Localization analyses of RPW8 homologs suggest that extrahaustorial membrane-targeting (EHM) may be an ancestral feature of the RPW8 family and that the EHM induced by powdery mildew is likely synthesized de novo in Arabidopsis.
Abstract
Upon penetration of the host cell wall, the powdery mildew fungus develops a feeding structure named the haustorium in the invaded host cell. Concomitant with haustorial biogenesis, the extrahaustorial membrane (EHM) is formed to separate the haustorium from the host cell cytoplasm. The Arabidopsis resistance protein RPW8.2 is specifically targeted to the EHM where it activates haustorium-targeted resistance against powdery mildew. RPW8.2 belongs to a small family with six members in Arabidopsis (Arabidopsis thaliana). Whether Homologs of RPW8 (HR) 1 to HR4 are also localized to the EHM and contribute to resistance has not been determined. Here, we report that overexpression of HR1, HR2, or HR3 led to enhanced resistance to powdery mildew, while genetic depletion of HR2 or HR3 resulted in enhanced susceptibility, indicating that these RPW8 homologs contribute to basal resistance. Interestingly, we found that N-terminally YFP-tagged HR1 to HR3 are also EHM-localized. This suggests that EHM-targeting is an ancestral feature of the RPW8 family. Indeed, two RPW8 homologs from Brassica oleracea tested also exhibit EHM-localization. Domain swapping analysis between HR3 and RPW8.2 suggests that sequence diversification in the N-terminal 146 amino acids of RPW8.2 probably functionally distinguishes it from other family members. Moreover, we found that N-terminally YFP-tagged HR3 is also localized to the plasma membrane and the fungal penetration site (the papilla) in addition to the EHM. Using this unique feature of YFP-HR3, we obtained preliminary evidence to suggest that the EHM is unlikely derived from invagination of the plasma membrane, rather it may be mainly synthesized de novo.
Powdery mildew is a common plant disease caused by biotrophic fungal pathogens in the order of Erysiphales. An infectious powdery mildew pathogen is able to penetrate the host cell wall and develop a feeding structure called the haustorium in the host epidermal cell for deriving water and nutrients for parasitic growth on the plant surface. Conceivably, the host-adaptation process is gradual and long, as the fungus has to overcome multilayered, spatiotemporally distinct plant defense mechanisms. In most cases, attempted penetration of the plant cell wall by any powdery mildew fungus induces cell wall-apposition (a structure called the papilla) underneath the penetration point (Bushnell and Bergquist, 1975; Aist, 1976). For a nonadapted fungus, further ingression is halted by the so-called penetration resistance mainly manifested by papilla formation. Previous studies revealed several genetic components involved in penetration resistance (Collins et al., 2003; Lipka et al., 2005; Stein et al., 2006; Campe et al., 2016). Interestingly, consistent with the notion that papilla formation is a conserved cell wall-based defense mechanism (Meyer et al., 2009; Wen et al., 2011), overexpression of PMR4, the callose synthase largely responsible for the callose deposited in the papilla resulted in complete resistance to well-adapted powdery mildew (Ellinger et al., 2013), further validating the defense function of the papilla.
Well-adapted powdery mildew, by definition, must have overcome penetration resistance of the host. Such a fungus must be able to penetrate the host cell wall and differentiate a haustorium from the tip of an appressorium. Earlier cytological studies revealed that though formed physically within the invaded cell, the haustorium is separated from the host cell cytoplasm by an interfacial membrane termed the extrahaustorial membrane (EHM; Gil and Gay, 1977; Bushnell and Gay, 1978; Roberts et al., 1993). Together with the extrahaustorial matrix, the EHM represents the host-pathogen interface where fungal effectors are translocated into the host cell, and nutrients from the host cell are transported into the haustorium. Concomitant with the biogenesis of the EHM, a physical barrier named the neckband is thought to form and seal the fungal plasma membrane (PM) in the haustorial neck with the invaginated host PM to prevent the apoplastic escape of materials from the extrahaustorial matrix (Heath, 1976; Gil and Gay, 1977).
In response to haustorial invasion, plants mount postpenetration resistance to constrain the haustorium or limit its function. At the cellular or subcellular level, postpenetration resistance is manifested by the formation of a haustorial encasement and focal accumulation of H2O2 around the haustorium, with occasional collapse of the invaded cell (i.e. hypersensitive response; Meyer et al., 2009; Wang et al., 2009; Wen et al., 2011). Both the papilla and the haustorial encasement are enriched for callose (β-1,3-glucan) and these two structures appear to be spatiotemporally connected (Meyer et al., 2009). At the molecular level, apart from resistance activated by immune receptors upon recognition of specific pathogen effectors (Halterman et al., 2001; Zhou et al., 2001; Yahiaoui et al., 2004; Jordan et al., 2011), plants have also evolved other forms of resistance mechanisms. For example, the Arabidopsis (Arabidopsis thaliana) atypical R proteins RPW8.1 and RPW8.2 confer postpenetration resistance to all infectious powdery mildew strains tested (Xiao et al., 2001). RPW8.1 and RPW8.2 belong to a small gene family in Arabidopsis and Brassica with the progenitor being a HOMOLOG of RPW8 3 (HR3)-like gene (Xiao et al., 2004). We recently revealed that RPW8.2 [also RPW8.1 when expressed in the haustorium-invaded cells (Ma et al., 2014)] is specifically targeted to the EHM where it enhances formation of the haustorial encasement and promotes onsite H2O2 accumulation and/or triggers hypersensitive response (Wang et al., 2009). Specific targeting of RPW8.2 to the EHM requires two basic residue-enriched motifs in RPW8.2 and engages an EHM-oriented specific trafficking pathway (Wang et al., 2013; Kim et al., 2014). Thus, the EHM is the subcellular locale for launching RPW8.2-mediated, haustorium-targeted defenses. These observations demonstrate that the EHM is not only a site for cross-border trafficking but also a critical host-pathogen battleground. Whether other members of the RPW8 family play roles in postpenetration resistance and/or are localized to the EHM have not been determined.
In this study, through genetic and localization analyses we showed that three Arabidopsis RPW8 homologs (AtHR1 to AtHR3) and two representative Brassica oleracea RPW8 homologs (BoHRa and BoHRb) are probable EHM resident proteins and all but HR4 likely contribute to basal resistance against powdery mildew pathogens. Unexpectedly, we found that HR3 tagged with YFP at its N terminus is also localized to the PM and the papilla. Using the triple localization feature of YFP-HR3, we obtained preliminary evidence to suggest that the EHM is unlikely derived from invagination of the PM, rather it may be mainly synthesized de novo.
