The crystal structure, kinetics assay, molecular docking, and temporal and spatial expression of PviPRX9, as well as its coexpression with other genes, revealed a role of PviPRX9 in lignification.
Abstract
Class III peroxidases (CIIIPRX) catalyze the oxidation of monolignols, generate radicals, and ultimately lead to the formation of lignin. In general, CIIIPRX genes encode a large number of isozymes with ranges of in vitro substrate specificities. In order to elucidate the mode of substrate specificity of these enzymes, we characterized one of the CIIIPRXs (PviPRX9) from switchgrass (Panicum virgatum), a strategic plant for second-generation biofuels. The crystal structure, kinetic experiments, molecular docking, as well as expression patterns of PviPRX9 across multiple tissues and treatments, along with its levels of coexpression with the majority of genes in the monolignol biosynthesis pathway, revealed the function of PviPRX9 in lignification. Significantly, our study suggested that PviPRX9 has the ability to oxidize a broad range of phenylpropanoids with rather similar efficiencies, which reflects its role in the fortification of cell walls during normal growth and root development and in response to insect feeding. Based on the observed interactions of phenylpropanoids in the active site and analysis of kinetics, a catalytic mechanism involving two water molecules and residues histidine-42, arginine-38, and serine-71 was proposed. In addition, proline-138 and gluntamine-140 at the 137P-X-P-X140 motif, leucine-66, proline-67, and asparagine-176 may account for the broad substrate specificity of PviPRX9. Taken together, these observations shed new light on the function and catalysis of PviPRX9 and potentially benefit efforts to improve biomass conservation properties in bioenergy and forage crops.
As a perennial grass adapted to many regions in North America, switchgrass (Panicum virgatum) has been targeted as a model grass for biofuel (Vogel and Mitchell, 2008). Switchgrass requires minimal agricultural input and, thus, can be sustainably grown on marginal croplands (Saathoff et al., 2013). Continuous improvement of the yield and quality of switchgrass is a pressing need to meet the U.S. national goal for replacing a portion of petroleum gasoline with biofuel (Perlack and Stokes, 2011). One plausible approach for achieving this goal is to manipulate plant lignin biosynthesis, leading to changes in lignin content and cell wall composition, which could significantly improve biofuel yields (Dien et al., 2009). Lignin is an aromatic polymer of plant cell walls that accounts for 20% to 30% of all terrestrial plant biomass (Vanholme et al., 2010; Fernández-Perez et al., 2015; Voxeur et al., 2015). Lignin is cross-linked to cell wall hemicellulose, resulting in both structural and protective fortification of plant cells (Wagner et al., 2007). However, lignin also is a major source of recalcitrance for the conversion of herbaceous biomass into biofuels (Dien et al., 2009). Thus, optimizing lignin levels in biomass will help to mitigate the negative impact of lignin in the biochemical conversion process of lignocellulose to bioethanol or, alternatively, will help increase the energy content of herbaceous biomass (Scully et al., 2016). However, there are still many unsolved questions associated with lignification, such as the functions of key enzymes involved in lignin biosynthesis, the mechanisms by which the lignin polymer grows, and how specific monomeric units get incorporated into the growing polymer (Ostergaard et al., 2000; Vanholme et al., 2010; Mansfield et al., 2012; Fernández-Perez et al., 2015).
The majority of monomeric units comprising the lignin polymer are the aromatic hydroxycinnamyl alcohols (monolignols) p-coumaryl alcohol, coniferyl alcohol, and sinapyl alcohol (Freudenberg, 1965; Vanholme et al., 2010; Barros et al., 2015; Fernández-Perez et al., 2015). Current evidence suggests that two different enzyme families, class III plant peroxidases (CIIIPRXs; EC 1.11.1.7 ) and laccases (EC 1.10.3.2), are responsible for monolignol oxidation and, ultimately, the oxidative radical coupling of lignin monomeric units (Ostergaard et al., 2000; Zhao et al., 2013; Wang et al., 2015). However, in the case of CIIIPRXs, which have broad substrate specificities and large numbers of CIIIPRX isozymes encoded in plant genomes, it is a very challenging task to decipher the biological functions that an individual CIIIPRX may play in cell wall physiology (Veitch, 2004a).
PRXs constitute a multigene family, with most plant PRXs belonging to class I and class III (Veitch, 2004b; Almagro et al., 2009). Class I PRXs, which contain ascorbate peroxidase (the only class I PRX detectable in plants), cytochrome c peroxidase, and catalase peroxidase, are intracellular peroxidases without signal peptides, disulfide bridges, or structural calcium ions. They are typically located in the chloroplasts, cytosol, mitochondria, and peroxisomes.
CIIIPRXs comprise all plant secretory peroxidases and have distinguishing attributes from the larger peroxidase superfamily. In the peroxidase database PeroxiBase (Fawal et al., 2013; http://peroxibase.toulouse.inra.fr/, accessed September 6, 2016), there are as many as 12,800 PRXs collected from more than 2,500 organisms available. Based on the sequences obtained from high-quality ESTs, approximately 400 PRX ESTs were identified from switchgrass and more than half of these belong to CIIIPRXs (Tobias et al., 2008; Saathoff et al., 2013). During plant growth, CIIIPRXs play a dual role in both cell wall stiffening and loosening (Francoz et al., 2015). In the presence of aromatic cell wall compounds (e.g. monolignols, cinnamic acids, aromatic amino acids, etc.) and the cosubstrate hydrogen peroxide (H2O2), CIIIPRXs catalyze the oxidation of the aromatic lignin monomers to generate monolignol radicals, leading to oxidative radical coupling and, ultimately, a growing lignin polymer. This results in cross-linking of cell wall components, decreased elasticity of the cell wall, and suspension of cell elongation (Marjamaa et al., 2009). With reverse genetic techniques, the functions of CIIIPRXs in Arabidopsis (Arabidopsis thaliana) cell wall lignification have been demonstrated (Lee et al., 2013). In addition, studies suggested that down-regulation of an anionic CIIIPRX, PrxA3a, in transgenic aspen (Populus sieboldii × Populus gradidentata) reduced the lignin content and modified the lignin composition (Li et al., 2003). Recent advances in genomics and next-generation sequencing have facilitated the global analysis of peroxidase transcripts in different organs, growing stages, and populations of switchgrass (Saathoff et al., 2013). However, the large numbers of CIIIPRXs in the switchgrass genome and their potential diversity of functions make it challenging to determine the functions of an individual CIIIPRX isozyme.
In this study, we have comprehensively characterized a switchgrass CIIIPRX, PviPRX9, through determining its crystal structure, examining enzyme kinetics toward phenylpropanoids, and investigating PviPRX9 expression patterns to understand its specific roles in lignification.
RESULTS
Overall Structure
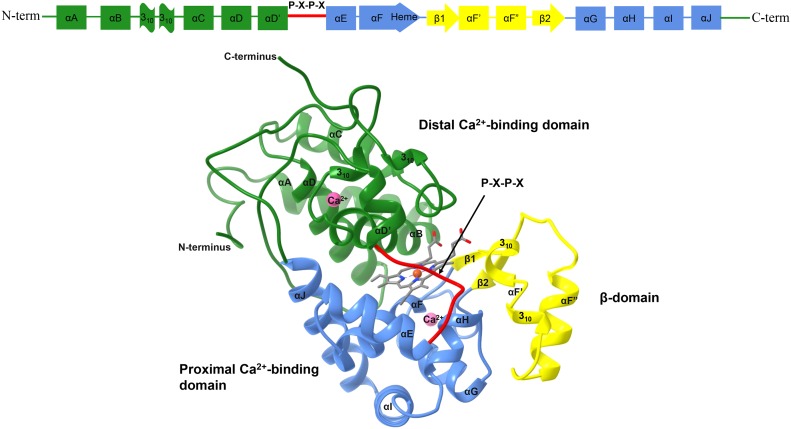
Our crystal structure suggested that PviPRX9 should be monomeric (31 kD) in solution, as indicated in its crystal packing of limited intermolecular interaction. To confirm the crystallographic monomeric state of PviPRX9, the structure was submitted to PDBePISA (Krissinel and Henrick, 2007). The resulting analysis indicated no significant interface interactions between monomers and implied a monomeric state of PviPRX9. The overall fold of PviPRX9 consisted of 13 α-helices and two β-strands, which was similar to those observed among other CIIIPRXs. PviPRX9 displayed three distinct domains (Fig. 1), two Ca2+-binding domains and the β-domain. The distal Ca2+-binding domain started at the N terminus and was composed of the first five α-helices and two 310-helices. The proximal Ca2+-binding domain started at αE, continued through αF (heme-binding helix), and ran up to the first β-strand (β1) of the β-domain (Fig. 1). The β-domain was connected to the proximal Ca2+-binding domain via an antiparallel β-sheet. Starting with β1, the β-domain ran through two 310-helices (named αF′ by convention), αF, and back through β2 (Fig. 1). Then, αG through αF form the rest of the proximal Ca2+-binding domain, with the C terminus wrapped back up to the distal Ca2+-binding domain. The structure of PviPRX9 also displayed four disulfide bonds, which are the conserved features among class III peroxidases (Ostergaard et al., 2000; Henriksen et al., 2001; Watanabe et al., 2010). Two of the four conserved disulfides were in the proximal Ca2+-binding domain, and the other two disulfides were in the distal Ca2+-binding domain. In PviPRX9, the two distal side disulfide linkages were established between sulfhydryl groups of Cys-11 and Cys-89 and between Cys-44 and Cys-49. The two proximal side disulfide linkages were established between Cys-95 and Cys-288 and between Cys-174 and Cys-198.
Figure 1.
Ribbon diagram representing the global structure of PviPRX9 with bound heme (gray) dividing the lower proximal Ca2+-binding domain (blue) containing the His-167 ligand, the upper distal Ca2+-binding domain (green), and the β-domain (yellow). The secondary structural elements were laid out as one-dimensional bars and are indicated following the convention of PRX. Ca2+ ions are depicted with pink balls. Molecular graphics images were produced using the Chimera package (Pettersen et al., 2004).
Calcium-Binding Sites, Heme Environment, Active Site, and Substrate-Binding Pocket
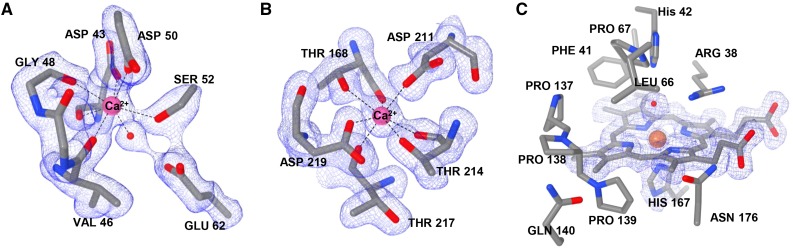
The difference electron density map (Fo − Fc) clearly showed two Ca2+ ions from the early stage of the refinement, which individually belonged to the proximal and distal domains of PviPRX9. The distal Ca2+ was coordinated by Asp-43, Val-46, Gly-48, Asp-50, Ser-52, and a water molecule (Fig. 2A). The proximal Ca2+ was coordinated by Thr-168, Asp-211, Thr-214, Thr-217, and Asp-219 (Fig. 2B). Those two Ca2+-binding domains were split by the heme cofactor that occupies a central location within PviPRX9 and all class III peroxidases characterized to date (Fig. 1). The distal domain above the central heme formed the enzyme active site of PviPRX9, and the proximal domain below the heme included the proximal His ligand (His-167) coordinated to the heme iron through its Nε2 atom (Fig. 2C). The carboxyl side chain of Asp-236 was within hydrogen bond distance to the Nδ1 atom of His-167. The heme propionate groups were stabilized by establishing a salt bridge with Arg-31 and hydrogen bonds with Ser-35, Ser-71, Gln-171, Gln-173, and Asn-176.
Figure 2.
Ca2+ and heme iron coordination. A, The distal Ca2+ coordinated by the side chains of Asp-43, Asp-50, and Ser-52, backbone carbonyls of Val-46, Gly-48, and Asp-43, and a water molecule. B, The proximal Ca2+ coordinated by the side chains of Thr-168, Asp-211, Thr-214, and Asp-219 and the backbone carbonyls of Thr-168, Thr-214, and Thr-217. C, The active site showing the catalytic His-42, ligand-binding Arg-38, heme and the fifth heme iron ligand His-167, and the P-X-P-X motif of PviPRX9 (137PPPQ140). Images were rendered with Chimera version 1.11.
