A new DII reporter indicates a low level of auxin signaling during telophase and G1 in maize.
Abstract
A sensitive and dynamically responsive auxin signaling reporter based on the DII domain of the INDOLE-3-ACETIC ACID28 (IAA28, DII) protein from Arabidopsis (Arabidopsis thaliana) was modified for use in maize (Zea mays). The DII domain was fused to a yellow fluorescent protein and a nuclear localization sequence to simplify quantitative nuclear fluorescence signal. DII degradation dynamics provide an estimate of input signal into the auxin signaling pathway that is influenced by both auxin accumulation and F-box coreceptor concentration. In maize, the DII-based marker responded rapidly and in a dose-dependent manner to exogenous auxin via proteasome-mediated degradation. Low levels of DII-specific fluorescence corresponding to high endogenous auxin signaling occurred near vasculature tissue and the outer layer and glume primordia of spikelet pair meristems and floral meristems, respectively. In addition, high DII levels were observed in cells during telophase and early G1, suggesting that low auxin signaling at these stages may be important for cell cycle progression.
Auxins are tryptophan or independently derived compounds that regulate many aspects of plant development (McSteen, 2010; Salehin et al., 2015). In maize (Zea mays), auxin contributes to inflorescence development (Galli et al., 2015), tassel branching (Gallavotti et al., 2008a), and root structure (Jansen et al., 2012; Zhang et al., 2014). The currently available auxin reporter in maize uses the synthetic promoter element DR5 to drive the expression of an endoplasmic reticulum retained Red Fluorescent Protein (DR5rev::mRFP-er, DR5) in response to increased auxin. Although it is an informative tool for determining relative auxin response in vivo (Gallavotti et al., 2008b), DR5 responds independently to brassinosteroids and therefore may not reliably and specifically report auxin signaling alone (Nakamura et al., 2003). In addition, DR5 is transcriptionally activated upon auxin perception and specific signal transduction, and therefore DR5 levels reflect the transcriptional output of the auxin signaling pathway. On the other hand, DII levels directly depend on inputs into the auxin signaling pathway such as auxin concentration and F-box coreceptor accumulation in each cell (Brunoud et al., 2012). Therefore, we modified an Arabidopsis DII-based auxin responsive reporter (Brunoud et al., 2012) for use in maize.
In Arabidopsis (Arabidopsis thaliana), the TRANSPORT INHIBITOR RESPONSE 1/AUXIN SIGNALING F-BOX proteins (TIR1/AFBs) bind directly to auxins and mediate degradation of the AUXIN/INDOLE-3-ACETIC ACID (AUX/IAA) repressors (Dharmasiri et al., 2005; Gray et al., 2001; Kepinski and Leyser, 2005; for review, see Salehin et al., 2015). Different AUX/IAA proteins share a highly conserved region called domain II (DII), or degron, which is the minimum region required for its auxin-mediated degradation (Moss et al., 2015; Tan et al., 2007). The translational fusion of the DII region of the IAA responsive protein AtIAA28 with the yellow fluorescent protein VENUS and a nuclear localization signal (NLS) has been used as a sensitive and quantitative auxin responsive reporter in Arabidopsis (Brunoud et al., 2012). DII degradation dynamics are influenced both by auxin accumulation and TIR1/AFB concentration. Differences in DII signal reflect differences in input signal into the auxin signaling pathway. When the F-box coreceptors are uniformly distributed, variations in DII fluorescence intensity can be indicative of differences in auxin concentration (Vernoux et al., 2011).
We generated and characterized maize transgenic plants expressing DII-VENUS-NLS (herein called DII) as a marker for auxin signaling. A mutated version, mDII-VENUS-NLS (mDII), which does not bind TIR1 upon auxin perception (Brunoud et al., 2012; Tan et al., 2007), was used as a control. The DII marker responded in a dose- and time-dependent manner to auxin via proteasome-mediated degradation. Furthermore, the DII signal was reduced in the leaf vasculature and in glume primordia of floral meristems, but a strong DII signal was observed in a “collar” of cells surrounding the floral primordia, potentially marking a transition site in lateral organ outgrowth. In addition, DII effectively reported differences in auxin signaling during the cell cycle. DII signal was higher during telophase and early G1 and lower during late interphase and the transition to mitosis. These observations support a role for auxin signaling in cell cycle transitions and suggest that DII will be useful for testing mechanisms of auxin signaling in maize.
RESULTS
Transgenic maize plants expressing the DII reporter under the transcriptional control of the maize ubiquitin promoter (DII-VENUS-NLS, DII) were generated as well as plants expressing a mutated version of DII (mDII-VENUS-NLS, mDII) that is incapable of auxin-dependent ubiquitin-mediated degradation (Tan et al., 2007). Both transgenic lines grew as well as nontransformed plants and were morphologically similar (Supplemental Fig. S3).
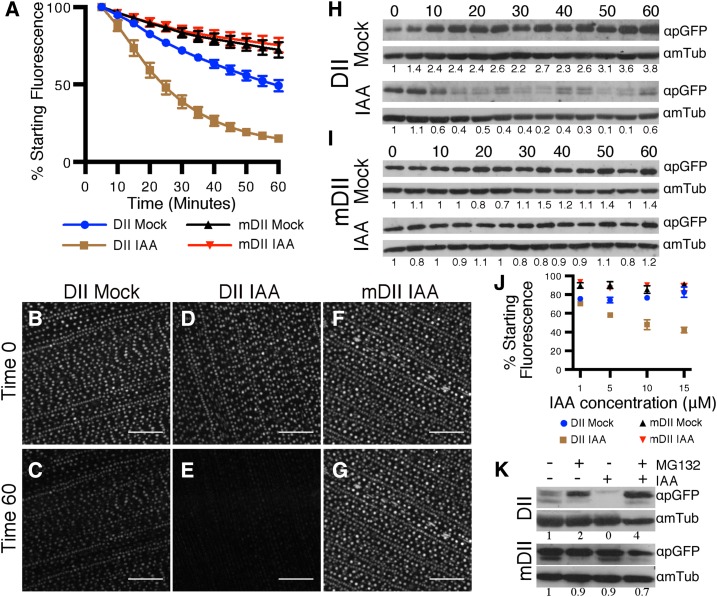
Time-lapse imaging demonstrated that DII samples treated with auxin showed a reduction in DII-specific nuclear fluorescence. A 0.5-cm2 piece from the base of the developing leaf blade (Supplemental Fig. S1A) was excised, and the nuclear fluorescence intensity was analyzed by confocal microscopy in mock-treated (0.05% dimethyl sulfoxide [DMSO]) and auxin-treated (10 μm IAA) samples. DII fluorescence intensity values of >4,000 adaxial epidermal nuclei per treatment were measured from images captured at 5-min intervals from a 1-h time-lapse recording. Nuclear fluorescence intensity values were displayed as percentages compared to the intensity at time zero. Normalized nuclear fluorescence intensity values (Fig. 1A) correspond to the mean value of five biological replicates. Under these conditions, we observed that more than 50% (52.1 ± 4.4%) of the fluorescence intensity measured for DII signal was lost in the first 25 min of IAA treatment, whereas mock treatment resulted in an approximately 20% decrease (23% ± 1.8%; Fig. 1A). At the end of the time-lapse, we observed a reduction of approximately 85% (84.9 ± 1.9%) DII fluorescence intensity upon IAA treatment. In contrast, after 60 min of mock treatment, we observed a reduction of approximately 50% (50.7 ± 15.1%; Fig. 1, A−E). The relative fluorescence loss measured for the control version mDII at the end of the 60-min time-lapse was approximately 25% for both mock- and IAA-treated leaves (27.6 ± 6.3% and 24.5 ± 6.3%, respectively; Fig. 1, A, F, and G). Loss of fluorescence in mDII and mock-treated DII plants was due to a combination of nuclear movement and photobleaching (Supplemental Fig. S2). Photobleaching was more severe in DII-treated samples, because the exposure time during time-lapse imaging for DII was longer (approximately 400 ms) than for mDII (approximately 150 ms).