RESULTS
RPW8 Homologs Contribute to Basal Resistance against Powdery Mildew
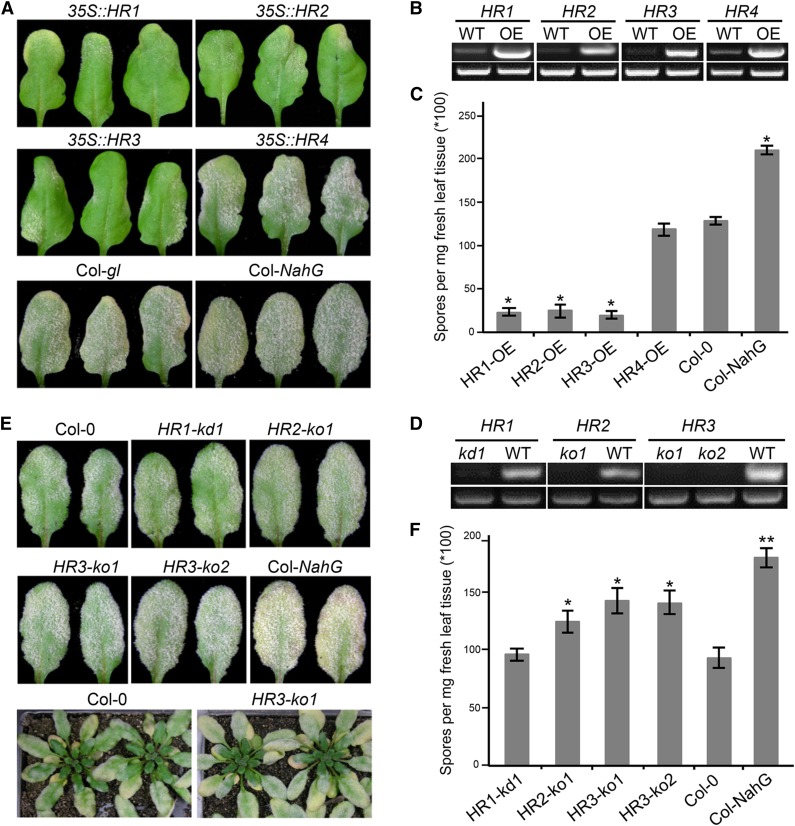
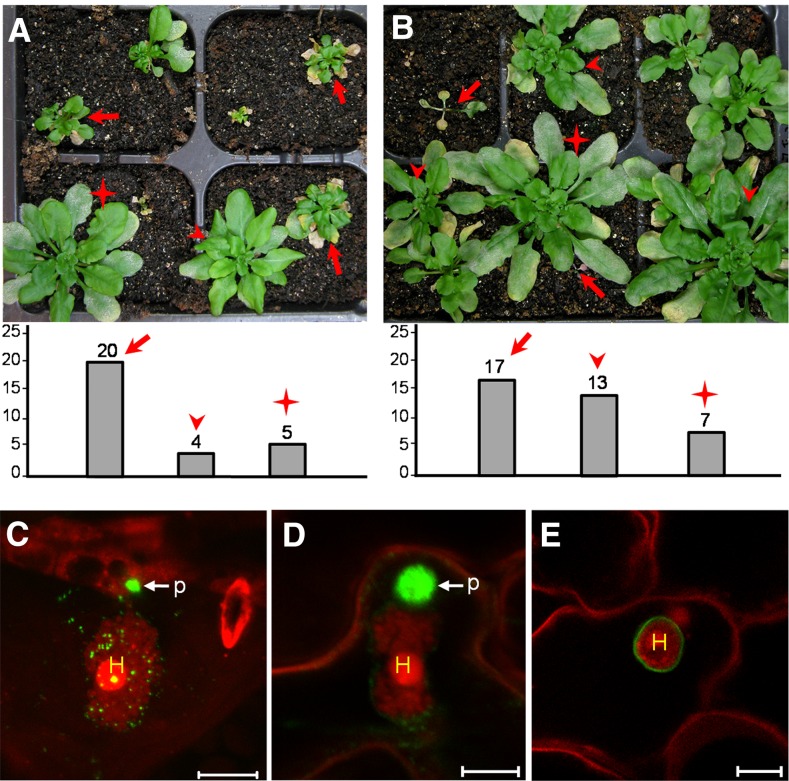
The RPW8 gene locus in Arabidopsis Ms-0 accession contains RPW8.1 and RPW8.2 and three homologs of RPW8, designated HR1 (At3g50450), HR2 (At3g50460), and HR3 (At3g50470; Xiao et al., 2001). The powdery mildew-susceptible accession Col-0 lacks RPW8.1 and RPW8.2, but contains HR4 (At3g50480) in the same location, along with HR1, HR2, and HR3 (Xiao et al., 2001, 2004). Because Col-0 mutants including those that are defective in salicylic acid (SA)-signaling display enhanced disease susceptibility to powdery mildew (Xiao et al., 2005), we reasoned that Col-0 is still capable of mounting a certain level of SA-dependent and perhaps SA-independent basal resistance. To test if HR1 to HR4 play a role in basal resistance in Col-0, we first overexpressed HR1, HR2, HR3, and HR4 using the 35S promoter together with the native 5′ regulatory sequence (496 bp for HR3, 1000 bp for the rest), a demonstrated strategy shown to effectively achieve higher expression of RPW8.1 and RPW8.2 (Orgil et al., 2007), in Col-gl (Col-0 containing the glabrous mutation gl1-1). For each DNA construct, >30 T1 transgenic plants were generated and five independent homozygous lines in the T3 or T4 generation were tested with a well-adapted powdery mildew isolate Golovinomyces cichoracearum (Gc) UCSC1. Plants of all tested lines transgenic for 35S::HR1, 35S::HR2, or 35S::HR3 displayed obvious enhanced diseases resistance compared to Col-gl, whereas plants transgenic for 35S::HR4 were as susceptible as Col-gl (Fig. 1, A and C). Reverse-transcription (RT)-PCR confirmed that expression levels of the four RPW8 homologs were much higher in the transgenic lines than the respective endogenous genes in Col-gl (Fig. 1B). These results suggest that HR1, HR2, and HR3, but not HR4, may contribute to basal resistance in Col-0 against powdery mildew.
Figure 1.
RPW8 homologs contribute to basal resistance to powdery mildew. A, Disease reaction phenotypes of representative leaves of T3 Col-gl lines overexpressing (i.e. 35S plus NP) HR1, HR2, HR3, or HR4 infected with Gc UCSC1. Col-gl and Col-nahG were used as control. Pictures were taken at 12 dpi. B, RT-PCR analysis of representative overexpression lines and Col-gl using gene-specific primers (top panel). UBC21 was used as control (bottom panel). cDNA was synthesized using total RNA prepared from uninfected plants. PCR was done with 24 (UBC21) or 30 (other genes) cycles. ImageJ (NIH) was used to estimate band intensities. C, Quantitative assay of disease susceptibility. Asterisks indicate significance at P < 0.01 compared with Col-gl based on Student’s t test. Data represent means ± se (n = 4) from one of three independent experiments. D, RT-PCR analysis of gene knockout or knockdown T-DNA insertion lines. Top panels (left to right): HR1, HR2, or HR3 using gene-specific primers within Exon 1 upstream of all T-DNA insertions. Bottom panels (all): UBC21. Infected leaves at 3 dpi were used for RNA extraction and cDNA synthesis. PCR was done with 22 (UBC21) or 30 (other genes) cycles. ImageJ was used to estimate band intensities. E, Representative infected leaves or plants of indicated mutant and control lines at 12 dpi. F, Quantitative assay of disease susceptibility at 12 dpi. Data represent means ± se (n = 4) from one of three independent experiments. Student’s t test was used to examine if the difference relative to Col-0 is statistically significant (* P < 0.05; ** P < 0.01).
