The genome-scale metabolic model of a nitrogen-fixing filamentous cyanobacterium helps to understand inter- and intracellular metabolic interactions and contributes to engineering strategies.
Abstract
Anabaena sp. PCC 7120 is a nitrogen-fixing filamentous cyanobacterium. Under nitrogen-limiting conditions, a fraction of the vegetative cells in each filament terminally differentiate to nongrowing heterocysts. Heterocysts are metabolically and structurally specialized to enable O2-sensitive nitrogen fixation. The functionality of the filament, as an association of vegetative cells and heterocysts, is postulated to depend on metabolic exchange of electrons, carbon, and fixed nitrogen. In this study, we compile and evaluate a comprehensive curated stoichiometric model of this two-cell system, with the objective function based on the growth of the filament under diazotrophic conditions. The predicted growth rate under nitrogen-replete and -deplete conditions, as well as the effect of external carbon and nitrogen sources, was thereafter verified. Furthermore, the model was utilized to comprehensively evaluate the optimality of putative metabolic exchange reactions between heterocysts and vegetative cells. The model suggested that optimal growth requires at least four exchange metabolites. Several combinations of exchange metabolites resulted in predicted growth rates that are higher than growth rates achieved by only considering exchange of metabolites previously suggested in the literature. The curated model of the metabolic network of Anabaena sp. PCC 7120 enhances our ability to understand the metabolic organization of multicellular cyanobacteria and provides a platform for further study and engineering of their metabolism.
Cyanobacteria are ubiquitous photosynthetic organisms found in almost every habitat on Earth, including hot springs and Antarctic rocks, as well as the fur of some sloths (Aiello, 1985). Cyanobacteria are highly diverse in terms of morphology: Some species are filamentous, others are unicellular or can form aggregates, several species are capable of nitrogen fixation in differentiated heterocysts, and some form motile hormogonia or spore-like akinetes (Flores and Herrero, 2010; Singh and Montgomery, 2011). In their natural environment, cyanobacteria are often an integral part of complex ecosystems with other species from all three domains of life (Stewart et al., 1983; Adams, 2000; Adams and Duggan, 2008). Several species build up thick microbial mats in extreme environments (Reysenbach et al., 1994) or composite with fungal filaments to form lichens (Rikkinen et al., 2002), while others live inside their symbiotic plant hosts (Adams, 2000). In case of the aquatic Azolla caroliniana, a small water fern, a filamentous, heterocyst-forming cyanobacterium is found within the ovoid cavities in the plant’s leaves, maintaining a mutually beneficial symbiotic relationship with the plant. This symbiont, Anabaena azollae, provides fixed nitrogen to the fern and, in return, receives carbon sources and a protected environment from Azolla (Hill, 1977; Lechno-Yossef and Nierzwicki-Bauer, 2002). The highly productive Azolla-Anabaena symbiosis has long been recognized as a cheap and effective biofertilizer of tropical rice paddies, and more recently it has been successfully applied in temperate climate as well (Wagner, 1997; Bocchi and Malgioglio, 2010). Outside of its plant host, the free-living form of Anabaena azollae has significant contribution to the carbon and nitrogen economy of tropical soils as well, forming microbial communities with other nitrogen-fixing cyanobacteria (Singh, 1950). When living freely, however, Anabaena azollae only develops 5% to 10% of its cell to heterocysts. This frequency increases up to 25% to 30%, when the symbiosis is extended to also include rice. This higher rate of nitrogen fixation is the result of an adjustment to provide sufficient nitrogen for all three species, i.e. the cyanobacterium, the fern, and the cocultivated rice (de Macale and Vlek, 2004). Anabaena sp. PCC 7120 strain, an isolated and sequenced form of Anabaena azollae, shows the same developmental pattern of a single heterocyst for every 10 to 20 vegetative cells (Kumar et al., 2010; Ehira, 2013) and acts as a representative model organism of the free-living cyanobacterium. To mimic the productivity of the symbiotic form, Anabaena sp. PCC 7120 has recently been modified to increase the expression of the HetR protein controlling heterocyst frequency and thus to enhance the organism’s potential as a nitrogen biofertilizer. The resulting mutant strain has been reported for its ability to cater rice seedlings with beneficial levels of nitrogen in short-term hydroponic experiments (Chaurasia and Apte, 2011). In order to utilize such biochemical traits in designed applied processes, it becomes important to understand community behavior and metabolic interactions in natural and simple ecosystems where these feature.
In fact, Anabaena sp. PCC 7120 can alone be argued to form such a very simple yet incompletely understood “community” of cells with multiple metabolic states and interdependent metabolic exchange. Under diazotrophic conditions, approximately every tenth vegetative cell irreversibly transforms into a heterocyst to provide a low-oxygen environment for the nitrogenase enzyme to function (Golden and Yoon, 2003). This enzyme is responsible for the conversion of atmospheric molecular nitrogen into ammonia in a highly energy-expensive reaction, consuming chemical energy stored in 16 molecules of ATP and eight electrons carried by ferredoxin molecules for every molecule of nitrogen assimilated. Furthermore, the nitrogenase is irreversibly inactivated by oxygen, which makes oxygenic photosynthesis and nitrogen fixation incompatible processes (Fay, 1992). Therefore, these specialized heterocyst cells undergo a series of changes to minimize the level of internal oxygen, including the deposition of two additional envelope layers around the cell and the degradation of photosystem II and carboxysomes (Wolk et al., 2004; Nicolaisen et al., 2009; Awai et al., 2010). As a result, heterocysts are dependent on vegetative cells as a source of electrons and carbon (Kumar et al., 2010). In return, vegetative cells obtain fixed nitrogen (Meeks and Elhai, 2002). Heterocysts and vegetative cells are therefore mutually interdependent, showing the features of a very simple “ecosystem.” This “ecosystem” is a suitable and simple model to simulate and elucidate community metabolism. In this study, our aim was to understand the metabolic interactions between the two cell types within the filament of Anabaena sp. PCC 7120 and to reveal the underlying reaction network enabling such a relationship. We therefore reconstructed a genome-scale metabolic network of Anabaena sp. PCC 7120 incorporating both cell types. The metabolic reconstruction allowed us to perform an exhaustive computational analysis of possible exchange metabolites and to rank exchange metabolites according to evolutionary optimality criteria, in particular growth yield of the filament. To benchmark our reconstruction, we compared growth states of Anabaena sp. PCC 7120 under both photoautotrophic and mixotrophic conditions, either consuming a combined nitrogen source or growing diazotrophically. Our stoichiometric model is represented using the systems biology markup language (Hucka et al., 2003) and is being analyzed by a constraint-based optimization approach (Price et al., 2003; Steuer et al., 2012). To our best knowledge, this reconstruction is the first extensively curated, genome-scale model for Anabaena sp. PCC 7120. It is also the first complete reconstruction for heterocyst metabolism and among the first attempts to simulate a simple multicellular organism at genome scale.
RESULTS AND DISCUSSION
A Manually Curated Stoichiometric Model of Anabaena sp. PCC 7120
The reconstruction process follows the protocol of Thiele and Palsson (2010) and is detailed in the “Materials and Methods.” In brief, based on the available genome annotation (Kaneko et al., 2001; Peterson et al., 2001) and reaction databases (Kanehisa et al., 2004; Caspi et al., 2012), gene-protein-reaction relationships were established. Elementally balanced biochemical reactions were sorted into six intracellular compartments (cytosol, thylakoid lumen, carboxysome, cytoplasmic membrane, thylakoid membrane, and periplasmic space) in order to simulate the growth of vegetative cells on a combined nitrogen source (single-cell model).
The single cell reconstruction contains a total of 777 metabolites interconnected via 804 enzymatic and 14 spontaneous reactions, as well as 79 transport reactions between intracellular compartments or the external space (Fig. 1). Ninety-nine of the unique metabolic reactions have not previously been reported in databases for Anabaena sp. PCC 7120 (Kanehisa et al., 2004; Caspi et al., 2012) but were acquired from the primary literature for this organism. In addition, 60 reactions were associated with their gene candidates here for the first time following a BLAST-based homology search. However, no candidate genes could be identified for a total of 36 reactions, of which 24 are essential for growth (see “Materials and Methods” for more details).
Figure 1.
Compartments considered in the two-cell model. The filamentous structure of Anabaena sp. PCC 7120 under diazotrophic conditions is represented by two supercompartments (a vegetative cell and a heterocyst) sharing certain metabolites via exchange reactions (red dashed line). Black arrows and numbers in italic indicate main fluxes of diazotrophic growth. Supercompartments are divided into (sub)compartments: Both cells contain a cytosol (light yellow body), a thylakoid lumen (green bands), a thylakoid membrane (dark green dashes), and a cytoplasmic membrane (blue dashes) and share a contiguous periplasmic space (white space delimited by the cytoplasmic and outer membranes). The vegetative cell also carries carboxysomes (orange hexagon). The brown body in the heterocyst is the nitrogen storage cyanophycin, not a separate compartment on its own. Numbers in parentheses indicate the number of reactions (including transport) associated with that compartment. Cell types and compartments are not to scale. BMG, biomass (growth); Cyt b6f, cytochrome b6f complex; Nase, nitrogenase; BMM, biomass (maintenance).
The reconstruction was examined for orphan reactions, that is, reactions that are disconnected from the remaining reactions. Such reactions (or reaction subsets) indicate misannotation or insufficiently known pathways. Orphan reactions cannot carry metabolic flux and therefore can either be kept or removed from the reconstruction without any observable impact on the flux balance analysis (FBA) solution. Here, a total of 98 orphan reactions were identified and moved to a separate file for later analysis (Supplemental File S5), all being more than two reaction steps away from the closest element within the network. Gaps that span two or less reaction steps were resolved by finding gene associations to the missing steps wherever possible. The gap-filling process is detailed in the “Materials and Methods” section.