Figure 1.
DII degradation dynamics. A, Relative nuclear fluorescence intensities (compared to time 0) of DII and mDII leaf samples treated with 10 μm IAA or 0.05% DMSO measured from confocal images. Data points represent mean values ± ses from five biological replicas. B to G, Maximum projection of Z-stacks of DII leaf image at time 0 (B) and after 1-h mock treatment (C; 0.05% DMSO), DII leaf image at time 0 (D) and after 1-h treatment with 10 μm IAA (E), mDII leaf image at time 0 (F) and after 1-h treatment with 10 μm IAA (G). H and I, Immunoblot of DII (H) and mDII (I) leaf samples after mock (0.05% DMSO, top) and IAA (10 μM, bottom) treatment sampled every 5 min for 1 h using anti-GFP antibody. Loading control was performed by stripping and reprobing with antitubulin antibody. Quantification of band intensity relative to tubulin band intensity is indicated below each band. J, Loss of DII and mDII nuclear fluorescence intensities after 20-min treatment using different IAA concentrations. Each intensity value was compared to fluorescence intensity at time zero to calculate mean ± se for three biological replicas. K, Immunoblot of protein extracts from DII (top) and mDII (bottom) primary roots after treatment with different combinations of MG132 (100 μM) and IAA (10 μM), as indicated. Loading control was performed by stripping and reprobing with antitubulin antibody.
Immunoblotting was used to test further whether the DII construct in maize responded to auxin. DII protein levels were measured during auxin treatment using a polyclonal anti-GFP antibody (Invitrogen). The developing leaf blade was cut into 0.5-cm2 pieces and placed on 10 mm MES (pH 5.7) solidified with 0.8% agar and supplemented with auxin (10 μm IAA) or mock treatment (0.05% DMSO; Supplemental Fig. S1E). Samples were collected every 5 min for 1 h. Mock-treated samples usually showed increasing DII protein accumulation toward the end of the 1-h time-lapse, but the relative change was somewhat variable among replicas (Fig. 1H). In contrast, IAA-treated pieces showed a robust but fluctuating reduction in signal after 10 to 15 min of treatment that remained low thereafter (Fig. 1H). For mDII samples, similar protein levels were observed for IAA- and mock-treated pieces throughout the experiment (Fig. 1I). These trends were similar across three independent experiments (Supplemental Figs. S4 and 5) and confirmed that DII protein was likely degraded in response to auxin.
We expected that DII degradation would be more pronounced upon treatment with higher IAA concentrations, as observed for DII in Arabidopsis (Brunoud et al., 2012). Confocal microscopy was used to image and measure the change in DII fluorescence intensity after treatment with increasing IAA concentrations (1, 5, 10, and 15 μM) for 20 min in 4-week-old maize plant leaf pieces, as described in Supplemental Figure S1A. Consistently, mock-treated DII fluorescence levels were reduced by approximately 20% after 20-min treatment (Fig. 1J). By contrast, DII levels responded in a dose-dependent manner to IAA. After 20-min treatment with 1 μm IAA, we observed a reduction of approximately 30% (29.6 ± 2.7%) of DII initial fluorescence (Fig. 1J). After 5 μm IAA treatment, the reduction was approximately 40% (41.8 ± 1.4%), while after 10 μm IAA treatment showed a fluorescence reduction of approximately 50% (52 ± 5.2%), similar to that observed in the time-lapse experiment (compare Figure 1A with 1J). Finally, treatment with 15 μm IAA resulted in the largest reduction in fluorescence, measured as approximately 60% (57.9 ± 3%) relative to the initial fluorescence. Mock DII, mock mDII, and IAA-treated mDII fluorescence intensity levels were similar regardless of the IAA concentrations applied (Fig. 1J). Together, these results indicated that DII responds to auxin in a dose-dependent manner.
AUX/IAA proteins are ubiquitinated upon auxin treatment by TIR1/AFBs and degraded via the proteasome in Arabidopsis (Gray et al., 2001). To determine whether DII degradation is proteasome dependent in maize, we assessed whether DII degradation was blocked by the proteasome inhibitor MG132 (Dharmasiri et al., 2005). DII accumulation was measured by immunoblotting total protein extracts from 3-d-old seedling roots grown in hydroponic media (10 mm MES, pH 5.7) pretreated for 3 h with proteasome inhibitor (100 μm MG132) or mock (DMSO) and then incubated for another hour with media supplemented with different combinations of auxin (10 μm IAA), MG132 100 μm, or the corresponding amount of DMSO. We observed higher DII accumulation in MG132-treated samples compared to mock-treated roots (Fig. 1K), indicating that blocking the proteasome activity results in higher stability of DII protein in the roots. Similar to leaf samples, treatment with 10 μm IAA triggered DII degradation (Fig. 1K). In contrast, application of IAA to MG132-pretreated roots did not cause DII degradation, because DII levels were similar to MG132-treated samples (Fig. 1K). mDII transgenic plants were treated with the same combinations of MG132 and IAA as the experiment for DII seedlings. mDII protein levels appeared unchanged under all experimental conditions (Fig. 1K). This experiment was repeated three times with similar results (Supplemental Fig. S6).