Next, we identified one T-DNA knockdown (kd) line (Salk_056764) for HR1 (designated HR1-kd1), one knockout (ko) line (Salk_093095) for HR2 (designated HR2-ko1) and two knockout lines (SALK _122954 and WiscDsLox420C08) for HR3 (designated HR3-ko1 and HR3-ko2) based on RT-PCR results (Fig. 1D and Supplemental Fig. S1). Plants of these mutant lines were inoculated with Gc UCSC1 along with plants of Col-0 and Col-NahG, a transgenic line expressing a bacterial SA hydrolase (Lawton et al., 1995). Except for HR1-kd1, the remaining three T-DNA lines exhibited a small but reproducible increase in fungal spores on the leaf surface 10-12 d postinoculation (dpi) when compared to Col-0, although they were not as susceptible as Col-NahG (Fig. 1, E and F). Disease quantification showed that plants of the HR2 and HR3 mutant lines produced approximately 25% to approximately 45% more fungal spores than Col-0 while Col-NahG supported nearly twice as many spores as Col-0 (Fig. 1F) at 12 dpi, indicating that genetic depletion of HR2 and HR3 results in enhanced disease susceptibility albeit at a lower degree compared to SA depletion by the SA hydrolase encoded by NahG. These results, together with the data from overexpression analyses, demonstrate that HR1, HR2, and HR3 probably contribute to basal resistance against powdery mildew.
Functional Diversification between RPW8.2 and HR3
In a recent study, we found that two R/K-R/K-X-R/K motifs comprise the core EHM-targeting signal (ETS) in RPW8.2 (Wang et al., 2013). Sequence analysis showed that while the first ETS has some variations, the second ETS is highly conserved among all the RPW8 family members in Arabidopsis and three RPW8 homologs from B. oleracea (Supplemental Fig. S2). Thus, we wondered if other Arabidopsis RPW8 family members are also EHM-resident proteins. RPW8.1-YFP has shown to be mainly expressed in mesophyll cells when expressed from its native promoter [NP; (Wang et al., 2007)]. Interestingly, when expressed from the RPW8.2 promoter, RPW8.1-YFP was also found to be EHM-localized (Ma et al., 2014). To examine subcellular localization of HR1 to HR4, each of these genes was translationally fused with YFP at the C terminus, and the fusion constructs were stably expressed in Col-gl by their respective NPs. Unexpectedly, we were unable to detect any fluorescent signal before or after inoculation with Gc UCSC1 from at least 30 independent T1 transgenic lines examined for each construct despite the detection of the transgene expression by RT-PCR in two representative lines per construct (not shown). Western blotting using an anti-GFP antibody recognizing YFP also revealed no protein accumulation in any of these lines (Supplemental Fig. S3A). While T1 plants transgenic for NP::HR1-YFP, NP::HR2-YFP, or NP::HR4-YFP appeared phenotypically normal in growth and development and were susceptible to powdery mildew (Fig. 2A), approximately 55% of T1 plants transgenic for NP::HR3-YFP exhibited stunted growth with leaf necrosis and varied degree of enhanced resistance to Gc UCSC1 (Fig. 2, B and C). These observations suggest that these fusion proteins cannot accumulate to levels detectable by confocal microscopy or western blotting, and that HR3-YFP is also toxic when stably expressed in Arabidopsis. To make sure that these fusion constructs are translated into proteins, we transiently expressed them in Nicotiana benthamiana leaves by agroinfiltration. Again, no YFP signal was detectable. We then transiently coexpressed each of the constructs with the coat protein of Turnip crinkle virus that also functions to suppress RNA silencing in host plants (Qu et al., 2003). Interestingly, except for NP::HR4-YFP, the remaining three constructs produced detectable, albeit weak, YFP signal. HR1-YFP and HR3-YFP appeared to be localized in the PM in epidermal cells, punctate spots were also seen around chloroplasts in mesophyll cells. However, HR2-YFP was mainly found in the ring structures surrounding chloroplasts in mesophyll cells (Supplemental Fig. S3B), which is reminiscent of RPW8.1-YFP’s localization in mesophyll cells (Wang et al., 2009). Moreover, consistent with cell death in Arabidopsis lines transgenic for NP::HR3-YFP, we found massive necrosis from the leaf section infiltrated with NP::HR3-YFP only (Supplemental Fig. S3C), demonstrating that HR3-YFP is capable of triggering cell death in both Arabidopsis and N. benthamiana. Taken together, these observations suggest that RPW8 homologs appear to be distinct from RPW8.2 in terms of protein function and accumulation.
Figure 2.
Expression of HR3-YFP results in necrotic cell death. A, Representative plants (five weeks old) or infected leaves (12 dpi) expressing indicated DNA constructs under control of their respective NPs in Col-0. More than 30 T1 lines for each construct were examined. Note that there was no obvious altered growth and defense phenotypes in any of the transgenic lines compared with Col-0. B, Representative T1 plants transgenic for NP::HR3-YFP with varied degrees of stunted growth and spontaneous cell death (red arrows). C, A group of T1 plants transgenic for NP::HR3-YFP at 10 dpi with Gc UCSC1 showing varied degree of resistance (indicated by red arrowheads) and susceptibility (indicated by yellow arrowheads). D, Representative T1 transgenic plants (upper panel) and total number of T1 plants with spontaneous leaf necrotic death (arrows) or without (arrowheads; lower panel) for the indicated DNA constructs.
