Abstract
miR156 and SPL transcription factors play various roles in conferring competence to flower in plants.
PHYSIOLOGY OF COMPETENCE TO FLOWER
Flowering occurs when the shoot apical meristem (SAM) transitions from forming vegetative organs to giving rise to flowers. This switch to flowering represents the most obvious transition in the growth of the shoot and initiates reproductive development. However, even during vegetative development, the plant shoot transitions through different phases, often referred to as juvenile and adult (Poethig, 1990). The vegetative organs formed sequentially on the flanks of the SAM can differ markedly during each of these phases; for example, the morphology, physiology, and epidermal characteristics of leaves formed during the juvenile phase differ from those formed later during the adult phase (Hackett, 1985; Poethig, 2003). The progressive and sequential transition through these phases was described initially in perennial species (Hackett, 1985) and more recently in detail in genetic model systems, particularly maize (Zea mays) and Arabidopsis (Arabidopsis thaliana; Poethig, 1990; Bergonzi and Albani, 2011; Huijser and Schmid, 2011). In Arabidopsis, the capacity of rosette leaves to form abaxial trichomes is considered to indicate the transition from juvenile to adult vegetative phase (Chien and Sussex, 1996; Telfer et al., 1997). In addition to these vegetative features, the propensity of plants to flower and initiate reproduction also increases with age, and older shoots are described as acquiring competence to flower. This process can be demonstrated most clearly in plants that show an obligate requirement for exposure to an environmental stimulus to undergo floral transition. Plants that have not yet acquired competence to flower will remain vegetative when exposed to stimuli such as photoperiod or vernalization, whereas competent plants exposed to the same environmental cue are induced to flower. Some of the first examples of this phenomenon were in perennial woody plants such as black currant (Ribes nigrum) or ivy (Hedera helix) and were reviewed extensively by Hackett (1985).
The extent to which vegetative phase change and competence to flower are causally interlinked is important in considering these processes. Early genetic and physiological experiments in maize exploited the teopod2 mutant to address these issues (Bassiri et al., 1992). This mutant shows a greatly extended juvenile vegetative phase but acquires competence to flower in response to exposure to short photoperiods at a similar stage in shoot development to wild-type plants (Bassiri et al., 1992). Therefore, this experimental approach suggested that vegetative phase change and competence to flower are not dependent on one another. Nevertheless, more recent work suggests that the underlying mechanisms controlling both processes are related, because microRNA156 (miR156) and its downstream targets the SQUAMOSA PROMOTER-BINDNG PROTEIN-LIKE (SPL) transcription factors control both vegetative phase change (Poethig, 2013) and competence to flower (Huijser and Schmid, 2011; Bergonzi et al., 2013; Zhou et al., 2013). In this short review, we focus on the emerging evidence that miR156/SPL control competence to flower as well as vegetative phase change and discuss their relationship to the growth regulator gibberellin GA. Finally, we describe other genetic systems that have been implicated in regulating competence to flower and discuss how these are related to miR156/SPL.
MIR156 AND MIR172 CONTRIBUTE TO THE CONTROL OF FLOWERING TIME
Many different factors influence the time after germination at which plants initiate flowering. Genetic analysis of flowering time in Arabidopsis defined several regulatory pathways controlling this transition, including those mediating responses to the seasonal environmental cues of photoperiod and vernalization and others influenced by endogenous factors such as the growth regulator GA (Fornara et al., 2010; Srikanth and Schmid, 2011; Andrés and Coupland, 2012). Therefore, to study specifically the basis of competence to flower, factors centrally involved in controlling age-related competence must be distinguished from those conferring environmental responses or participating in general endogenous pathways. During the last 10 years, interest has focused on miR156 and miR172, because their abundance is dependent on the age of the shoot and they influence flowering time. In this section, we briefly review the discovery of these microRNAs (miRNAs) and their involvement in controlling flowering time.
Initially, miR156 was identified in Arabidopsis by sequencing small RNAs predicted to be processed by DICER (Reinhart et al., 2002), and based on computational approaches, the targets of these miRNAs were identified as mRNAs of genes encoding SPL transcription factors (Rhoades et al., 2002). These miRNAs are encoded by eight loci in Arabidopsis (Morea et al., 2016); therefore, their activities were initially difficult to dissect by loss-of-function genetics. However, overexpression of miR156 has effects on leaf morphology and causes reduced apical dominance, shorter plastochron, and later flowering (Schwab et al., 2005). Furthermore, detailed analysis of vegetative phase change showed that overexpression of miR156 delayed the transition from juvenile to adult phase (Wu and Poethig, 2006; Wu et al., 2009). Similarly, in maize, the corngrass mutation, which extends the juvenile phase, is caused by the insertion of a retrotransposon upstream of a MIR156 precursor gene, leading to the overexpression of miR156 (Chuck et al., 2007).
More recently, the phenotypic effects of reducing miR156 activity were described. Overexpression of a MIM156 transgene was used to reduce miR156 activity through sequestration (Franco-Zorrilla et al., 2007), and these transgenic Arabidopsis plants showed opposite phenotypes to the miR156 overexpressors: accelerated vegetative phase transition and reduced number of leaves at flowering, which might be caused by a longer plastochron rather than by accelerated flowering time (Wang et al., 2009; Wu et al., 2009; Todesco et al., 2010). Furthermore, double mutant plants carrying T-DNA insertions in two of the miR156 precursor genes, MIR156a and MIR156c, also exhibited accelerated adult vegetative phase transition, flowered with fewer leaves, and flowered slightly earlier under long days (Yang et al., 2013; Yu et al., 2013).
Mature miR156 levels are lower in older than younger plants. In RNA samples extracted from whole young Arabidopsis plants or specifically from their apices, miR156 levels are higher than in samples derived from similar tissues of older plants (Wu and Poethig, 2006; Wang et al., 2009; Wu et al., 2009). Although miR156 is encoded by eight precursors, MIR156a and MIR56c were the only precursor genes whose transcripts were reduced in abundance between successive leaf primordia, and these are major contributors to the pool of mature 20-nucleotide miR156, although a 21-nucleotide form seems to be expressed from other precursors (Yang et al., 2013; Yu et al., 2013). These data suggest that, in Arabidopsis, progressive reduction of miR156 in leaves that develop successively on the shoot confers the gradual transition from juvenile to adult phase, while this miRNA also accelerates plastochron and causes variable but reproducible delays in flowering time.