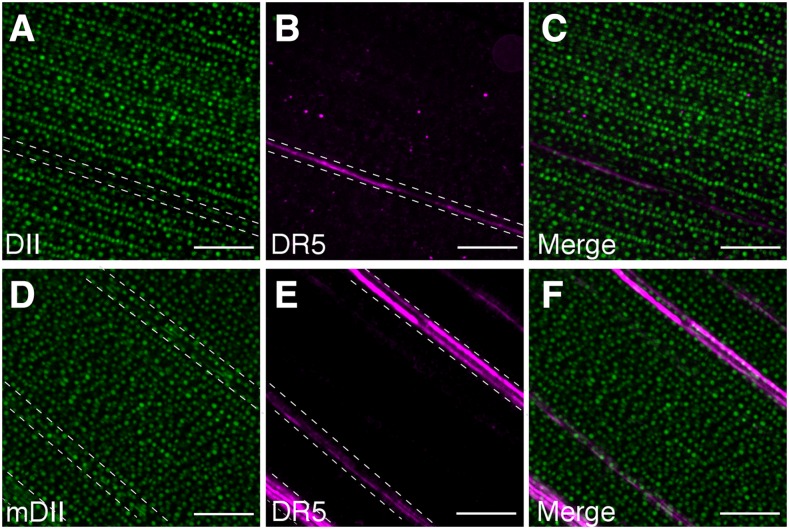
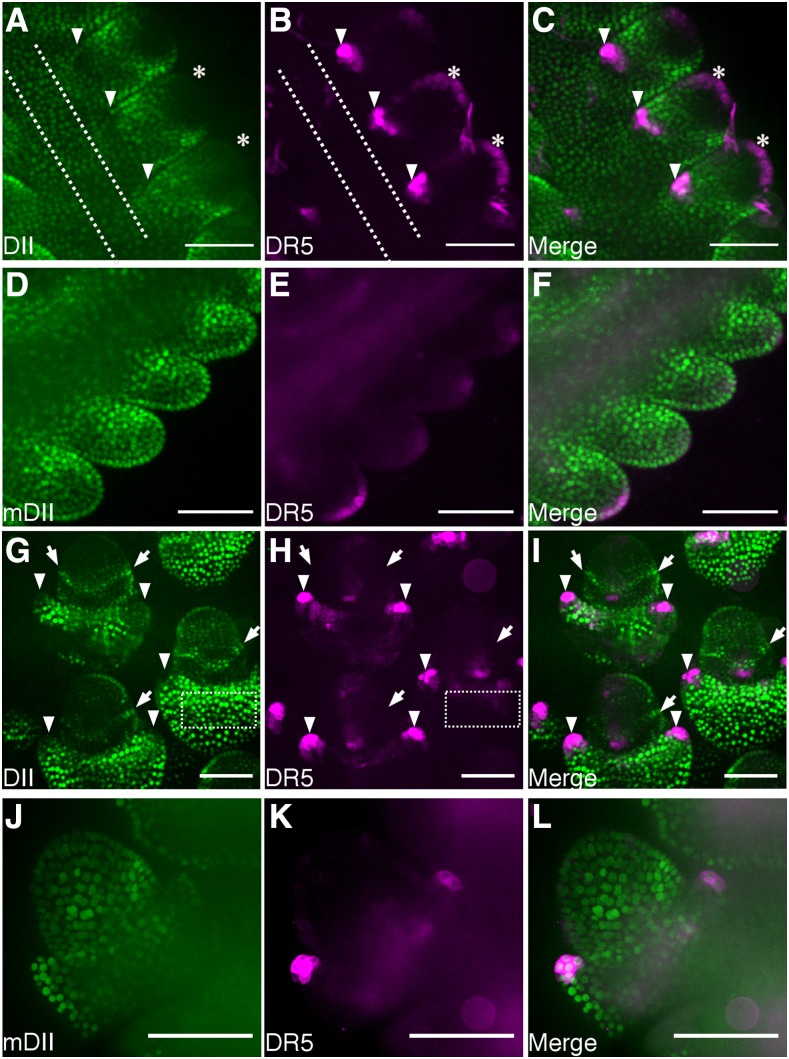
The DII-based auxin responsive marker was used to detect endogenous auxin signaling in maize by imaging tissues or structures for which auxin-responsive DR5 activity is well documented, such as near vasculature (Scarpella et al., 2006) or tassel branches (Gallavotti et al., 2008a). Plants simultaneously expressing DII or mDII and DR5 (Gallavotti et al., 2008b) were generated by crossing lines carrying the transgenes. DII-specific fluorescence signal near vasculature was lower by approximately 30% (28% ± 2.1%) compared to nearby epidermal cells (Fig. 2A). In contrast, DR5 fluorescence was approximately 400% higher (370.3 ± 28%) near vasculature than nearby epidermal cells (Fig. 2B). Therefore, DII and DR5 were virtually opposite in signal distribution (Fig. 2C). By contrast, mDII fluorescence was ubiquitous (Fig. 2D) and not locally reduced near DR5-labeled vasculature (Fig. 2, E and F). Together, these results reinforce the efficacy of the DII marker and show that both DII and DR5 identify higher auxin signaling in vascular cells and overlying epidermal cells. To assess DII localization in developing tassels, 5-week-old plants were dissected to reveal tassels coexpressing DII and DR5, and spikelet pair meristems and floral meristems were imaged (Supplemental Fig. S7). In spikelet pair meristems, DII fluorescence was reduced to approximately 70% in the outer region and in the glume primordia, compared to DII signal in the central region of the tassel (65.7 ± 5.1% and 72.4 ± 4.8%, respectively; Fig. 3A; central region indicated with dotted lines in Fig. 3, A and B). In contrast, DR5 signal was increased >300% (336.6 ± 8.6% and 564.6 ± 19.3%) in the L1 layer and the glume primordia, respectively, compared to the central region (Fig. 3B). In floral meristems, low DII signal was measured in the tip of glume primordia as approximately 60% (56.3 ± 0.7%) of the fluorescence measured in the central region of the glume outside the floral meristem (Fig. 3G; an example of one central region of the glume is indicated with a dotted-line rectangle in Fig. 3, G and H). DR5 signal however, was increased in tips of glume primordia approximately 500% (496.9 ± 7.9%) compared to the central region of the glume around the floral meristem (Fig. 3H). Again, DII and DR5 showed essentially opposite fluorescence patterns in developing tassels, as expected (Fig. 3, C and I). Intriguingly, we observed a DII-specific “collar” around floral meristems that suggests there may be a ring of cells with low auxin signaling or perception that might be important for floral meristem development (Fig. 3, G and I; Supplemental Fig. S8). It is possible that the observed gradient in auxin signaling in the meristem could mark the incipient primordia initials. The control mDII fluorescence was not excluded from areas with DR5 expression but instead was observed throughout the tassel, both in spikelet pair meristems (Fig. 3, D–F) and more developed floral meristems (Fig. 3, J–L). Together, these observations suggest an auxin-signaling gradient, as reported by DII accumulation in the axillary meristems and lateral organ boundaries such as the site of glume initials and the tip of developing glume primordia.
Figure 2.
DII and mDII signal near vasculature in the leaf blade. Maximum Z-stack projections of fluorescence of epidermal and vasculature cells from leaf sections of DII (A), DR5 (B), merged DR5 and DII (C), mDII (D), DR5 (E), and merged DR5 and mDII (F). Bar = 100 μm. For fluorescence intensity quantification, random positions within the dashed lines were taken and compared with fluorescence in the leaf epidermis (region outside of the dashed line region).
Figure 3.
DII and mDII signal in developing tassels. Fluorescence observed in secondary tassels excised from 5- to 6-week-old plants expressing DII (A and G) DR5 (B, E, H, and K), mDII (D and J), merged DII and DR5 (C and I), and merged mDII and DR5 (F and L). Imaged structures correspond to spikelet pair meristems (A–F) and floral meristems (G–I) and spikelet meristems (J–L). Bar = 100 μm. Asterisks indicate L1 layer. Arrowheads indicate glume primordia. Arrows indicate collar with high DII signal. Dotted lines in A and B and G and H indicate the central region of the tassel and the glume outside the spikelet meristem, respectively, that was used to quantify the fluorescence.