Along with the reconstructed biomass objective function (see below), biosynthesis routes for the different carotenoids that accumulate in Anabaena sp. PCC 7120 were updated based on literature (Albrecht et al., 1996; Takaichi et al., 2005; Takaichi and Mochimaru, 2007; Mochimaru et al., 2008; Graham and Bryant, 2009). The currently incomplete pathways for the synthesis of phycobilin, thiamine, and molybdopterin in the Kyoto Encyclopedia of Genes and Genomes (KEGG; Kanehisa et al., 2004) were also revised (Schluchter and Glazer, 1997; Gutzke et al., 2001; Ruiz et al., 2010; Biswas, 2011). In addition, the Suc metabolism of Anabaena sp. PCC 7120 was extended as compared to current database entries for this organism (Cumino et al., 2007; Marcozzi et al., 2009; Du et al., 2013). The reconstruction also includes a proposed pathway for the iron (III)-siderophore schizokinen based on the biosynthesis route of a similar siderophore, rhizobactin, from Sinorhizobium meliloti 1021 (Lynch et al., 2001; Nicolaisen et al., 2008; D. Malatinszky and P.R. Jones, unpublished data). In order to aid the analysis of the model, a visual representation of the metabolic network was created (Supplemental Files S6 and S7) using the software package Cytoscape (Shannon et al., 2003).
Two-Cell Model and Biomass Composition
In a separate model, the single-cell model is transformed into two supercompartments (Table I) to reflect the multicellular structure of the Anabaena sp. PCC 7120 filament under diazotrophic conditions (two-cell model). The two-cell model contains a total of 1797 reactions, including exchange between the two supercompartments (see Fig. 1 and Table I for selected modifications to the heterocyst supercompartment). Transport reactions across compartments and exchange between supercompartments are assumed bidirectional, independent of ATP, and unconstrained in contrast to transport reactions to the external space, which are defined as ATP-driven, unless evidence for a different driving mechanism was found in the literature (rows 1, 3, and 8 in Supplemental File S8).
Table I. Major differences between the two supercompartments (cell types) in the Anabaena sp. PCC 7120 model.
| Vegetative Cell | Heterocyst |
|---|---|
| Photosynthesis (PSI and PSII)a | Cyclic photophosphorylation (PSI only)a (Wolk et al., 2004) |
| Carboxysomes and Rubisco | Neither carboxysomes nor an active Rubisco (Madan and Nierzwicki-Bauer, 1993;Valladares et al., 2007) |
| No nitrogen fixation | Nitrogen fixation (nitrogenase) |
| GS-GOGAT cycleb | Fd-GOGAT missingb (Martín-Figueroa et al., 2000) |
| FNR at PSI produces NADPHc | FNR produces red. FdxH for nitrogenasec (Razquin et al., 1996) |
| cox1 cytochrome c oxidase | Contains cox2 and cox3 only (Valladares et al., 2003) |
PSI, Photosystem I; PSII, photosystem II. bFd-, Ferredoxin-dependent. cFdxH, Heterocyst-specific ferredoxin.
The vast majority of the reactions exist in both the vegetative cell and the heterocyst, although there are characteristic differences between the two supercompartments. Most importantly, only the vegetative cell is able to perform oxygenic photosynthesis via linear photophosphorylation, whereas only the heterocyst is capable of performing nitrogen fixation, using cyclic photophosphorylation on photosystem I. Reactions responsible for oxygen evolution in photosystem II were therefore deleted from the heterocyst supercompartment. Similarly, the inactive Rubisco-dependent carbon fixation was removed from the heterocyst, although other carbon fixation mechanisms may still be active (see below). In addition, nitrogen metabolism in the heterocyst lacks the expression of the Gln oxoglutarate aminotransferase (GOGAT) enzyme, but it may have an active nitrogenase in place. The physiological differences between the two supercompartments are listed in Table I, while the resulting differences in active reactions are described in Supplemental File S5.
The terminally differentiated heterocyst does not grow or undergo cell division; therefore, the objective function of the two-cell model is defined as the growth of the vegetative cell (row 5 in Supplemental File S8). To account for macromolecular turnover in the heterocyst (row 6 in Supplemental File S8), the biomass reaction in this supercompartment is constrained to a lower bound equal to 10% of the maximum biomass production in the vegetative cell (van Bodegom, 2007). Moreover, both supercompartments include an artificial ATP hydrolysis reaction to account for the energy requirement of growth-independent cell maintenance at a fixed flux rate (row 7 in Supplemental File S8). This flux rate is equal to 10% of total ATP consumption at maximum growth rate, similar to previous stoichiometric models (Feist et al., 2007; Nogales et al., 2012; Knoop et al., 2013). In the initial two-cell model, the two supercompartments are allowed to exchange four metabolites: Suc (Schilling and Ehrnsperger, 1985; Cumino et al., 2007; Nürnberg et al., 2015), Gln (Wolk et al., 1976; Thomas et al., 1977; Picossi et al., 2005), Glu (Martín-Figueroa et al., 2000), and 2-oxoglutarate (2-og; Böhme, 1998; row 12 in Supplemental File S8). The transport reactions for these metabolites are unconstrained, bidirectional, and provide direct exchange between supercompartments without the involvement of other compartments or the external space. In addition, any dilution occurring due to the size difference of the two cell types is not taken into account (rows 2 and 9 in Supplemental File S8). At optimal growth, the heterocyst supercompartment was found to supply sufficient fixed nitrogen for the growth of exactly 7.6 new vegetative cells, based on the nitrogen content of the vegetative cell in the biomass equation. This calculation determines the maximum number of growing vegetative cells a single heterocyst can support. However, already existing vegetative cells adjacent to the heterocyst require less nitrogen to remain functional, increasing this ratio to the range observed experimentally (i.e. 10–20 vegetative cells per heterocyst). At such ratios, the predicted growth rate drops gradually, reaching about 46% of the maximal rate when a single heterocyst sustains exactly 20 vegetative cells. It is worth noting that the growth rate predicted by the model does not predict actual cell number, but biomass accumulation rate. Therefore, in our reconstruction, we represented the Anabaena sp. PCC 7120 filament as a single vegetative supercompartment and a single heterocyst supercompartment, denoted as the two-cell model in the following (Fig. 1; row 4 in Supplemental File S8).
Figure 1 summarizes the main metabolic fluxes concerning carbon and nitrogen metabolism in the two-cell model under diazotrophic growth conditions. The vegetative cell fixes carbon via the Calvin cycle driven by photosynthesis and produces an excess of Suc from glyceraldehyde 3-phosphate and an excess of Glu synthesized by the GOGAT enzyme. The primary source of Glu is internally recycled 2-og and Gln of heterocyst origin. The excess Suc and Glu are exchanged for Gln and 2-og from the heterocyst. Gln is derived from Glu by incorporating ammonia from heterocystous nitrogen fixation to Glu from the vegetative cell. Energy (ATP) and electron (reduced ferredoxin) requirements of the nitrogenase reaction are mainly covered by cyclic photophosphorylation at photosystem I. The rest of the energy is provided by degrading Suc from the vegetative cell and spending its carbon content on cellular maintenance (Fig. 1).
There is very little information on the exact composition of Anabaena sp. PCC 7120 biomass in the literature (Table II). Therefore, the biomass equation constructed for Synechocystis sp. PCC 6803 by Nogales et al. (2012) and adjusted by Knoop et al. (2013) was used here and adapted to Anabaena sp. PCC 7120, complemented by sparsely available analytical data (Table II). Vegetative cells and heterocysts were assumed to share the same fractional composition comprising DNA, RNA, proteins, pigments, lipids, cell wall, inorganic ions, and the metabolic pool (row 10 in Supplemental File S8). The fractional composition given in Nogales et al. (2012) was left unchanged; however, some of the fractions were recalculated if data were available (Table II; row 11 in Supplemental File S8). The impact of variation in the biomass composition on the predicted growth rate was simulated and the results are shown in Supplemental File S9. Even a ±20% variation in any one component did not influence the predicted growth rate by more than ±3%.
Table II. Biomass composition used in the model.
Fractional composition was adopted from the Synechocystis sp. PCC 6803 model developed by Nogales et al. (2012) and Knoop et al. (2013). The definition and the reaction formula of the different fractions were adapted for Anabaena sp. PCC 7120, wherever specific analysis data were available.
| Fraction | Source of Data and Reference |
|---|---|
| Pigments | Analysis of carotenoid composition (Takaichi et al., 2005; Takaichi and Mochimaru, 2007; Mochimaru et al., 2008; Graham and Bryant, 2009) |
| DNA | Base abundance calculated from the genome sequence (Kaneko et al., 2001) |
| RNA | Base abundance calculated for annotated genes and weighted by RNAseq abundance (Kaneko et al., 2001; Flaherty et al., 2011) |
| Proteins | Amino acid abundance calculated and weighted by RNAseq abundance (Kaneko et al., 2001; Flaherty et al., 2011) |
| Lipids | Adopted from Synechocystis sp. PCC 6803 (Nogales et al., 2012; Knoop et al., 2013) |
| Cell wall | Adopted from Synechocystis sp. PCC 6803 |
| Inorganic ions | Adopted from Synechocystis sp. PCC 6803 |
| Pool fraction | Adopted from Synechocystis sp. PCC 6803a |
No genes could be identified for the biosynthesis of spermidine (Jantaro et al., 2003; Incharoensakdi et al., 2010).
Computational Characterization of the Reconstructed Metabolic Model
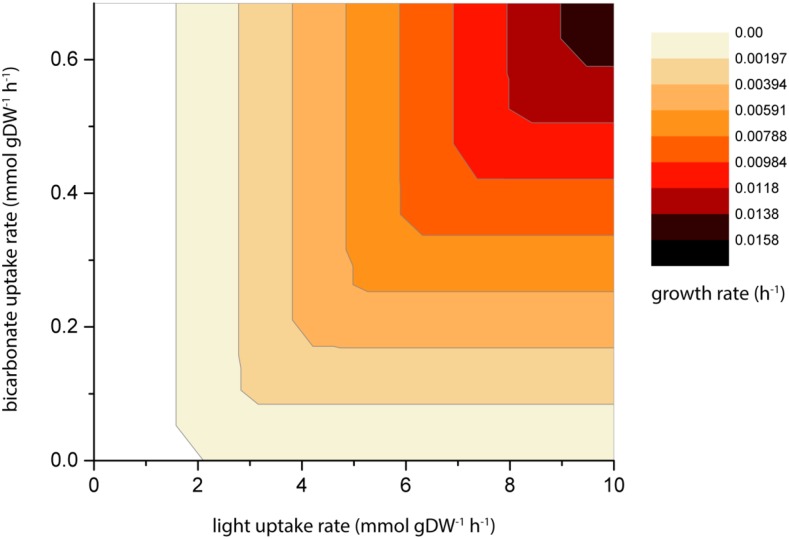
Simulations were run using the two-cell model under photodiazotrophic conditions with Glu, Gln, 2-og, and Suc as possible exchange metabolites, and bicarbonate, molecular dinitrogen and light as only external substrates. The uptake of the external substrates was constrained to upper bounds of 10 mmol g dry weight−1 h−1 each. Figure 2 shows the relationship between light intensity and bicarbonate uptake. The maximal growth rate is reached at 10 mmol g dry weight−1 h−1 photon flux (the upper bound) and the bicarbonate uptake rate of 0.68 mmol g dry weight−1 h−1.