As described for miR156, miR172 was identified initially by random sequencing of small RNAs (Park et al., 2002) but soon after was characterized by forward and reverse genetics (Aukerman and Sakai, 2003; Chen, 2004). Based on homology, miR172 was predicted to target mRNAs encoding a small set of transcription factors consisting of APETALA2 (AP2) and closely related proteins (Park et al., 2002). Reverse genetic approaches showed that overexpression of miR172 caused a floral phenotype similar to that of ap2 mutants and that a transgene expressing AP2 mRNA containing a disrupted miR172 recognition sequence caused severe floral defects related to those of agamous (ag) mutants, consistent with AP2 repressing AG transcription (Chen, 2004). Similarly, an early-flowering mutant, early activation tagged (eat), with floral defects was identified in a T-DNA activation tagging screen and shown to be caused by the overexpression of miR172 (Aukerman and Sakai, 2003). The latter result suggested that the AP2-like transcription factors targeted by miR172 are likely to be repressors of the floral transition, and this was confirmed in the same genetic screen by the recovery of the late-flowering activation tagged mutant target of eat1 (toe1), in which one of the AP2-like genes targeted by miR172 was overexpressed (Aukerman and Sakai, 2003). Loss-of-function genetic analysis of the six AP2-like transcription factors targeted by miR172 showed that they act redundantly to repress flowering and that the hextuple mutant in which all of the genes are inactivated is extremely early flowering (Mathieu et al., 2009; Yant et al., 2010). Based on misexpression studies and analysis of its binding sites, SCHLAFMÜTZE was proposed to inhibit flowering mainly by binding to and repressing the transcription of the floral promoter FLOWERING LOCUS T (FT) in the leaves (Mathieu et al., 2009). Thus, miR172 is an activator of flowering and floral development whose targets are mRNAs encoding AP2 and five other AP2-like transcription factors.
The transcription of miR172 precursors is regulated by the age of the plant and is part of the same network as the miR156/SPL module. The abundance of miR172 shows the opposite temporal pattern to the accumulation of miR156, so that in plants grown under long days, it is present at low levels 2 to 5 d after germination and increases progressively to accumulate at high levels around 16 d after germination (Aukerman and Sakai, 2003). Some of this increase is likely to be due to older plants forming flowers, where miR172 is highly expressed (Chen, 2004), but the miR172/AP2-like module also is involved in controlling vegetative phase change (Wu et al., 2009). The toe1 toe2 double mutant and plants overexpressing miR172 prematurely undergo the transition to adult vegetative phase. Finally, miR172 acts downstream of miR156/SPL, so that higher levels of miR156 lead to reduced expression of miR172, while SPL transcription factors, particularly SPL9 and SPL15 but probably also SPL2, SPL10, SPL11, and SPL13, bind directly to and activate the transcription of the miR172 precursor gene MIR172b (Wu et al., 2009; Hyun et al., 2016; Xu et al., 2016b). This interaction contributes to the inverse relationship of miR156 and miR172 abundance in apices of the same plants (Wu et al., 2009), and these patterns are strikingly conserved in distantly related species, including maize (Chuck et al., 2007). Although the details are not yet clear, whole-genome analyses suggest that the network of interactions among the miR156/SPL and miR172/AP2 modules is likely to be intricate and complex, because AP2 also binds directly to MIR172 genes as well as to MIR156 loci and levels of both miRNAs are altered in ap2 mutants (Yant et al., 2010).
In summary, reduction in miR156 levels as plants age allows increased expression of specific SPL transcription factors, and these in turn activate the transcription of MIR172 genes. The resulting inverse temporal expression patterns of the miRNAs confer their opposing effects on vegetative phase change and, presumably, flowering time.
miRNAs AND COMPETENCE TO FLOWER
The observations that the abundance of miR156 declines with the age of the plant and that it regulates flowering time through the repression of SPL transcription factors suggested that miR156 might play a central role in controlling competence to flower. The decline of miR156 levels and the expression of SPL transcription factors were correlated with the initiation of flowering of older Arabidopsis plants that were not exposed to promotive environmental signals such as long photoperiods (Wang et al., 2009). Also, miR156 overexpressor plants and spl9 spl15 double mutants were less sensitive to short exposures of 1 or 3 long days given 3 weeks after germination (Schwarz et al., 2008). However, whether this effect was due to impairing age-related competence to flower or more directly to reducing responsiveness to long days was not tested. Overall, testing competence to flower is difficult in Arabidopsis because it responds to inductive environmental cues extremely early after germination (Mozley and Thomas, 1995) and reference accessions such as Columbia do not exhibit an obligate requirement for these stimuli.
By contrast, flowering and the acquisition of competence are delayed in perennial Brassicaceae relatives of Arabidopsis, and analysis of their obligate vernalization response demonstrated that, in these systems, miR156 levels act as the timer in controlling competence to flower (Fig. 1; Bergonzi et al., 2013; Zhou et al., 2013). Some accessions of perennial Arabis alpina and Cardamine flexuosa exhibit an obligate vernalization response, and these flowered only if exposed to cold when several weeks old but remained vegetative if vernalized as younger plants (Wang et al., 2011; Bergonzi et al., 2013; Zhou et al., 2013). In each species, the stage in development that miR156 reached trough levels correlated with the time that the plant became sensitive to vernalization to induce flowering. Analysis of transgenic plants of these species supported a causal relationship between the down-regulation of miR156 and the acquisition of competence to flower in response to vernalization. In A. alpina, the overexpression of miR156 from the cauliflower mosaic virus (CaMV) 35S promoter prevented flowering in response to vernalization, whereas reduction of miR156 activity by overexpression of MIM156 caused plants to respond to vernalization sooner after germination (Bergonzi et al., 2013). Similarly, in C. flexuosa, miR156 and miR172 levels were found to be inversely related, as described above for other species, and overexpression of miR172 caused plants to flower without vernalization (Zhou et al., 2013). These experiments suggested that repression of miR156 and the resulting increase in expression of miR172 in older plants confer an age-related response to vernalization in C. flexuosa. Evidence for a role of miR172 also was obtained in A. alpina, because mutations in the ortholog of AP2 caused flowering without vernalization (Fig. 1; Bergonzi et al., 2013). Overall, these results support the idea that, in perennial Brassicaceae species, the acquisition of competence to flower in response to vernalization is conferred by age-related down-regulation of miR156.
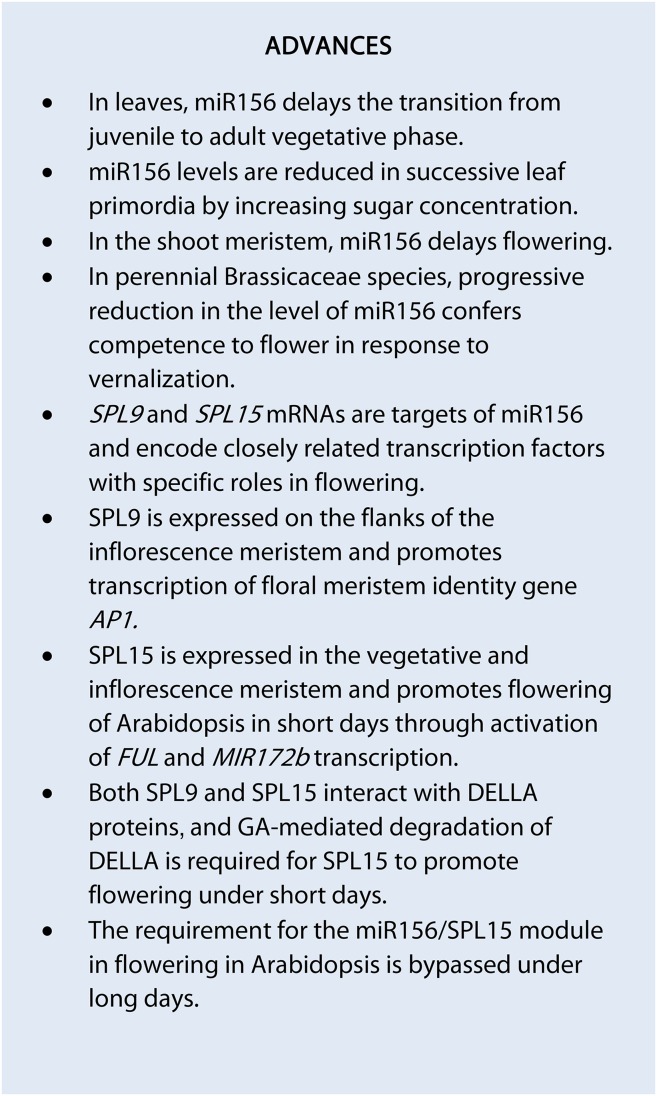
Figure 1.