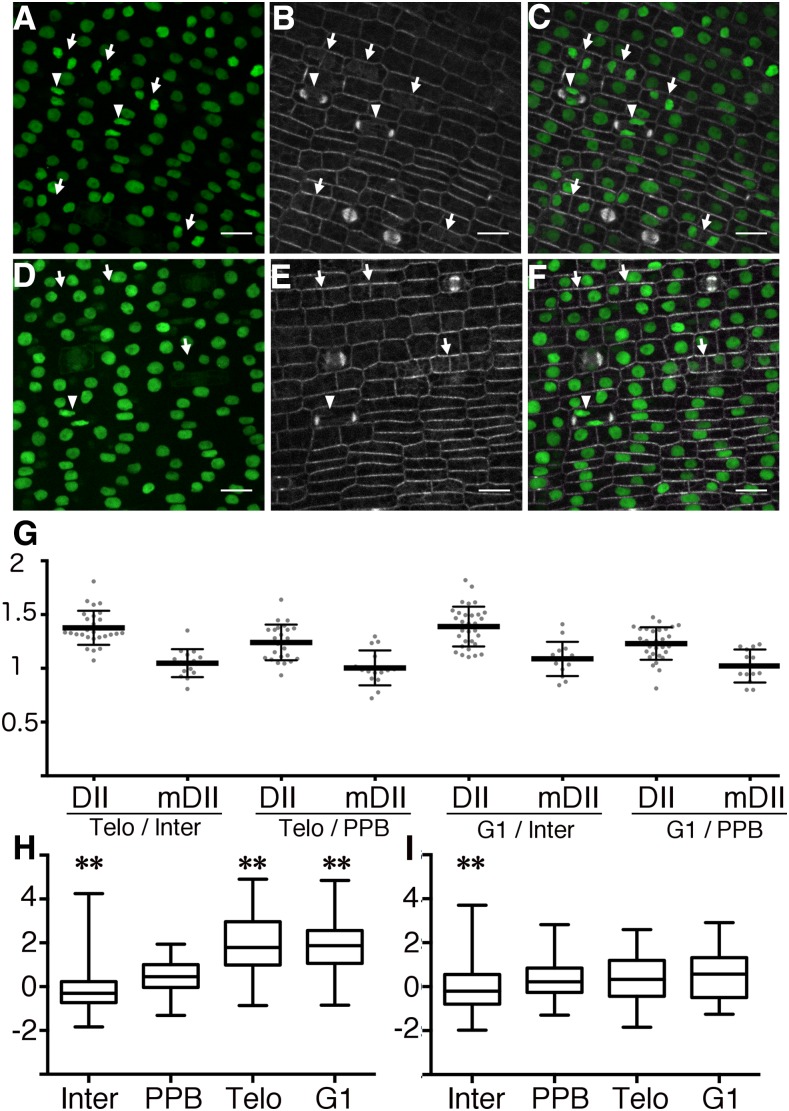
Auxins together with cytokinins are important regulators of cell cycle progression (for review, see Perrot-Rechenmann, 2010), so we hypothesized that individual cells would show differential auxin signaling along the cell cycle. Indeed, auxin has been shown to directly bind to and cause degradation of the cell cycle regulator F-Box S-Phase Kinase-Associated protein 2A (SKP2A; Jurado et al., 2010). We analyzed changes in DII fluorescence during the cell cycle of symmetrically dividing epidermal cells in maize. Plants expressing DII or mDII and a live cell marker for microtubules, CFP-TUBULIN (Mohanty et al., 2009), allowed identification of cell-cycle stages. For this analysis, “interphase cells” are broadly defined to include cells in late G1 or S or early G2, because these stages cannot be unambiguously identified by characteristic microtubule arrays. In contrast, distinct microtubule structures distinguish cells in late G2/prophase, metaphase, anaphase, telophase, and G1. Cells in late G2/prophase have preprophase bands (PPBs), cells in telophase contain phragmoplasts, and cells in early G1 have pairs of decondensing nuclei that are often closely appressed to the new cross wall (Kumagai et al., 2001; Rasmussen et al., 2013) or have a ring of microtubules nucleating from the nuclei (Ambrose and Wasteneys, 2011). DII levels were not measured in metaphase or anaphase cells, because DII during those stages is dispersed throughout the cytoplasm. Cells in telophase (n = 94; Fig. 4, B and E arrowheads) had approximately 21% and approximately 36% higher DII fluorescence compared to cells with PPBs (n = 124) and cells in interphase (n = 3,468; Fig. 4, A–C and G), respectively. Similar values were observed when we compared cells in early G1 (n = 172; Fig. 4, B and E, arrows) with cells in interphase (37% higher fluorescence intensities) and with cells with PPB (22% higher fluorescence intensities; Fig. 4G). In contrast, mDII fluorescence across different cell stages was similar between cells with PPBs (n = 88), cells in telophase (n = 90), cells in G1 (n = 191), and cells in interphase (n = 2,926; Fig. 4, D–F and G). Normalized values (Z-score values) of DII fluorescence showed a higher median signal for cells in telophase (1.79) and G1 (1.87) than for cells in interphase (−0.31) and cells with a PPB (0.44; Fig. 4H). In contrast, normalized values of mDII fluorescence showed a similar median in all cell stages (interphase: −0.2, PPB: 0.22, telophase: 0.33, G1: 0.57; Fig. 4I). Moreover, the distribution of normalized values of DII fluorescence in telophase and G1 was significantly different than the one observed for cells with a PPB (P < 0.0001 using the nonparametric Kruskal–Wallis test; Fig. 4H). On the other hand, the distribution of the mDII-specific fluorescence normalized values measured for cells in telophase and G1 did not differ statistically from the one observed for cells with a PPB (P-value = 0.62 Kruskal–Wallis test; Fig. 4I). In addition, we analyzed the fluorescence of each nucleus compared with the brightest nucleus in each micrograph and then separated them by cell-cycle type. Similar to the Z-score-based analysis, statistically significant differences were observed between DII samples in cells with PPBs, and cells in telophase and early G1. In contrast, mDII samples with PPBs or in telophase or G1 were not significantly different from each other (Supplemental Fig. S9).
Figure 4.
DII and mDII accumulation during different cell cycle stages. A and D, Maximum projection of Z-stacks showing the fluorescence of DII (A) and mDII (D) in leaf epidermal cells. B and E, Microtubule live-cell marker CFP-TUBULIN (Mohanty et al., 2009) in the corresponding epidermal cells in A and D, respectively. C and F, Merged images of leaf epidermal cells coexpressing CFP-TUBULIN and DII (C) or mDII (F). Bar = 10 μm. Arrowheads indicate cells in telophase, and arrows indicate cells in G1. G, Graph of the ratios between the mean fluorescence values of nuclei in different cell stages expressing DII or mDII, as indicated. H, Distribution of Z-scores based on fluorescence intensity of DII measured from signal observed in cells in interphase (n = 2,229), PPB (n = 74), telophase (n = 58), and G1 (n = 129). I, Distribution of Z-scores based on fluorescence intensity of mDII measured from signal observed in cells in interphase (n = 1,510), PPB (n = 48), telophase (n = 63), and G1 (n = 63). G1, Cells in early G1; Inter, interphase; PPB, cells with PPB; Telo, telophase. The Kruskal-Wallis test was used to measure significance. Interphase cells were excluded from comparative analysis.