The volume of the substrate-binding pocket of PviPRX9 estimated by the CASTp server (Dundas et al., 2006) was 487.9 Å3. In addition to the other conserved residues among most CIIIPRXs, such as Arg-38, Phe-41, and His-42, the substrate-binding pocket was composed of three major features (Fig. 2C). (1) One side of the substrate-binding pocket above the heme was established by the P-X-P-X motif, located between the fifth (αD′) and sixth (αE) α-helices. This P-X-P-X motif of PviPRX9 consisted of Pro-137, Pro-138, and Pro-139 followed by Gln-140 (Figs. 1 and 2C). (2) Another side of the substrate-binding pocket was constituted by the residues at the tail of β1-strand, Gln-173, Leu-175, and Asn-176 (Figs. 1 and 2C). (3) The top and third side of the substrate-binding pocket was established by Arg-38, Phe-41, His-42, Leu-66, and Pro-67 (Figs. 1 and 2C). The heme group was positioned at the bottom of this substrate-binding pocket. The crystal structure of PviPRX9 indicated that the heme iron was in a mixed occupancy of oxidation states, probably due to x-ray exposure during data collection, as shown before among crystal structures of heme proteins (Berglund et al., 2002).
Structural Alignment of PviPRX9 with Other CIIIPRXs
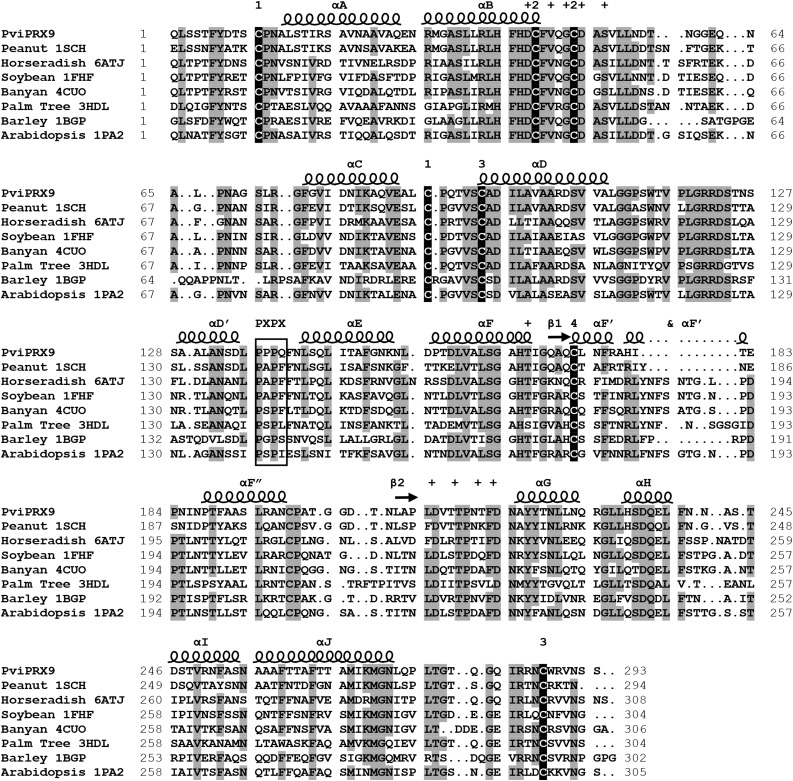
Structural sequence alignment of PviPRX9 was performed with its homologous peroxidases obtained from the Protein Data Bank (PDB) with a threshold identity value of 49% or higher (Fig. 3). The structure of peanut (Arachis hypogaea) PRX (PDB: 1SCH) had the highest sequence identity of 72% followed by horseradish (Armoracia lapathifolia) PRX C (PDB:1ATJ) with an identity of 51%, soybean (Glycine max) PRX (PDB: 1FHF), banyan (Ficus benghalensis) PRX (PDB: 4CUO), and palm tree (Roystonea regia) PRX (PDB: 3HDL) with identities of 50%, and barley (Hordeum vulgare) PRX (PDB: 1BGP) along with Arabidopsis PRX (PDB: 1PA2) with sequence identities of 49%. The areas of greatest conservation across these CIIIPRXs mainly included the residues for maintaining the peroxidase fold and catalytic activity. Among the 10 residues responsible for coordinating Ca2+ ions, eight of them were completely conserved across all CIIIPRXs examined (Fig. 3). In one of those remaining two residues, the Ca2+-coordinating residue Thr-168 in the proximal domain of PviPRX9 was conservatively substituted to Ser in the banyan PRX, while a Thr residue was maintained in all the other CIIIPRXs examined. However, high heterogeneity was observed for Thr-217, which participates in the coordination of Ca2+ ion at the proximal domain only via its backbone carboxyl group (Fig. 3).
Figure 3.
Structural alignment of PviPRX9 with seven CIIIPRXs from the Research Collaboratory for Structural Bioinformatics PDB. Secondary structural elements were labeled with helices indicated by spirals and β-sheets indicated by arrows. The secondary structure was letter mapped according to the system laid out by Schuller et al. (1996). Residues coordinating with the two conserved Ca2+ atoms are indicated with plus signs. The conserved P-X-P-X motif is outlined with a box. The Cys residues participating in conserved disulfides are numbered according to the corresponding Cys. Amino acids with 75% or greater identity in the alignment are highlighted in gray, and the conserved Cys residues are highlighted in black.
The eight Cys residues that participated in the four disulfides were completely conserved among all CIIIPRXs examined. Full conservation also was observed for the fifth heme iron ligand His-167 in the proximal Ca2+-binding domain and Arg-38 and His-42 in the distal Ca2+-binding domain. Additionally, the Pro repeat motif P-X-P-X was present in all CIIIPRXs examined. Significantly, only PviPRX9 had this motif as 137P-P-P-Q140 among compared CIIIPRXs. In peanut, horseradish, and soybean, Pro-138 was substituted by an Ala. In banyan, palm tree, and Arabidopsis, residue 138 was replaced by a Ser. For PviPRX9, a polar Gln-140 was located at the surface-exposed entrance to the substrate-binding pocket, which was substituted to a hydrophobic amino acid such as Pro or Phe in all other CIIIPRXs examined except barley PRX, where a polar Ser was found. At the top of the substrate-binding pocket, Leu-66 from a loop of the distal Ca2+-binding domain of PviPRX9 was substituted with Gly in peanut, Phe in horseradish, Ala in banyan and barley, Ile in palm tree, and Gly in Arabidopsis.
Significant variation also was observed for residues that constituted the opposite side of the P-X-P-X motif in the substrate-binding pocket, such as Gln-173, Leu-175, and Asn-176. Gln-173 was maintained in peanut, horseradish, and banyan, was substituted to an Arg in both soybean and Arabidopsis, and was substituted to His in palm tree and barley. Leu-175 in PviPRX9 was substituted to Thr in peanut, Arg in horseradish, Ser in soybean, palm tree, and barley, Gln in banyan, and Gly in Arabidopsis. Asn-176 in PviPRX9 was substituted to Ala in peanut, Phe in horseradish, Thr in soybean, Phe in banyan, Ser in palm tree, Ser in barley, and Val in Arabidopsis.
Enzyme Kinetic Assays
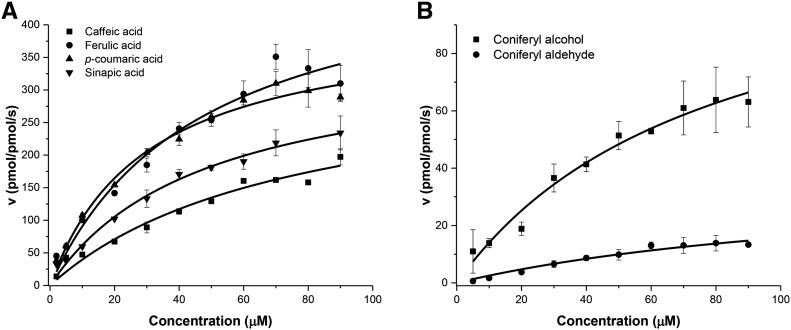
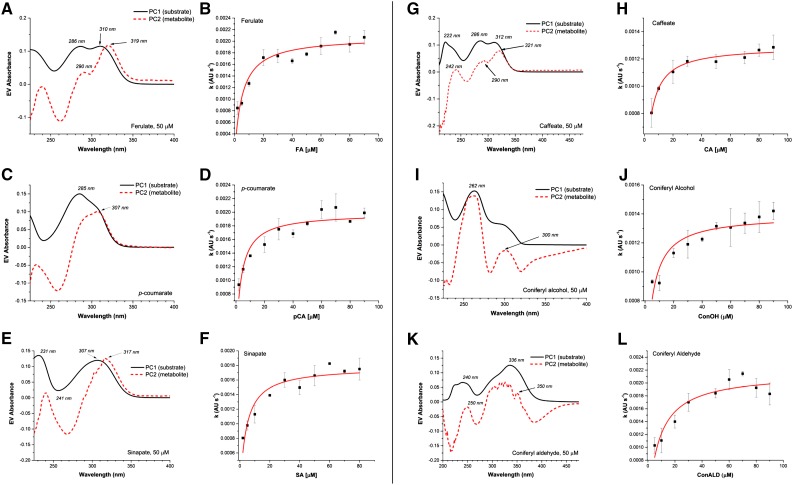
To characterize the substrate preference for PviPRX9, we performed steady-state kinetics with ferulate, p-coumarate, sinapate, caffeate, coniferyl alcohol, and coniferyl aldehyde (Fig. 4). As shown in Table I, the kcat values, the turnover numbers, were 518.3 s−1 for ferulate, 410.7 s−1 for p-coumarate, 357.9 s−1 for sinapate, 337.5 s−1 for caffeate, 124.5 s−1 for coniferyl alcohol, and 37.1 s−1 for coniferyl aldehyde. The catalytic efficiency (kcat/Km) of p-coumarate gave the highest value at 13.7 s−1 µm−1, followed by the value for ferulate at 11 s−1 µm−1, sinapate at 7.5 s−1 µm−1, caffeate at 4.5 s−1 µm−1, coniferyl alcohol at 1.6 s−1 µm−1, and coniferyl aldehyde at 0.3 s−1 µm−1.
Figure 4.
Steady-state initial rates are plotted versus reducing substrate concentrations. A, Carboxylates were varied from 2 to 90 µm, and the concentration of H2O2 was held constant at 500 µm. B, Coniferyl alcohol and coniferyl aldehyde were varied from 5 to 90 µm, and the concentration of H2O2 was held constant at 500 µm. Plots were generated using OriginPro 2016, and final graphs were generated with Microsoft Excel 2015.
Table I. Kinetic parameters for the catalytic reaction of PviPRX9 with constant H2O2 concentration obtained through monitoring the disappearance of substrates.
The turnover number kcat was found using the nonbinding buffer MES at pH 6 with 1 mm Ca2+.
| Substrate | Substrate Disappearance |
SVD/ALS |
ΔGbinding | ||
|---|---|---|---|---|---|
| Km | kcat | kcat/Km | Km | ||
| µm | s−1 | s−1 µm−1 | µm | kcal mol−1 | |
| Ferulate | 47.2 ± 11.9 | 518.3 ± 60.2 | 11.0 | 5.1 ± 1.1 | −6.4 |
| p-Coumarate | 29.9 ± 4.3 | 410.7 ± 21.9 | 13.7 | 3.4 ± 0.8 | −5.9 |
| Sinapate | 47.9 ± 7.4 | 357.9 ± 26.7 | 7.5 | 3.8 ± 0.9 | −7.2 |
| Caffeate | 75.3 ± 23.0 | 337.5 ± 57.5 | 4.5 | 3.1 ± 0.3 | −6.4 |
| Coniferyl alcohol | 78.7 ± 19.3 | 124.5 ± 17.3 | 1.6 | 3.3 ± 0.6 | −6.7 |
| Coniferyl aldehyde | 136.5 ± 59.5 | 37.1 ± 11.0 | 0.3 | 6.1 ± 1.1 | −6.2 |
Using a combined approach of singular value decomposition (SVD) coupled with alternating least square (ALS) analysis, we monitored product formation to gain a better estimation of binding affinities (Fig. 5; Table I). However, kcat could not be computed with this approach, since the optical absorbance for those metabolites was unknown. The lowest Km value result from SVD/ALS product formation was for caffeate at 3.1 µm, and the highest Km value was observed for coniferyl aldehyde at 6.1 µm (Table I). The SVDs for the different monolignyl compounds and the corresponding saturation curves also were calculated (Fig. 5). The principal component of the spectrum assigned to the metabolite overlapped with the parent compound in all cases, with a shift in the λmax of approximately 10 to 15 nm for the carboxylates. In the case of coniferyl alcohol and coniferyl aldehyde, the spectra of the metabolites largely overlapped with the parent compounds, which possibly explains the linearity observed when substrate disappearance was used for rate measurements. For coniferyl alcohol, caffeate, and ferulate, the spectra of the products were similar to those obtained by Rasmussen and coworkers (1995) after adding excess amounts of H2O2. For p-coumarate, the spectrum of the product was analogous to the spectrum reported by Bakovic and Dunford (1993) monitoring the reaction for extended times. The time-dependent traces for metabolite formation showed linearity over the entire time scale (60 s) and were subsequently fitted with a linear function to compute the apparent velocity (Supplemental Fig. S1). The corresponding plots showed saturating profiles, with calculated affinities (Table I) lower than the ones obtained with the substrate-disappearance method.