Figure 2.
Predicted optimal growth rates of Anabaena sp. PCC 7120 as a function of light and bicarbonate. Darker colors represent higher growth rates (see legend on the right). Photon requirement of cell maintenance is represented by a white area on the left side of the contour plot. DW, Dry weight.
To obtain insight into the properties of the two-cell model and to test to what extent model-based predictions coincide with known metabolic exchange fluxes, we evaluated the two-cell model under photodiazotrophic conditions (Fig. 3).
Figure 3.
Growth rates predicted as a function of different transport reactions under diazotrophic conditions. A, Impact of light availability on growth rate when both cell types (dotted red line) or only the vegetative cell (solid blue line) harvest photons. B, Photon uptake by the heterocyst in combination with optimal light harvesting in the vegetative cell. C, Bicarbonate uptake by the two transport reactions (sodium symport, solid blue line; active transport, dotted red line) in the vegetative cell. D, Bicarbonate uptake by the heterocyst. E, Exchange of Suc if the Gln-to-Glu ratio is unbound (red dotted line) or fixed to 1 (black dashed line). F, Exchange of Glu and Gln at ratios fixed to 1 (black dashed line) or left unbound (solid blue and dotted red lines). Red double arrows on E and F show the direction of exchange. VC, Vegetative cell; HC, heterocyst; DW, dry weight.
The minimum photon requirement to cover non-growth-associated maintenance costs without supporting growth (x-intercept) is shown in Figure 3A. This requirement is equal to a photon flux of 6 mmol g dry weight−1 h−1, when light is harvested by the vegetative cell only (Fig. 3A, solid blue line). At the upper bound of the vegetative cell’s photon uptake (10 mmol g dry weight−1 h−1), the growth rate is 0.006 h−1 (Fig. 3A, solid blue line; Fig. 3B, y-intercept) that increases up to the maximum (0.0144 h−1) by the heterocyst’s contribution to light harvesting. In the case when both supercompartments harvest light (Fig. 3A, dotted red line), the energy contribution by the heterocyst lowers the requirement from the vegetative cell by about 2.3 mmol g dry weight−1 h−1. This contribution by the heterocyst via cyclic photophosphorylation saturates at approx. 4.2 mmol g dry weight−1 h−1, over which the proton gradient through the thylakoid membrane is replenished by secondary reactions in the electron transport chain without the synthesis of additional ATP (Fig. 3B). In contrast, light or carbon uptake by the heterocyst alone cannot support growth of the vegetative cell (data not shown). On the other hand, any of the two bicarbonate transporters in the vegetative cell can provide sufficient carbon for growth, although the maximum rate is lower for the ATP-driven transport (Fig. 3C, dotted red line). When forcing bicarbonate uptake over the optimum, however, both transport reactions have a negative effect on growth, as light becomes limiting (Fig. 3C, both lines). According to Figure 3D, maximal growth can be achieved even at zero bicarbonate uptake by the heterocyst, suggesting that the vegetative cell alone can fix sufficient amount of carbon (via Rubisco in the carboxysome) to reach maximum growth rate. Moreover, if bicarbonate uptake in the heterocyst is enforced (Fig. 3D, both curves), the carbon is not utilized for growth (Fig. 3D, straight horizontal lines on the left side of each curve) but rather recycled via the C4 dicarboxylic acid cycle and released as carbon dioxide (not shown). The recycling capacity depletes around 1.8 and 2.6 mmol g dry weight−1 h−1 bicarbonate over the active transport and the symport, respectively. Nonetheless, it is still unclear whether in reality heterocysts utilize this C4 route to fix their own carbon (Popa et al., 2007), although cyanobacteria have been described to assimilate about 20% of the total fixed CO2 in the form of C4 acids (Owttrim and Colman, 1988; Luinenburg and Coleman, 1992, 1993). Notably, experimental evidence suggests that the main source of carbon for heterocysts is likely to be Suc (Böhme, 1998; Martín-Figueroa et al., 2000; Kumar et al., 2010; Nürnberg et al., 2015). In contrast, the model predicts a very low flux for Suc at the optimal growth rate (Fig. 3E, red dotted line). In the same simulation, Glu is transferred to the heterocyst at a 2-fold higher rate than that of Gln, moving in the reverse direction. This is possible because Gln and Glu exchange were optimized as independent reactions and suggests that Glu is partially utilized as a carbon source in the heterocyst. However, Suc becomes the primary source of carbon in the heterocyst (also increasing its flux by 4 times) if the Gln-Glu exchange ratio is fixed to 1, while the growth optimum decreases by only about 7% (Fig. 3, E and F, dashed black lines; row 13 in Supplemental File S8). Further tests of the model comparing growth rates on different carbon and nitrogen sources can be found in Supplemental File S10.
Experimental Evaluation of Model Predictions on Carbon Source Utilization by the Single-Cell Model
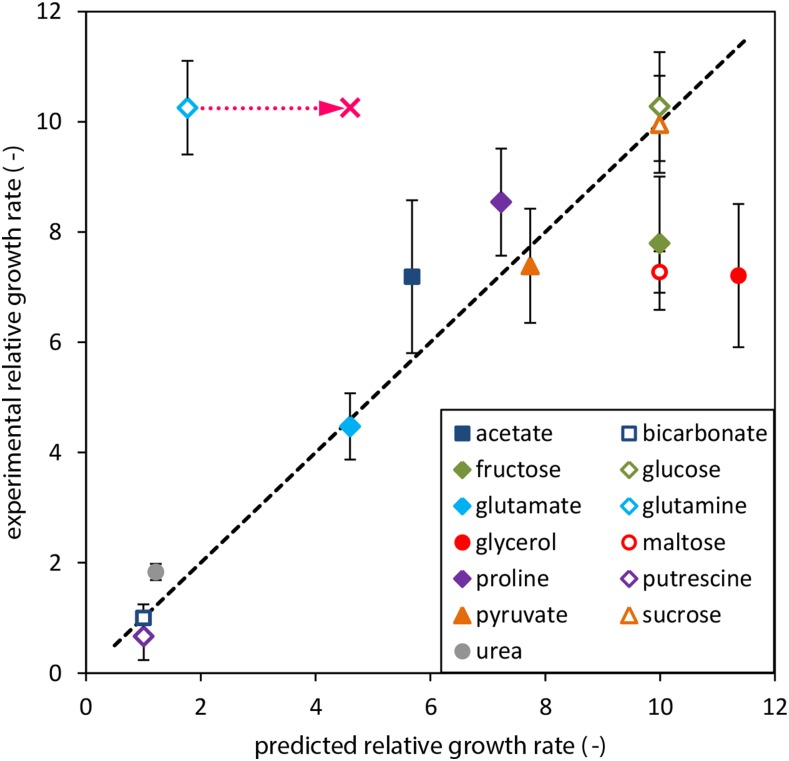
Experimental and predicted mixotrophic growth rates on a variety of carbon sources were compared relative to autotrophic growth on bicarbonate, using nitrate as the sole nitrogen source. In the presence of nitrate, no heterocysts are expected to form; the following growth rate calculations were therefore performed using the single-cell model. Tested carbon sources included bicarbonate, urea, sugars (Glc, Fru, Suc, and maltose), a sugar alcohol (glycerol), fermentation products (pyruvate and acetate), amino acids (Glu, Gln, and Pro), and a polyamine (putrescine). For comparative reasons, each uptake reaction was assumed to hydrolyze one molecule of ATP and was constrained to transport equal moles of carbon with each substrate (row 14 in Supplemental File S8). Light harvesting and bicarbonate and nitrate uptake were constrained to an upper bound of 10 mmol g dry weight−1 h−1. The two datasets of relative growth rates in exponential phase are shown in Figure 4.
Figure 4.
Correlation between experimental and predicted mixotrophic growth rates in the exponential phase on 12 different carbon sources, relative to autotrophic growth on bicarbonate. The pink cross and dotted arrow show the shift in predicted growth rate on Gln if ammonia is being excreted. The black dashed line highlights the correlation between experimental and computational datasets. Error bars depict ± sd of three biological replicates.
Most data points show a good fit to the trend, except for glycerol, some sugars, and Gln. The latter amino acid supported the second highest growth rate in the experiments, exceeded only by Glc. In contrast, simulated growth rate on Gln was just above the control bicarbonate. Interestingly, the model predicted the same growth rate for Gln and Glu when ammonia excretion was allowed, resulting in a 2.6-fold increase of growth rate on Gln. This suggests that the 2-fold higher molar nitrogen content of Gln makes this amino acid a stoichiometrically unfavored substrate compared to Glu, which can only be overcome by the removal of excess nitrogen. By excreting nitrogen, the model compensates for the suboptimal C/N ratio of Gln and shifts the intracellular C/N ratio to the same level as with Glu (supplemental figure S3 in Supplemental File S10, green squares). The adjustment via addition of ammonia excretion only brings predicted Gln halfway to experimental levels, and the contrasting difference between the two datasets therefore remains incompletely explained. In addition, excretion of ammonia under diazotrophic conditions has not yet been observed experimentally. However, it may be possible that the excess nitrogen is being deposited to nitrogen storage (i.e. cyanophycin), rather than being lost via excretion.
Among the sugar compounds, datasets for Glc and Suc showed good correlation, while Fru and maltose underperformed in wet-lab experiments. In fact, the four sugars showed essentially the same growth rate in simulations, due to high stoichiometric similarities between the metabolic pathways of these substrates (further discussed for supplemental figure S1 in Supplemental File S10). In silico, growth on glycerol resulted in the highest growth rate, whereas experimental growth on Glc achieved the highest rate. Glc and glycerol are both metabolized to glyceraldehyde 3-phosphate, but via two different pathways. We note that suboptimal growth on glycerol as sole carbon source, as compared to predictions using an FBA-based model, was also previously observed for Escherichia coli K-12 (Ibarra et al., 2002). For E. coli, adaptive evolution resulted in an increased growth rate on glycerol, in good agreement with model-predicted values (Ibarra et al., 2002). For this model, a variety of reasons might be responsible for the discrepancy, such as the thermodynamic properties of the dehydrogenation step to dihydroxyacetone phosphate or lack of appropriate NAD(P)(H) balancing. Nonetheless, the results in Figure 4 show that the constraint-based model provides a reasonable prediction of mixotrophic growth rates on different carbon sources, thereby justifying the use of the stoichiometric model to evaluate the feasibility and optimality of potential exchange reactions in more detail.