Age-related responsiveness to vernalization controlled by the miR156/SPL and miR172/AP2-like modules in A. alpina, a perennial relative of Arabidopsis. Top row, Young plants that have not achieved competence to flower are exposed to vernalization. In the shoot apex, miR156 is broadly expressed across the SAM and leaves, and the abundance of miR156 remains high during vernalization. Flowering does not occur during vernalization. After vernalization, miR156 is down-regulated and SPL-encoding genes are expressed, but flowering does not proceed until the plants are vernalized. The AP2 ortholog PEP2 is expressed at all stages throughout the plant and is shown here at the SAM. Bottom row, Older plants that have achieved competence are exposed to vernalization. The age-related down-regulation of miR156 occurred, allowing the expression of SPL-encoding genes at the shoot apex. The level of miR172 is increased markedly at the SAM during vernalization, presumably through the activity of SPLs. The increase of miR172 at the SAM inhibits the accumulation of the floral repressor PEP2 in the center of the meristem. Flowering is induced during vernalization, and floral meristem identity genes such as LFY are expressed. In another perennial Brassicaceae species, C. flexuosa, the miR172 level was shown to be increased during the growth of adult plants even before vernalization. The miR172/AP2 module also plays a role in the floral meristem formed on the flanks of the SAM to determine the identity of developing floral organs. This figure is based on data from Bergonzi et al. (2013) and Zhou et al. (2013).
SPL TRANSCRIPTION FACTORS THAT REGULATE FLOWERING TIME
SPL transcription factors were identified originally in a biochemical screen for proteins that bind to the promoter of SQUAMOSA, a gene that encodes a MADS box protein involved in floral development of Antirrhinum majus (Klein et al., 1996). They were originally called SQUAMOSA-BINDING PROTEINs (SBPs), and the abundance of the mRNAs encoding SBP1 and SBP2 increased as the plant aged, they were expressed prior to SQUAMOSA, and they exhibited specific spatial expression patterns in the SAM and floral primordia (Klein et al., 1996). Genes encoding proteins related to SBPs were then isolated from Arabidopsis and named SPLs (Cardon et al., 1997). Subsequently, this class of transcription factor was identified in all species examined in the green plant lineage and is defined by a 79-amino acid highly conserved region that represents the DNA-binding domain (Klein et al., 1996; Guo et al., 2008). The SPL family is comprised of 16 genes in Arabidopsis, and 11 of these were reported to contain miRNA recognition sites (Guo et al., 2008). Of these 11 SPLs, SPL3/SPL4/SPL5 and SPL9/SPL15, which represent two clades in the family, contain recognition sequences for miR156 and have been associated most strongly with the floral transition. Therefore, we focus particularly on these five members of the family.
The SPL3/SPL4/SPL5 genes have a simpler structure than other members of the family and encode only two exons (Cardon et al., 1999). In this group, the miR156 recognition sequence is located in the 3′ untranslated region (Rhoades et al., 2002; Wu and Poethig, 2006; Gandikota et al., 2007). The mRNAs of all three genes are strongly expressed in inflorescences (Fig. 2; Cardon et al., 1999). SPL4 and SPL5 mRNAs are strongly increased in abundance under inductive long days through the activity of the photoperiodic flowering pathway and have distinct spatial patterns of expression, with SPL4 mRNA being expressed mainly in the rib meristem and later in floral primordia while SPL5 mRNA is expressed on the flanks of the inflorescence meristem and in floral primordia (Fig. 2; Cardon et al., 1999; Schmid et al., 2003; Jung et al., 2012; Torti et al., 2012). SPL3 mRNA has been detected widely in the meristem, shoot, and flowers (Cardon et al., 1997; Wang et al., 2009; Wu et al., 2009) and in vegetative meristems (Wu et al., 2009) and has been reported to be up-regulated during the floral transition (Wang et al., 2009). Furthermore, miR156 inhibits the translation of SPL3 mRNA when expressed from the CaMV 35S promoter (Wu and Poethig, 2006; Gandikota et al., 2007). Several reverse genetic experiments based on overexpression and biochemical analyses suggest that SPL3/SPL4/SPL5 contribute to the promotion of flowering. Transgenic plants carrying a fusion of the SPL3 open reading frame to the CaMV 35S promoter were early flowering (Cardon et al., 1997), which was enhanced if the miRNA recognition sequence was removed from the transgene, and these plants also prematurely underwent the transition from juvenile to adult vegetative phases (Wu and Poethig, 2006; Gandikota et al., 2007). Similar phenotypes to those for SPL3 were observed when SPL4 and SPL5 were overexpressed without their miR156 recognition sites, supporting the idea that these genes show functional redundancy (Wu and Poethig, 2006). However, recent analysis demonstrated that an spl3 spl4 spl5 triple mutant was not delayed in flowering time compared with wild-type plants, although it did form slightly higher numbers of cauline leaves. Thus, these genes were proposed not to promote the floral transition but to act relatively late during the flowering process in conferring floral identity on the developing primordium (Xu et al., 2016b). The mechanism by which SPL3/SPL4/SPL5 confer floral meristem identity might involve the direct activation of genes involved in the early stages of floral development. In transgenic plants expressing from the CaMV 35S promoter, an SPL3-GFP fusion protein lacking the miR156 binding site in the 3′ untranslated region or GFP-SPL3 expressed from the endogenous SPL3 promoter, the fusion protein was found to bind directly to the promoters of LEAFY (LFY), APETALA1 (AP1), and FRUITFULL (FUL) and the CaMV 35S transgene caused increased expression of the target genes (Yamaguchi et al., 2009). These genes also were increased in expression in transgenic plants overexpressing from the CaMV 35S promoter SPL4 or SPL5 transcripts lacking the miR156-binding site (Yamaguchi et al., 2009). Also, in the apices of spl3 spl4 spl5 triple mutants grown for 11 long days, LFY and AP1 transcripts were present at lower levels than in wild-type plants (Xu et al., 2016b). Thus, the expression patterns of SPL3, SPL4, and SPL5, their target genes, and the phenotypic effects of their mutation suggest that they act late in the flowering process to contribute to the early steps in floral development.