DISCUSSION
A DII-based auxin-responsive tool was generated expressing the nuclear localized DII domain from AtIAA28 fused to a yellow fluorescent protein (Brunoud et al., 2012) under the transcriptional control of the maize ubiquitin promoter and was further characterized in maize. The DII marker rapidly degrades in response to exogenous auxin as assessed using both live-imaging and immunoblot analysis in maize leaves (Fig. 1, A–I), although the observed degradation dynamic varies when using these two different techniques. DII-specific signal in leaf epidermal cells was reduced by 50% after 25 min of treatment and reached basal levels after 1 h of auxin treatment. When immunoblotting was used to detect DII accumulation in leaves over 1 h of auxin treatment, basal levels were reached after 15 min of treatment. This difference in dynamics can be due to differences in either sensitivity of the technique or in cell types used in these experiments. For time-lapse analysis, only epidermal cells in leaves with a 3- to 5-mm ligule were analyzed by microscopy, whereas for immunoblot analysis, a leaf piece from leaves with a 5- to 8-mm ligule containing multiple cell types and layers was sampled. These different cell types or developmental stages could have variable amounts and types of auxin receptors. In Arabidopsis, rigorous analysis of expression levels of the F-box coreceptors as well as the AUX/IAA proteins and ARFs was determined in the shoot apical meristem to characterize the inputs into the auxin signaling pathway (Vernoux et al., 2011). In the shoot apical meristem, TIR1, AFB1, and AFB5 were primarily responsible for auxin-mediated degradation of AUX/IAAs. TIR1 had lower expression in the central zone, while AFB1 and AFB5 were ubiquitously expressed in the central zone, indicating spatial variability in F-box coreceptor expression (Vernoux et al., 2011). Determining the expression in vivo of the TIR1/AFB is critical, because they have different AUX/IAA degradation dynamics in vitro. Use of different auxin receptors changed AUX/IAA degradation dynamics in a synthetic yeast system: full-length IAA28 half-life was 33.1 min when TIR1 was used for auxin perception, whereas when AFB2 was used, IAA28 half-life was 20.5 min (Havens et al., 2012). In Arabidopsis plants, IAA28 levels decreased 50% after 14.8 min of exogenous auxin treatment (Dreher et al., 2006).
In addition to differential degradation of AUX/IAAs in response to different coreceptors, regions outside the DII domain also influence proteolysis dynamics, although they are dependent on an intact proteasome (Dreher et al., 2006; Moss et al., 2015). For example, degradation dynamics of AtIAA28 DII domain used in this work and in Brunoud et al., 2012 was slower than that observed for whole-length IAA28 of Arabidopsis (Havens et al., 2012). Recent work shows that IAA1 ubiquitination and degradation were not dependent on a Lys residue anywhere within the IAA1 protein, since Lys removal did not impair IAA1 degradation. This raises the possibility that IAA1 may be degraded via another proteasome dependent mechanism (Gilkerson et al., 2015). Together, this indicates that while DII is degraded in response to auxin, other factors such as F-box coreceptor concentration or regions outside DII domain may contribute to DII protein stability. Here, we show that DII was degraded in a dose-dependent manner with increasing IAA concentrations (Fig. 1J). Moreover, we show that DII degradation was blocked by MG132, indicating protein turnover requires an intact proteasome (Fig. 1K). Despite heterologous expression of IAA28 DII from Arabidopsis in maize, we observed reproducible degradation dynamics that were similar to degradation dynamics reported in Arabidopsis roots (Brunoud et al., 2012). Regardless of the potential impact of other regions on AUX/IAA protein stability and the exact mechanism that triggers DII-degradation upon auxin perception, the DII auxin response reporter characterized in this work can be used as a tool to monitor spatial and temporal differences in input signaling.
Auxin concentration gradients in plant tissues are important for the function of auxin as a developmental regulator (Benjamins and Scheres, 2008; Ikeda et al., 2009; Petersson et al., 2009). DII was used to assess endogenous levels of auxin signaling inputs in maize plants, and we observed mostly opposite patterns of DR5 and DII in vasculature-associated tissue of leaves and in glume primordia of developing tassels (Figs. 2, A–C and 3, G–I). However, an advantage of the DII marker is that it clearly indicates regions with auxin signaling input minima. Interestingly, DII accumulation pattern in the floral meristems of developing tassels revealed a collar of cells that accumulated high levels of DII, which indicates a minimal auxin zone surrounded by regions with higher auxin signaling inputs might be important for floral organ formation (Fig. 3, G and I; Supplemental Fig. S8). This might indicate low auxin levels in a boundary region of floral organ primordia, although further work will be necessary to determine whether these auxin signaling input minima play a specific role in floral development. Other examples of auxin minima that have a critical role in development have been identified using the DII marker. Auxin minima identified using the DII marker line are observed at the leaf axil boundary and are required for axillary meristem formation in both tomato and Arabidopsis (Wang et al., 2014a, 2014b). Localized auxin production in leaf axils decreased axillary meristem formation, while reduced auxin perception increased axillary meristem formation (Wang et al., 2014a). In addition to an important role in axillary meristem development, auxin minima on the adaxial leaf primodia are observed transiently during leaf polarity establishment in Arabidopsis (Qi et al., 2014). In each of these cases, polar auxin transport is critical for the establishment of a local auxin minimum and subsequent development. Polar auxin transport is also essential for axillary meristem development in maize (Gallavotti et al., 2008a, 2008b). Similarly, auxin minima in pericycle cells along the root are crucial for proper lateral root formation in Arabidopsis (Dubrovsky et al., 2011). Intriguingly, similar to the shoot apical meristem in Arabidopsis (Vernoux et al., 2011), low DII was observed at the tip of the spikelet pair meristem, although DR5 signal was also low. This potentially suggests that low auxin transcriptional response rather than accumulation may explain the lack of DR5 accumulation. Therefore, although DII and DR5 showed an overall opposite distribution as expected, specific features of auxin signaling, particularly transient areas of low auxin accumulation or perception, were highlighted using the DII marker.
Prior studies suggested that cell cycle progression is controlled by both cytokinins and auxins, with auxin promoting transitions from G2 to M and G1 to S (for review, see Perrot-Rechenmann, 2010). Proper cell cycle regulation is critical for the growth of any organism. It is important for plant cells to be able to reenter the cell cycle during development, since organogenesis in plants takes place during the whole life cycle. Intimate links between cell cycle progression and auxin have been demonstrated. Specifically, auxin promotes cell cycle progression in the pericycle as an early step of lateral root formation in Arabidopsis (Himanen et al., 2002) and maize (von Behrens et al., 2011; Yu et al., 2016, 2015). In the developing maize leaf, increased concentrations of auxins and cytokinins were found in the rapidly dividing zone of the leaf in regions prior to cell expansion (Nelissen et al., 2012). Maize lateral root initiation in response to nitrate occurred through localized auxin accumulation and corresponding expression of cell-cycle promoting factors and degradation of cell-cycle inhibitors (Yu et al., 2016). Auxin specifically binds and triggers the degradation of the SKP2A, which is important for the stability of the cell division transcription factors E2FC/DBP demonstrating that auxin directly modulates cell-cycle progression (Jurado et al., 2010). In addition, auxin regulates transcription of cell-cycle control genes (Himanen et al., 2002; Jansen et al., 2012; Martínez-de la Cruz et al., 2015; Roudier et al., 2003), and the transcription of auxin-responsive genes fluctuates during cell cycle progression (Breyne et al., 2002; Menges et al., 2005). Therefore, it is likely that auxin accumulation and signaling are dynamic during cell cycle progression. However, to our knowledge, high DII signal (reflecting low auxin concentration or signaling input) during telophase and G1 has not been previously reported. Using lines expressing DII and CFP-TUBULIN, we identified a significant increase in DII-specific fluorescence (approximately 30%), and thus reduction in auxin signaling inputs, in dividing leaf epidermal cells in late telophase and early G1 (Fig. 4). The observed increase in auxin signaling inputs after early G1 might be associated with stabilizing transcriptional regulators in preparation for the G2/M transition. These results suggest that the auxin-mediated aspects of both symmetric and asymmetric division (Himanen et al., 2002; Yu et al., 2016) may have common features.