Stoichiometric Evaluation of Metabolite Exchange within the Filament
Following the characterization of the curated single-cell model, we sought to obtain insight into the stoichiometric optimality of the metabolic exchange between vegetative cells and heterocysts. Notably, a systematic analysis of the stoichiometric and energetic implications of different metabolites in intraspecies cellular exchange is challenging to carry out experimentally, while being feasible in silico. From previous studies, Suc was proposed to act as the sole source of electrons and carbon for heterocysts (Curatti et al., 2002; Golden and Yoon, 2003; Cumino et al., 2007), and Gln was suggested to serve as a nitrogen carrier and Glu as carbon skeleton for ammonia incorporation (Flores and Herrero, 2010; Kumar et al., 2010). In addition, the lack of GOGAT in heterocysts was postulated to result in the accumulation and subsequent transport of 2-og into the vegetative cells (Böhme, 1998; Martín-Figueroa et al., 2000). These compounds and other central carbon metabolites, including some amino acids that may be involved in intercellular nitrogen exchange (Montesinos et al., 1995), were included as potential exchange metabolites in the two-cell model. In addition to amino acids, ammonia was also investigated as an alternative carrier of nitrogen. Therefore, in total, twelve different metabolites were considered as exchange metabolites, and all possible combinations were comprehensively evaluated with respect to the predicted maximal growth rate, resulting in 4096 combinations. Earlier simulations suggested that the ratio of Gln and Glu exchange may be constrained to unity (Fig. 3), and therefore this case (as an additional constraint) was also included. Results were plotted as a distribution chart against the number of exchange reactions involved in each solution (Fig. 5A). Selected exchange metabolite combinations (Fig. 5B) showing the highest growth rates by involving the least number of exchange metabolites are further evaluated in Figures 6 and 7, highlighting the major metabolic pathways and reactions involved in the solution.
Figure 5.
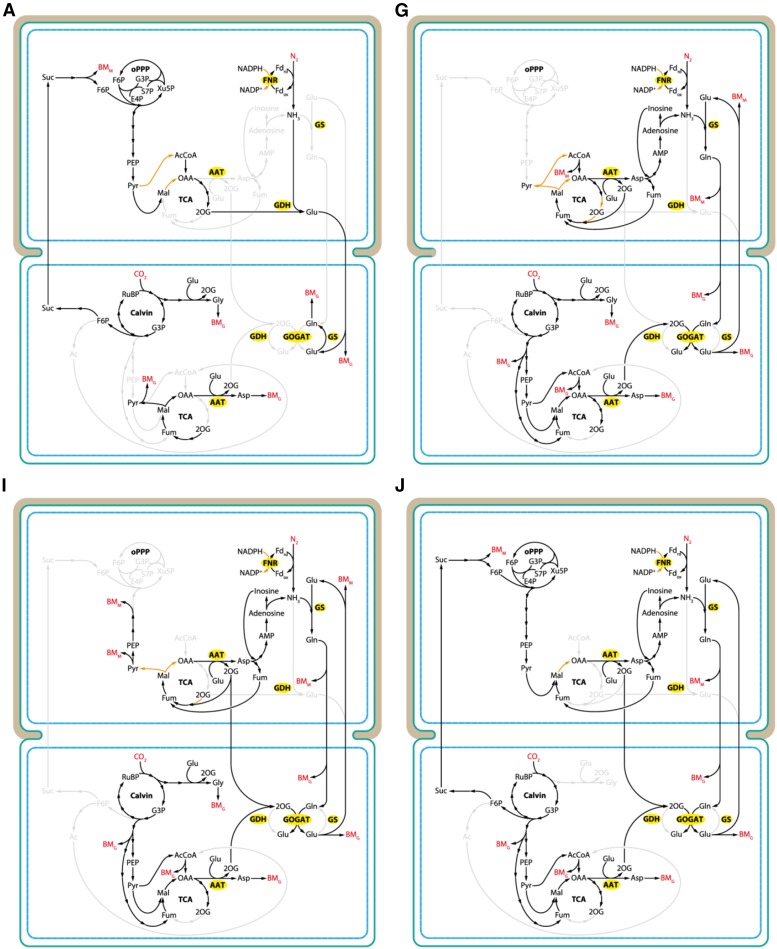
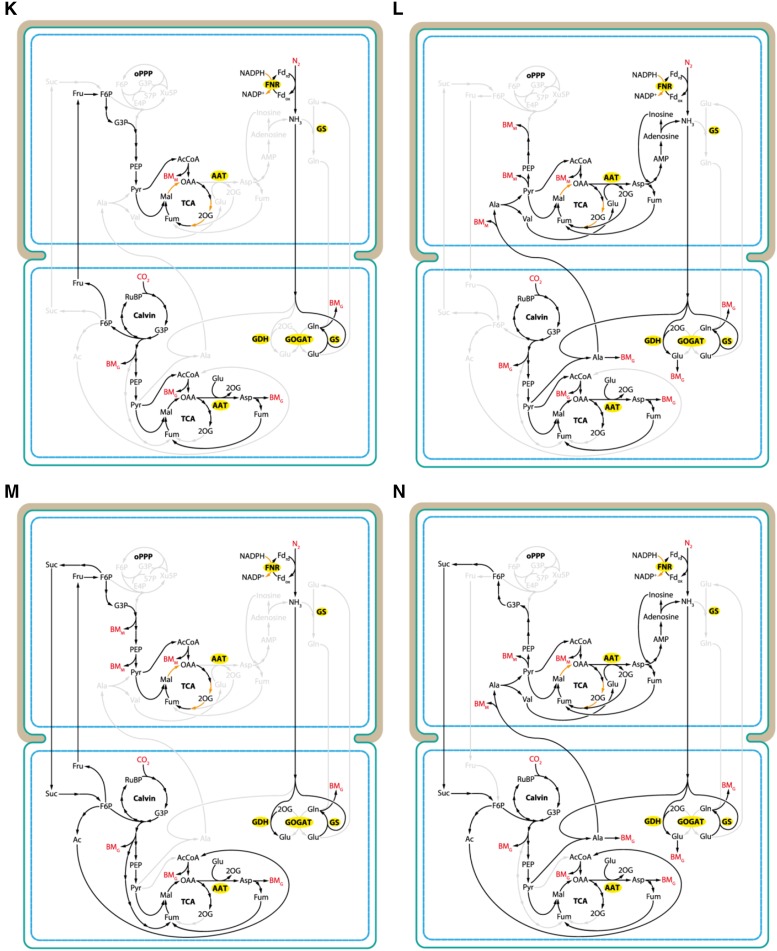
Predicted growth rates in response to the number of intercellular exchange reactions. A, Each colored area represents the distribution of nonzero solutions with a different set of exchange reactions. In case of yellow and green, Gln-to-Glu ratio was fixed to 1. The ratio was unbound for cases red and teal. Yellow and red areas evaluate the effect of ammonia exchange. Blue and purple areas highlight solutions where Suc was consumed by the heterocyst. The growth rate for the four exchange metabolites suggested in the literature (i.e. Suc, 2-og, Glu, and Gln) is highlighted by a red circle (Gln-to-Glu ratio unbound) and a blue square (ratio fixed to 1). All simulations were performed using the two-cell model. suc, Suc; nh3, ammonia; gln, l-Gln; glu, l-Glu; 2og, 2-oxoglutarate; fru, Fru; glc, Glc; pyr, pyruvate; ala, l-Ala; his, l-His; orn, l-Orn; cyp, cyanophycin monomer. B, Zoomed-in section from A showing growth rates for selected exchange metabolite combinations. Letters refer to cases in C and the corresponding cases in Figures 6 and 7. C, Combination of a maximum of four reactions exchanging Suc, Gln, Glu, or 2-oxoglutarate (2OG). Arrows indicate the metabolites exchanged and the direction of the exchange (uptake by the vegetative cell and the heterocyst is represented by green and beige colors, respectively). Black bars show growth rate in each case. . VC, Vegetative cell; HC, heterocyst.
Figure 6.
Main metabolic fluxes connecting exchange metabolites Suc, Gln, Glu, and 2-og. Panels are labeled according to cases on Figure 5, B (blue dots) and C, and show increasing growth rates from case A to case J. Exchange reactions are as follows: A, +Suc −Glu; G, +Glu −Gln; I, +Glu −Gln −2-og; J, +Suc +Glu −Gln −2-og. Ratio of Glu to Gln was unbound. Compounds in red indicate a sink or uptake of an external metabolite. Orange arrows in the heterocyst highlight reactions providing electrons (NADPH) for nitrogenase. BMG, Biomass (growth); BMM, biomass (maintenance). Enzyme names are highlighted in yellow. Upper cells, Heterocysts; lower cells, vegetative cells. oPPP, oxidative pentose-phosphate pathway; BMM, biomass (maintenance); G3P, glyceraldehyde 3-phosphate; E4P, erythrose 4-phosphate; S7P, sedoheptulose 7-phoshate; Xu5P, xylulose 5-phosphate; PEP, phosphoenolpyruvate; Pyr, pyruvate; AAT, aspartate aminotransferase; Mal, malate; Fum, fumarate; OAA, oxaloacetate; RuBP, ribulose 1,5-bisphosphate; BMG, biomass (growth).
Figure 7.
Main metabolic fluxes connecting exchange metabolites Suc, Fru, Ala, and ammonia. Panels are labeled according to cases on Figure 5B (red dots) and show increasing growth rates from case K to case N. Exchange reactions are as follows: K, +Fru −NH3; L +Ala −NH3; M, +Fru −Suc −NH3; N, +Ala −Suc −NH3. Ratio of Glu to Gln was unbound. Compounds in red indicate a sink or uptake of an external metabolite. Orange arrows in the heterocyst highlight reactions providing electrons (NADPH) for nitrogenase. BMG, Biomass (growth); BMM, biomass (maintenance). Enzyme names are highlighted in yellow. Upper cells, Heterocysts; lower cells, vegetative cells. Ac, acetate; AcCoa, acetyl-coenzyme A; AMP, adenosine monophosphate.