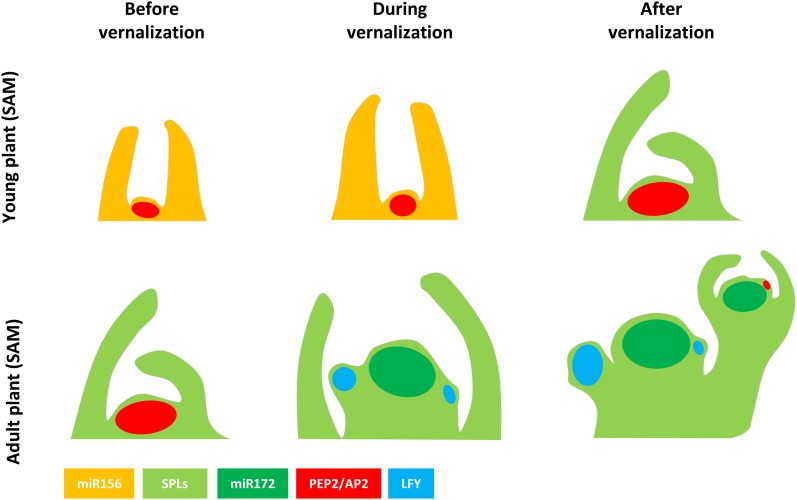
Figure 2.
Spatially distinct roles of SPL genes in the Arabidopsis SAM. Left, Two closely related genes, SPL9 and SPL15, are expressed before floral induction in leaves and the SAM, respectively. SPL9 is expressed in leaves, where, in adult plants after reduction in miR156, it participates in the accumulation of miR172 to promote the transition to adult leaf morphology. Middle, During floral induction under short days, the accumulation of miR172 and mRNA of the floral activator FUL at the SAM requires the function of SPL15. During and after floral induction, SPL9 mRNA appears on the flanks of the meristem and the protein activates the floral identity gene AP1 in cooperation with DELLA and LFY. SPL3, SPL4, and SPL5 mRNAs are expressed at the shoot apex. Right, After floral induction, SPL15 is expressed in the floral meristem and the inflorescence meristem. SPL3, SPL4, and SPL5 are expressed in specific patterns at the apex. See text for references.
The closely related SPL9 and SPL15 genes represent one clade within the Arabidopsis SPL family and have distinct functions in floral induction and floral development (Schwarz et al., 2008; Wang et al., 2009; Yamaguchi et al., 2014; Hyun et al., 2016). Although they are closely related paralogues, the SPL9 and SPL15 genes exhibit distinct mRNA expression patterns in the shoot meristem (Fig. 2; Wang et al., 2009; Wu et al., 2009; Hyun et al., 2016). SPL9 mRNA appears not to be expressed in the shoot meristem prior to the floral transition but rises on the flanks of the meristem during the transition before the emergence of floral primordia (Hyun et al., 2016). In vegetative plants, SPL9 mRNA is expressed in leaf primordia and leaves (Wang et al., 2009; Wu et al., 2009; Hyun et al., 2016). SPL15 mRNA is expressed in the vegetative meristem prior to floral induction, throughout the meristem during induction, and in the inflorescence meristem (Hyun et al., 2016). During the vegetative phase, SPL15 mRNA is present in leaves in a more restricted pattern than SPL9 mRNA (Hyun et al., 2016). Gain-of-function transgenic alleles of SPL9 and SPL15, in which the miRNA recognition site is mutated without affecting the protein sequence, both cause early flowering (Wang et al., 2009; Hyun et al., 2016), and the SPL9 transgene causes early transition to the vegetative adult phase (Wu et al., 2009). Also, a dominant ethyl methanesulfonate-induced mutation of SPL15 that affected the miR156-binding site caused premature transition from the juvenile to the adult vegetative phase and reduced cell size in leaves (Usami et al., 2009). These experiments, which rely on gain-of-function approaches, suggest that SPL9 and SPL15 have similar functions in controlling flowering time and the vegetative phase transition. However, loss-of-function alleles of SPL9 and SPL15 also have been described and seem to distinguish between the activities of the genes (Schwarz et al., 2008; Wu et al., 2009; Hyun et al., 2016). The spl15 single mutant and the spl9 spl15 double mutant were slightly later flowering than wild-type plants under long days (Schwarz et al., 2008). However, under noninductive short days, the spl15 mutant and the spl9 spl15 double mutant showed severe late flowering, whereas the spl9 mutant did not (Hyun et al., 2016). This phenotype of spl15 was much weaker and more variable in the studies of Xu et al. (2016b), suggesting that additional environmental variables such as light quality or intensity that have not yet been identified play an important role in determining the phenotype of the mutant. When observed, the late-flowering phenotype of spl15 under short days was similar but not quite as severe as that of 35S:miR156 plants, suggesting that most of the late flowering caused by the overexpression of miR156 is through the inhibition of SPL15 activity (Hyun et al., 2016). The spl9 mutant also showed delayed transition to the adult vegetative phase that was enhanced in the spl9 spl15 double mutant, which also showed reduced leaf plastochron (Schwarz et al., 2008; Wu et al., 2009). Thus, the expression patterns and loss-of-function phenotypes of SPL9 and SPL15 suggest that SPL15 plays the larger role in floral induction and is particularly important under noninductive short days, whereas SPL9 plays a more significant role in vegetative phase change and acts in floral primordia after the floral induction.
SPECIFIC FUNCTIONS OF SPL9 AND SPL15 IN CONTROLLING FLOWERING AND THEIR REGULATION BY GA
In the floral primordium, SPL9 has specific roles in the activation of genes required for early flower development (Yamaguchi et al., 2014). Analysis of DNA binding showed that SPL9 binds to functionally important regions in the AP1 promoter (Wang et al., 2009; Yamaguchi et al., 2014), and a constitutively expressed chemically inducible form of SPL9 increases AP1 transcription synergistically with inducible LFY (Yamaguchi et al., 2014). In addition, analysis of transgenic plants expressing miR156-resistant SPL9 mRNA showed that SPL9 binds to FUL and SOC1 (Wang et al., 2009), which, considering the expression pattern of SPL9, also might be most relevant in wild-type plants during the early stages of floral development. In addition to being regulated by miR156 at the posttranscriptional level, SPL9 also is regulated at the posttranslational level by DELLA proteins (Yu et al., 2012; Zhang et al., 2007; Porri et al., 2012; Box 1). These proteins interact directly with and regulate the activity of transcription factors and are degraded in the presence of GA, providing a mechanism by which this plant growth regulator controls gene expression (Xu et al., 2014). A series of experiments indicated that SPL9 recruits the DELLA protein REPRESSOR OF ga1-3 (RGA) to the AP1 promoter and that DELLA binding enhances the ability of SPL9 to activate AP1 transcription in the floral primordium (Box 1; Yamaguchi et al., 2014). By contrast, the interaction of RGA and SPL9 represses the ability of SPL9 to activate the transcription of SOC1 and MIR172b (Yu et al., 2012). Thus, the effect of the interaction between SPL9 and DELLA appears to differ among target genes or tissues, leading to the activation of transcription of some targets such as AP1 and the repression of others including SOC1 and MIR172b (Yu et al., 2012; Yamaguchi et al., 2014).