Results presented in this work demonstrate the suitability of the DII-based auxin responsive reporter in maize to assess dynamic auxin input signaling. DII nuclear localization and rapid temporal response to auxin simplifies live imaging, quantification, and the study of auxin signaling both in individual cells and across tissues. Although the recent combination of DII with mDII in one plasmid, R2D2, has increased the utility of the reporter in Arabidopsis (Liao et al., 2015), the DII marker alone is highly valuable in maize, particularly considering the mostly ubiquitous expression of mDII driven by the ubiquitin promoter. DII responded to both exogenous IAA applied to leaf pieces and roots as well as endogenous auxin signaling in leaves, roots, and developing tassels. The important role of auxin in maize development (Forestan and Varotto, 2012; Galli et al., 2015; McSteen, 2010; von Behrens et al., 2011; Yu et al., 2016; Zhang et al., 2014) and during the cell cycle (Del Pozo and Manzano, 2014; Himanen et al., 2002) highlights the utility of the DII marker.
MATERIALS AND METHODS
Construction of the DII and mDII Maize Lines
The DII and mDII reporters were amplified using primers AtIAA28DII and N7SpeI (Supplemental Table S1) from the original Arabidopsis (Arabidopsis thaliana) plasmids (Brunoud et al., 2012). These fragments were cloned by BamHI/SpeI restriction digest into pAL1004, a binary vector modified from pTF101.1 (Frame et al., 2002) and pAHC25 (Christensen and Quail, 1996). The maize (Zea mays) Ubi-1 promoter-GUS-Nos terminator cassette from pAHC25 was subcloned into pTF101.1, and GUS was replaced with a customized MCS (BamHI-XmaI-HindIII-SpeI-SacI). After sequence verification (UC Davis Sequencing), the DII and mDII vectors were introduced into Agrobacterium tumefaciens EHA101 to transform the maize hybrid HiII by the Plant Transformation Facility (Iowa State University). The transformants were screened by microscopy for DII or mDII signal and crossed to B73.
Growth Conditions and Genotyping
Plants were grown under standard greenhouse conditions and selected by application of 0.2 g/L glufosinate with 0.05% Tween20. Glufosinate-resistant plants were used in all experiments. Both mDII and DII segregated with ratios consistent with insertion in a single locus. Plants expressing DII or mDII together with DR5 or CFP-TUBULIN (GRMZM2G164696) were generated by crossing and confirmed by genotyping using primers listed in Supplemental Table S1. Transgenes were maintained as heterozygotes by backcrossing to inbred line B73 or into other lines expressing transgenes. Experiments were performed using T2 or later generation plants. PCR was performed using KOD Polymerase (EMD-Millipore) according to the manufacturer’s instructions with the addition of 7% DMSO.
Microscopy and Image Analysis
All microscopy was performed using a spinning-disk confocal system (Solamere Technology, Inc) with an inverted Eclipse TE stand (Nikon), a Yokagawa W1 spinning disk (Yokagawa), and EM-CCD camera (Hamamatsu 9100c). The following Nikon objectives were used: 40× water immersion lens (1.15 NA), 20× (0.75 NA), or 10× (0.45 NA). The water immersion lens was used with perfluorcarbon immersion liquid (RIAAA-678, Cargille). The stage contains both a Piezo Z (ASI) and 3 axis DC servo motor controller to allow fully automated time-lapse imaging that is managed by Micromanager software (www.micromanager.org). The solid-state lasers used emit at 561, 514, and 445 (Obis from 40-100 mW). All emission filters are from Chroma Technology. For DII and mDII, a 514 laser with emission filter 540/30 was used. For CFP-TUBULIN imaging, a 445 laser with emission filter 480/40 was used. For DR5 imaging, a 561 laser with emission filter 620/60 was used. Figures were assembled in GNU image manipulation program (GIMP, https://www.gimp.org/).
Leaves from 4-week-old glufosinate-resistant plants were dissected to excise 0.5-cm2 leaf pieces (Supplemental Fig. S1A). The leaf pieces were placed on microscope slides adding 10 to 15 μL of 10 mm MES (pH 5.7) supplemented with 0.05% DMSO (mock-treatment) or 10 μm IAA. Z-stack images with 10 μm depth were taken for six different positions every 5 min for 1 h. Care was taken to avoid regions near the cut surface, as described (Rasmussen, 2016). In all of the following experiments, any Z stack image that did not capture the whole nucleus was excluded from further analysis. Images were analyzed using FIJI software (http://fiji.sc/wiki/index.php/Fiji). Z stacks were compressed into one image using maximum intensity projections, corrected for drift with “StackReg.” Background was removed using “background subtraction rolling 50.” Next, binary files were generated corresponding to the first time point. The “Analyze particles” tool was used to select individual nuclei from binary files (Supplemental Fig. S1B–D), Particle selection was used to measure individual nuclear fluorescence values throughout the time-lapse. Prism (Graph Pad) was used to analyze and plot the data.
Different cell cycle stages were identified by imaging microtubule structures using a previously reported CFP-TUBULIN line in maize (Mohanty et al., 2009). The presence of a PPB and phragmoplast indicated cells were in late G2 or prophase and telophase, respectively. Once mitosis was completed, early G1 stage was identified by the proximity of the nuclei of the daughter cells to the newly formed cell wall as well as a characteristic microtubule array. One distinguishing feature of a G1 stage cell are microtubules radiating from the newly formed nucleus (Ambrose and Wasteneys, 2011). Those cell stages were clearly distinguishable from the cells in late G1, S, or early G2, which we grouped as “interphase” cells. Measurement of DII or mDII levels during cell cycle stages was performed as follows. The mean value of the fluorescence of cells in telophase was divided by the mean fluorescence values for cells with PPBs or interphase cells in each micrograph. In addition, mean florescence values for cells in early G1 were also divided by mean fluorescence values of cells with PPBs or cells in interphase in each micrograph. Average values of the calculated ratios of all different micrographs belonging to different biological replica are plotted in Figure 4G. Moreover, mean intensity fluorescence levels were measured and normalized by calculating a Z-score value for each micrograph to allow comparisons between separate images. Distribution of Z-score values as well as median and variance for each cell-cycle stage group was plotted in Figure 4, H and I. The statistical significance of the distributions for the different cell stages was performed by the nonparametric Kruskal-Wallis test (Prism). Interphase was excluded from the comparisons between cell cycle stages because of variability that may be due to differential expression of the promoter (because variability was seen in both mDII samples and DII samples). In addition, we used an alternative method of analyzing that data by making ratios using raw fluorescence values per each micrograph instead of using Z scores. The highest fluorescence signal was used as the denominator so all the values lie between zero and one.
To estimate loss of fluorescence due to nuclear movement we analyzed mDII time-lapse images because mDII does not respond to either exogenous or endogenous auxin. First, we determined whether nuclear movement was dependent on auxin treatment by assessing colocalization of nuclei during the time-lapse using the Coloc2 tool in FIJI. Nuclear movement was not altered by auxin treatment (Supplemental Fig. S2A). Next, we assessed how much nuclear movement caused apparent loss of fluorescence during 1-h time-lapse by comparing binary mask position at each time point in the experiment. Each time-lapse experiment uses the first time point to generate the binary mask. Subsequent masks may not perfectly align due to nuclear movement, as indicated by detected shifts between the first and last mask (Supplemental Fig. S2B). To assess how much fluorescence is lost from nuclear movement, we compared the average fluorescence calculated from the binary mask generated at time 0 to a binary mask generated at each time point. The relative difference calculated from three different mock- and auxin-treated replicas was plotted (Supplemental Fig. S2B).