Figure 5.
Metabolite kinetics of PviPRX9. A, Plots of the ferulate substrate spectrum and the product spectrum showing a clear overlap of wavelengths. B, Michaelis-Menten plot for ferulate product formation. C, Plots of the p-coumarate substrate spectrum and the product spectrum showing wavelength overlap. D, Michaelis-Menten plot for p-coumarate product formation. E, Plots of the sinapate substrate spectrum and the product spectrum showing wavelength overlap. F, Michaelis-Menten plot for sinapate product formation. G, Plots of the caffeate substrate spectrum and the product spectrum showing wavelength overlap. H, Michaelis-Menten plot for caffeate product formation. I, Plots of the coniferyl alcohol substrate spectrum and the product spectrum showing wavelength overlap. J, Michaelis-Menten plot for coniferyl alcohol product formation. K, Plots of the coniferyl aldehyde substrate spectrum and the product spectrum showing wavelength overlap. L, Michaelis-Menten plot for coniferyl aldehyde product formation. The Michaelis-Menten metabolite plot y axis units are arbitrary units (AU) per second. Plots were generated using SVD/ALS with Mathcad and OriginPro 2016.
o-Phenylenediamine (OPD), a common peroxidase substrate, was used to compare the activity of PviPRX9 with the horseradish peroxidase. The specific activity of PviPRX9 for the substrate OPD was 23.4 ± 0.778 µmol s−1 mg−1, while for the commercially available horseradish peroxidase, the specific activity for OPD was 27.4 ± 0.643 µmol s−1 mg−1.
Molecular Docking
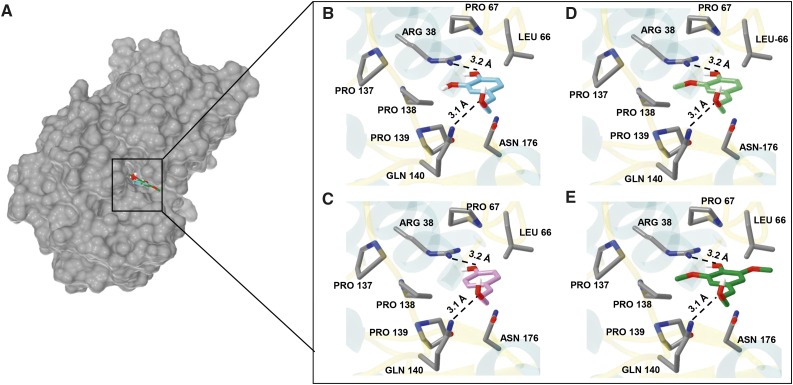
In order to rationalize the observed profile of kinetic efficiency among tested phenylpropanoids, molecular docking was performed with the compounds used in the kinetics study (Trott and Olson, 2010). All compounds were found to dock in more than one pose with favorable docked binding affinity (Fig. 6; Table II). To choose the most appropriate pose, the crystal structure of PviPRX9 with docked compounds was overlaid with the structure of the ferulate:cyanide:horseradish PRX C ternary complex (PDB: 7ATJ; Henriksen et al., 1999). For the compounds, the poses most closely corresponding to the location of ferulate in the ternary complex were selected, and the binding poses for the alcohol compounds are shown in Figure 6. The docked ferulate with PviPRX9 showed its phenolic oxygen establishing a hydrogen bond with Arg-38 Nη2 at a distance of 3.2 Å and a propenyl oxygen 3 Å from the Nε2 of Gln-140. Ferulate had a predicted binding affinity of −6.4 kcal mol−1 (Table II). For the compounds coniferyl alcohol and coniferyl aldehyde, the predicted binding affinities were −6.7 and −6.4 kcal mol−1. For docked sinapate, sinapyl aldehyde, and sinapyl alcohol, the phenolic oxygen was positioned at a hydrogen-bonding distance of 3.2 Å from Arg-38 Nη2. The propenyl oxygen of sinapate and sinapyl aldehyde was 3 Å from the Nε2 of Gln-140. For sinapyl alcohol, the propenyl oxygen was 3.1 Å from the Nε2 of Gln-140. Predicted docked binding affinities were −7.2 kcal mol−1 for sinapate, −7.1 kcal mol−1 for sinapyl aldehyde, and −7.5 kcal mol−1 for sinapyl alcohol (Table II). The docking results for caffeate, caffeyl aldehyde, and caffeyl alcohol placed the 4′ phenolic oxygen of caffeate and caffeyl aldehyde at 2.8 Å from Arg-38 Nη2 and that of caffeyl alcohol at 3.2 Å. The propenyl oxygens of the respective carboxylate, alcohol, and aldehyde were 3.2, 3.1, and 3.3 Å from the Nε2 of Gln-140, with the predicted docked binding affinities −6.4, −6.2, and −6.2 kcal mol−1. For p-coumarate, p-coumaryl aldehyde, and p-coumaryl alcohol, the compounds docked with phenolic oxygen at 2.8, 2.8, and 3.2 Å from Arg-38 Nη2, respectively. The propenyl oxygens of the respective carboxylate, aldehyde, and alcohol were 3.3, 3.8, and 3.1 Å from the Nε2 of Gln-140, and the docked binding affinities were −5.9, −5.9, and −5.9 kcal mol−1 (Table II).
Figure 6.
Active site of PviPRX9 with docked substrates coniferyl alcohol, p-coumaryl alcohol, sinapyl alcohol, and caffeyl alcohol. A, Surface representation of PviPRX9 showing the binding pocket with docked monolignols. B, Caffeyl alcohol is pictured in cyan oriented with the meta phenolic oxygen 3.2 Å from Arg-38 Nη2, and the propenyl oxygen was 3.1 Å from the Nε2 of Gln-140. C, p-Coumaryl alcohol is pictured in purple docked with phenolic oxygen oriented 3.2 Å from Arg-38 Nη2, and the propenyl oxygen was 3.1 Å from the Nε2 of Gln-140. D, Coniferyl alcohol is shown in lime green docked with the phenolic oxygen position within the hydrogen-bonding distance of 3.2 Å from Arg-38 Nη2, and the propenyl oxygen was 3.1 Å from the Nε2 of Gln-140. E, Sinapyl alcohol (forest green) docked with the phenolic oxygen 3.2 Å from Arg-38 Nη2, and the propenyl oxygen was 3.1 Å from the Nε2 of Gln-140. Molecular graphics images were produced using the Chimera package (Pettersen et al., 2004).
Table II. Binding energies calculated for molecular docking.
The ΔG of binding for phenylpropanoids was calculated by the molecular docking program AutoDock Vina for the most probable poses.
| Reducing Substrate | ΔGbinding |
|---|---|
| kcal mol−1 | |
| Ferulate | −6.4 |
| Coniferyl alcohol | −6.7 |
| Coniferyl aldehyde | −6.2 |
| Sinapate | −7.2 |
| Sinapic alcohol | −7.5 |
| Sinapic aldehyde | −7.1 |
| p-Coumarate | −5.9 |
| p-Coumaryl alcohol | −5.9 |
| p-Coumaryl aldehyde | −5.9 |
| Caffeate | −6.4 |
| Caffeic alcohol | −6.2 |
| Caffeic aldehyde | −6.2 |
PviPRX9 Expression
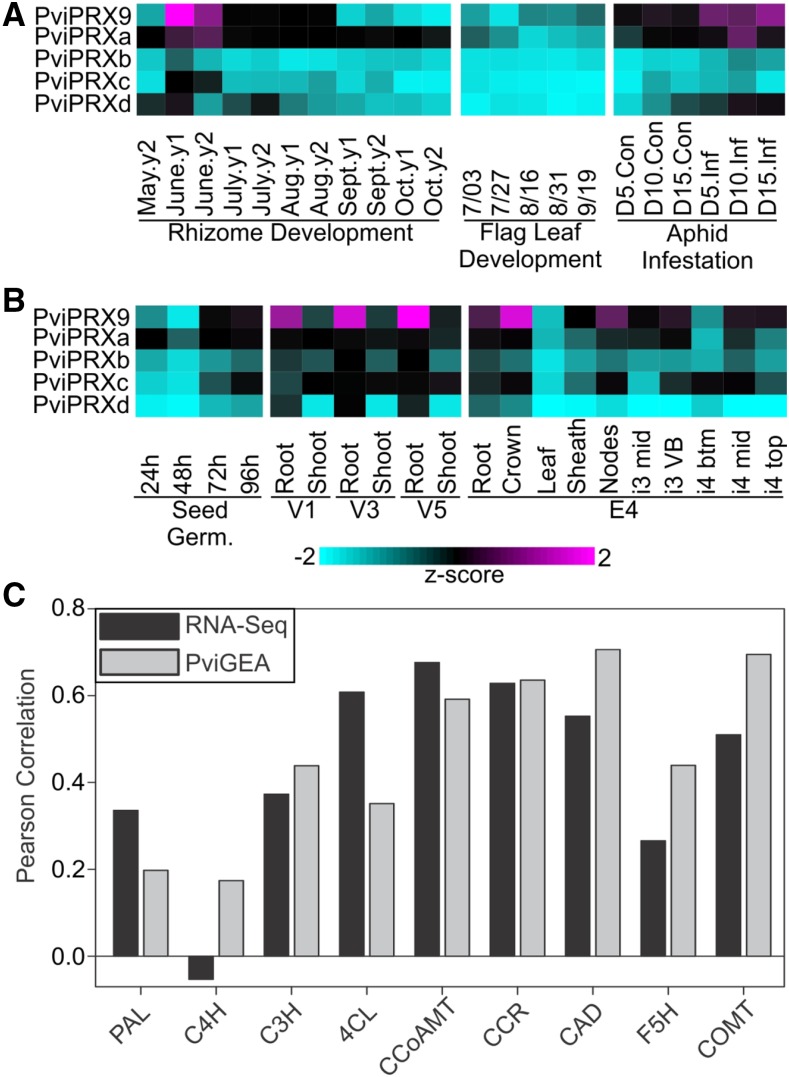
A 2-year study of switchgrass rhizome development (N.A. Palmer, A.J. Saathoff, E.D. Scully, C.M. Tobias, P. Twigg, S. Madhavan, M. Schmer, R. Cahoon, S.E. Sattler, S.J. Edmè, R.B. Mitchell, G. Sarath, unpublished data; accession no. SRX1601466), a single-year study of switchgrass flag leaf development (Palmer et al., 2015), and a greenhouse study investigating the transcriptional response of switchgrass seedlings to wheat aphid (Schizaphis graminum) infestation (T. Donze-Reiner et al., unpublished data; accession no. SRX1600826) were analyzed with RNA sequencing (RNA-Seq). The expression patterns of PviPRX9 were used to detect other peroxidases that shared a comparable expression profile with a correlation of expression of 0.75 or greater in existing RNA-Seq and microarray data sets (Fig. 7). This search yielded four other switchgrass peroxidase-encoding genes labeled as PviPRXa to PviPRXd for convenience (PviPRXa, Pavir.2NG016500; PviPRXb, Pavir.3KG134300; PviPRXc, Pavir.3NG190800; and PviPRXd, Pavir.5NG345000). Across all data sets, PviPRX9 was most highly expressed, followed by PviPRXa. PviPRXb to PviPRXd were invariably expressed at lower levels in the RNA-Seq data sets described above.
Figure 7.