SPL15 promotes the floral transition under noninductive short days. Fluorescent protein fusions to SPL15 expressed from endogenous regulatory sequences accumulated in the meristem and were regulated directly by miR156 (Hyun et al., 2016), suggesting that the protein acts in the meristem to promote flowering. In agreement with this conclusion, the level of FT mRNA, which is the output of flowering pathways that act in the leaves, was the same in spl9 spl15 double mutant and wild-type plants (Hyun et al., 2016). Also, although the overexpression of SPL9 from heterologous promoters in leaves did promote FT transcription and early flowering, the effect was less strong than when SPL9 was expressed in the shoot meristem (Wang et al., 2009). Therefore, in wild-type plants, these SPLs probably act exclusively in the meristem to promote flowering. SPL15 binds directly to the FUL and miR172b genes and is required for their activation in the shoot meristem under short days. Genetic experiments in which GA was depleted from the shoot meristem by the overexpression of a GA catabolic enzyme supported the idea that the interaction of RGA with SPL15 prevents the activation of SPL15 target genes such as MIR172b and FUL (Hyun et al., 2016; Box 1). These results are consistent with the roles of SPL15 and GA in promoting the floral transition under short days (Wilson et al., 1992; Hyun et al., 2016) and suggest that one way in which GA promotes flowering under short days is by stimulating the degradation of DELLA, allowing SPL15 to activate its target genes in the meristem. Thus, among Arabidopsis SPL transcription factors, SPL15 seems to play the major role in the floral transition. Interestingly, the mechanism by which SPL15 activates the transcription of its target genes involves cooperativity with known regulators of flowering, particularly the MADS box transcription factor SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) (Fig. 3), which acts early during the floral transition (Borner et al., 2000; Samach et al., 2000).
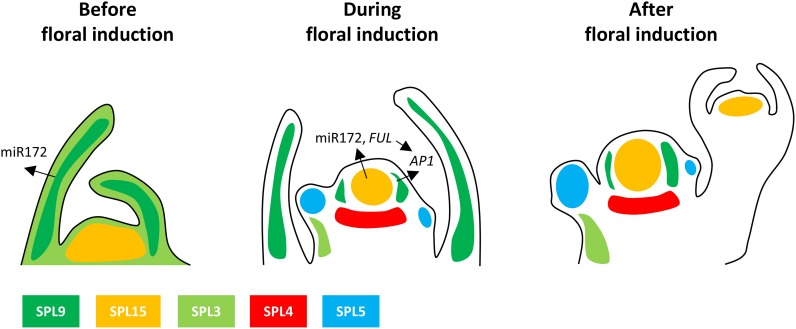
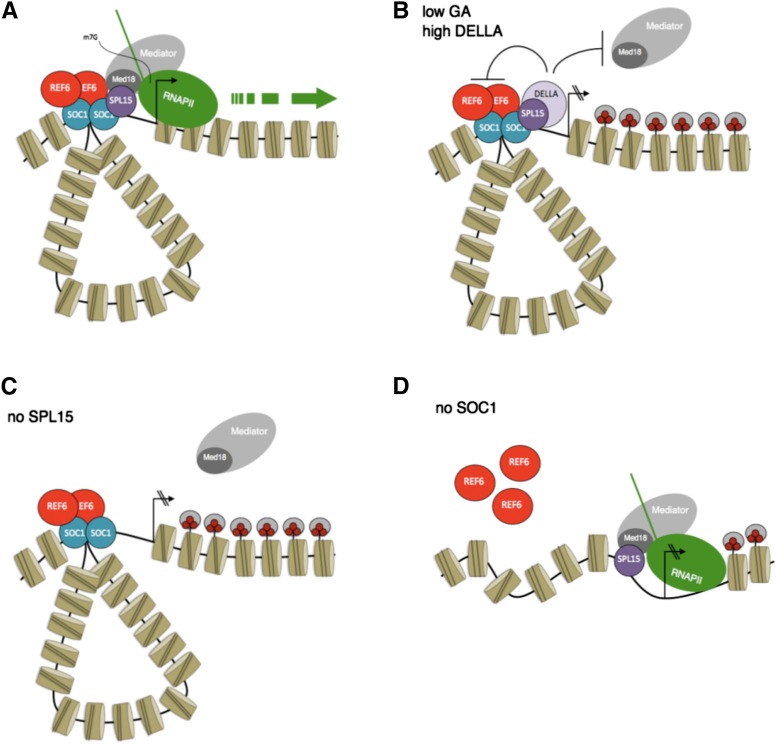
Figure 3.
Mechanism by which SPL15 activates its target genes FUL and MIR172b during floral induction under short days. A, Activation of transcription requires SPL15 and the MADS box transcription factor SOC1. SOC1 recruits the trimethylation of Lys-27 on histone 3 (H3K27me3) demethylase REF6, allowing the removal of repressive chromatin marks from the target gene. SPL15 interacts with the Mediator complex to promote transcription through RNA Pol II. B, If GA levels are low at the meristem, DELLA protein levels are high. DELLA interacts with SPL15 at the target gene promoter, preventing the interaction of SPL15 with the Mediator complex. No transcription occurs. C, In the absence of SPL15, SOC1 binds to the target gene and REF6 is recruited, but no transcription occurs. The Mediator complex is not recruited to the target gene. D, In the absence of SOC1, SPL15 binds to the target gene but REF6 is not recruited. The repressive chromatin mark H3K27me3 remains on the gene, and no transcription occurs. Data in this figure are from Hyun et al. (2016).
The genetic and molecular analyses of SPL9 and SPL15 clearly implicate them in the early stages of reproductive development and suggest that, if miR156 acts to regulate competence to flower, as suggested by the experiments in perennial Brassicaceae species, then it likely does so by repressing the activity of SPL15 and probably SPL9. Also, the observation that DELLAs and GA act to regulate SPL9 and SPL15 activity is consistent with early observations that this growth regulator is implicated in vegetative phase change (Chien and Sussex, 1996; Telfer et al., 1997; Poethig, 2003) as well as floral induction under noninductive conditions (Wilson et al., 1992; Hyun et al., 2016).