To estimate the maximum loss of fluorescence due to photobleaching, we used the longest exposure times (400 ms) corresponding to those used during DII time-lapse on mDII leaf piece, because their nuclei do not respond to auxin (Supplemental Fig. S2C). A leaf piece expressing mDII was imaged every 15 s 12 times with 400-ms exposure (165 s total time lapse) to provide the same amount of photobleaching that occurs during 1-h time-lapse for DII. Exposure time for time-lapse imaging of mDII (150 ms; Fig. 1) was shorter than the one used for DII imaging (400 ms), because the fluorescence signal was much brighter in mDII samples than in DII samples. Therefore, this is an accurate estimate for DII photobleaching and an overestimate of loss-of-fluorescence due to photobleaching for mDII samples.
Sample Preparation for Immunoblot Analysis
Leaf pieces were incubated on 0.8% agar 10 mm MES (pH 5.7) supplemented with 0.05% DMSO (mock) or 10 μm IAA (Supplemental Fig. S1E). Samples were harvested every 5 min for 1 h and frozen in liquid nitrogen. For the proteasome-dependent DII degradation experiment (Fig. 1K), maize seeds were germinated for 2 d on wet paper towels and then transferred to liquid 10 mm MES (pH 5.7) for 24 h. Media was replaced with 10 mm MES (pH 5.7) supplemented with 100 μm MG132 or the corresponding control amount of DMSO and incubated for 3 h. Then media was replaced with 10 mm MES (pH 5.7) supplemented with 10 μm IAA with or without 100 μm MG132, or with the corresponding amount of DMSO for control samples. Proteins were extracted in TBS buffer supplemented with 10 mm β-mercaptoethanol, 100 μm MG132, 1 mm phenylmethylsulfonyl fluoride protease inhibitor (Life Technologies), 1% Protease Inhibitor Cocktail (Sigma), 0.2% SDS, and 1% NP-40. Then 5 μg of protein extracts was separated on a 12% acrylamide SDS-PAGE gel. DII and mDII were detected by immunoblotting with a polyclonal anti-GFP antibody (Life Technologies, A-6455). As loading control, membranes were stripped and reprobed using monoclonal antialpha-Tubulin antibody (Life Technologies, 32-2500).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers pAL004(Ubi-nos)DII (KY313415) and pAL004(Ubi-nos)mDII (KY313416).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Procedure for sample collection, drug treatment, and image analysis.
Supplemental Figure S2. Nuclear fluorescence loss due to nuclear movement and photobleaching during time-lapse experiment.
Supplemental Figure S3. Growth of plants expressing the DII or mDII transgene.
Supplemental Figure S4. Other biological replicas related to the experiment shown in Figure 1H.
Supplemental Figure S5. Other biological replicas related to the experiment shown in Figure 1I.
Supplemental Figure S6. Degradation of DII depends on functional proteasome.
Supplemental Figure S7. Photograph of a developing tassel of 5- to 6-d-old maize plants grown under greenhouse conditions.
Supplemental Figure S8. DII and mDII signal in floral meristems belonging to developing tassels.
Supplemental Figure S9. Distribution of ratios for DII- and mDII-specific fluorescence intensity values at different cell-cycle stages.
Supplemental Table S1. Primers used in this study.
Supplementary Material
Acknowledgments
The Plant Transformation Facility at Iowa State University is gratefully acknowledged for generating maize transgenic plant lines under MCB number 1027445 (University of Wyoming).
Glossary
- DMSO
dimethyl sulfoxide
- PPB
preprophase band
Footnotes
L.Z.A. was supported by a MARC U-STAR training grant from the National Institutes of Health (T34GM062756). C.G.R. gratefully acknowledges funding from NSF-MCB150484 (University of California, Riverside), USDA-NIFA-CA-R-BPS-5108-H (University of California, Riverside), and DEB #1052051 (University of Wyoming).
Articles can be viewed without a subscription.
References
- Ambrose C, Wasteneys GO (2011) Cell edges accumulate gamma tubulin complex components and nucleate microtubules following cytokinesis in Arabidopsis thaliana. PLoS One 6: e27423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamins R, Scheres B (2008) Auxin: the looping star in plant development. Annu Rev Plant Biol 59: 443–465 [DOI] [PubMed] [Google Scholar]
- Breyne P, Dreesen R, Vandepoele K, De Veylder L, Van Breusegem F, Callewaert L, Rombauts S, Raes J, Cannoot B, Engler G, et al. (2002) Transcriptome analysis during cell division in plants. Proc Natl Acad Sci USA 99: 14825–14830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunoud G, Wells DM, Oliva M, Larrieu A, Mirabet V, Burrow AH, Beeckman T, Kepinski S, Traas J, Bennett MJ, et al. (2012) A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature 482: 103–106 [DOI] [PubMed] [Google Scholar]
- Christensen AH, Quail PH (1996) Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res 5: 213–218 [DOI] [PubMed] [Google Scholar]
- Del Pozo JC, Manzano C (2014) Auxin and the ubiquitin pathway. Two players-one target: the cell cycle in action. J Exp Bot 65: 2617–2632 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M (2005) The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445 [DOI] [PubMed] [Google Scholar]
- Dreher KA, Brown J, Saw RE, Callis J (2006) The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell 18: 699–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky JG, Napsucialy-Mendivil S, Duclercq J, Cheng Y, Shishkova S, Ivanchenko MG, Friml J, Murphy AS, Benková E (2011) Auxin minimum defines a developmental window for lateral root initiation. New Phytol 191: 970–983 [DOI] [PubMed] [Google Scholar]
- Forestan C, Varotto S (2012) The role of PIN auxin efflux carriers in polar auxin transport and accumulation and their effect on shaping maize development. Mol Plant 5: 787–798 [DOI] [PubMed] [Google Scholar]
- Frame BR, Shou H, Chikwamba RK, Zhang Z, Xiang C, Fonger TM, Pegg SEK, Li B, Nettleton DS, Pei D, et al. (2002) Agrobacterium tumefaciens-mediated transformation of maize embryos using a standard binary vector system. Plant Physiol 129: 13–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A, Barazesh S, Malcomber S, Hall D, Jackson D, Schmidt RJ, McSteen P (2008a) sparse inflorescence1 encodes a monocot-specific YUCCA-like gene required for vegetative and reproductive development in maize. Proc Natl Acad Sci USA 105: 15196–15201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A, Yang Y, Schmidt RJ, Jackson D (2008b) The relationship between auxin transport and maize branching. Plant Physiol 147: 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli M, Liu Q, Moss BL, Malcomber S, Li W, Gaines C, Federici S, Roshkovan J, Meeley R, Nemhauser JL, et al. (2015) Auxin signaling modules regulate maize inflorescence architecture. Proc Natl Acad Sci USA 112: 13372–13377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkerson J, Kelley DR, Tam R, Estelle M, Callis J (2015) Lysine residues are not required for proteasome-mediated proteolysis of the auxin/indole acidic acid protein IAA1. Plant Physiol 168: 708–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M (2001) Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414: 271–276 [DOI] [PubMed] [Google Scholar]