None of the exchange metabolites investigated here, including ammonia and Gln, were able to individually allow growth of the filament (Fig. 1). Any combination of two exchange metabolites did not result in maximal growth rates. As the best-ranked combination of two exchange reactions, export of ammonia and import of Ala (from the heterocysts’ perspective) resulted in 70% of the maximal growth rate (Fig. 5B, case L). Moreover, although a total of twelve metabolites were allowed to exchange, no more than 10 were ever chosen in any combination by the model to provide a feasible solution and nonzero growth. The maximal growth rate was reached by combinations of seven reactions, with only slight improvements over the combinations of four reactions. Flux distributions were, to variable extent, different in each case.
At maximal growth rates, regardless of the number of exchange reactions involved or the Gln-Glu ratio, Ala was consumed by the heterocyst and nitrogen was transferred as ammonia. Furthermore, Suc was produced and excreted, rather than consumed, by the heterocyst. When Suc was consumed (blue and purple areas), Gln, Glu, and 2-og were transported, as suggested by literature. However, the growth rate was on average 30% higher if Suc was exported, independent of the Gln-Glu ratio. Ammonia was favored over Gln for the transfer of nitrogen throughout the simulation, increasing growth rates by 5% to 7% when the Gln-to-Glu ratio was fixed (Fig. 5A, yellow area) and by 3% to 7% when it was unbound (Fig. 5A, red area). When only four reactions were allowed, inclusion of ammonia increased growth rate by 38% over the reference (Fig. 5A, blue square) if the Gln-to-Glu ratio was fixed, and by 34% over the reference (Fig. 5A, red circle) if unbound. Those four exchange metabolites suggested in literature, i.e. Suc, Gln, Glu, and 2-og were evaluated in more detail by looking at the flux distribution of each possible combination individually (Fig. 5, B, blue line, and C). Only 10 out of the 16 exchange combinations allowed growth, with +Suc and −Glu giving the lowest growth rate (in the following, + and − signs preceding metabolite names denote uptake and excretion by the heterocyst, respectively). In this case (Fig. 6, case A) an incomplete tricarboxylic acid cycle (TCA cycle) is driven by Suc originated from the vegetative cell to produce 2-og by isocitrate dehydrogenase and then convert it to Glu by Glu dehydrogenase (GDH), incorporating ammonia fixed by nitrogenase. Eventually, electrons required by nitrogenase are also derived partially from Suc via pyruvate. The Glu produced is transferred back to the vegetative cell to serve as a source of assimilable nitrogen. This nitrogen is directly incorporated into different amino acids and, finally, biomass (Fig. 6, case A). Growth rate increases in ascending order when −Glu +2-og (Fig. 5C, case B), +Suc −Gln (Fig. 5C, case C), and Gln +2-og (Fig. 5C, case D) are combined. Interestingly, pairing Gln with Glu (−Gln +Glu; Fig. 5C, case G) improves growth rate the most significantly among all the combinations of two reactions. About 78% of Glu is used to incorporate ammonia into Glu by Gln synthetase (GS) and is sent back to the vegetative cell as Gln. The rest of the Glu fuels the second half of the TCA cycle via Asp transaminase. In the vegetative cell, GOGAT instead of GS becomes active and produces Glu for filament growth and for the heterocyst. The 2-og required by GOGAT is the product of a broken TCA cycle and Asp transaminase converting oxaloacetate to Asp in the vegetative cell (Fig. 6, case G). Addition of 2-og exchange increases growth rate by another 7% (Fig. 5C), allowing the heterocyst to recycle some of the Glu and return it to the vegetative cell as 2-og, independent of the exchange of nitrogen (Fig. 6, case I). This carbon transfer from the heterocyst in the form of 2-og becomes higher if Suc is also allowed to exchange (Fig. 6, case J). In this case, Suc is used to run only the second half of the TCA cycle without consuming any 2-og in the pathway and sending the majority of 2-og to the vegetative cell. In the absence of an active first half of the TCA cycle, the primary source of electrons for nitrogenase becomes malate dehydrogenase, also involving transhydrogenase (Fig. 6, case J, orange arrow in the upper cell). Transhydrogenase shuffles electrons from NADH produced in the malate dehydrogenase reaction to NADPH. In all cases discussed above (Fig. 6, cases A, G, I, and J), the main provider of reduced ferredoxin for nitrogenase is ferredoxin-NADP+ reductase (FNR) transferring electrons from NADPH to ferredoxin.
The highest increase in growth rate due to the addition of one more reaction occurred from two to three (Fig. 5B, red line and circles), using ammonia as the carrier of fixed nitrogen. The lowest growth rate among those with only two reactions was achieved when ammonia exchanged for Fru (Fru, Fig. 7, case K). Ammonia in the vegetative cell is assimilated solely by GS, and the resulting Gln is incorporated to other amino acids and biomass. The source of Fru is the Calvin cycle, and Fru is converted to pyruvate in the heterocyst. Pyruvate eventually forms oxaloacetate to maintain the TCA cycle, providing NADPH for the reverse reaction at FNR producing reduced ferredoxin for nitrogenase (Fig. 7, case K). As discussed above, exchange of ammonia for Ala (Fig. 7, case L) gave the highest growth rate among all the combinations of only two reactions. Ammonia in this case is assimilated by both GS and GDH in the vegetative cell with significantly higher contribution from the first enzyme, while GOGAT remains inactive. In addition, ammonia is also incorporated to pyruvate, forming Ala that is then transferred back to the heterocyst. The nitrogen carried by Ala is converted eventually back to ammonia via Asp and adenosine.
Allowing exchange of Suc, growth rate improves by about 30% (Fig. 5B, case M) compared to the case when only +Fru and −NH3 were exchanging (Fig. 5B, case K). Also, the model increases Fru exchange 100 times and sets Suc flux accordingly. In other words, Fru and Suc exchange fluxes are closely equivalent for carbon content, except for some Fru consumed by the heterocyst to run the TCA cycle. Stoichiometrically, the net outcome of this cycling of Fru to Suc in the heterocyst and Suc to Fru in the vegetative cell is ultimately one mole of ATP for every mole of Fru by the reverse reaction of fructokinase in the vegetative cell. It is unclear, however, if such metabolite concentrations to shift the fructokinase reaction kinetics to the direction of ATP generation can ever occur in a vegetative cell. It is also worth noting that in most cases the model favored, when available, exchange of carbon sources other than Suc or transported Suc toward the vegetative cell. In case of the fastest growing among those using only three reactions (case N), the increase in growth rate due to the addition of Suc exchange is about 36% compared to case L (Fig. 5B), which uses only two reactions (+Ala −NH3). Similar to that, case N has no active GOGAT in the vegetative cell while actively assimilating ammonia via GS, GDH, and Ala dehydrogenase producing Ala. The higher exchange rate of Ala compared to case L increases the flux over the second half of the TCA cycle as well (generating more reducing equivalents), allowing higher nitrogen fixation rate and, ultimately, more vegetative cell biomass. The carbon content of Ala is recycled through pyruvate and Fru 6-phosphate and sent back to the vegetative cell in the form of Suc.
CONCLUSION
As yet, stoichiometric reconstructions of cyanobacterial metabolism have mainly focused on unicellular nondiazotrophs (Saha et al., 2012; Knoop et al., 2013) or nonheterocystous diazotrophs (Resendis-Antonio et al., 2007; Saha et al., 2012; Vu et al., 2012). Here, we present a curated genome-scale stoichiometric model of a filamentous heterocystous nitrogen-fixing cyanobacterium with two distinct cell types.
During the reconstruction, process 60 genes and proteins have been newly annotated and associated with a metabolic function based on sequence homology and, in a few cases, experimental observations. In addition, a total of 36 gene candidates have been proposed to fill essential and nonessential metabolic gaps in the biochemical network of Anabaena sp. PCC 7120. Moreover, the extensive manual curation of every reaction in the reconstruction and the design of a detailed interactive network map (Supplemental File S6) allowed us to identify and eliminate inconsistent reactions that represented roughly 30% of the total reactions found in current metabolic databases for this organism.
The model correctly predicted the vegetative cell-to-heterocyst ratio under diazotrophic conditions, showing that the heterocyst supercompartment (a single heterocyst) can supply the formation of 7.6 vegetative cells at maximum growth rate. This ratio was increased at the expense of vegetative cell growth rate, decreasing growth rate to 46% percent of the maximum to sustain 20 vegetative cells by a single heterocyst. Nitrogen uptake rate in the heterocyst was not limited by the upper bound of its current constraint (i.e. 10 mmol g dry weight−1 h−1) or by light, but instead, it was limited by the amount of carbon skeleton (Glu) the vegetative cell was able to provide for the incorporation of ammonia. From a stoichiometric point of view, the heterocyst can be forced to produce even more fixed nitrogen from more Glu, at the expense of vegetative cell growth rate.
Growth simulations on 13 different carbon sources suggested that Glc and Suc are among the highest yielding substrates, followed by pyruvate, Pro, acetate, and Glu. These results were confirmed by growth rate experiments, which showed a good overall correlation between the two datasets.
Based on these results, all possible combinations of 12 potential exchange metabolites were evaluated in the two-cell model with respect to stoichiometric optimality. Within the simulations, no more than 10 exchange reactions were active simultaneously, and the maximum growth rate was achieved by the combination of only seven exchange reactions. A minimum of two exchange metabolites were always required for growth, and the exchange of two metabolites already results in 70% of the maximum growth rate. The best-ranked two-metabolite scenario was exchange of ammonia for Ala in the heterocyst. In general, when ammonia was transferred, growth rates were consistently higher by about 3% to 7% than the growth rates obtained using Gln as the nitrogen carrier. In case of four reactions, inclusion of ammonia increased growth rate by 34% to 38%, while Suc was excreted by the heterocyst rather than being consumed. When the heterocyst utilized Suc as a source of electrons and carbon, however, Gln, Glu, and 2-og were selected by the model as the best combination for growth, in a good agreement with the literature (Wolk et al., 1976; Thomas et al., 1977; Schilling and Ehrnsperger, 1985; Böhme, 1998; Martín-Figueroa et al., 2000; Picossi et al., 2005; Cumino et al., 2007). Notably, the rate of Suc transport was closer to reported figures (Nürnberg et al., 2015) when the exchange of Gln to Glu was fixed to a 1:1 ratio. The model correctly predicted the importance of Suc in the vegetative cell-heterocyst system, when Gln, Glu, and 2-og were also exchanged. However, in many cases higher growth rates were achieved when Suc was transferred back to the vegetative cell rather than being consumed by the heterocyst, suggesting that stoichiometrically this direction of Suc exchange is more optimal. In other cases combining the exchange of Fru and Suc (among other metabolites), the model found a way to ultimately transfer ATP from the heterocyst to the vegetative cell. This transfer utilizing the reverse reaction of fructokinase may be thermodynamically feasible at product concentrations 100 times higher than that of Fru, although there is no evidence that such concentrations occur in a living cell. Nevertheless, it is important to evaluate the consequences of different exchange metabolite combinations, even if the results do not match all experimental observations. In fact, only a model can provide exact answers to stoichiometric optimality, which may be a key in understanding an organism's complex metabolic network.