REGULATION OF THE MIR156 TIMER
The temporal regulation of vegetative phase change and competence to flower described above ultimately relies on the gradual reduction of miR156 levels. This process is widely conserved in higher plants (Wu and Poethig, 2006; Chuck et al., 2007; Bergonzi et al., 2013; Zhou et al., 2013), but the precise age-related mechanisms by which miR156 levels are regulated remain unclear. Eight genes encode miR156 in Arabidopsis (Rhoades et al., 2002; Morea et al., 2016). The precise spatial and temporal expression patterns of these precursor genes have not been described, and it remains unclear, for example, which are expressed in the meristem and what their temporal patterns of expression are. However, two of them, MIR156A and MIR156C, are highly expressed in the shoot of young plants and express most of the mature miR156 detected at this stage (Yang et al., 2013; Yu et al., 2013). The abundance of these precursor RNAs falls in successive leaf primordia and shows a similar regulation to mature miR156 (Yang et al., 2013). Thus, the temporal pattern of reduction in miR156 levels appears to be conferred at least in large part by transcriptional regulation of these precursors, and this conclusion was further supported by analyzing fusions of the regulatory sequences of these precursor genes to the GUS marker gene (Xu et al., 2016a).
The transcriptional down-regulation of MIR156A and MIR156C is regulated by chromatin modification. H3K27me3 is a chromatin mark associated with the repression of transcription (Derkacheva and Hennig, 2014), and this mark accumulates to higher levels on the MIR156A and MIR156C genes in apices of 5-week-old plants than of 1- or 2-week-old plants (Xu et al., 2016a). Deposition of this mark on these genes involves the SWINGER methyl transferase and the chromatin remodeler PICKLE, which was shown previously to associate with genes rich in H3K27me3 modification (Zhang et al., 2012). Both of these proteins regulate vegetative phase change, bind directly to the MIR156 precursor genes, and contribute to the accumulation of H3K27me3 on MIR156A and MIR156C (Xu et al., 2016a). However, these proteins are part of the general enzyme machinery that contributes to H3K27me3 deposition across the genome, and the mechanisms by which they are recruited to MIR156A and MIR156C in an age-dependent manner are unclear.
However, several experiments suggest that endogenous sugar levels may act earlier in the process to repress the transcription of MIR156 precursor genes (Wahl et al., 2013; Yang et al., 2013; Yu et al., 2013), perhaps by increasing deposition of the H3K27me3 mark. MIR156 transcriptional repression in vegetative phase change was shown to be promoted by a signal produced in leaf primordia (Yang et al., 2011). The possible role of sugars as this signal was then tested in a range of genetic and physiological experiments, based on classical work suggesting that sugars regulate the maturation of the shoot (Goebel, 1908). Application of exogenous sucrose reduced miR156 levels and specifically the transcription of MIR156A and MIR156C. Also, Suc and glucose levels were higher in older plants, therefore showing an inverse relationship to miR156, and mutants exhibiting impaired rates of photosynthesis had higher levels of miR156 as well as delayed transition to the adult vegetative phase (Yang et al., 2013; Yu et al., 2013). These and related experiments suggest that the higher levels of sugar, particularly Suc but also Glc, in older plants contribute to the down-regulation of MIR156 gene transcription to accelerate transition to the adult phase. However, it remains unclear from these experiments whether sugar also acts as the timer in the shoot meristem to regulate the floral transition.
A further series of genetic experiments implicated another sugar, trehalose 6-phosphate (T6P), in the repression of miR156 levels during flowering (Gómez et al., 2010; Wahl et al., 2013). T6P is present at low concentrations in plant cells and is proposed to act as a signaling molecule rather than to have a function in primary metabolism (Lunn et al., 2006). Mutations impairing TREHALOSE 6-PHOSPHATE SYNTHASE (TPS) are embryo lethal (Gómez et al., 2010), but if this defect is complemented with a transgene active in embryos, then the resulting plants are viable and late flowering (Gómez et al., 2010; Wahl et al., 2013). In these plants, miR156 levels were up to 8 times higher than in the wild type, suggesting that this is one of the causes of the late-flowering phenotype. Consistent with this interpretation, the SPL3, SPL4, and SPL5 genes were expressed at lower levels in tps mutants. These results suggest that T6P signaling might be important in regulating MIR156 transcription as part of the sugar-signaling pathway. However, in tps mutants, other flowering time genes were altered in expression, and as described above, roles for SPL3, SPL4, and SPL5 in flowering time control have not been established, so the mechanism and extent to which T6P controls flowering time through miR156 regulation still require elucidation.
OTHER GENETIC SYSTEMS CONTROLLING COMPETENCE TO FLOWER
In addition to the miR156/SPL module, other genetic systems have been proposed to contribute to the age at which plants become sensitive to environmental cues that induce flowering. Notable among these are the TEMPRANILLO (TEM) transcription factors that repress the response to photoperiod in young Arabidopsis plants. TEM1 and TEM2 are members of the RAV transcription factor family and contain two DNA-binding domains related to those of AP2 and B3 (Castillejo and Pelaz, 2008). TEM1 binds directly to the promoter of FT and to exons of genes encoding GA biosynthetic enzymes to repress their transcription and thereby delay floral induction (Castillejo and Pelaz, 2008; Osnato et al., 2012). Furthermore, TEM1 mRNA abundance falls abruptly between 8 and 10 d after germination under long days (Castillejo and Pelaz, 2008). The timing of this reduction correlates with a strong increase in FT mRNA and to enhanced sensitivity of the plants to long photoperiods for floral induction (Castillejo and Pelaz, 2008; Sgamma et al., 2014). These results suggest that TEM genes act mainly in young plants to block the response to long days and that reduction in their expression contributes to the acquisition of competence to flower in response to photoperiod. Furthermore, this system appears to be evolutionarily conserved, because the mRNA of a TEM ortholog from A. majus also was reduced in abundance at the time at which plants became sensitive to photoperiod to induce flowering (Sgamma et al., 2014). How the levels of TEM1 and TEM2 mRNA are reduced with age is unknown, but in Arabidopsis, exposure to long days reduces TEM1 and TEM2 mRNA levels (Osnato et al., 2012), suggesting that the repressive effects of these transcription factors on flowering may be regulated directly by environmental conditions rather than, or as well as, by endogenous mechanisms associated with aging of the plant.
Finally, the floral repressor TERMINAL FLOWER1 (TFL1) extends the phase during which plants are insensitive to inductive cues. In perennial A. alpina, TFL1 activity blocked activation of the floral meristem identity gene LFY during the vernalization of young plants (Wang et al., 2011). Reduction of TFL1 expression by RNA interference allowed LFY transcription and flowering to occur during the vernalization of young plants, in a similar way to that in 35S:MIM156 plants (Wang et al., 2011; Bergonzi et al., 2013). These data, together with the work described above on miR156/SPL function in A. alpina, suggest that the repression of flowering by TFL1 is required to block the flowering of young plants and that this can be overcome later through the action of SPL transcription factors. TFL1 is proposed to interact with the bZIP transcription factor FD and, thereby, repress transcription (Hanano and Goto, 2011), so it is possible that TFL1 and SPL transcription factors have common target genes and that whether flowering proceeds is determined by the relative abundance of each class of protein.