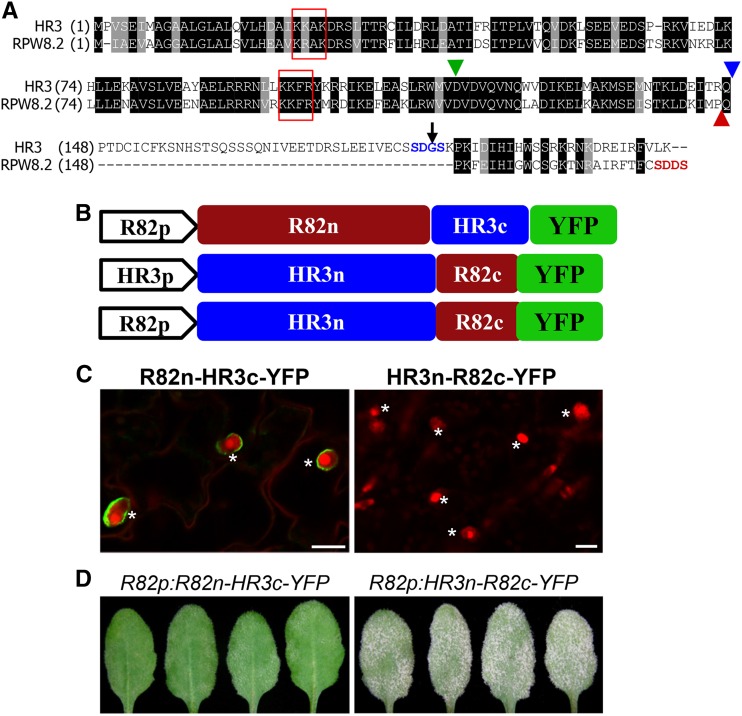
Given that an HR3-like gene is the probable progenitor of the RPW8 gene family (Xiao et al., 2004), we investigated how RPW8.2 might have functionally diversified from HR3 via the following three aspects. First, we expressed HR3-YFP from the RPW8.2 promoter (R82P) in the Col-gl background. We found that approximately 86% (42 of 49) of transgenic T1 plants showed strong or lethal cell death (Fig. 2D), suggesting that the RPW8.2 promoter is stronger than that of HR3. Again, YFP signal was not detected in any of these transgenic lines even before onset of cell death. Second, because substitution of Asp at 116 to Gly (D116G) in RPW8.2 abolishes RPW8.2-triggered hypersensitive response and resistance (Orgil et al., 2007; Wang et al., 2009) and D116 is conserved in HR3 (Fig. 3A), we thus made the D116G mutation in HR3 and stably expressed the HR3D116G-YFP fusion protein from the HR3 promoter. We found that this mutation did not affect HR3-YFP–triggered cell death (Fig. 2D) despite that the fusion protein remained undetectable. This suggests that hypersensitive response cell death activated by RPW8.2 is mechanistically distinct from the necrotic cell death caused by HR3-YFP. Third, because both RPW8.1 and RPW8.2 contain a shorter C terminus compared to HR3 (Xiao et al., 2004) and the C terminus SDDS of RPW8.2 appears to be required or full scale interaction with 14-3-3 (Yang et al., 2009), we made an HR3 variant with a truncation of the C-terminal 26 amino acids (ΔCt26) and the G186 to D substitution to make an SDDS C terminus as seen in RPW8.2 such that the resultant HR3ΔCt26/G186D mutant most structurally resembles RPW8.2 (Fig. 3A). Also, in another construct we removed the C-terminal 65 amino acids of HR3 (HR3ΔCt65) to see if it can make the fusion protein more stable. However, in both cases, cell death still occurred in respective T1 transgenic plants without detectable YFP signal (Fig. 2D). Based on these results we reasoned that the N-terminal portion of HR3 (amino acid 1 to 147) is responsible for (1) inappropriate cell death activated by HR3-YFP and (2) lack of protein accumulation at a detectable level by microscopy when the C-terminal portion is perturbed (i.e. due to fusion with a fluorescent protein). We speculated that these properties of HR3 might have been lost in RPW8.2 as part of its functional diversification from an HR3-like progenitor.
Figure 3.
Sequence and function diversification of RPW8.2 from HR3. A, Protein sequence alignment between HR3 and RPW8.2. The blue arrowhead indicates the position (amino acid 147) in HR3 for recombination with the C terminus of RPW8.2, which is indicated by a red arrowhead. The green arrowhead indicates Asp-116 in RPW8.2, which is conserved in HR3. The black arrow indicates Gly-186, which is mutated to Asp in the HR3ΔCt26/G186D mutant. The two core EHM-targeting motifs are boxed in red. B, Schematic illustration of the chimeric R82n-HR3c and HR3n-R82c genes in fusion with YFP driven either by the RPW8.2 or HR3 promoter. HR3n = HR31-147, R82n = RPW8.21-146, HR3c = HR3148-213, and R82c = RPW8.2147-174. C, Representative images showing expression and localization of the indicated fusion proteins. Note that HR3n-R82c-YFP was undetectable. Bar, 20 μm. D, Phenotypes of the indicated genotypes. Representative leaves from infected plants at 12 dpi were shown to reflect the functionality of the chimeric genes. Plants transgenic for HR3p::HR3n-R82c-YFP were also susceptible (not shown). *, Haustoria.
To further test the above speculation, we made chimeric constructs by swapping the N- and C-terminal domains of RPW8.2 and HR3, which are designated R82n (amino acids 1 to 146)-HR3c (148 to 213), and HR3n (1 to 147)-R82c (147–174) as shown in Figure 3, A and B. Interestingly, R82n-HR3c-YFP was nicely localized to the EHM (Fig. 3C) and conferred resistance to Gc UCSC1 (Fig. 3D) in a similar manner as RPW8.2-YFP, whereas HR3n-R82c-YFP was still undetectable and lost ability to trigger cell death or resistance. These results suggest that (1) R82n is essential for protein stability, defense and EHM-localization of RPW8.2 while R82c is replaceable, which is compatible with results from our recent mutational analysis of RPW8.2 (Wang et al., 2009); (2) R82c may be able to suppress HR3n-mediated inappropriate cell death and/or that HR3n-R82c-YFP cannot accumulate to a level sufficient for triggering cell death. However, a more detailed structure-function analysis between HR3 and RPW8.2 is needed to further define the molecular and evolutionary mechanisms underlying the functional diversification between these two proteins.
Because HR3 has more likely retained the original conserved function of the HR3-like progenitor gene (Xiao et al., 2004), whereas RPW8.2 has apparently evolved novel resistance function, we also wondered if HR3 is required for RPW8.2-mediated resistance. To test this, we introduced the HR3-ko1 allele to S5, a Col-gl transgenic for a single copy of a genomic fragment containing RPW8.1 and RPW8.2 and their NPs by crossing. We found that S5/HR3-ko1 plants were as resistant as S5 plants (Supplemental Fig. S4), indicating that HR3 is dispensable for RPW8.2-activated resistance.
All RPW8 Homologs Are Likely EHM-Resident Proteins
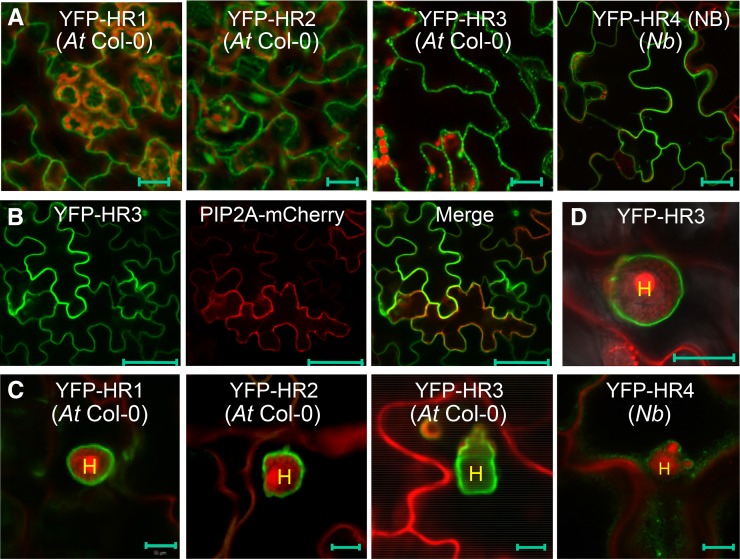
From the above analysis, we were unable to determine if the RPW8 homologs are also EHM-resident proteins due to the unknown intrinsic constraints associated with C-terminal YFP tagging. Neither did we succeed in detection of internally YFP-tagged HR3 by microscopy (not shown). Previously we reported that N-terminally YFP-tagged RPW8.2 (YFP-RPW8.2) was also targeted to the EHM, despite its loss of function in defense (Wang et al., 2010). We thus made N-terminally YFP-tagged DNA constructs for HR1, HR2, HR3, and HR4 and stably expressed them from the 35S promoter in Col-gl. While YFP signal was still not detectable from any of over 40 transgenic lines expressing YFP-HR4, YFP signal from the remaining three fusion constructs appeared to be distributed in the cytoplasm and possibly in the PM (Fig. 4A). To see if the YFP-HR4 fusion protein can be made, we transiently expressed the construct in N. benthamiana leaves and detected YFP signal along the cell wall possibly in the PM (Fig. 4A). These results suggest that N-terminally YFP-tagged HR4 is unable to accumulate in stable transgenic Arabidopsis plants, which is different from those of the other family members. To determine if the observed YFP signal from YFP-HR3 was indeed in the PM, we transiently coexpressed YFP-HR3 with an Arabidopsis PM aquaporin AtPIP2A in fusion with mCherry (Nelson et al., 2007) in N. benthamiana leaf cells. We found that YFP signal nicely colocalized with mCherry signal (Fig. 4B), indicating that N-terminally YFP-HR3 is localized to the PM.