PviPRX9 expression analysis and relationship to the expression levels of genes associated with lignin biosynthesis. A, RNA-Seq-based transcript abundances of PviPRX9 and four additional peroxidases with correlated expression (PviPRXa to PviPRXd) in diverse switchgrass tissues shown as z-scores. Data are shown for rhizome development based on RNA-Seq experiments with rhizomes collected from field-grown plants of cv Summer over 2 years; flag leaf development; and aphid infestation with RNA-Seq analysis of tissues collected from uninfested (Con) and aphid-infested (Inf) cv Summer plants at 5, 10, and 15 d (D) post infestation. B, Microarray-based transcript abundances from PviGEA for PviPRX9 and four additional peroxidases with correlated expression (PviPRXa to PviPRXd) in multiple tissues and developmental stages shown as z-scores. Data are shown for four time points during seed germination (Seed Germ.); root and shoot tissue at three vegetative growth stages (V1, V3, and V5); and multiple tissues at the stem elongation 4 (E4) stage: root, crown, leaf (pooled leaves from E4 tiller), sheath (pooled sheaths from E4 tiller), nodes (pooled nodes from E4 tiller), the middle of the third internode (i3 mid), vascular bundles from the third internode (i3 VB), bottom one-fifth of the fourth internode (i4 btm), middle one-fifth of the fourth internode (i4 mid), and top one-fifth of the fourth internode (i4 top). Respective data sets and bioinformatic routines used for these analyses are given in “Materials and Methods.” Magenta indicates high expression and cyan indicates low expression. C, Correlation of transcript abundances between PviPRX9 and genes associated with monolignol biosynthesis in the data sets described in A and B. PAL, Phe ammonia lyase; C4H, cinnamate 4-hydroxylase; C3H, p-coumarate 3-hydroxylase; 4CL, 4-coumarate:CoA ligase; CCoAMT, caffeoyl-CoA o-methyltransferase; CCR, cinnamoyl-CoA reductase; CAD, cinnamoyl alcohol dehydrogenase; F5H, ferulate 5-hydroxylase; COMT, caffeic acid o-methyltransferase.
During rhizome development, PviPRX9 showed peak expression in June, a period of active rhizome and plant growth. Intermediate PviPRX9 expression was observed in July and August, when plants enter their reproductive phase and rhizome growth slows. PviPRX9 expression was down-regulated significantly during harvests, corresponding to the onset of aerial senescence (September) and the onset of winter dormancy (October; Fig. 7A). In minimally lignified tissues such as flag leaves, the expression of PviPRX9 was low (Fig. 7A). PviPRXa was expressed moderately in these data sets, and peak expression in rhizomes also was detected in the June harvests (Fig. 7A). The patterns for the more minimally expressed PviPRXb to PviPRXd genes were comparable to that of PviPRX9 in rhizomes and flag leaves (Fig. 7A).
In response to aphid infestation across a 15-d time series, PviPRX9 was significantly induced in infested plants relative to uninfested control plants, with an expression peak on day 15 after infestation (Fig. 7A). PviPRX9 is part of a syntenic cluster of peroxidase genes present in the genomes of all C3 and C4 grasses that were induced variably in response to aphid feeding in switchgrass, sorghum (Sorghum bicolor), and foxtail millet (Setaria italica; Scully et al., 2016). PviPRXa expression also was induced by aphid herbivory, although peak expression was noticed on day 10 after infestation. Up-regulation of PviPRXb to PviPRXd expression in aphid-infested plants was not as distinct. The patterns of expression of PviPRX9 in rapidly growing tissues, and potentially in response to aphid herbivory, could indicate a role in lignification. It can be expected that other peroxidases, conceivably those identified here, could play a role in switchgrass cell wall lignification as well.
Expression of these five peroxidases also was investigated in the switchgrass gene expression atlas database (PviGEA [http://switchgrassgenomics.noble.org/]; Fig. 7B; Zhang et al., 2013). The expression level of PviPRX9 was minimal during the first 48 h of seed germination and then increased in the next 48 h. During early vegetative growth stages (V1–V5), PviPRX9 had higher expression in root tissue and lower expression in young shoot tissues (Fig. 7B). During the stem elongation 4 (E4) growth stage, PviPRX9 had the highest expression in crown tissue, followed by pooled node tissue, while the expression in leaf and sheath tissues was lower. Interestingly, a striking expression gradient was observed within a single internode, with the bottom of internode 4 having low expression and the middle and top of the same internode having significantly higher expression (Fig. 7B). The patterns of expression of PviPRXa to PviPRXd were essentially similar to those observed for PviPRX9, except that PviPRXd was expressed at a low level in internodes (Fig. 7B).
The correlation of expression between PviPRX9 and genes encoding monolignol biosynthesis enzymes was calculated by mining the RNA-Seq and microarray data sets (Fig. 7C). With the exception of C4H, PviPRX9 had comparable expression correlations with each gene associated with the monolignol biosynthesis pathway in both RNA-Seq and microarray data sets. The highest correlations in the RNA-Seq data set occurred with CCoAMT, CCR, 4CL, CAD, and COMT (0.676, 0.628, 0.608, 0.552, and 0.51, respectively), while the microarray data set had the best correlations with CAD, COMT, CCR, and CCoAMT (0.706, 0.685, 0.636, and 0.592, respectively). Correlations for PviPRXa to PviPRXd were in the same range of 0.5 to 0.7. Together, these data reinforce the suggested role of PviPRX9 in the lignification of switchgrass tissues, although the potential roles for PviPRXa to PviPRXd and other as yet uncharacterized peroxidases in lignification cannot be excluded.
Analysis of Switchgrass Genomic Data for Peroxidases
To find other full-length CIIIPRX annotated genes, the switchgrass genome (version 3.1; https://phytozome.jgi.doe.gov) was mined for the presence of the peroxidase PFAM domain (PF00141). This search yielded 333 loci encoding proteins containing the peroxidase domain. Of these 333 loci, 24 encoded putative ascorbate peroxidases, while the remaining 309 encoded putative CIIIPRXs. In comparison, peroxidase domain searches of several grasses yielded 164 proteins in Panicum halli, 154 in sorghum, 169 in foxtail millet, 166 in Setaria viridis, 158 in rice (Oryza sativa), and 158 in Brachypodium distachyon. Based on the tetraploid nature of the switchgrass genome relative to the diploid grasses, these numbers indicate that a majority of the switchgrass peroxidases were found using the PFAM search. However, it is likely that pseudogenes and incomplete sequences exist in the current switchgrass genome annotation.
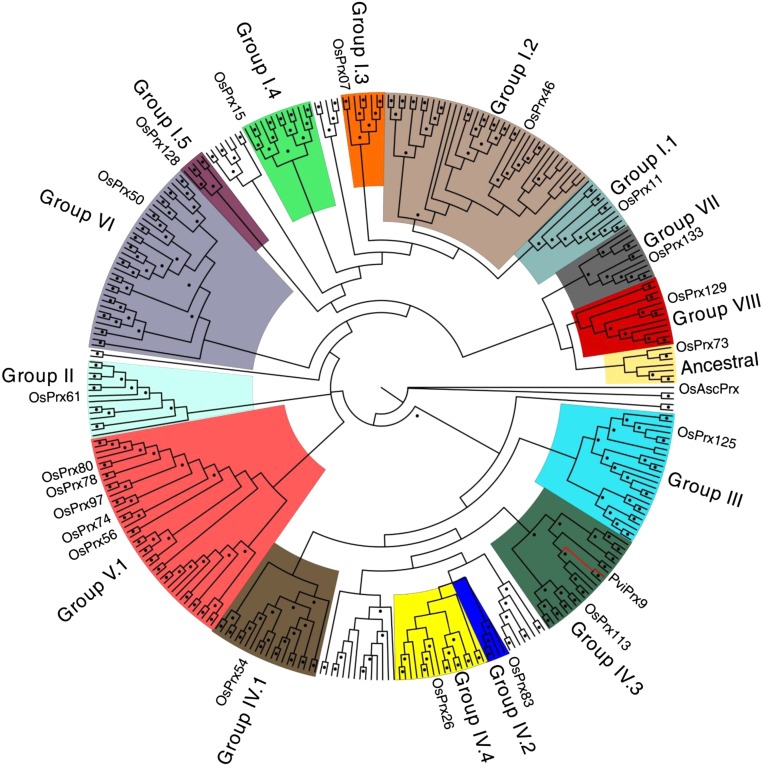
A maximum likelihood phylogenetic tree for all full-length class III PviPRXs in the switchgrass genome (version 3.1) was constructed to examine the evolutionary relationships of PviPRX9 with other PviPRXs. PviPRXs were assigned to evolutionary groups based on phylogenetic proximity with previously characterized rice peroxidases (Passardi et al., 2004). The bootstrap consensus tree is shown in Figure 8. Based on branching points and bootstrap values, the 300 PviPRXs were grouped into nine major evolutionary clades labeled groups I through VIII and ancestral. Among these clades, PviPRX9 was positioned in group IV.3.
Figure 8.
Maximum likelihood tree of all full-length CIIIPRXs in switchgrass. Maximum likelihood analysis was performed using the program Garli with 500 bootstrap pseudoreplicates. PviPRXs were assigned to evolutionary groups based on a phylogenetic grouping with previously assigned rice peroxidases. Dots indicate nodes with bootstrap support of 50 or greater. PviPRX9 was allocated in group IV.3 with the node in red.
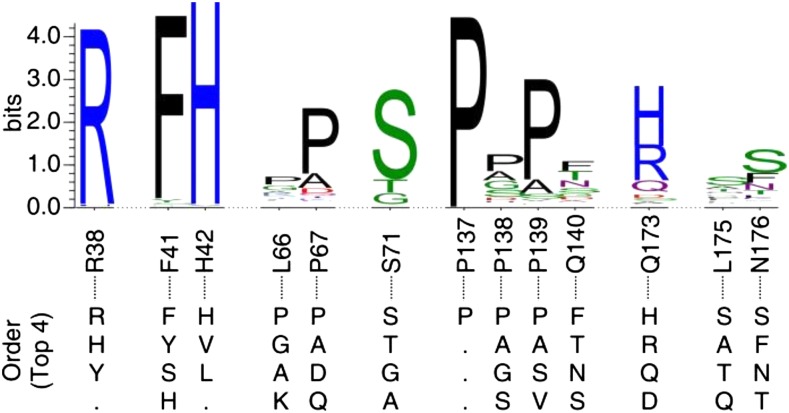
Within the substrate-binding pocket, both conservation and variation were observed among amino acids. Across all PviPRXs, the residues with proposed catalytic roles along with several residues in the substrate-binding pocket, such as Arg-38, Phe-41, His-42, and Pro-137 surrounding the substrate-binding pocket, showed a high degree of conservation (Fig. 9). Several residues associated with the substrate-binding pocket showed higher degrees of variation, including Leu-66, Pro-67, Ser-71, Pro-138, Pro-139, Gln-173, Leu-175, and Asn-176 (Fig. 9).
Figure 9.
Sequence logos of the residues constituting the substrate-binding pocket in all switchgrass CIIIPRXs. The top four amino acids at each position for all switchgrass CIIIPRXs were included below at their matching positions. Position labels were based on the sequence of PviPRX9. This figure was generated by WebLogo 3 (http://weblogo.threeplusone.com). The distributions of amino acids found in all positions are given in Supplemental Table S2.
DISCUSSION
Substrate Specificity
Previous studies in other plants showed that CIIIPRXs preferentially catalyzed the coniferyl and p-coumaryl compounds over sinapyl compounds or vice versa (Rasmussen et al., 1995; Nielsen et al., 2001; Gabaldón et al., 2006; Shigeto et al., 2014). For example, the CIIIPRX ATP A2 from Arabidopsis exhibited at least 1 magnitude of order higher catalytic activity with coniferyl and p-coumaryl than sinapyl compounds (Nielsen et al., 2001), while CIIIPRXs from Zinnia elegans showed higher catalytic activity with the sinapyl compounds compared with coniferyl and p-coumaryl compounds (Gabaldón et al., 2006). In contrast, our results suggested that the catalytic efficiency values of PviPRX9 for all three substrates were within 1 order of magnitude (Fig. 4; Table I), suggesting that PviPRX9 catalyzed all those compounds with similar efficiencies. For p-coumarate, the catalytic efficiency was highest at 13.7 s−1 µm−1, followed closely by ferulate and sinapate at 80.3% and 54.8%, respectively. For turnover number, ferulate exhibited the highest kcat of 518.3 s−1, followed by p-coumarate, sinapate, caffeate, and coniferyl alcohol with relative activities of 79.2%, 69.1%, 65.1%, and 24% respective to ferulate. Coniferyl aldehyde had the lowest relative activity respective to ferulate at 7.16%. The kinetics results for substrate disappearance showed that there was no strong preference of PviPRX9 observed for any specific methoxy or hydroxyl substituent pattern on the phenyl ring among the compounds examined, suggesting that PviPRX9 might have a rather broad substrate specificity. Similarly, in the case of Km values obtained from our product formation analysis, PviPRX9 showed a change of only 3 µm from highest to lowest Km (Fig. 5; Table I), suggesting a similar binding affinity across the compounds examined. As a whole, our kinetics data suggest that PviPRX9 has a broad substrate specificity, which may reflect its function in oxidizing all the physiologically relevant subunits of lignin polymer with similar ability.