Our model not only provides a comprehensively curated blueprint for the genome-scale metabolic network of Anabaena sp. PCC 7120, but also serves as an important computational tool that may allow the design of engineering strategies for the most studied nitrogen-fixing cyanobacterium. Even though Anabaena sp. PCC 7120 may primarily be suitable for laboratory research, it is a highly suited first target organism to assess proof-of-principle engineering strategies toward the sustainable production of combined nitrogen (Chaurasia and Apte, 2011) or other important bioproducts (Heyer and Krumbein, 1991).
In future work, the model would greatly benefit from the inclusion of a more precise biomass equation, specific for Anabaena sp. PCC 7120 both under diazotrophic and nondiazotrophic conditions. Such an update of the current biomass equation to reflect the actual elemental composition of the organism's biomass may resolve the discrepancy between the computationally predicted and experimentally observed growth with Gln.
MATERIALS AND METHODS
Metabolic Reconstruction of Anabaena sp. PCC 7120
A comprehensive protocol for the generation of high-quality genome-scale models was followed here (Thiele and Palsson, 2010) to reconstruct the metabolic network of Anabaena sp. PCC 7120. Assumptions made during the reconstruction process are collected in Supplemental File S8.
The complete genome sequence and gene annotation of Anabaena sp. PCC 7120 are available in several databases (Kaneko et al., 2001; Kanehisa et al., 2004; Nakao et al., 2010). The most commonly used annotation was acquired from the curated RefSeq Genome Database of the National Center for Biotechnology Information (NCBI; Tatusova et al., 2014) and juxtaposed to a recent independent annotation from the J. Craig Venter Institute (Peterson et al., 2001) to pair a meaningful function and a protein product to as many genes as possible. The metabolic function of proteins derived from genomic data were collected from biochemical repositories (Kanehisa et al., 2004; Magrane and Consortium, 2011; Caspi et al., 2012) and primary literature (Supplemental File S3). A systematic, automated algorithm to predict novel gene-protein reaction associations for cyanobacteria was also considered (Krishnakumar et al., 2013) but not used here due to the large number of contradictions to the other sources and experimental data.
All the information gathered above was mapped onto general metabolic pathways drawn for the KEGG database (Kanehisa et al., 2004) and compared with the data therein. The most recent metabolic reconstruction of the unicellular cyanobacterium Synechocystis sp. PCC 6803 (Knoop et al., 2013) was also mapped for comparison and to serve as a template during the identification of critical metabolic functions and gaps. The majority of these gaps were resolved either by finding genes in primary literature or by identifying novel gene/protein candidates based on sequence homology (light and dark pink bands in Fig. 8, respectively). Homology-based searches were performed using the BLASTX engine (Altschul et al., 1990) on the NCBI Anabaena sp. PCC 7120 proteome against gene sequences of reviewed protein entries from the UniProt Knowledgebase (Magrane and Consortium, 2011) to identify best hits. The best hits were verified by a BLASTX search against NCBI RefSeq protein database filtered for cyanobacterial entries (Supplemental File S4). A few gaps, otherwise unresolved, were resolved by artificially adding the corresponding metabolic reaction to the model to allow the biosynthesis of key metabolites (e.g. l-Met and l-Asn; light green bands in Fig. 8) or complete missing steps of close-to-compete pathways (dark green bands in Fig. 8). Gaps with over two missing reaction steps were treated differently. In most cases, such gaps indicate orphan reactions and appear in sparsely represented pathways due to misannotation. Lacking any connection to the rest of the network, these orphan reactions do not carry metabolic flux in an FBA solution and were therefore removed from the reconstruction (dark gray bands in Fig. 8). Dead-end metabolites not consumed by any reaction in the network were resolved by sink reactions artificially added to the model (yellow bands in Fig. 8, also including the biomass equations).
Figure 8.
Comparison between Anabaena sp. PCC 7120 and Synechocystis sp. PCC 6803 (Knoop et al., 2013) stoichiometric models and their improvement over the KEGG database (Kanehisa et al., 2004). Metabolic gaps were resolved by either adapting reactions from literature (light pink bands) or by identifying new gene candidates (dark pink bands). A number of gaps could not be associated with any gene in Anabaena sp. PCC 7120 (light and dark green bands). Some reactions in the KEGG database were omitted due to inconsistent coenzyme usage or incomplete reaction formula (light gray band) or due to the lack of connection to the rest of the network (orphan reactions, dark gray band).
The equations for each metabolic reaction were adopted either from public databases (Kanehisa et al., 2004; Caspi et al., 2012) or from the Synechocystis sp. PCC 6803 model (Knoop et al., 2013). Reaction thermodynamics, in terms of reaction directionality, were either acquired from the MetaCyC database (Caspi et al., 2012) or calculated using eQuilibrator, an online calculator to estimate reactions’ Gibbs free energy change (Flamholz et al., 2012). Also, every reaction was evaluated for mass and charge balance using eQuilibrator and adjusted if necessary. Those reactions not found in public resources were set bidirectional. Coenzyme dependencies (NAD+, NADP+, and quinones) in the KEGG database were adapted for Anabaena sp. PCC 7120 wherever primary biochemical evidence was available in the literature or left unchanged (light gray band in Fig. 8).
The stoichiometric model for Anabaena sp. PCC 7120 was generated in two formats: as a single-cell model including photosynthesis and carbon-concentrating reactions for nondiazotrophic growth in a vegetative cell (Supplemental File S2), and as a two-cell model setting every reaction to the right supercompartment (either a heterocyst or a vegetative cell) under diazotrophic conditions (Supplemental File S1). Simulations described in the “Results and Discussion” section, unless otherwise noted, were performed on the two-cell model considering six intracellular compartments (cytoplasm, cytoplasmic membrane, thylakoid lumen, thylakoid membrane, carboxysome, and periplasmic space) and the external medium (Fig. 1).
Stoichiometric Simulations and Model Evaluation
All simulations were run using the COBRA toolbox version 2 with Gurobi Optimizer 5.6.0 as the solver in MATLAB R2103b environment (MATLAB, 2011; Schellenberger et al., 2011; Gurobi Optimization, 2013). All FBA optimizations were calculated using a parameter setting to minimize the taxicab norm. In addition, bicarbonate uptake rate was constrained to an upper bound of 10 mmol g dry weight−1 h−1 in phototrophic conditions for both cell types. Simulations with the single-cell model were run on nitrate, as the nitrogen source up to 10 mmol g dry weight−1 h−1, unless otherwise noted. The two-cell model was set to the uptake of molecular nitrogen with an upper limit of 10 mmol g dry weight−1 h−1. When comparing single-cell growth on combined nitrogen to the two-cell model under diazotrophic conditions, both models were constrained for an equivalent total photon uptake of 10 mmol g dry weight−1 h−1. The optimal distribution of the 10 mmol g dry weight−1 h−1 photons among the two supercompartments was 7 and 3 mmol g dry weight−1 h−1 in the vegetative cell and the heterocyst, respectively. In all other simulations using the two-cell model, photon uptake in both supercompartments was constrained to 10 mmol g dry weight−1 h−1. For the evaluation of exchange metabolites between the heterocyst and the vegetative cell, unconstrained bidirectional diffusion was included to the model for each metabolite.
ATP-driven transport reactions have been added to the single-cell model for each carbon and nitrogen source except for molecular nitrogen and ammonia that were exchanged via simple diffusion. In case of mixo- and heterotrophic simulations, nitrogen was supplied solely by nitrate, whereas nitrogen sources were compared on bicarbonate in autotrophic conditions. In heterotrophic simulations, both photon and bicarbonate uptake rates were set to zero. Under mixotrophic conditions, photon and bicarbonate uptake reactions were constrained to 10 mmol g dry weight−1 h−1. Both carbon and nitrogen uptake fluxes have been constrained to carry a maximum of 10 mmol g dry weight−1 h−1 carbon and nitrogen source, respectively. For example, upper bound of the bicarbonate transport reaction (a single carbon atom) was set to 10 mmol g dry weight−1 h−1, whereas Gln uptake was set to 2 mmol g dry weight−1 h−1 (five carbon atoms).
Growth Rate Experiments
Wild-type Anabaena sp. strain PCC 7120 was grown in triplicates at 30°C, with continuous illumination from cool-white LED lamps at 60 μE m−2 s−1, on a rotary shaker at 200 rpm and in sterile 100-mL Erlenmeyer flasks containing 30 mL BG-11 medium (Rippka et al., 1979) until approximately OD730 = 1 measured in a 1-cm cuvette. Culture health was evaluated by reading the absorbance spectrum between 300 and 800 nm. The spectra indicated no differences in pigment composition of the biological replicates. The three replicates were then mixed to minimize biological variation. Cells from the mixed wild-type culture were harvested by centrifugation at 3000g, washed in fresh BG-11, and resuspended to the original volume in fresh BG-11.
Each of the BG-11C media containing one of the organic carbon sources were prepared from the same BG-11 standard medium described elsewhere (Rippka et al., 1979), by replacing bicarbonate for the corresponding organic substrate encapsulating equimolar carbon with 5 mm Glc, and filter sterilized. For example, Fru (six carbon atoms per molecule) and glycerol (three carbon atoms per molecule) were set to a final concentration of 5 mm and 10 mm, respectively. The washed and resuspended cyanobacterial culture was diluted 10 times in 3 mL of each BG-11C media and dispensed into three wells of untreated 6-well flat-bottom microtiter plates in a prerandomized fashion. Plates were covered with a sterile lid, wrapped into Parafilm, and incubated under the same conditions as the original shake flask cultures. Cyanobacterial growth and health was observed for up to 8 d by optical density measurements in a Tecan M200 Pro plate reader at 730 nm for culture density and 440 nm (absorbance maximum of chlorophyll a) for cellular health. The two sets of growth curves acquired at the different wavelengths showed good correlation, and therefore only readings at 730 nm were evaluated thereafter. Growth rates were determined for the exponential phase from the growth curves provided on supplemental figure S2 in Supplemental File S10.