Figure 4.
Subcellular localization of YFP-HR1, YFP-HR2, YFP-HR3, and YFP-HR4. Except for YFP-HR4, which was transiently expressed in leaves of N. benthamiana, the rest were stably expressed in Arabidopsis Col-0 (At Col-0). A, Representative images showing cytoplasmic and likely plasma membrane localization of indicated fusion proteins expressed from the 35S promoter in leaves of Arabidopsis transgenic plants (for HR1, HR2 and HR3) or agroinfiltrated N. benthamiana plants (for HR4). B, A representative image showing colocalization of YFP-HR3 with AtPIP2A-mCherry in N. benthamiana epidermal cells when transiently coexpressed. C, Representative images showing likely EHM-localization of indicated fusion proteins expressed from the 35S promoter at 2 dpi with Gc UCSC1. D, EHM-localization of YFP-HR3 expressed from the HR3 NP in At Col-0 at 2 dpi with Gc UCSC1. Bar, 20 μm in A; 50 μm in B; 10 μm in C and D. H, Haustorium; Nb, N. benthamiana.
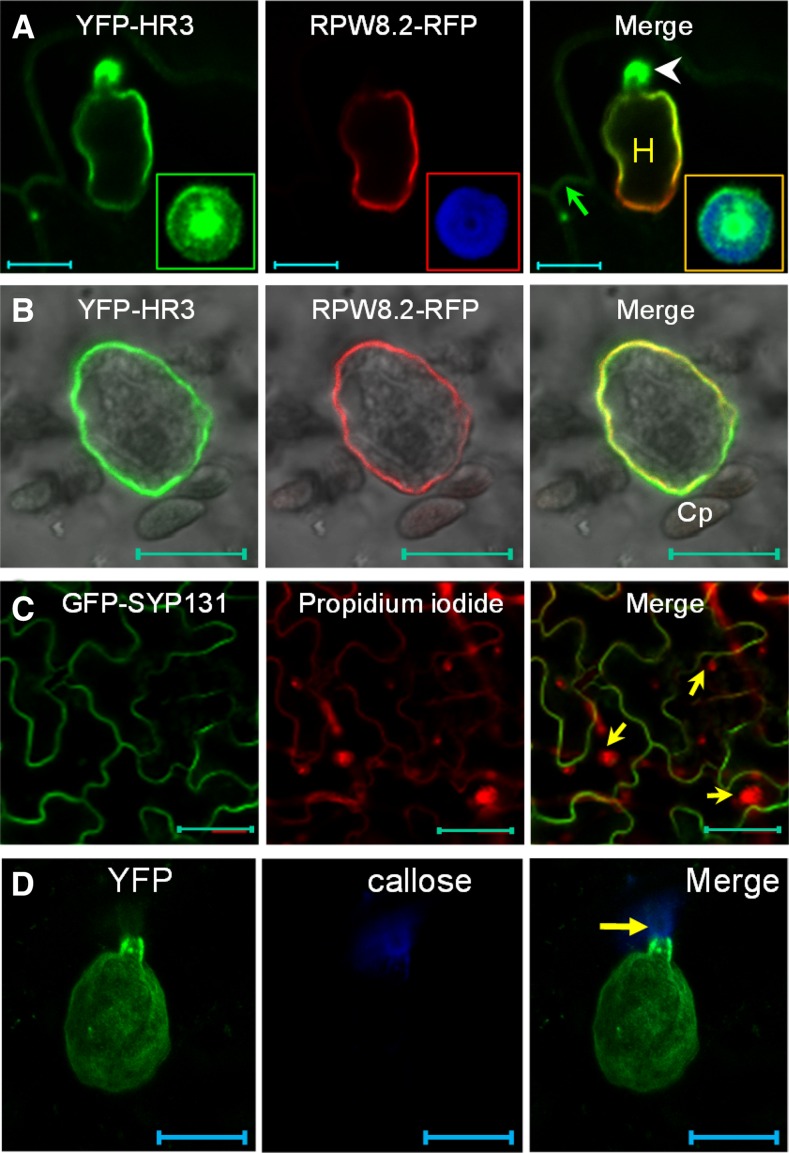
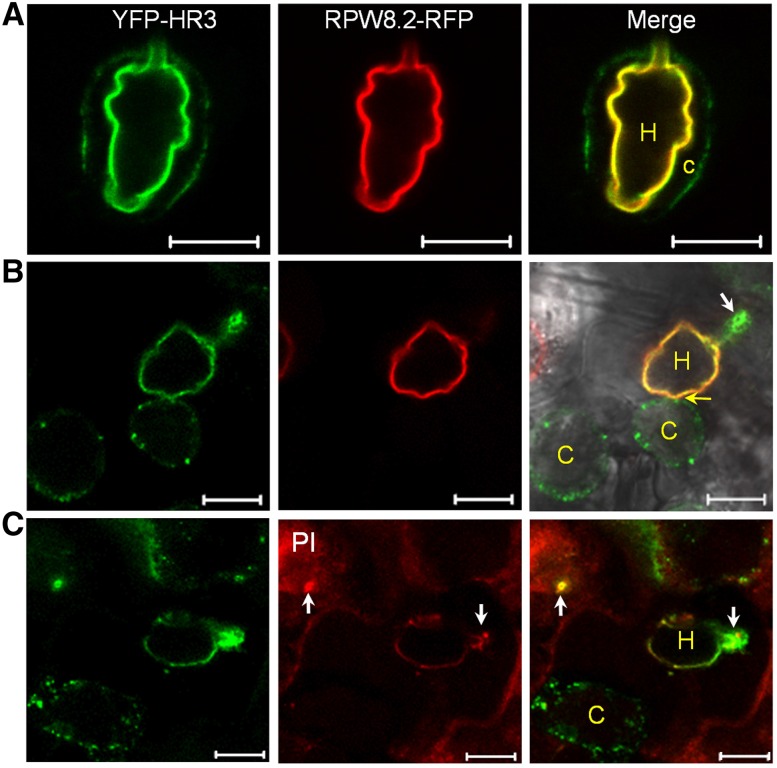
We then inoculated respective transgenic lines with Gc UCSC1 to examine the localization of these fusion proteins in epidermal cells invaded by haustoria. Interestingly, we found that YFP-HR1, YFP-HR2, YFP-HR3 appeared to be preferentially or exclusively localized to the EHM in epidermal cells invaded by haustoria (Fig. 4C), similar to our observation with YFP-RPW8.2 (Wang et al., 2010). The EHM-localization of YFP-HR3 is confirmed by precise colocalization with RPW8.2-RFP at the EHM (Fig. 5A). Moreover, YFP-HR3 signal remained detectable at the EHM of isolated haustorial complexes extracted from infected leaves coexpressing YFP-HR3 and R82-RFP (Fig. 5B). To exclude the possibility that EHM-localization was caused by overexpression of the fusion protein, we also generated transgenic lines expressing YFP-HR3 from the RPW8.2 promoter. As expected, expression of YFP-HR3 was mostly confined to haustorium-invaded epidermal cells and YFP signals were mostly or exclusively found in the EHM (Fig. 4D). To test if YFP-HR4 is also localized to the EHM, we inoculated YFP-HR4 expressing N. benthamiana leaves with Gc UCSC1, which is weakly infectious on N. benthamiana (Xiao et al., 2003). We also observed YFP signal as small punctae around the haustorium as well as in the cytoplasm (Fig. 4C). All T1 transgenic plants expressing these fusion proteins (> 20 for each construct) were as susceptible as Col-gl to Gc-UCSC1 (not shown), agreeing with the nonfunctional nature of these fusion proteins in defense as seen for YFP-RPW8.2 (Wang et al., 2010). To rule out the possibility that membrane proteins may be targeted by default to the EHM in the haustorium-invaded cells during the biogenesis of the EHM, we expressed the PM-localized membrane protein SYP131 (Uemura et al., 2004) tagged with YFP from the RPW8.2 promoter in Col-gl. YFP-SYP131 showed exclusive PM localization in haustorium-invaded cells (Fig. 5C), indicating that EHM-targeting of the YFP-tagged RPW8 homologs is attributable to the properties of these proteins rather than a consequence of activation of a default EHM-oriented trafficking pathway.