To explain the reasons that cause the difference of substrate specificities among CIIIPRXs, one compelling hypothesis is that the conformation of the substrate-binding pocket was responsible for imparting the broad substrate specificity to CIIIPRXs (Abelskov et al., 1997). In addition, the structural characteristics of the substrate-access channel have been proposed to be responsible for CIIIPRX’s preference for oxidizing coniferyl and p-coumaryl or sinapyl compounds (Henriksen et al., 1998). Their diminished ability to oxidize sinapyl compounds has been explained as due to steric overlap between conserved Pro-139 (horseradish peroxidase C numbering) and the extra methoxy substituent (Nielsen et al., 2001), which would necessitate the need for a separate enzyme responsible for the oxidation of sinapyl compounds or an elaborate oxidation state shuttle through p-coumaryl compounds (Shigeto et al., 2014). However, recent studies have shown that some peroxidases with the conserved Pro-139, such as those from Z. elegans and Arabidopsis, have an undiminished or even increased capability to oxidize sinapyl compounds (Gabaldón et al., 2006; Shigeto et al., 2014).
As shown in Figure 9, some amino acids constituting the substrate-binding pocket are located within highly variable regions among switchgrass CIIIPRXs. Our structural and sequence analysis revealed that PviPRX9 has three separate domains that merge at the substrate-binding pocket (Figs. 1–3). At the interface of the three domains, αB in the distal Ca2+-binding domain offers the catalytic amino acid His-42 and the phenylpropanoid-binding amino acid Arg-38 (Fig. 2C). Additionally, Leu-66 and Pro-67 of the loop region (65ALPXXXS71) in the distal Ca2+-binding domain makes up the top of the substrate-binding pocket (Fig. 2C). On the other hand, the P-X-P-X motif as 137P-P-P-Q140 comprises the side of the substrate-binding pocket that links the distal Ca2+-binding domain to the proximal Ca2+-binding domain. Opposite to this P-X-P-X motif, the side chains of Gln-173 and Asn-176 in the β-domain constitute the other side of the substrate-binding pocket (Fig. 2C). Based on the observed substitution pattern among examined CIIIPRXs, significant alteration in hydrophobicity occurs within the substrate-binding pocket, along with the pattern of potential hydrogen bond network residues. Both the degree of change and the specific location within the substrate-binding pocket to which these changes occur could lead to a wide range of substrate specificity among the approximately 300 switchgrass CIIIPRXs.
PviPRX9 contained 137P-P-P-Q140 on one side of the binding pocket following the general P-X-P-X motif observed in other CIIIPRXs; thus, it was expected to have significantly lower activity toward sinapate compared with ferulate and p-coumarate based on previous studies (Nielsen et al., 2001). The docked position for sinapate into the substrate-binding pocket of PviPRX9 was shifted away from the P-X-P-X motif due to the second methoxy substituent. However, this predicted repositioning of sinapate within the binding pocket of PviPRX9 did not decrease the activity of the enzyme toward this substrate as drastically as would be expected. Through our molecular docking and structural analysis, the unique presence of Gln-140 located at the entrance of the binding pocket was found to establish a hydrogen bond with the propenyl oxygen of the substrates ranging from 2.1 to 3.1 Å depending on the compound. Therefore, being consistent with the apparent binding affinities conferred by the kinetics parameter Km and the estimated binding affinity from the docking study, all the compounds examined for PviPRX9 exhibited similar binding affinity and binding pose. In the case of horseradish peroxidase C, Gln at the fourth position of the P-X-P-X motif is substituted to Phe; thus, it lacks a hydrogen bond with the substrate’s propenyl oxygen at the binding pocket entrance. Therefore, we hypothesized that 137P-P-P-Q140 and Leu-66 could be the major structural motifs responsible for PviPRX9’s broad specificity with small hierarchical preference. Work is in progress to test the hypothesis with site-directed mutagenesis followed by enzyme activity assays.
Catalytic Reaction Mechanism
The overall mechanism of PviPRX9 for the phenylpropanoid oxidation consists of four steps, starting from its resting state. Our crystal structure suggests that the catalytic center of PviPRX9 possessed a penta-coordinated Fe(III) ligated to the four nitrogens of the porphyrin and the proximal His, with a noncoordinating water molecule positioned distal to the porphyrin Fe(III) center.
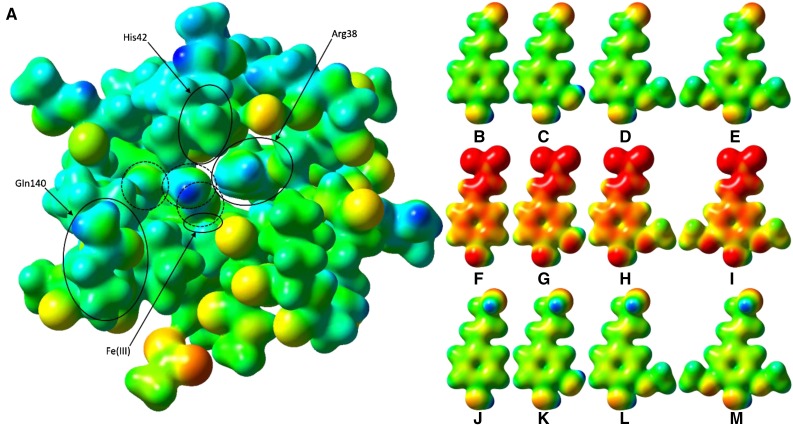
The electrostatic potential surface of the PviPRX9 active site optimized by semiempirical theory highlighted the points at which the substrates interact with the enzyme once bound (Fig. 10A). The oxygen-iron distance of the noncoordinating water molecule in the crystal structure was optimized to 2.06 Å, which resulted in being 2.14 Å away from the Hε atom of Arg-38. A second water molecule observed in the crystal structure appears to donate a hydrogen bond to the peptide carbonyl of Pro-137, orienting its second hydrogen atom down toward Nβ of the heme and accepting a hydrogen bond from a third water molecule. This third crystallographic water molecule accepted a hydrogen bond from the Hη2 atom of Arg-38. A previous study hypothesized that a water molecule bound by the P-X-P-X motif was the sole proton-shuttling mechanism (Henriksen et al., 1999). However, our optimized structure of PviPRX9 showed that the water molecule bound by Pro-137 is too far away (4.24 Å) and at an improper angle (its hydrogen atom is not in the plane of the imidazole ring) for deprotonation by His-42, necessitating a second water molecule positioned below His-42, which is conveniently generated by the heterolytic cleavage of H2O2. The highly conserved amino acid among CIIIPRXs, Arg-38, helps drive the reaction forward by electrostatic stabilization of the hydroxide and phenolate intermediates of the first water and the phenol, respectively. The phenol is quickly eliminated by the formation of the hydroxycinnamyl radical.
Figure 10.
Electrostatic potential surfaces of PviPRX9 and phenylpropanoids. A, Heme (hexcoordinate, high spin), Arg-31 (as methylguanidinium), Ser-35 (as ethanol), Arg-38 (as n-propylguanidinium), Phe-41 (as toluene), His-42 (as 5-methyl-1H-imidazole; mostly obscured by Leu-66), Ala-65 (as ACE) to Arg-73 (as NMA), Gly-170 (as formamide), Asp-135 (as ACE) to Phe-141 (as NMA), Ala-172 (as ACE) to Phe-177 (as NMA), His-167 (as 5-methyl-1H-imidazole), Asp-326 (as acetate ion), and six water molecules; Cys-174 was mutated to Gly to reduce computational cost. ACE and NMA are acetyl and N-methyl capping groups on the N and C termini, respectively, of each stretch of multiple residues. The three water molecules circled with dashed lines were, from right to left, the water involved in the first proton transfer, a noncatalytic water that occupies the nascent binding site of the hydroxycinnamyl substrate phenol functional group, and the water released upon H2O2 binding to the heme. The latter water molecule was regenerated by heterolytic cleavage of H2O2 and involved in catalytic proton transfer. B, p-Coumaryl aldehyde. C, Caffeyl aldehyde. D, Coniferyl aldehyde. E, Sinapyl aldehyde. F, p-Coumarate. G, Caffeate. H, Ferulate. I, Sinapate. J, p-Coumaryl alcohol. K, Caffeyl alcohol. L, Coniferoyl alcohol. M, Sinapyl alcohol. The active site and ligand electrostatic potentials, mapped on their corresponding self-consistent field densities, are shown on a potential scale of −2.7 × 10−1 hartrees (red) to +2.97 × 101 hartrees (blue) for the active site and −1 × 10−1 (red) to 3 × 101 (blue) for the ligands. This figure was generated using GaussView 3.09.
To analyze any complementarity between the molecular geometry and the electrostatic potential of the active site and its possible substrates, the electrostatic potential surfaces of 12 ligands (p-coumaryl aldehyde, p-coumarate, p-coumaryl alcohol, caffeyl aldehyde, caffeate, caffeyl alcohol, coniferyl aldehyde, ferulate, coniferyl alcohol, sinapyl aldehyde, sinapate, and sinapyl alcohol) were calculated. As shown in Figure 10, B to M, the substrates were similar within each class. When the three classes are all plotted on the same potential scale of −0.1 hartrees (red) to +0.3 hartrees, the aldehydes (Fig. 10, B–E) and alcohols (Fig. 10, J–M) were largely similar, whereas the hydroxycinnamates (Fig. 10, F–I) had their −1 formal charge partially delocalized throughout the molecule but concentrated predominantly in the carboxylate end. The gas-phase molecular geometry of the aldehydes and acids was completely planar, whereas that of the alcohols was not, averaging C6-C7-C8-O and C7-C8-O-H dihedral angles of 119.9° and −54.0°, respectively, between the four of them. The nonplanar geometry and rotatable bonds between the propenyl and 2° alcohol functional groups could allow alcohols an advantage in binding, as they would be better able to position themselves to hydrogen bond with the side chain of Gln-140 while still remaining coplanar to the Hη2 of Arg-38.
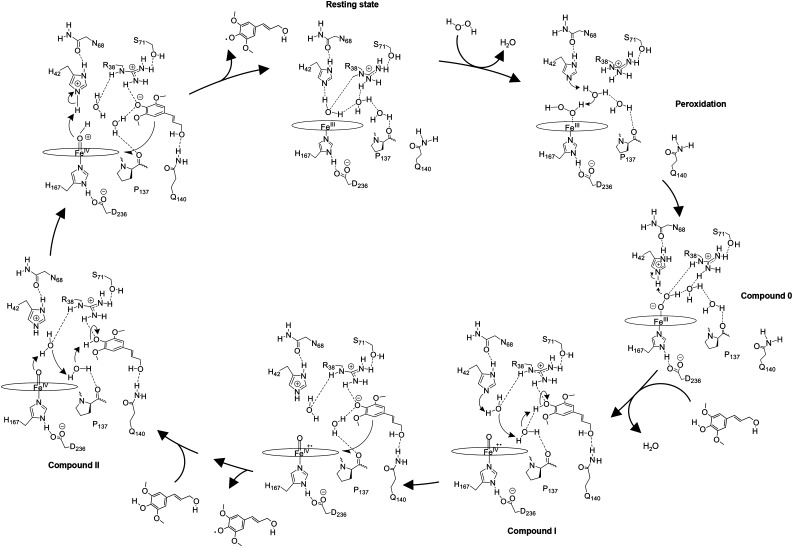
In the first step of the catalytic reaction mechanism, H2O2 displaces the water at the distal side by coordinating to the Fe(III) of the heme (Fig. 11). His-42 deprotonates the Fe-coordinating oxygen of H2O2 through a water-mediated proton shuttle to form Fe(III)-hydroperoxo-heme (compound 0; Baek and Van Wart, 1989; Miller et al., 1994; Vidossich et al., 2010). In the second step, protonation of the distal oxygen results in heterolytic cleavage of the hydroperoxy O-O bond, generating a first water and the formation of the Fe(IV)-oxo heme π-cation radical (compound I; Hiner et al., 2002). In the third step, the hydroxycinnamyl substrate binds to the enzyme with hydrogen bond formation between its phenolic oxygen and Hη2 of Arg-38, its phenolic hydrogen and the water molecule bound to Pro-137, and its propenyl oxygen and the amide of Gln-140 (Poulos and Kraut, 1980; Henriksen et al., 1999). His-42 abstracts a proton from the water generated from H2O2, which deprotonates the water molecule bound to Pro-137, which then deprotonates the phenol of the hydroxycinnamyl substrate. The deprotonated hydroxycinnamyl substrate then transfers an electron to the heme, resulting in the formation of a hydroxycinnamyl radical, which diffuses away, and Fe(IV)-oxo heme (compound II). In the last step, a second hydroxycinnamyl substrate binds to the enzyme, as in step 3. Upon binding, the hydroxycinnamyl substrate is deprotonated through a water-mediated proton shuttle, protonating the oxo ligand of the heme to form an Fe(IV)-hydroxo heme. Then, the deprotonated hydroxycinnamyl substrate transfers an electron to the heme and His-42 protonates the hydroxo ligand of the Fe(IV)-hydroxo heme, generating a second water molecule and the second hydroxycinnamyl radical that diffuses out from the pocket, regenerating the resting-state peroxidase.