VARIATION IN COMPETENCE TO FLOWER AMONG ANNUALS AND PERENNIALS
The acquisition of competence to flower is usually strongly delayed in perennials, whereas annuals can flower rapidly after germination. This delay in perennials allows the plant to produce more biomass and axillary meristems prior to reproduction and, thereby, likely increases the possibility of surviving flowering and reproducing the following year (Bergonzi and Albani, 2011). Annual and perennial life history can diverge rapidly during evolution, suggesting that the genetic system conferring competence to flower also can change relatively quickly (Bergonzi et al., 2013; Zhou et al., 2013). By contrast, the miR156/SPL system appears to be ancient and present in all flowering plants (Morea et al., 2016). In the Brassicaceae, this discrepancy is proposed to be explained by increased dependency on the miR156/SPL system for flowering in perennials, whereas annuals evolve genetic mechanisms that bypass the requirement for SPLs during flowering. For example, in Arabidopsis, there is a strong requirement for SPL15 to promote flowering under noninductive short days, whereas in long days, this requirement is bypassed so that spl15 mutants have a very mild phenotype under these conditions (Hyun et al., 2016). Therefore, the balance of quantitative activities of different flowering pathways can explain how the time taken to acquire competence is more important in determining the flowering time of some species than others. Similarly, the evolution in annuals of pathways that bypass the requirement for miR156/SPL to induce flowering can explain how the miR156/SPL module is present and similarly expressed in annuals and perennials, but annuals do not show a strong requirement for the acquisition of competence to flower. The recent progress in defining closely related annual and perennial experimental systems that differ in competence phenotypes provides a means of understanding how these bypass pathways evolve and how their activities vary quantitatively.
Glossary
- SAM
shoot apical meristem
- miRNA
microRNA
- H3K27me3
trimethylation of Lys-27 on histone 3
- T6P
trehalose 6-phosphate
Footnotes
This work was supported by the ERC through Hylife, by the Deutsch Forschungsgemeinschaft (grant no. SPP1530), and by the Max Planck Society.
Articles can be viewed without a subscription.
References
- Andrés F, Coupland G (2012) The genetic basis of flowering responses to seasonal cues. Nat Rev Genet 13: 627–639 [DOI] [PubMed] [Google Scholar]
- Aukerman MJ, Sakai H (2003) Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassiri A, Irish EE, Poethig RS (1992) Heterochronic effects of Teopod 2 on the growth and photosensitivity of the maize shoot. Plant Cell 4: 497–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergonzi S, Albani MC (2011) Reproductive competence from an annual and a perennial perspective. J Exp Bot 62: 4415–4422 [DOI] [PubMed] [Google Scholar]
- Bergonzi S, Albani MC, Ver Loren van Themaat E, Nordström KJ, Wang R, Schneeberger K, Moerland PD, Coupland G (2013) Mechanisms of age-dependent response to winter temperature in perennial flowering of Arabis alpina. Science 340: 1094–1097 [DOI] [PubMed] [Google Scholar]
- Borner R, Kampmann G, Chandler J, Gleissner R, Wisman E, Apel K, Melzer S (2000) A MADS domain gene involved in the transition to flowering in Arabidopsis. Plant J 24: 591–599 [DOI] [PubMed] [Google Scholar]
- Cardon G, Höhmann S, Klein J, Nettesheim K, Saedler H, Huijser P (1999) Molecular characterisation of the Arabidopsis SBP-box genes. Gene 237: 91–104 [DOI] [PubMed] [Google Scholar]
- Cardon GH, Höhmann S, Nettesheim K, Saedler H, Huijser P (1997) Functional analysis of the Arabidopsis thaliana SBP-box gene SPL3: a novel gene involved in the floral transition. Plant J 12: 367–377 [DOI] [PubMed] [Google Scholar]
- Castillejo C, Pelaz S (2008) The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr Biol 18: 1338–1343 [DOI] [PubMed] [Google Scholar]
- Chen X. (2004) A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303: 2022–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien JC, Sussex IM (1996) Differential regulation of trichome formation on the adaxial and abaxial leaf surfaces by gibberellins and photoperiod in Arabidopsis thaliana (L.) Heynh. Plant Physiol 111: 1321–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G, Cigan AM, Saeteurn K, Hake S (2007) The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat Genet 39: 544–549 [DOI] [PubMed] [Google Scholar]
- Derkacheva M, Hennig L (2014) Variations on a theme: Polycomb group proteins in plants. J Exp Bot 65: 2769–2784 [DOI] [PubMed] [Google Scholar]
- Fornara F, de Montaigu A, Coupland G (2010) SnapShot: control of flowering in Arabidopsis. Cell 141: 550–550.e2 [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA, Paz-Ares J (2007) Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 39: 1033–1037 [DOI] [PubMed] [Google Scholar]
- Gandikota M, Birkenbihl RP, Höhmann S, Cardon GH, Saedler H, Huijser P (2007) The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J 49: 683–693 [DOI] [PubMed] [Google Scholar]
- Goebel K. (1908) Einleitung in die Experimentelle Morphologie der Pflanzen. BG Teubner, Leipzig, Germany [Google Scholar]
- Gómez LD, Gilday A, Feil R, Lunn JE, Graham IA (2010) AtTPS1-mediated trehalose 6-phosphate synthesis is essential for embryogenic and vegetative growth and responsiveness to ABA in germinating seeds and stomatal guard cells. Plant J 64: 1–13 [DOI] [PubMed] [Google Scholar]
- Guo AY, Zhu QH, Gu X, Ge S, Yang J, Luo J (2008) Genome-wide identification and evolutionary analysis of the plant specific SBP-box transcription factor family. Gene 418: 1–8 [DOI] [PubMed] [Google Scholar]
- Hackett WP. (1985) Juvenility, maturation, and rejuvenation in woody plants. Hortic Rev 7: 109–155 [Google Scholar]
- Hanano S, Goto K (2011) Arabidopsis TERMINAL FLOWER1 is involved in the regulation of flowering time and inflorescence development through transcriptional repression. Plant Cell 23: 3172–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijser P, Schmid M (2011) The control of developmental phase transitions in plants. Development 138: 4117–4129 [DOI] [PubMed] [Google Scholar]
- Hyun Y, Richter R, Vincent C, Martinez-Gallegos R, Porri A, Coupland G (2016) Multi-layered regulation of SPL15 and cooperation with SOC1 integrate endogenous flowering pathways at the Arabidopsis shoot meristem. Dev Cell 37: 254–266 [DOI] [PubMed] [Google Scholar]
- Jung JH, Ju Y, Seo PJ, Lee JH, Park CM (2012) The SOC1-SPL module integrates photoperiod and gibberellic acid signals to control flowering time in Arabidopsis. Plant J 69: 577–588 [DOI] [PubMed] [Google Scholar]
- Klein J, Saedler H, Huijser P (1996) A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol Gen Genet 250: 7–16 [DOI] [PubMed] [Google Scholar]
- Lunn JE, Feil R, Hendriks JHM, Gibon Y, Morcuende R, Osuna D, Scheible WR, Carillo P, Hajirezaei MR, Stitt M (2006) Sugar-induced increases in trehalose 6-phosphate are correlated with redox activation of ADPglucose pyrophosphorylase and higher rates of starch synthesis in Arabidopsis thaliana. Biochem J 397: 139–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J, Yant LJ, Mürdter F, Küttner F, Schmid M (2009) Repression of flowering by the miR172 target SMZ. PLoS Biol 7: e1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morea EGO, da Silva EM, Silva GFFE, Valente GT, Rojas CHB, Vincentz M, Nogueira FTS (2016) Functional and evolutionary analyses of the miR156 and miR529 families in land plants. BMC Plant Biol 16: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozley D, Thomas B (1995) Developmental and photobiological factors affecting photoperiodic induction in Arabidopsis thaliana Heynh Landsberg erecta. J Exp Bot 46: 173–179 [Google Scholar]