Figure 5.
YFP-HR3 is localized to the plasma membrane, the EHM, and the papilla. A, Representative image showing that YFP-HR3 is localized to the PM (arrow), the EHM (labeled by RPW8.2-RFP), and the papilla (arrowhead). Inset is a close-up view of the transverse section of a fungal penetration site where both YFP-HR3 and callose focally accumulate. Note the heterogeneous distribution of YFP-HR3. B, Representative isolated haustorial complex with an intact EHM labeled by both YFP-HR3 and RPW8.2-RFP. C, PM-localization of GFP-SYP131 in the epidermal cells containing haustoria (arrows). D, Maximum projection of a Z-stack image Z-stack image showing YFP-RPW8.2 is localized to the EHM but not in the papilla where callose is deposited (arrow). Bar, 10 μm in A, B, and D; 50 μm in C. Cp, Chloroplast.
YFP-HR3 Is Localized to the Papilla
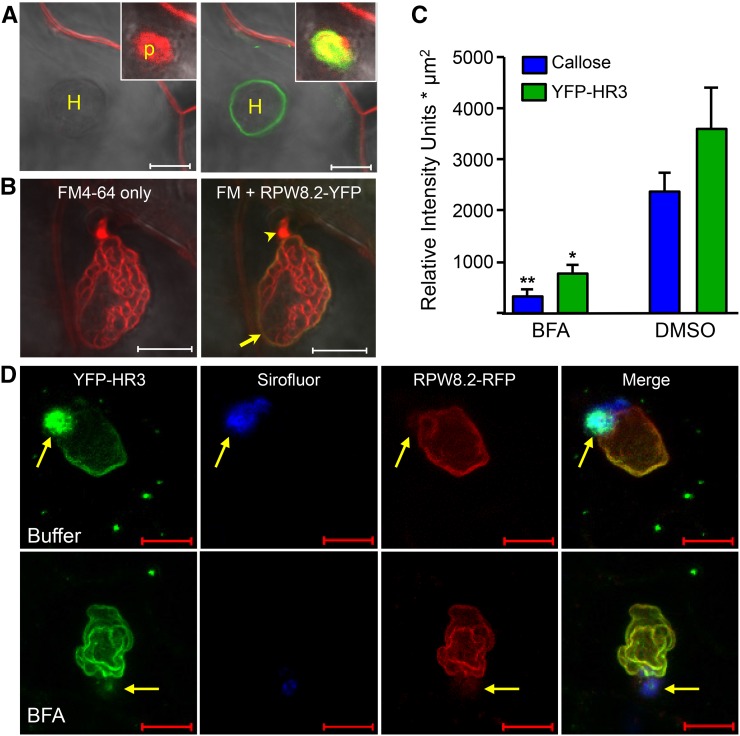
Intriguingly, we also noticed that YFP-HR3 showed focal accumulation around the fungal penetration site where a callosic papilla is formed (Fig. 5A). This localization pattern is reminiscent of PEN1/SYP121, a syntaxin involved in penetration (Collins et al., 2003; Assaad et al., 2004). To determine the precise localization of YFP-HR3 in relation to the papilla, we stained callose in the papilla by Sirofluor and found that YFP-HR3 was not precisely colocalized with callose, as it was more concentrated in the middle and the rim of the bull’s-eye-shaped callosic papilla (inset in Fig. 5A). Interestingly, we never observed signal from RPW8.2-YFP (RFP) or YFP (RFP)-RPW8.2 in the papilla (Fig. 5, A and D). These observations suggest that papilla-localization of YFP-HR3 is attributable to HR3 and may reflect a cellular function of HR3 that is distinct from RPW8.2.
Homologs of RPW8 exist in Brassica species (Xiao et al., 2004) but their biological functions have not been determined. To test if EHM- and papilla localization is indeed an ancestral feature of the RPW8 protein family, we made and expressed YFP fusion (both at the N- and C terminus) constructs for two RPW8 homologs from B. oleracea, BoHRa and BoHRb (Xiao et al., 2004) from the RPW8.2 promoter in Arabidopsis. We found that over 65% T1 lines (> 40) transgenic for BoHRa-YFP or BoHRb-YFP had greatly reduced plant stature with spontaneous cell death lesions and powdery mildew resistance (Fig. 6, A and B), suggesting that these Brassica homologs probably contribute to basal resistance against powdery mildew. Interestingly, unlike HR3-YFP, YFP signal from BoHRa-YFP, albeit weaker compared with RPW8.2-YFP, was detectable in powdery mildew-invaded cells as puncta mostly around the haustorium, with strong accumulation in the fungal penetration site (Fig. 6C). However, we never observed strong diffuse YFP signal from BoHRa-YFP at the EHM, highlighting a clear difference from RPW8.2-YFP. As expected, all the T1 lines (>30) transgenic for YFP-BoHRa or YFP-BoHRb were suscepible to Gc UCSC1 (not shown) and the fusion proteins were also found in the papilla (Fig. 6D) and the EHM (Fig. 6E). These observations further support that the EHM-targeting and papilla-location feature had likely evolved in RPW8 homologs before the separation of Brassica and Arabidopsis. Considering the gradual adaptation process of powdery mildew, it seems likely that during the Arabidopsis-powdery mildew coevolutionary conflict, RPW8.2 has gained EHM-focused localization while losing its papilla-localization.
Figure 6.