Figure 11.
Proposed catalytic reaction mechanism of PviPRX9. In the first step, H2O2 displaces a water molecule from the active site in its resting state and reacts with His-42 to generate a hydroperoxide molecule via a water-mediated proton transfer. Next, the hydroperoxide molecule coordinates Fe(III) heme to generate Fe(III)-hydroperoxo heme (compound 0). In the second step, the hydroperoxo ligand is protonated by His-42 to generate first water and an Fe(IV)-oxo heme π-cation radical (compound I). Then, a monolignol binds to PviPRX9, displacing a water. His-42 deprotonates the monolignyl 4-hydroxy through a water-mediated proton-shuttling mechanism, generating the negatively charged monolignol that donates an electron to compound I to generate the first monolignol radical and Fe(IV)-oxo heme (compound II). In the third step, a second monolignol binds and is deprotonated through a water-mediated proton shuttle. Next, an electron is transferred to the heme from the deprotonated monolignol and His-42 protonates the Fe(IV)-hydroxo heme, generating the second water molecule along with the second monolignol radical, which diffuses from the pocket, returning PviPRX9 to its resting state.
Functional Diversity among Switchgrass CIIIPRXs
The observed heterogeneity among the noncatalytic residues within the substrate-binding pocket of the switchgrass CIIIPRX family reveals potential diversity in their substrate specificities. For example, a nonpolar-to-polar substitution in the P-X-P-X motif will produce significant changes in hydrophobicity, which could result in alterations in specificity favorable toward substrates with a more preferable hydrophobicity. In addition, observed substitutions in the P-X-P-X motif, such as Pro to Glu, Asp, Gln, Ala, or Ser, could result in changes to the hydrophobic and electrostatic character of the substrate-binding pocket. The observed heterogeneity also includes changes for the residues constituting the substrate entrance site, altering from hydrophobic to polar or charged and the length of the side chains. Additional polar or charged residues at the solvent-exposed surface of the binding pocket would likely allow for preferential affinity to those compounds with polar or charged tail groups, such as the phenylpropanoids examined in this study. The heterogeneity among switchgrass CIIIPRXs within their substrate-binding pocket also indicates diversity in the shape and size of the binding pocket. For example, Ser at the position of Gln-140 could lead to a specificity change toward compounds with longer linear carbon chains than the phenylpropanoids examined in this study. In addition, at the top of the pocket, several alterations are observed, such as Leu to Gly, which would result in easier access to the guanidinium group on Arg-38.
CONCLUSION
This study, to our knowledge for the first time, characterized the crystal structure, a substrate-specificity binding pocket, and the plausible catalytic reaction mechanisms of a switchgrass CIIIPRX, PviPRX9. The crystal structure, kinetics experiments, molecular docking, as well as temporal, spatial, and insect-induced expression of PviPRX9 revealed the function of PviPRX9 in lignification by binding and oxidizing phenylpropanoids. Significantly, our study suggests that PviPRX9 has the ability to oxidize most of the tested compounds with rather similar efficiencies, which reflects its role in the fortification of cell walls during normal growth and root development, and in response to insect feeding, where ectopic lignification often is observed. An enzyme such as PviPRX9 with broad substrate specificity could conceivably utilize any available monolignol substrate. The crystal structure of PviPRX9 reveals a substrate-binding pocket encapsulated by all three domains: (1) one loop at the distal Ca2+-binding domain positions with one hydrophobic residue (Leu-66) at the top of the substrate-binding pocket; (2) P-P-P-Q in between the proximal and distal Ca2+-binding domains that dictates a shape of the pocket and allows Gln-140 to form a hydrogen bond with the incoming substrate; and (3) Leu-175 and Asn-176 at the beginning portion of the β-domain that could interact with the functional groups of bound substrate. Substitution of those amino acids within the substrate-binding region can be postulated to lead to changes in substrate affinity and result in the diversification of CIIIPRX function after gene duplication.
MATERIALS AND METHODS
Culture
Escherichia coli strain Rosetta was transfected with a pET 30a vector containing the switchgrass (Panicum virgatum) gene PviPRX9 (Pavir.2NG638900). Luria-Bertani medium was inoculated with transformed E. coli cells and incubated at 37°C. Cultures were induced with isopropyl β-D-1-thiogalactopyranoside at 0.5 mm after they had reached an optical density at 600 nm of 0.6 to 0.7, followed by incubation at 25°C for 18 h. Cells were then harvested and collected by centrifugation at 5,000 rpm. Cell pellets were frozen for later purification.
Purification and Refolding
Refolding of the peroxidase was achieved by modifying the previously published protocols (Smith et al., 1990; Shigeto et al., 2014). Frozen cell pellets were thawed in lysis buffer consisting of 20 mm Tris-HCl and 1% (v/v) Triton X-100 at pH 9. Resuspended cells were sonicated five times and then centrifuged at 16,000 rpm for 30 min. The supernatant was discarded, and pellets were resuspended in a wash buffer consisting of 20 mm Tris-HCl, 5% (v/v) glycerol, and 300 mm NaCl at pH 9, followed by centrifugation at 15,000g for 30 min. This wash step was repeated. Next, the washed pellet was resuspended in buffer A (20 mm Tris-HCl, 20 mm Bis-Tris propane, 7.8 m urea, and 50 mm CaCl2 at pH 9) followed by centrifugation at 15,000g for 30 min. The resulting supernatant was applied to a nickel-nitrilotriacetic acid agarose (Ni-NTA) gravity flow column preequilibrated with buffer A. The Ni-NTA column was washed with 4 column volumes of buffer A. Next, the Ni-NTA column was washed with 4 column volumes of 20 mm Tris-HCl, 20 mm Bis-Tris propane, 7.8 m urea, and 300 mm NaCl at pH 6.3. The recombinant peroxidase was eluted from the Ni-NTA column with 20 mm Tris-HCl, 20 mm Bis-Tris, 10 mm citrate, 7.8 m urea, and 300 mm NaCl at pH 4.5 and collected for refolding.
The solution containing unfolded purified protein was first buffer exchanged to pH 9 with a buffer consisting of 20 mm Tris-HCl, 20 mm Bis-Tris, 10 mm citrate, 7.8 m urea, and 300 mm NaCl. Next, the peroxidase was buffer exchanged into 20 mm Tris-HCl, 20 mm Bis-Tris, 4 m urea, 10 mm CaCl2, 49.2 μm heme, 0.25 mm reduced glutathione (GSH), and 0.45 mm oxidized glutathione (GSSG), pH 9, at 4°C, followed by buffer exchange into 20 mm Tris-HCl, 20 mm Bis-Tris, 2 m urea, 100 mm CaCl2, 49.2 μm heme, 0.25 mm GSH, and 0.45 mm GSSG, pH 9. Next, urea was removed by dialyzing against 20 mm Tris-HCl, 20 mm Tris, 10 mm CaCl2, 49.2 μm heme, 0.25 mm GSH, and 0.45 mm GSSG at pH 8.5. The solubilized, refolded protein was finally buffer exchanged to 20 mm Tris and 5% glycerol, pH 8.5, for purification on a Resource Q column (GE Healthcare). Peroxidase was eluted from the Resource Q column by the linear gradient elution with buffer A (20 mM Tris-HCl containting 5% glycerol, pH 8.5) and buffer B (20 mM Tris-HCl, 5% glycerol, and 2 M NaCl, pH 8.5). 20 mm Tris containing 5% glycerol, pH 8.5, 20 mm Tris, 5% glycerol, and 2 m NaCl, pH 8.5. Fractions were collected and analyzed by SDS-PAGE. The fractions that absorbed at both 280 and 403 nm along with expected molecular weights by SDS-PAGE were combined and concentrated for further analysis.
Protein Crystallization and Structure Determination
PviPRX9 crystals were grown at 4°C with the hanging-drop vapor diffusion method. The mother liquor consisted of 2 m (NH4)2SO4 and was mixed 1:1 with purified and concentrated PviPRX9 at 5 mg mL−1 in 20 mm Tris (pH 8) and equilibrated against a mother liquor reservoir. Crystals were cryoprotected with a solution of 20% glycerol and 2 m (NH4)2SO4 before data collection at 100 K. PviPRX9 crystallized in the P212121 space group with the unit cell dimensions a = 53.854 Å, b = 59.432 Å, c = 96.874 Å, α = 90°, β = 90°, γ = 90°. Data were collected at the Advanced Light Source beamline 8.2.1 with a wavelength of 1 Å. The software package HKL2000 was used for diffraction data processing (Otwinowski and Minor, 1997). Phasing was done by molecular replacement with the search model peanut (Arachis hypogaea) PRX (PDB: 1SCH) and using PHENIX Phaser (Schuller et al., 1996; Adams et al., 2010). Refinement and model building were done using PHENIX and Coot (Adams et al., 2010; Emsley et al., 2010). The diffraction data statistics are listed in Table III. Crystallographic data and coordinates (PDB ID: 5TWT) were deposited in the PDB.
Table III. Crystallographic data of PviPRX9.
Numbers in parentheses refer to the highest resolution shell.
| Data Collection | PvPRX9 |
|---|---|
| PDB identifier | 5TWT |
| Space group | P212121 |
| Cell dimensions | |
| a, b, c (Å) | 53.854, 59.432, 96.874 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution (Å) | 37.55–1.30 |
| Wavelength (Å) | 1.00 |
| Asymmetric unit | 1 |
| Rsym | 0.094 (0.617) |
| I/σI | 6.2 (2.1) |
| CC1/2 | 0.991 (0.818) |
| Completeness | 97.14 (95.3) |
| Redundancy | 4.0 |
| Refinement | |
| Resolution (Å) | 37.55–1.30 |
| Unique reflections | 75,730 |
| Rwork/Rfree | 0.1346/0.1453 |
| Root-mean-square deviations | |
| Root-mean-square deviation bonds (Å) | 0.007 |
| Root-mean-square deviation angles (°) | 0.92 |
| Ramachandrans (%) | |
| Favored | 98.3 |
| Outliers | 0 |
| No. of atoms | 4,759 |
| Protein and ligand | 4,312 |
| Water | 447 |
Structure-Based Sequence Alignment
Structural sequence alignment of PviPRX9 was performed with homologous peroxidases that were obtained from the PDB. Peroxidase sequences used for structure-based sequence alignment had a sequence identity of 49% or higher. For sequence alignment and structure superposition root-mean-square deviations, the MatchMaker tool in the molecular viewing and analysis program UCSF Chimera was used (Pettersen et al., 2004). Default settings including the Needleman-Wunsch alignment algorithm and the Blosum-62 matrix were used to superimpose the structures. Sequence alignment from the structure superposition was set to iterate until converged. The sequence alignment (Fig. 3) was generated with the program BioEdit (Hall, 1999) with structural annotation added by hand.
Enzyme Kinetic Assays
The recombinant PviPRX9 enzyme was prepared at 1,000× the required reaction concentration in 50 mm MES containing 1 mm CaCl2 at pH 6. For specific activity comparisons, horseradish peroxidase type II (P-8250; Sigma-Aldrich) was rehydrated to 10 mg mL−1 in buffer used for the assay (50 mm MES and 1 mm CaCl2, pH 6). The concentration of peroxidase used for kinetic experiments was determined using the extinction coefficient ε of 100 mm−1 cm−1 at 403 nm (Nielsen et al., 2001). The enzyme reaction concentration for steady-state kinetics used was 100 pm. The specific activity assay enzyme concentration used after optimization was 1 nm, while the substrate OPD was 40 mm and H2O2 was 5 mm. For steady-state kinetics, reducing substrates were made fresh right before the experiments and ranged in concentration from 10 to 90 µm. For reactions with variable reducing substrate concentrations, H2O2 concentration was held constant at 500 µm. Higher concentrations of reducing substrate were not used to measure initial rates because of their high absorbance, as observed previously (Abelskov et al., 1997). Total reaction volume was 1 mL, and data were recorded for 60 s at 5-s intervals. Spectra were collected with an Agilent 8453 UV-Vis spectrophotometer (Agilent Technologies).