- Osnato M, Castillejo C, Matías-Hernández L, Pelaz S (2012) TEMPRANILLO genes link photoperiod and gibberellin pathways to control flowering in Arabidopsis. Nat Commun 3: 808. [DOI] [PubMed] [Google Scholar]
- Park W, Li J, Song R, Messing J, Chen X (2002) CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol 12: 1484–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig RS. (1990) Phase change and the regulation of shoot morphogenesis in plants. Science 250: 923–930 [DOI] [PubMed] [Google Scholar]
- Poethig RS. (2003) Phase change and the regulation of developmental timing in plants. Science 301: 334–336 [DOI] [PubMed] [Google Scholar]
- Poethig RS. (2013) Vegetative phase change and shoot maturation in plants. Curr Top Dev Biol 105: 125–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porri A, Torti S, Romera-Branchat M, Coupland G (2012) Spatially distinct regulatory roles for gibberellins in the promotion of flowering of Arabidopsis under long photoperiods. Development 139: 2198–2209 [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP (2002) MicroRNAs in plants. Genes Dev 16: 1616–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP (2002) Prediction of plant microRNA targets. Cell 110: 513–520 [DOI] [PubMed] [Google Scholar]
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616 [DOI] [PubMed] [Google Scholar]
- Schmid M, Uhlenhaut NH, Godard F, Demar M, Bressan R, Weigel D, Lohmann JU (2003) Dissection of floral induction pathways using global expression analysis. Development 130: 6001–6012 [DOI] [PubMed] [Google Scholar]
- Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D (2005) Specific effects of microRNAs on the plant transcriptome. Dev Cell 8: 517–527 [DOI] [PubMed] [Google Scholar]
- Schwarz S, Grande AV, Bujdoso N, Saedler H, Huijser P (2008) The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol Biol 67: 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgamma T, Jackson A, Muleo R, Thomas B, Massiah A (2014) TEMPRANILLO is a regulator of juvenility in plants. Sci Rep 4: 3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth A, Schmid M (2011) Regulation of flowering time: all roads lead to Rome. Cell Mol Life Sci 68: 2013–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer A, Bollman KM, Poethig RS (1997) Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development 124: 645–654 [DOI] [PubMed] [Google Scholar]
- Todesco M, Rubio-Somoza I, Paz-Ares J, Weigel D (2010) A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet 6: e1001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torti S, Fornara F, Vincent C, Andrés F, Nordström K, Göbel U, Knoll D, Schoof H, Coupland G (2012) Analysis of the Arabidopsis shoot meristem transcriptome during floral transition identifies distinct regulatory patterns and a leucine-rich repeat protein that promotes flowering. Plant Cell 24: 444–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usami T, Horiguchi G, Yano S, Tsukaya H (2009) The more and smaller cells mutants of Arabidopsis thaliana identify novel roles for SQUAMOSA PROMOTER BINDING PROTEIN-LIKE genes in the control of heteroblasty. Development 136: 955–964 [DOI] [PubMed] [Google Scholar]
- Wahl V, Ponnu J, Schlereth A, Arrivault S, Langenecker T, Franke A, Feil R, Lunn JE, Stitt M, Schmid M (2013) Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science 339: 704–707 [DOI] [PubMed] [Google Scholar]
- Wang JW, Czech B, Weigel D (2009) miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138: 738–749 [DOI] [PubMed] [Google Scholar]
- Wang R, Albani MC, Vincent C, Bergonzi S, Luan M, Bai Y, Kiefer C, Castillo R, Coupland G (2011) Aa TFL1 confers an age-dependent response to vernalization in perennial Arabis alpina. Plant Cell 23: 1307–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RN, Heckman JW, Somerville CR (1992) Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol 100: 403–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS (2009) The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138: 750–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Poethig RS (2006) Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133: 3539–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Liu Q, Yao T, Fu X (2014) Shedding light on integrative GA signaling. Curr Opin Plant Biol 21: 89–95 [DOI] [PubMed] [Google Scholar]
- Xu M, Hu T, Smith MR, Poethig RS (2016a) Epigenetic regulation of vegetative phase change in Arabidopsis. Plant Cell 28: 28–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Hu T, Zhao J, Park MY, Earley KW, Wu G, Yang L, Poethig RS (2016b) Developmental functions of miR156-regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes in Arabidopsis thaliana. PLoS Genet 12: e1006263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi A, Wu MF, Yang L, Wu G, Poethig RS, Wagner D (2009) The microRNA-regulated SBP-box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev Cell 17: 268–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N, Winter CM, Wu MF, Kanno Y, Yamaguchi A, Seo M, Wagner D (2014) Gibberellin acts positively then negatively to control onset of flower formation in Arabidopsis. Science 344: 638–641 [DOI] [PubMed] [Google Scholar]
- Yang L, Conway SR, Poethig RS (2011) Vegetative phase change is mediated by a leaf-derived signal that represses the transcription of miR156. Development 138: 245–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Xu M, Koo Y, He J, Poethig RS (2013) Sugar promotes vegetative phase change in Arabidopsis thaliana by repressing the expression of MIR156A and MIR156C. eLife 2: e00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant L, Mathieu J, Dinh TT, Ott F, Lanz C, Wollmann H, Chen X, Schmid M (2010) Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell 22: 2156–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Cao L, Zhou CM, Zhang TQ, Lian H, Sun Y, Wu J, Huang J, Wang G, Wang JW (2013) Sugar is an endogenous cue for juvenile-to-adult phase transition in plants. eLife 2: e00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Galvão VC, Zhang YC, Horrer D, Zhang TQ, Hao YH, Feng YQ, Wang S, Schmid M, Wang JW (2012) Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA promoter binding-like transcription factors. Plant Cell 24: 3320–3332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Bishop B, Ringenberg W, Muir WM, Ogas J (2012) The CHD3 remodeler PICKLE associates with genes enriched for trimethylation of histone H3 lysine 27. Plant Physiol 159: 418–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Schwarz S, Saedler H, Huijser P (2007) SPL8, a local regulator in a subset of gibberellin-mediated developmental processes in Arabidopsis. Plant Mol Biol 63: 429–439 [DOI] [PubMed] [Google Scholar]
- Zhou CM, Zhang TQ, Wang X, Yu S, Lian H, Tang H, Feng ZY, Zozomova-Lihová J, Wang JW (2013) Molecular basis of age-dependent vernalization in Cardamine flexuosa. Science 340: 1097–1100 [DOI] [PubMed] [Google Scholar]