Brassica RPW8 homologs are also localized to the EHM and the papilla. A and B, Representative powdery mildew-infected T1 plants expressing BoHRa-YFP (A) or BoHRb-YFP (B) from the RPW8.2 promoter (upper panel) and total number of T1 transgenic plants from each indicated category (lower panel). Arrows indicate plants with spontaneous cell death and resistance to powdery mildew (disease reaction score, DR = 0 to 1; Xiao et al., 2005); arrowheads indicate plants displaying enhanced mildew resistance without spontaneous cell death (DR = 1 to 2); asterisks indicate plants susceptible to powdery mildew (DR = 2 to 3 or 3). Pictures were taken at 9 dpi. C and D, Representative images showing accumulation of BoHRa-YFP (C) or YFP-BoHRa (D) in the papilla. E, Representative image showing EHM-localization of YFP-BoHRa. Bar, 10 μm in C to E.
The EHM Is Fixed at the Haustorial Neck and Physically Separable from the Plasma Membrane and the Papilla
Triple localization (i.e. the EHM, the PM and the papilla) of YFP-HR3, regardless of its relevance to the physiological functions of HR3, may provide a unique tool to interrogate the origin and biogenesis of the EHM and how these membrane compartments might be physically connected with or separated from each other. To test if the EHM is separable from the PM and the papilla, we made transgenic plants coexpressing YFP-HR3 from the 35S promoter and RPW8.2-RFP from the RPW8.2 promoter. Constitutive expression of YFP-HR3 allows PM-labeling by YFP-HR3 before powdery mildew infection, and possible direct lateral diffusion or translocalization of YFP-HR3 from the PM to the EHM during haustorial biogenesis. Leaf sections inoculated with Gc UCSC1 were prepared at 3 dpi and subjected to plasmolysis (incubation with 0.5M NaCl). Interestingly, in some cells, the shrunk cytoplasm with the YFP-HR3-labeled PM was pulled off the cell wall but got stuck at the haustorial neck, wrapping around the haustorial complex with an EHM labeled by both YFP-HR3 and RPW8.2-RFP (Fig. 7A). In other cells, the shrunk cytoplasm with YFP-HR3-labeled PM was nearly or completely torn off the haustorial complex; however, YFP-HR3 localized at the papilla and the EHM remained unaffected (Fig. 7, B and C). These observations demonstrate that the PM is physically separable from the EHM and the papilla, and support that (1) there indeed exists a physical barrier (i.e. the neckband) at the haustorial neck (Gil and Gay, 1977) that fixes and separates the PM from the EHM, and (2) the papilla is an extracellular membrane compartment likely formed via exosome secretion (Meyer et al., 2009). It is also important to note that despite that the cytoplasm in most affected cells was shrunk >5 times during plasmolysis, the size and shape of most haustorial complexes did not seem to have significant changes (Fig. 7), suggesting that the EHM and the fungal cell wall in particular may be rigid and resistant to high osmotic pressure.
Figure 7.
The EHM is physically separable from the PM and the papilla. Gc UCSC-infected leaves expressing YFP-HR3 (from 35S) and RPW8.2-RFP (from the RPW8.2 promoter) were subjected to plasmolysis (0.5 M NaCl). A, Representative confocal image taken at 10 min after treatment. Note that the cytoplasm with YFP-HR3-labeled PM was shrunk around the seemingly unaffected haustorium labeled by both YFP-HR3 and RPW8.2-RFP. B and C, Representative confocal images taken at 30 min after treatment showing that the cytoplasm with YFP-HR3-labeled PM was nearly (B) or completely (C) detached from the haustorial complex with an intact EHM labeled by YFP-HR3 and RPW8.2-RFP. Note that YFP-HR3 localized to the papilla stained by PI (arrows in C) was also unaffected. Bar, 10 μm in A to C. C, Cytoplasm; H, haustorium.
YFP-HR3 Is Targeted to the Papilla and the EHM via Distinct Trafficking Pathways
To determine if YFP-HR3 is targeted to the papilla and the EHM via the same or distinct intracellular trafficking pathways, we first tested if there is lateral diffusion and/or an intracellular trafficking pathway between the PM and the EHM. FM4-64 is a lipophilic dye that labels the PM and tracks PM-derived endosomes to various subcellular compartments in a time-dependent manner in approximately 30 min (Bolte et al., 2004). While FM4-64 (20 μM in 0.002% Silwet L-77 aqueous solution) nicely labeled the PM in 5 min to 10 min after application of the dye and then was found to be colocalized with YFP-HR3 in the papilla in approximately 20 min, FM4-64 did not label the EHM in 45 min when all endosomal compartments should be labeled (Fig. 8A). However, if vacuum-infiltrated into the leaf tissue, FM4-64 could immediately stain the EHM (Fig. 8B). Given that the haustorial membrane lobes and the haustorial neck in particular were more strongly stained by FM4-64 (Fig. 8B), we speculate that the haustorial neckband as the presumable diffusion barrier is probably disrupted by vacuum infiltration, resulting in diffusion of the FM4-64 into the extrahaustorial matrix from the apoplast. This observation suggests that while membrane lipids or proteins from the PM can reach the papilla likely via an endosomal trafficking pathway, there is no or little (undetectable) lateral diffusion or endosomal trafficking from the PM to the EHM.
Figure 8.
Papilla-accumulation of YFP-HR3 is BFA sensitive while its targeting to the EHM is not. A, Surface-applied FM4-64 (20 μM) in 0.002% Silwet L-77 stained the PM and the papilla in 20 min (image on the left) but did not stain the EHM labeled by YFP-HR3 in 45 min (image on the right). B, When vacuum-infiltrated into leaf tissue (see “Materials and Methods”), FM4-64 could stain the EHM (arrow) labeled by RPW8.2-YFP as well as the haustorial membrane lobes. Notably, the haustorial neck is often more strongly stained (arrowhead). C, BFA inhibits focal accumulation of YFP-HR3 and callose to the papilla. Quantification of relative fluorescence intensity for YFP-HR3 and Sirofluor-stained callose at the papilla in confocal images. Data represent means ± se (n = 6) from one of two independent experiments. Student’s t test was used to compare the differences between treatments (* indicates P < 0.01, ** indicates P < 0.001). D, Representative images showing localization of YFP-HR3 to the PM, the papilla, and the EHM at 45 hpi in epidermal cells infiltrated with 1% DMSO buffer (upper panel) or 300 μM BFA (lower panel) at approximately 6 hpi from the base of the leaf. The EHM was labeled by RPW8.2-RFP and the callose was stained with Sirofluor. Note that both YFP-HR3 and callose were diminished in the papilla due to BFA treatment, but YFP-HR3 and RPW8.2-RFP at the EHM were unaffected. Bar, 10 μm in A to E. H, Haustorium; p, papilla.
We then determined if YFP-HR3’s papilla- and the EHM-localization can be inhibited by the same pharmacological inhibitor. It has been shown that Brefeldin A (BFA), a fungal toxin widely used to block multiple intracellular vesicle trafficking pathways, can inhibit focal accumulation of both GFP-PEN1 and callose deposition to the papilla (Nielsen et al., 2012). We treated leaves of plants transgenic for 35S::YFP-HR3 and R82p::RPW8.2-RFP with BFA (300 μM) and found that while EHM localization of YFP-HR3 and RPW8.2-RFP appeared unaffected, both focal accumulation of YFP-HR3 and callose to the papilla were similarly diminished (Fig. 8, C and D). These observations indicate that EHM-oriented protein targeting and papilla-focused protein accumulation are realized via two distinct trafficking pathways.