Analysis of Steady-State Kinetics Data Using Substrate Disappearance and SVD/ALS
Substrate disappearance kinetics showed linearity within the first 30 to 40 s, which were subsequently used to calculate initial rates using the slope found with a linear fitting function. Then, initial rates versus reducing substrate concentration were fitted using a Michaelis-Menten equation. Both linear and nonlinear fitting were performed in the Origin 2015 software package (OriginLab). Initial rates were measured as substrate disappearance using ε of 16 mm−1 cm−1 at 310 nm for ferulate (Nielsen et al., 2001), ε of 15.2 mm−1 cm−1 at 320 nm for caffeate (Rasmussen et al., 1995), ε of 19.2 mm−1 cm−1 at 308 nm for p-coumarate (Nielsen et al., 2001), ε of 21.3 mm−1 cm−1 at 308 nm for sinapate (Nielsen et al., 2001), ε of 13.8 mm−1 cm−1 for coniferyl alcohol at 264 nm (Nielsen et al., 2001), and ε of 20.6 mm−1 cm−1 at 341 nm for coniferyl aldehyde (Barceló et al., 2001). This approach was initially used in order to estimate both the affinity (Km) and turnover (kcat) of our enzyme preparation; however, a nonsaturating behavior was observed in most of the substrates. This is due to the formation of a spectrally active metabolite in the same region of the absorbance spectrum, as reported previously by several authors (Rasmussen et al., 1993, 1995; Quiroga et al., 2001). In principle, a better kinetic analysis can be performed using stopped-flow spectrophotometry and considering the initial few seconds of substrate disappearance (Abelskov et al., 1997). Rasmussen and coworkers (1993) estimated the molecular absorptivity of the products using an excess of H2O2 until substrate exhaustion, a strategy later applied by Barceló and Pomar (2001). In both cases, the chemical instability of the radical metabolites prevents a reliable optical characterization.
An alternative approach is to rely on data deconvolution using a combined approach of SVD coupled with ALS analysis. This multivariate strategy (called SVD/ALS here) was adopted to deconvolute the time-dependent evolution of substrate disappearance/product formation during reaction. In matrix language, the number of transient species (components) in the observable Y, consisting of i spectra collected at i successive delay times and j wavelengths, is estimated analyzing the chemical rank of the matrix using SVD (Maeder and Neuhold, 2007). To overcome spectral overlapping, advantage can be taken by using evolving factor analysis (EFA), which performs successive SVD on gradually increasing submatrices in the time direction, adding a row at a time from the top of the matrix to the bottom (forward EFA) and vice versa (backward EFA; Maeder, 1987; Maeder and Neuhold, 2007). The emergence and evolution of the singular values can thus be followed individually with increasing time and reverse sequence (i.e. their disappearance can be observed with increasing time). Under the assumption that the evolving system studied is sequential, an estimation of the window concentration profiles can be provided combining the forward and backward traces. The final estimate was obtained through ALS, which iteratively optimizes the profile of the matrix C (concentration) using an initial guess of concentrations from EFA and then explicitly computes the shapes of the profiles using a nonnegativity concentration constraint (Maeder and Neuhold, 2007). Spectra were collected and analyzed using Matlab (The Mathworks). Before multivariate analysis, Savitzky-Golay smoothing and baseline correction were performed on all the recorded spectra. SVD, EFA, and ALS were carried out with Matlab programs written by the authors. Two components with their corresponding kinetic and spectral eigenvectors were considered as significantly above baseline.
Molecular Docking
p-Coumarate, caffeate, ferulate, sinapate, and their respective aldehydes and alcohols were docked into the PM7-optimized structure using AutoDock Vina (Trott and Olson, 2010). The ligands were generated from the QM-optimized geometries, and their 3-phenylprop-2-enyl dihedral angles were fixed for docking. Both PviPRX9 and the 12 ligands were prepped for docking using AutoDock tools (Morris et al., 2009). The 3-phenylprop-2-enyl dihedral angles of the phenylpropanoids were fixed during the docking procedure.
Expression Profiling of PviPRX9 and Correlation with Monolignol Biosynthesis Genes
Expression patterns for PviPRX9 and four peroxidases sharing expression profiles (correlation of expression ≥ 0.75) and monolignol biosynthesis genes were analyzed in RNA-Seq and microarray data sets in order to create a tissue-specific and developmental expression profile for PviPRX9 and similarly expressed peroxidase-encoding genes. RNA-Seq data sets consisted of a 2-year survey of rhizome development (Palmer et al., unpublished data), a single-year survey of flag leaf development (Palmer et al., 2015), and an investigation of transcriptional responses to aphid infestation (Donze-Reiner et al., unpublished data). Microarray data sets were compiled in PviGEA (http://switchgrassgenomics.noble.org/), where PviPRX9 is annotated as AP13CTG06599 (Zhang et al., 2013). Expression values were converted to z-scores prior to heat map generation. Expression values for genes involved in monolignol biosynthesis also were obtained from the same data sets in order to analyze the correlation of expression between PviPRX9 and genes encoding enzymes associated with monolignol biosynthesis.
Maximum Likelihood-Based Phylogenetic Analysis and Classification of Full-Length CIIIPRXs from Switchgrass
CIIIPRX-encoding loci (333) were identified in the switchgrass genome (version 3.1; https://phytozome.jgi.doe.gov) based on annotation with the PFAM peroxidase domain (PF00141). Of the 333 peroxidase loci, 24 encoded putative ascorbate peroxidases and were removed from further analyses. Of the remaining 309 peroxidase-encoding loci, nine were incomplete genes and did not contain either a start or a stop codon and also were removed from subsequent analyses. Amino acid sequences for the remaining 300 full-length PviPRXs were retrieved from the latest version of the switchgrass genome and aligned to representative sequences from nine major peroxidase evolutionary groups identified previously in rice (Oryza sativa) using MUSCLE as implemented in MEGA 5 (Edgar, 2004; Passardi et al., 2004; Tamura et al., 2011). Bootstrap consensus maximum likelihood trees were constructed and compiled as described previously (Scully et al., 2016). Switchgrass peroxidases were assigned to evolutionary groups based on phylogenetic similarity to the rice peroxidases (Supplemental Table S1). The PFAM peroxidase domain was used to query several other grass genomes available through the Joint Genome Institute phytozome database.
Optimization and Electrostatic Potential Surface Generation of the PviPRX9 Resting State
The structure of PviPRX9 was prepared for modeling of its resting state (sextet spin state) by replacing the crystallographic water coordinating heme with a water molecule, reducing the structure and crystallographic solvent, and optimizing the complete hydrogen-bonding network using the PDB Prep Wizard in Schrödinger Maestro (Sastry et al., 2013; Schrödinger, 2016). The hydrogen atoms of the reduced and hydrogen bonding-optimized model were then optimized in CPU+GPU MOPAC2016 (Maia et al., 2012; Stewart, 2016) at the PM7 level of theory (Stewart, 2013) along with unconstrained optimization of three active site waters, the modeled crystallographic water, the water previously identified as catalytic, and the water molecule that docking suggests occupies the site into which the phenol functional group of the hydroxycinnamyl substrates is bound. A single-point calculation at the CAM-B3LYP level of theory (Yanai et al., 2004) using a double-ζ correlation-consistent basis set and a pseudopotential for iron (Dolg, 2005; K.A. Peterson, personal communication) and 3-21G basis sets (Gordon et al., 1982) for hydrogen, carbon, nitrogen, and oxygen was performed in Gaussian 09 (Frisch et al., 2009). Six points bohr−1 total electron density and electrostatic potential grids were then generated from the single-point self-consistent field density by the Gaussian 09 cubegen utility and mapped in GaussView 3.09 (Frisch, 2004) as the electrostatic potential on the electron density at an iso value of 0.02 electrons bohr−3.
Quantum Mechanics Optimization and Electrostatic Potential Surface Generation of Phenylpropanoids
p-Coumarate, caffeate, ferulate, sinapate, and their respective aldehydes and alcohols were optimized in Gaussian 09 at the CAM-B3LYP level of theory using double-ζ correlation-consistent basis sets (cc-pVDZ) with augmented functions (aug-cc-pVDZ) on oxygen (Peterson and Dunning, 2002). Each molecule was optimized in its s-trans-form with the 4-phenol group set to the syn orientation. The optimal dihedral angle for the propenol group of each monolignol was found by performing a relaxed potential energy scan at the same level of theory. The aldehydes and acids were optimized in the Cs point group, and the alcohols were optimized in the C1 point group. Following confirmation, via frequency calculation at the CAM-B3LYP/(aug)-cc-pVDZ level of theory, that each optimized molecule was in its lowest energy geometry, a single-point calculation at the CAM-B3LYP level of theory with triple-ζ basis sets (cc-pVTZ for hydrogen and carbon and aug-cc-pVTZ for oxygen) was performed on each. Twelve points bohr−1 total electron density and electrostatic potential grids were generated from the single-point triple-ζ self-consistent field densities by the Gaussian 09 cubegen utility and mapped in GaussView 3.09 as the electrostatic potential on the electron density at an iso value of 0.02 electrons bohr−3.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Example of SVD/ALS analysis of time-dependent spectra obtained through the reaction of PviPRX9 with ferulic acid (50 μm).
Supplemental Table S1. Family assignments of peroxidases identified in the switchgrass genome (version 3.1).
Supplemental Table S2. Frequency of amino acids at each position in Figure 9.
Supplementary Material
Glossary
- PDB
Protein Data Bank
- SVD
singular value decomposition
- ALS
alternating least square
- OPD
o-Phenylenediamine
- RNA-Seq
RNA sequencing
- PviGEA
switchgrass gene expression atlas database
- Ni-NTA
nickel-nitrilotriacetic acid agarose
- GSH
reduced glutathione
- GSSG
oxidized glutathione
- EFA
evolving factor analysis
Footnotes
This work was supported by the National Science Foundation (grant nos. DBI 0959778 and CHE 118359), the National Institutes of Health (grant no. 1R01GM11125401), and Murdock Charitable Trust (to C.K.), and the U.S. Department of Agriculture (National Institute of Food and Agriculture grant no. 2011–67009–30096 to G.S. and Agricultural Research Service CRIS project no. 5440–21000–030–00D).