Acknowledgments
We would like to thank Henning Knoop for his support to D.M. in making an early reconstruction, and for discussion and confirmation of some of the simulations.
Supplemental Data
The following supplemental materials are available.
Supplemental File S1. The reconstructed network of Anabaena sp. PCC 7120 under diazotrophic conditions (two-cell model) in a COBRA-compatible SBML format (XML).
Supplemental File S2. The reconstructed network of Anabaena sp. PCC 7120 under nondiazotrophic conditions (single-cell model) in a COBRA-compatible SBML format (XML).
Supplemental File S3. List of genes, reactions, and reconstruction data for both the single-cell and the two-cell model of Anabaena sp. PCC 7120.
Supplemental File S4. List of genes and reactions annotated here for Anabaena sp. PCC 7120.
Supplemental File S5. Reactions different in the two supercompartments; orphan reactions and KEGG reaction entries omitted from the working reconstruction.
Supplemental File S6. Interactive metabolic map of the reconstruction in Cytoscape 2.8.3 format (CYS).
Supplemental File S7. Matlab script and map definition files to export COBRA flux distributions and overlay onto the Cytoscape map (ZIP).
Supplemental File S8. An exhaustive list of assumptions made during the reconstruction process.
Supplemental File S9. Robustness of the model to changes in biomass composition.
Supplemental File S10. Growth rate comparisons on different carbon and nitrogen sources.
Supplemental Figure S1. Predicted growth rates in mix- and heterotrophic conditions compared to autotrophic growth on bicarbonate.
Supplemental Figure S2. Preferential order of carbon sources for mixotrophic growth of An abaena sp. PCC 7120 on combined nitrogen source (nitrate).
Supplemental Figure S3. Comparison of nitrogen sources in phototropic growth.
Supplementary Material
Footnotes
This work was supported by a Biotechnology and Biological Sciences Research Council grant (BB/N003608/1) and the People Programme (Marie Curie Actions) of the European Union’s Seventh Framework Programme FP7/2007-2013/ under REA grant agreement number 317184 (D.M.). This material reflects only the authors’ views and the European Union is not liable for any use that may be made of the information contained therein. R.S. is supported by the “e:Bio—Innovationswettbewerb Systembiologie” initiative (CyanoGrowth, FKZ 0316192).
[CC-BY]: Article free via Creative Commons CC-BY 4.0 license.
References
- Adams DG. (2000) Symbiotic interactions. In Whitton BA, Potts M, eds, The Ecology of Cyanobacteria: Their Diversity in Time and Space. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 523–561 [Google Scholar]
- Adams DG, Duggan PS (2008) Cyanobacteria-bryophyte symbioses. J Exp Bot 59: 1047–1058 [DOI] [PubMed] [Google Scholar]
- Aiello A. (1985) Sloth hair: unanswered questions. In Montgomery GG, ed, The Evolution and Ecology of Armadillos, Sloths, and Vermilinguas. Smithsonian Institution Press, Washington, DC, pp 213–218 [Google Scholar]
- Albrecht M, Linden H, Sandmann G (1996) Biochemical characterization of purified zeta-carotene desaturase from Anabaena PCC 7120 after expression in Escherichia coli. Eur J Biochem 236: 115–120 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Awai K, Lechno-Yossef S, Wolk CP (2010) Heterocyst envelope glycolipids. In Wada H, Murata N, eds, Lipids in Photosynthesis, Vol 30 Springer, Dordrecht, The Netherlands, pp 179–202 [Google Scholar]
- Biswas A. (2011) Identification and characterization of enzymes involved in the biosynthesis of different phycobiliproteins in cyanobacteria. PhD thesis. University of New Orleans, New Orleans, LA [Google Scholar]
- Bocchi S, Malgioglio A (2010) Azolla-Anabaena as a biofertilizer for rice paddy fields in the Po Valley, a temperate rice area in Northern Italy. Int J Agron 2010: 152158 [Google Scholar]
- Böhme H. (1998) Regulation of nitrogen fixation in heterocyst-forming cyanobacteria. Trends Plant Sci 3: 346–351 [Google Scholar]
- Caspi R, Altman T, Dreher K, Fulcher CA, Subhraveti P, Keseler IM, Kothari A, Krummenacker M, Latendresse M, Mueller LA, et al. (2012) The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res 40: D742–D753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaurasia AK, Apte SK (2011) Improved eco-friendly recombinant Anabaena sp. strain PCC7120 with enhanced nitrogen biofertilizer potential. Appl Environ Microbiol 77: 395–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumino AC, Marcozzi C, Barreiro R, Salerno GL (2007) Carbon cycling in Anabaena sp. PCC 7120. Sucrose synthesis in the heterocysts and possible role in nitrogen fixation. Plant Physiol 143: 1385–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curatti L, Flores E, Salerno G (2002) Sucrose is involved in the diazotrophic metabolism of the heterocyst-forming cyanobacterium Anabaena sp. FEBS Lett 513: 175–178 [DOI] [PubMed] [Google Scholar]
- de Macale M, Vlek PG (2004) The role of Azolla cover in improving the nitrogen use efficiency of lowland rice. Plant Soil 263: 311–321 [Google Scholar]
- Du W, Liang F, Duan Y, Tan X, Lu X (2013) Exploring the photosynthetic production capacity of sucrose by cyanobacteria. Metab Eng 19: 17–25 [DOI] [PubMed] [Google Scholar]
- Ehira S. (2013) Transcriptional regulation of heterocyst differentiation in Anabaena sp. strain PCC 7120. Russ J Plant Physiol 60: 443–452 [Google Scholar]
- Fay P. (1992) Oxygen relations of nitrogen fixation in cyanobacteria. Microbiol Rev 56: 340–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feist AM, Henry CS, Reed JL, Krummenacker M, Joyce AR, Karp PD, Broadbelt LJ, Hatzimanikatis V, Palsson BØ (2007) A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol Syst Biol 3: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty BL, Van Nieuwerburgh F, Head SR, Golden JW (2011) Directional RNA deep sequencing sheds new light on the transcriptional response of Anabaena sp. strain PCC 7120 to combined-nitrogen deprivation. BMC Genomics 12: 332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flamholz A, Noor E, Bar-Even A, Milo R (2012) eQuilibrator—the biochemical thermodynamics calculator. Nucleic Acids Res 40: D770–D775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores E, Herrero A (2010) Compartmentalized function through cell differentiation in filamentous cyanobacteria. Nat Rev Microbiol 8: 39–50 [DOI] [PubMed] [Google Scholar]
- Golden JW, Yoon HS (2003) Heterocyst development in Anabaena. Curr Opin Microbiol 6: 557–563 [DOI] [PubMed] [Google Scholar]
- Graham JE, Bryant DA (2009) The biosynthetic pathway for myxol-2′ fucoside (myxoxanthophyll) in the cyanobacterium Synechococcus sp. strain PCC 7002. J Bacteriol 191: 3292–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurobi Optimization (2013) Gurobi Optimizer Reference Manual. http://www.gurobi.com [Google Scholar]
- Gutzke G, Fischer B, Mendel RR, Schwarz G (2001) Thiocarboxylation of molybdopterin synthase provides evidence for the mechanism of dithiolene formation in metal-binding pterins. J Biol Chem 276: 36268–36274 [DOI] [PubMed] [Google Scholar]
- Heyer H, Krumbein WE (1991) Excretion of fermentation products in dark and anaerobically incubated cyanobacteria. Arch Microbiol 155: 284–287 [Google Scholar]
- Hill DJ. (1977) The role of Anabaena in the Azolla-Anabaena symbiosis. New Phytol 78: 611–616 [Google Scholar]
- Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, Arkin AP, Bornstein BJ, Bray D, Cornish-Bowden A, et al. (2003) The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics 19: 524–531 [DOI] [PubMed] [Google Scholar]
- Ibarra RU, Edwards JS, Palsson BO (2002) Escherichia coli K-12 undergoes adaptive evolution to achieve in silico predicted optimal growth. Nature 420: 186–189 [DOI] [PubMed] [Google Scholar]
- Incharoensakdi A, Jantaro S, Raksajit W, Mäenpää P (2010) Polyamines in cyanobacteria: biosynthesis, transport and abiotic stress response. In Méndez-Vilas A, ed, Current Research, Technology and Education Topics in Applied Microbiology and Microbial Biotechnology, Vol 1 Formatex, Badajoz, Spain, pp 23–32 [Google Scholar]
- Jantaro S, Mäenpää P, Mulo P, Incharoensakdi A (2003) Content and biosynthesis of polyamines in salt and osmotically stressed cells of Synechocystis sp. PCC 6803. FEMS Microbiol Lett 228: 129–135 [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M (2004) The KEGG resource for deciphering the genome. Nucleic Acids Res 32: D277–D280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko T, Nakamura Y, Wolk CP, Kuritz T, Sasamoto S, Watanabe A, Iriguchi M, Ishikawa A, Kawashima K, Kimura T, et al. (2001) Complete genomic sequence of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res 8: 205–213, 227–253 [DOI] [PubMed] [Google Scholar]
- Knoop H, Gründel M, Zilliges Y, Lehmann R, Hoffmann S, Lockau W, Steuer R (2013) Flux balance analysis of cyanobacterial metabolism: the metabolic network of Synechocystis sp. PCC 6803. PLOS Comput Biol 9: e1003081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar S, Durai DA, Wangikar PP, Viswanathan GA (2013) SHARP: genome-scale identification of gene-protein-reaction associations in cyanobacteria. Photosynth Res 118: 181–190 [DOI] [PubMed] [Google Scholar]
- Kumar K, Mella-Herrera RA, Golden JW (2010) Cyanobacterial heterocysts. Cold Spring Harb Perspect Biol 2: a000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechno-Yossef S, Nierzwicki-Bauer S (2002) Azolla-Anabaena symbiosis. In Rai A, Bergman B, Rasmussen U, eds, Cyanobacteria in Symbiosis. Springer, Dordrecht, The Netherlands, pp 153–178 [Google Scholar]
- Luinenburg I, Coleman JR (1992) Identification, characterization and sequence analysis of the gene encoding phosphoenolpyruvate carboxylase in Anabaena sp. PCC 7120. J Gen Microbiol 138: 685–691 [DOI] [PubMed] [Google Scholar]