- Havens KA, Guseman JM, Jang SS, Pierre-Jerome E, Bolten N, Klavins E, Nemhauser JL (2012) A synthetic approach reveals extensive tunability of auxin signaling. Plant Physiol 160: 135–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen K, Boucheron E, Vanneste S, de Almeida Engler J, Inzé D, Beeckman T (2002) Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell 14: 2339–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y, Men S, Fischer U, Stepanova AN, Alonso JM, Ljung K, Grebe M (2009) Local auxin biosynthesis modulates gradient-directed planar polarity in Arabidopsis. Nat Cell Biol 11: 731–738 [DOI] [PubMed] [Google Scholar]
- Jansen L, Roberts I, De Rycke R, Beeckman T (2012) Phloem-associated auxin response maxima determine radial positioning of lateral roots in maize. Philos Trans R Soc B Biol Sci 367: 1525–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurado S, Abraham Z, Manzano C, López-Torrejón G, Pacios LF, Del Pozo JC (2010) The Arabidopsis cell cycle F-box protein SKP2A binds to auxin. Plant Cell 22: 3891–3904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S, Leyser O (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451 [DOI] [PubMed] [Google Scholar]
- Kumagai F, Yoneda A, Tomida T, Sano T, Nagata T, Hasezawa S (2001) Fate of nascent microtubules organized at the M/G1 interface, as visualized by synchronized tobacco BY-2 cells stably expressing GFP-tubulin: time-sequence observations of the reorganization of cortical microtubules in living plant cells. Plant Cell Physiol 42: 723–732 [DOI] [PubMed] [Google Scholar]
- Liao C-Y, Smet W, Brunoud G, Yoshida S, Vernoux T, Weijers D. 2015. Reporters for sensitive and quantitative measurement of auxin response. Nat Methods 12: 207–210, 2 p following 210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-de la Cruz E, García-Ramírez E, Vázquez-Ramos JM, Reyes de la Cruz H, López-Bucio J (2015) Auxins differentially regulate root system architecture and cell cycle protein levels in maize seedlings. J Plant Physiol 176: 147–156 [DOI] [PubMed] [Google Scholar]
- McSteen P. (2010) Auxin and monocot development. Cold Spring Harb Perspect Biol 2: a001479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M, de Jager SM, Gruissem W, Murray JAH (2005) Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J 41: 546–566 [DOI] [PubMed] [Google Scholar]
- Mohanty A, Luo A, DeBlasio S, Ling X, Yang Y, Tuthill DE, Williams KE, Hill D, Zadrozny T, Chan A, et al. (2009) Advancing cell biology and functional genomics in maize using fluorescent protein-tagged lines. Plant Physiol 149: 601–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss BL, Mao H, Guseman JM, Hinds TR, Hellmuth A, Kovenock M, Noorassa A, Lanctot A, Villalobos LIAC, Zheng N, et al. (2015) Rate motifs tune auxin/indole-3-acetic acid degradation dynamics. Plant Physiol 169: 803–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Higuchi K, Goda H, Fujiwara MT, Sawa S, Koshiba T, Shimada Y, Yoshida S (2003) Brassinolide induces IAA5, IAA19, and DR5, a synthetic auxin response element in Arabidopsis, implying a cross talk point of brassinosteroid and auxin signaling. Plant Physiol 133: 1843–1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelissen H, Rymen B, Jikumaru Y, Demuynck K, Van Lijsebettens M, Kamiya Y, Inzé D, Beemster GTS (2012) A local maximum in gibberellin levels regulates maize leaf growth by spatial control of cell division. Curr Biol 22: 1183–1187 [DOI] [PubMed] [Google Scholar]
- Perrot-Rechenmann C. (2010) Cellular responses to auxin: division versus expansion. Cold Spring Harb Perspect Biol 2: a001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersson SV, Johansson AI, Kowalczyk M, Makoveychuk A, Wang JY, Moritz T, Grebe M, Benfey PN, Sandberg G, Ljung K (2009) An auxin gradient and maximum in the Arabidopsis root apex shown by high-resolution cell-specific analysis of IAA distribution and synthesis. Plant Cell 21: 1659–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J, Wang Y, Yu T, Cunha A, Wu B, Vernoux T, Meyerowitz E, Jiao Y (2014) Auxin depletion from leaf primordia contributes to organ patterning. Proc Natl Acad Sci USA 111: 18769–18774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen CG. (2016) Using live-cell markers in maize to analyze cell division orientation and timing. In M-C Caillaud, ed, Plant Cell Division. Springer, New York, pp 209–225. [DOI] [PubMed] [Google Scholar]
- Rasmussen CG, Wright AJ, Müller S (2013) The role of the cytoskeleton and associated proteins in determination of the plant cell division plane. Plant J 75: 258–269 [DOI] [PubMed] [Google Scholar]
- Roudier F, Fedorova E, Lebris M, Lecomte P, Györgyey J, Vaubert D, Horvath G, Abad P, Kondorosi A, Kondorosi E (2003) The Medicago species A2-type cyclin is auxin regulated and involved in meristem formation but dispensable for endoreduplication-associated developmental programs. Plant Physiol 131: 1091–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehin M, Bagchi R, Estelle M (2015) SCFTIR1/AFB-based auxin perception: mechanism and role in plant growth and development. Plant Cell 27: 9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpella E, Marcos D, Friml J, Berleth T (2006) Control of leaf vascular patterning by polar auxin transport. Genes Dev 20: 1015–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LIA, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446: 640–645 [DOI] [PubMed] [Google Scholar]
- Vernoux T, Brunoud G, Farcot E, Morin V, Van den Daele H, Legrand J, Oliva M, Das P, Larrieu A, Wells D, et al. (2011) The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol Syst Biol 7: 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Behrens I, Komatsu M, Zhang Y, Berendzen KW, Niu X, Sakai H, Taramino G, Hochholdinger F (2011) Rootless with undetectable meristem 1 encodes a monocot-specific AUX/IAA protein that controls embryonic seminal and post-embryonic lateral root initiation in maize. Plant J 66: 341–353 [DOI] [PubMed] [Google Scholar]
- Wang Q, Kohlen W, Rossmann S, Vernoux T, Theres K (2014a) Auxin depletion from the leaf axil conditions competence for axillary meristem formation in Arabidopsis and tomato. Plant Cell 26: 2068–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang J, Shi B, Yu T, Qi J, Meyerowitz EM, Jiao Y (2014b) The stem cell niche in leaf axils is established by auxin and cytokinin in Arabidopsis. Plant Cell 26: 2055–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P, Baldauf JA, Lithio A, Marcon C, Nettleton D, Li C, Hochholdinger F (2016) Root type-specific reprogramming of maize pericycle transcriptomes by local high nitrate results in disparate lateral root branching patterns. Plant Physiol 170: 1783–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P, Eggert K, von Wirén N, Li C, Hochholdinger F (2015) Cell type-specific gene expression analyses by RNA sequencing reveal local high nitrate-triggered lateral root initiation in shoot-borne roots of maize by modulating auxin-related cell cycle regulation. Plant Physiol 169: 690–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Paschold A, Marcon C, Liu S, Tai H, Nestler J, Yeh C-T, Opitz N, Lanz C, Schnable PS, et al. (2014) The Aux/IAA gene rum1 involved in seminal and lateral root formation controls vascular patterning in maize (Zea mays L.) primary roots. J Exp Bot 65: 4919–4930 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.