References
- Abelskov AK, Smith AT, Rasmussen CB, Dunford HB, Welinder KG (1997) pH dependence and structural interpretation of the reactions of Coprinus cinereus peroxidase with hydrogen peroxide, ferulic acid, and 2,2′-azinobis(3-ethylbenzthiazoline-6-sulfonic acid). Biochemistry 36: 9453–9463 [DOI] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almagro L, Gómez Ros LV, Belchi-Navarro S, Bru R, Ros Barceló A, Pedreño MA (2009) Class III peroxidases in plant defence reactions. J Exp Bot 60: 377–390 [DOI] [PubMed] [Google Scholar]
- Baek HK, Van Wart HE (1989) Elementary steps in the formation of horseradish peroxidase compound I: direct observation of compound 0, a new intermediate with a hyperporphyrin spectrum. Biochemistry 28: 5714–5719 [DOI] [PubMed] [Google Scholar]
- Bakovic M, Dunford HB (1993) Kinetics of the oxidation of p-coumaric acid by prostaglandin H synthase and hydrogen peroxide. Biochemistry 32: 833–840 [DOI] [PubMed] [Google Scholar]
- Barceló AR, Pomar F (2001) Oxidation of cinnamyl alcohols and aldehydes by a basic peroxidase from lignifying Zinnia elegans hypocotyls. Phytochemistry 57: 1105–1113 [DOI] [PubMed] [Google Scholar]
- Barros J, Serk H, Granlund I, Pesquet E (2015) The cell biology of lignification in higher plants. Ann Bot (Lond) 115: 1053–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund GI, Carlsson GH, Smith AT, Szöke H, Henriksen A, Hajdu J (2002) The catalytic pathway of horseradish peroxidase at high resolution. Nature 417: 463–468 [DOI] [PubMed] [Google Scholar]
- Dien BS, Sarath G, Pedersen JF, Sattler SE, Chen H, Funnell-Harris DL, Nichols NN, Cotta MA (2009) Improved sugar conversion and ethanol yield for forage sorghum (Sorghum bicolor L. Moench) lines with reduced lignin contents. BioEnergy Res 2: 153–164 [Google Scholar]
- Dolg M. (2005) Improved relativistic energy-consistent pseudopotentials for 3d-transition metals. Theor Chem Acc 114: 297–304 [Google Scholar]
- Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J (2006) CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res 34: W116–W118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawal N, Li Q, Savelli B, Brette M, Passaia G, Fabre M, Mathé C, Dunand C (2013) PeroxiBase: a database for large-scale evolutionary analysis of peroxidases. Nucleic Acids Res 41: D441–D444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Pérez F, Vivar T, Pomar F, Pedreño MA, Novo-Uzal E (2015) Peroxidase 4 is involved in syringyl lignin formation in Arabidopsis thaliana. J Plant Physiol 175: 86–94 [DOI] [PubMed] [Google Scholar]
- Francoz E, Ranocha P, Nguyen-Kim H, Jamet E, Burlat V, Dunand C (2015) Roles of cell wall peroxidases in plant development. Phytochemistry 112: 15–21 [DOI] [PubMed] [Google Scholar]
- Freudenberg K. (1965) Lignin: its constitution and formation from p-hydroxycinnamyl alcohols. Science 148: 595–600 [DOI] [PubMed] [Google Scholar]
- Frisch M (2004) GaussView, version 3. Gaussian, Wallingford, CT [Google Scholar]
- Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, et al (2009) Gaussian 09, revision C.01. Gaussian, Wallingford, CT [Google Scholar]
- Gabaldón C, López-Serrano M, Pomar F, Merino F, Cuello J, Pedreño MA, Barceló AR (2006) Characterization of the last step of lignin biosynthesis in Zinnia elegans suspension cell cultures. FEBS Lett 580: 4311–4316 [DOI] [PubMed] [Google Scholar]
- Gordon MS, Binkley JS, Pople JA, Pietro WJ, Hehre WJ (1982) Self-consistent molecular-orbital methods. 22. Small split-valence basis sets for second-row elements. J Am Chem Soc 104: 2797–2803 [Google Scholar]
- Hall TA. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41: 95–98 [Google Scholar]
- Henriksen A, Mirza O, Indiani C, Teilum K, Smulevich G, Welinder KG, Gajhede M (2001) Structure of soybean seed coat peroxidase: a plant peroxidase with unusual stability and haem-apoprotein interactions. Protein Sci 10: 108–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksen A, Smith AT, Gajhede M (1999) The structures of the horseradish peroxidase C-ferulic acid complex and the ternary complex with cyanide suggest how peroxidases oxidize small phenolic substrates. J Biol Chem 274: 35005–35011 [DOI] [PubMed] [Google Scholar]
- Henriksen A, Welinder KG, Gajhede M (1998) Structure of barley grain peroxidase refined at 1.9-Å resolution: a plant peroxidase reversibly inactivated at neutral pH. J Biol Chem 273: 2241–2248 [DOI] [PubMed] [Google Scholar]
- Hiner AN, Raven EL, Thorneley RN, García-Cánovas F, Rodríguez-López JN (2002) Mechanisms of compound I formation in heme peroxidases. J Inorg Biochem 91: 27–34 [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372: 774–797 [DOI] [PubMed] [Google Scholar]
- Lee Y, Rubio MC, Alassimone J, Geldner N (2013) A mechanism for localized lignin deposition in the endodermis. Cell 153: 402–412 [DOI] [PubMed] [Google Scholar]
- Li Y, Kajita S, Kawai S, Katayama Y, Morohoshi N (2003) Down-regulation of an anionic peroxidase in transgenic aspen and its effect on lignin characteristics. J Plant Res 116: 175–182 [DOI] [PubMed] [Google Scholar]
- Maeder M. (1987) Evolving factor analysis for the resolution of overlapping chromatographic peaks. Anal Chem 59: 527–530 [Google Scholar]
- Maeder M, Neuhold YM (2007) Practical Data Analysis in Chemistry. Elsevier Science [Google Scholar]
- Maia JD, Urquiza Carvalho GA, Mangueira CP Jr, Santana SR, Cabral LA, Rocha GB (2012) GPU linear algebra libraries and GPGPU programming for accelerating MOPAC semiempirical quantum chemistry calculations. J Chem Theory Comput 8: 3072–3081 [DOI] [PubMed] [Google Scholar]
- Mansfield SD, Kim H, Lu F, Ralph J (2012) Whole plant cell wall characterization using solution-state 2D NMR. Nat Protoc 7: 1579–1589 [DOI] [PubMed] [Google Scholar]
- Marjamaa K, Kukkola EM, Fagerstedt KV (2009) The role of xylem class III peroxidases in lignification. J Exp Bot 60: 367–376 [DOI] [PubMed] [Google Scholar]
- Miller MA, Shaw A, Kraut J (1994) 2.2 Å structure of oxy-peroxidase as a model for the transient enzyme: peroxide complex. Nat Struct Biol 1: 524–531 [DOI] [PubMed] [Google Scholar]
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30: 2785–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen KL, Indiani C, Henriksen A, Feis A, Becucci M, Gajhede M, Smulevich G, Welinder KG (2001) Differential activity and structure of highly similar peroxidases: spectroscopic, crystallographic, and enzymatic analyses of lignifying Arabidopsis thaliana peroxidase A2 and horseradish peroxidase A2. Biochemistry 40: 11013–11021 [DOI] [PubMed] [Google Scholar]
- Ostergaard L, Teilum K, Mirza O, Mattsson O, Petersen M, Welinder KG, Mundy J, Gajhede M, Henriksen A (2000) Arabidopsis ATP A2 peroxidase: expression and high-resolution structure of a plant peroxidase with implications for lignification. Plant Mol Biol 44: 231–243 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Palmer NA, Donze-Reiner T, Horvath D, Heng-Moss T, Waters B, Tobias C, Sarath G (2015) Switchgrass (Panicum virgatum L) flag leaf transcriptomes reveal molecular signatures of leaf development, senescence, and mineral dynamics. Funct Integr Genomics 15: 1–16 [DOI] [PubMed] [Google Scholar]
- Passardi F, Longet D, Penel C, Dunand C (2004) The class III peroxidase multigenic family in rice and its evolution in land plants. Phytochemistry 65: 1879–1893 [DOI] [PubMed] [Google Scholar]
- Perlack R, Stokes B (2011) Billion-Ton Update: Biomass Supply for a Bioenergy and Bioproducts Industry. Oak Ridge National Laboratory, Oak Ridge, TN [Google Scholar]
- Peterson KA, Dunning TH (2002) Accurate correlation consistent basis sets for molecular core-valence correlation effects: the second row atoms Al-Ar, and the first row atoms B-Ne revisited. J Chem Phys 117: 10548–10560 [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Poulos TL, Kraut J (1980) The stereochemistry of peroxidase catalysis. J Biol Chem 255: 8199–8205 [PubMed] [Google Scholar]
- Quiroga M, de Forchetti SM, Taleisnik E, Tigier HA (2001) Tomato root peroxidase isoenzymes: kinetic studies of the coniferyl alcohol peroxidase activity, immunological properties and role in response to salt stress. J Plant Physiol 158: 1007–1013 [Google Scholar]
- Rasmussen CB, Bakovic M, Welinder KG, Dunford HB (1993) Unique reaction of a barley peroxidase with hydrogen peroxide. FEBS Lett 321: 102–105 [DOI] [PubMed] [Google Scholar]
- Rasmussen CB, Dunford HB, Welinder KG (1995) Rate enhancement of compound I formation of barley peroxidase by ferulic acid, caffeic acid, and coniferyl alcohol. Biochemistry 34: 4022–4029 [DOI] [PubMed] [Google Scholar]
- Saathoff AJ, Donze T, Palmer NA, Bradshaw J, Heng-Moss T, Twigg P, Tobias CM, Lagrimini M, Sarath G (2013) Towards uncovering the roles of switchgrass peroxidases in plant processes. Front Plant Sci 4: 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sastry GM, Adzhigirey M, Day T, Annabhimoju R, Sherman W (2013) Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des 27: 221–234 [DOI] [PubMed] [Google Scholar]
- Schrödinger L (2016) Schrödinger Suite 2016-2 Protein Preparation Wizard: Maestro, version 10.2. Schrödinger, New York [Google Scholar]
- Schuller DJ, Ban N, Huystee RB, McPherson A, Poulos TL (1996) The crystal structure of peanut peroxidase. Structure 4: 311–321 [DOI] [PubMed] [Google Scholar]
- Scully E, Donze-Reiner T, Wang H, Eickhoff T, Baxendale F, Twigg P, Kovacs F, Heng-Moss T, Sattler S, Sarath G (2016) A clade of syntenic peroxidases are expressed in C4 bioenergy grasses in response to feeding by greenbugs (Schizaphis graminum). Funct Plant Biol 43: 1134–1148 [DOI] [PubMed] [Google Scholar]
- Shigeto J, Nagano M, Fujita K, Tsutsumi Y (2014) Catalytic profile of Arabidopsis peroxidases, AtPrx-2, 25 and 71, contributing to stem lignification. PLoS ONE 9: e105332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AT, Santama N, Dacey S, Edwards M, Bray RC, Thorneley RN, Burke JF (1990) Expression of a synthetic gene for horseradish peroxidase C in Escherichia coli and folding and activation of the recombinant enzyme with Ca2+ and heme. J Biol Chem 265: 13335–13343 [PubMed] [Google Scholar]
- Stewart J (2016) MOPAC2016. Stewart Computational Chemistry, Colorado Springs, CO [Google Scholar]
- Stewart JJ. (2013) Optimization of parameters for semiempirical methods. VI. More modifications to the NDDO approximations and re-optimization of parameters. J Mol Model 19: 1–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobias CM, Sarath G, Twigg P, Lindquist E, Pangilinan J, Penning BW, Barry K, McCann MC, Carpita NC, Lazo GR (2008) Comparative genomics in switchgrass using 61,585 high-quality expressed sequence tags. Plant Genome 1: 111–124 [Google Scholar]
- Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31: 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R, Demedts B, Morreel K, Ralph J, Boerjan W (2010) Lignin biosynthesis and structure. Plant Physiol 153: 895–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veitch NC. (2004a) Horseradish peroxidase: a modern view of a classic enzyme. Phytochemistry 65: 249–259 [DOI] [PubMed] [Google Scholar]
- Veitch NC. (2004b) Structural determinants of plant peroxidase function. Phytochem Rev 3: 3–18 [Google Scholar]
- Vidossich P, Fiorin G, Alfonso-Prieto M, Derat E, Shaik S, Rovira C (2010) On the role of water in peroxidase catalysis: a theoretical investigation of HRP compound I formation. J Phys Chem B 114: 5161–5169 [DOI] [PubMed] [Google Scholar]
- Vogel KP, Mitchell KB (2008) Heterosis in switchgrass: biomass yield in swards. Crop Sci 48: 2159–2164 [Google Scholar]
- Voxeur A, Wang Y, Sibout R (2015) Lignification: different mechanisms for a versatile polymer. Curr Opin Plant Biol 23: 83–90 [DOI] [PubMed] [Google Scholar]
- Wagner A, Ralph J, Akiyama T, Flint H, Phillips L, Torr K, Nanayakkara B, Te Kiri L (2007) Exploring lignification in conifers by silencing hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyltransferase in Pinus radiata. Proc Natl Acad Sci USA 104: 11856–11861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Bouchabke-Coussa O, Lebris P, Antelme S, Soulhat C, Gineau E, Dalmais M, Bendahmane A, Morin H, Mouille G, et al. (2015) LACCASE5 is required for lignification of the Brachypodium distachyon Culm. Plant Physiol 168: 192–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe L, de Moura PR, Bleicher L, Nascimento AS, Zamorano LS, Calvete JJ, Sanz L, Pérez A, Bursakov S, Roig MG, et al. (2010) Crystal structure and statistical coupling analysis of highly glycosylated peroxidase from royal palm tree (Roystonea regia). J Struct Biol 169: 226–242 [DOI] [PubMed] [Google Scholar]
- Yanai T, Tew DP, Handy NC (2004) A new hybrid exchange-correlation functional using the Coulomb-attenuating method (CAM-B3LYP). Chem Phys Lett 393: 51–57 [Google Scholar]
- Zhang JY, Lee YC, Torres-Jerez I, Wang M, Yin Y, Chou WC, He J, Shen H, Srivastava AC, Pennacchio C, et al. (2013) Development of an integrated transcript sequence database and a gene expression atlas for gene discovery and analysis in switchgrass (Panicum virgatum L.). Plant J 74: 160–173 [DOI] [PubMed] [Google Scholar]
- Zhao Q, Nakashima J, Chen F, Yin Y, Fu C, Yun J, Shao H, Wang X, Wang ZY, Dixon RA (2013) Laccase is necessary and nonredundant with peroxidase for lignin polymerization during vascular development in Arabidopsis. Plant Cell 25: 3976–3987 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.