- Luinenburg I, Coleman JR (1993) Expression of Escherichia coli phosphoenolpyruvate carboxylase in a cyanobacterium. Functional complementation of Synechococcus PCC 7942 ppc. Plant Physiol 101: 121–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch D, O’Brien J, Welch T, Clarke P, Cuív PO, Crosa JH, O’Connell M (2001) Genetic organization of the region encoding regulation, biosynthesis, and transport of rhizobactin 1021, a siderophore produced by Sinorhizobium meliloti. J Bacteriol 183: 2576–2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan AP, Nierzwicki-Bauer SA (1993) In situ detection of transcripts for ribulose-1,5-bisphosphate carboxylase in cyanobacterial heterocysts. J Bacteriol 175: 7301–7306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magrane M, UniProt Consortium (2011) UniProt Knowledgebase: a hub of integrated protein data. Database (Oxford) 2011: bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcozzi C, Cumino AC, Salerno GL (2009) Role of NtcA, a cyanobacterial global nitrogen regulator, in the regulation of sucrose metabolism gene expression in Anabaena sp. PCC 7120. Arch Microbiol 191: 255–263 [DOI] [PubMed] [Google Scholar]
- Martín-Figueroa E, Navarro F, Florencio FJ (2000) The GS-GOGAT pathway is not operative in the heterocysts. Cloning and expression of glsF gene from the cyanobacterium Anabaena sp. PCC 7120. FEBS Lett 476: 282–286 [DOI] [PubMed] [Google Scholar]
- MATLAB (2011) MATLAB and Statistics Toolbox Release R2011b. MathWorks, Natick, MA [Google Scholar]
- Meeks JC, Elhai J (2002) Regulation of cellular differentiation in filamentous cyanobacteria in free-living and plant-associated symbiotic growth states. Microbiol Mol Biol Rev 66: 94–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochimaru M, Masukawa H, Maoka T, Mohamed HE, Vermaas WFJ, Takaichi S (2008) Substrate specificities and availability of fucosyltransferase and β-carotene hydroxylase for myxol 2′-fucoside synthesis in Anabaena sp. strain PCC 7120 compared with Synechocystis sp. strain PCC 6803. J Bacteriol 190: 6726–6733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montesinos ML, Herrero A, Flores E (1995) Amino acid transport systems required for diazotrophic growth in the cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol 177: 3150–3157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao M, Okamoto S, Kohara M, Fujishiro T, Fujisawa T, Sato S, Tabata S, Kaneko T, Nakamura Y (2010) CyanoBase: the cyanobacteria genome database update 2010. Nucleic Acids Res 38: D379–D381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaisen K, Hahn A, Schleiff E (2009) The cell wall in heterocyst formation by Anabaena sp. PCC 7120. J Basic Microbiol 49: 5–24 [DOI] [PubMed] [Google Scholar]
- Nicolaisen K, Moslavac S, Samborski A, Valdebenito M, Hantke K, Maldener I, Muro-Pastor AM, Flores E, Schleiff E (2008) Alr0397 is an outer membrane transporter for the siderophore schizokinen in Anabaena sp. strain PCC 7120. J Bacteriol 190: 7500–7507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales J, Gudmundsson S, Knight EM, Palsson BO, Thiele I (2012) Detailing the optimality of photosynthesis in cyanobacteria through systems biology analysis. Proc Natl Acad Sci USA 109: 2678–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nürnberg DJ, Mariscal V, Bornikoel J, Nieves-Morión M, Krauß N, Herrero A, Maldener I, Flores E, Mullineaux CW (2015) Intercellular diffusion of a fluorescent sucrose analog via the septal junctions in a filamentous cyanobacterium. MBio 6: e02109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owttrim GW, Colman B (1988) Phosphoenolpyruvate carboxylase mediated carbon flow in a cyanobacterium. Biochem Cell Biol 66: 93–99 [Google Scholar]
- Peterson JD, Umayam LA, Dickinson T, Hickey EK, White O (2001) The comprehensive microbial resource. Nucleic Acids Res 29: 123–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picossi S, Montesinos ML, Pernil R, Lichtlé C, Herrero A, Flores E (2005) ABC-type neutral amino acid permease N-I is required for optimal diazotrophic growth and is repressed in the heterocysts of Anabaena sp. strain PCC 7120. Mol Microbiol 57: 1582–1592 [DOI] [PubMed] [Google Scholar]
- Popa R, Weber PK, Pett-Ridge J, Finzi JA, Fallon SJ, Hutcheon ID, Nealson KH, Capone DG (2007) Carbon and nitrogen fixation and metabolite exchange in and between individual cells of Anabaena oscillarioides. ISME J 1: 354–360 [DOI] [PubMed] [Google Scholar]
- Price ND, Papin JA, Schilling CH, Palsson BO (2003) Genome-scale microbial in silico models: the constraints-based approach. Trends Biotechnol 21: 162–169 [DOI] [PubMed] [Google Scholar]
- Razquin P, Fillat MF, Schmitz S, Stricker O, Böhme H, Gómez-Moreno C, Peleato ML (1996) Expression of ferredoxin-NADP+ reductase in heterocysts from Anabaena sp. Biochem J 316: 157–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resendis-Antonio O, Reed JL, Encarnación S, Collado-Vides J, Palsson BØ (2007) Metabolic reconstruction and modeling of nitrogen fixation in Rhizobium etli. PLOS Comput Biol 3: 1887–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reysenbach AL, Wickham GS, Pace NR (1994) Phylogenetic analysis of the hyperthermophilic pink filament community in Octopus Spring, Yellowstone National Park. Appl Environ Microbiol 60: 2113–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikkinen J, Oksanen I, Lohtander K (2002) Lichen guilds share related cyanobacterial symbionts. Science 297: 357. [DOI] [PubMed] [Google Scholar]
- Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY (1979) Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol 111: 1–61 [Google Scholar]
- Ruiz M, Bettache A, Janicki A, Vinella D, Zhang C-C, Latifi A (2010) The alr2505 (osiS) gene from Anabaena sp. strain PCC7120 encodes a cysteine desulfurase induced by oxidative stress. FEBS J 277: 3715–3725 [DOI] [PubMed] [Google Scholar]
- Saha R, Verseput AT, Berla BM, Mueller TJ, Pakrasi HB, Maranas CD (2012) Reconstruction and comparison of the metabolic potential of cyanobacteria Cyanothece sp. ATCC 51142 and Synechocystis sp. PCC 6803. PLoS One 7: e48285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberger J, Que R, Fleming RM, Thiele I, Orth JD, Feist AM, Zielinski DC, Bordbar A, Lewis NE, Rahmanian S, et al. (2011) Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox v2.0. Nat Protoc 6: 1290–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling N, Ehrnsperger K (1985) Cellular differentiation of sucrose metabolism in Anabaena variabilis. Z Naturforsch C 40c: 776–779 [Google Scholar]
- Schluchter WM, Glazer AN (1997) Characterization of cyanobacterial biliverdin reductase. Conversion of biliverdin to bilirubin is important for normal phycobiliprotein biosynthesis. J Biol Chem 272: 13562–13569 [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RN. (1950) Reclamation of “usar” lands in India through blue-green algae. Nature 165: 325–326 [Google Scholar]
- Singh SP, Montgomery BL (2011) Determining cell shape: adaptive regulation of cyanobacterial cellular differentiation and morphology. Trends Microbiol 19: 278–285 [DOI] [PubMed] [Google Scholar]
- Steuer R, Knoop H, Machné R (2012) Modelling cyanobacteria: from metabolism to integrative models of phototrophic growth. J Exp Bot 63: 2259–2274 [DOI] [PubMed] [Google Scholar]
- Stewart WD, Rowell P, Rai AN (1983) Cyanobacteria-eukaryotic plant symbioses. Ann Microbiol (Paris) 134B: 205–228 [DOI] [PubMed] [Google Scholar]
- Takaichi S, Mochimaru M (2007) Carotenoids and carotenogenesis in cyanobacteria: unique ketocarotenoids and carotenoid glycosides. Cell Mol Life Sci 64: 2607–2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaichi S, Mochimaru M, Maoka T, Katoh H (2005) Myxol and 4-ketomyxol 2′-fucosides, not rhamnosides, from Anabaena sp. PCC 7120 and Nostoc punctiforme PCC 73102, and proposal for the biosynthetic pathway of carotenoids. Plant Cell Physiol 46: 497–504 [DOI] [PubMed] [Google Scholar]
- Tatusova T, Ciufo S, Fedorov B, O’Neill K, Tolstoy I (2014) RefSeq microbial genomes database: new representation and annotation strategy. Nucleic Acids Res 42: D553–D559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele I, Palsson BØ (2010) A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat Protoc 5: 93–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J, Meeks JC, Wolk CP, Shaffer PW, Austin SM (1977) Formation of glutamine from [13n]ammonia, [13n]dinitrogen, and [14C]glutamate by heterocysts isolated from Anabaena cylindrica. J Bacteriol 129: 1545–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valladares A, Herrero A, Pils D, Schmetterer G, Flores E (2003) Cytochrome c oxidase genes required for nitrogenase activity and diazotrophic growth in Anabaena sp. PCC 7120. Mol Microbiol 47: 1239–1249 [DOI] [PubMed] [Google Scholar]
- Valladares A, Maldener I, Muro-Pastor AM, Flores E, Herrero A (2007) Heterocyst development and diazotrophic metabolism in terminal respiratory oxidase mutants of the cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol 189: 4425–4430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bodegom P. (2007) Microbial maintenance: a critical review on its quantification. Microb Ecol 53: 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu TT, Stolyar SM, Pinchuk GE, Hill EA, Kucek LA, Brown RN, Lipton MS, Osterman A, Fredrickson JK, Konopka AE, et al. (2012) Genome-scale modeling of light-driven reductant partitioning and carbon fluxes in diazotrophic unicellular cyanobacterium Cyanothece sp. ATCC 51142. PLOS Comput Biol 8: e1002460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner G. (1997) Azolla: A review of its biology and utilization. Bot Rev 63: 1–26 [Google Scholar]
- Wolk CP, Ernst A, Elhai J (2004) Heterocyst metabolism and development. In Bryant D, ed, The Molecular Biology of Cyanobacteria, Vol 1 Springer, Dordrecht, The Netherlands, pp 769–823 [Google Scholar]
- Wolk CP, Thomas J, Shaffer PW, Austin SM, Galonsky A (1976) Pathway of nitrogen metabolism after fixation of 13N-labeled nitrogen gas by the cyanobacterium, Anabaena cylindrica. J Biol Chem 251: 5027–5034 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.