DISCUSSION
In this study, we showed that HR3, the probable progenitor of the RPW8 family, HR2, and possibly HR1 play a positive role in basal resistance and that EHM-localization is probably a conserved feature of the RPW8 family. Using HR3’s localization to the PM and papillae, we also collected evidence to suggest that the EHM is unlikely formed via PM invagination and the PM-to-EHM endocytic trafficking makes little or no contribution to the EHM biogenesis.
Our previous studies showed that RPW8.1 and RPW8.2 evolved recently in Arabidopsis after its separation from Brassica species, while RPW8 homologs more closely related to HR3 are found in B. oleracea and B. rapa and appear to be under purifying selection (Xiao et al., 2004; Orgil et al., 2007). Though anticipated, whether these RPW8 homologs are involved in basal resistance against powdery mildew remained undetermined. Through genetic and localization analyses we now have demonstrated that HR1, HR2, and HR3 contribute to basal resistance to powdery mildew and that all the Arabidopsis RPW8 family proteins including RPW8.1 (Ma et al., 2014) are probable EHM-residents. HR1, HR2, and HR3 probably serve a partially redundant function in basal resistance but this collective role cannot be accurately measured without a triple knockout mutant. Because these three genes are tandemly arrayed in an 8 Kb region, making double or triple mutants by genetic crossing is extremely difficult. We tried to silence HR1 and HR2 in the HR3-kd1 background but did not see any significant additional disease susceptibility (data not shown). Targeted mutagenesis using gene editing tools such CRISPR/Cas9 will be used to generate a triple mutant to assess the overall role of these three genes in the future. HR4 is believed to be the newest member of the RPW8 gene family, likely created from the deletion of RPW8.1 and RPW8.2 that resulted from a transposition event (Xiao et al., 2004). Hence, it is not a surprise to us that HR4 did not appear to contribute to basal resistance against powdery mildew and YFP-HR4 was not detectable in Arabidopsis. Interestingly, HR4 has been shown to be induced by powdery mildew (Chandran et al., 2011) and the beneficial fungus Trichoderma (Sáenz-Mata and Jiménez-Bremont, 2012), suggesting that HR4 is involved in biotic stress response. Unfortunately, no T-DNA lines for HR4 are available. Future targeted mutagenesis is required for understanding the biological function of HR4.
Taken together, it seems that RPW8.2’s capability in activating haustorium-targeted defense is not entirely new, but instead a capability that stemmed from and then surpassed that of the older family members. A critical question then is how RPW8.2-mediated resistance is mechanistically differentiated from the original function of the HR3-like progenitor (Xiao et al., 2004). Our present studies provided some insights into this question. First, RPW8.2-YFP can accumulates at high levels in the EHM, whereas HR3-YFP is undetectable, yet triggers massive cell death (Figs. 2, 3, and 6). This suggests that the C-tail of HR3 may serve as an autoinhibitory domain that needs to be intact for proper function of HR3, whereas this constraint is probably relaxed if not completely removed for RPW8.2. Second, because the chimeric protein R82n-HR3c-YFP but not HR3n-R82c-YFP behaved similarly as RPW8.2-YFP in defense and EHM-targeting (Fig. 3), we can conclude that sequence diversification in the N-terminal 147 amino acids between HR3 and RPW8 largely underscores their functional differences. A detailed mutational analysis is needed to identify key residues for improved defense activity in RPW8.2 or higher activity in triggering cell death in HR3 (when its C terminus is fused with YFP) from a total of 42 amino acid substitutions between these two proteins in this region (Fig. 3A). Third, YFP-HR3 but not YFP-RPW8.2 was found in the papilla, suggesting that HR3 might play a role in penetration resistance and a signal in HR3 for papilla localization might have been lost in RPW8.2, which seems to be reasonable as RPW8.2 apparently triggers haustorium-targeted defense at the postpenetration stage. It should be pointed out however that functional enhancement of RPW8.2 from an HR3-like gene may be partially attributable to the RPW8.2 promoter, which renders powdery mildew inducible, epidermal cell-specific expression of RPW8.2 (Wang et al., 2009) and showed higher activity when driving HR3-YFP expression as reflected by a higher proportion of T1 lines with lethal cell death (Fig. 2D). Thus, functional evolution of RPW8.2 from an HR3-like progenitor gene probably necessitated DNA sequence diversification in the 5′ regulatory region for enhanced, powdery mildew-inducible expression, and in the coding region for higher efficiency in defense activation while conserving the amino acids for EHM-targeting.
The origin and biogenesis of the EHM is of great interest to researchers in the field because of its apparent importance in understanding the host-pathogen interaction but unfortunately remains largely undetermined. The absence of eight PM-localized proteins in the EHM (Koh et al., 2005) and the presence of RPW8.2 exclusively in the EHM (Wang et al., 2009) suggest that the EHM and the PM are more likely two distinct membranes with different origins. However, theoretically, one cannot exclude the possibility that the EHM is initially derived from invagination of the PM when the haustorium develops from the tip of the fungal penetration peg inside the host epidermal cell, and then differentiates into a membrane distinct from the PM by selective exclusion of certain PM proteins and specific recruitment of other proteins such as RPW8.2. One of the difficulties we face when addressing this question is the lack of a good protein marker that is localized to the PM and the EHM at high levels. In this regard, we were lucky to find that YFP-HR3 is localized in the PM, the papilla, and the EHM (Fig. 5), because this localization feature could be very valuable for testing the above possibility and assessing possible interconnection between the three membrane compartments. Based on our results from YFP-HR3 localization and FM4-64 staining analyses, we suggest that the EHM is unlikely derived from PM invagination; rather, it is more likely synthesized de novo via exocytosis with possible contribution from other intracellular membrane compartments. Specifically, our above reasoning is supported by the following three aspects. First, we performed plasmolysis of haustorium-invaded epidermal cells expressing YFP-HR3 and RPW8.2-RFP and found that YFP-HR3 signal was virtually intact in the EHM throughout the plasmolysis and was separable from the shrunk cytoplasm with YFP-HR3-labeled PM (Fig. 7). This demonstrates that the EHM is indeed a membrane compartment distinct and physically separable from the PM, which allows isolation of individual haustorial complexes containing the EHM (Fig. 5B). Second, we found that PM-incorporated FM4-64 failed to reach the EHM in 45 min although it could reach the papilla in 20 min under conditions when the integrity of the haustorial complex is not affected (Fig. 8A). This indicates that there is a diffusion barrier (most likely attributable to the haustorial neckband) between the PM and the EHM, and implies that endocytic trafficking from the PM to the EHM either does not occur at all or is minimal. Indeed, when such a diffusion barrier is likely disrupted upon vacuum infiltration of a leaf section containing haustorial complexes in the presence of 0.002% surfactant Silwet L-77, FM4-64 can readily and immediately stain the EHM and the haustorial membrane (Fig. 8B). In line with this inference, an exocytosis pathway defined by a SNARE complex defined by Vamp721/Vamp722 and SNAP33 has recently been shown to be required for efficient targeting of RPW8.2 vesicles to the EHM (Kim et al., 2014). Third, consistent with the above results, we also found that YFP-HR3’s localization to the papilla is BFA sensitive, whereas EHM-targeting of YFP-HR3 (and RPW8.2-RFP) is BFA insensitive (Fig. 8, D and E). Papilla-localization of YFP-HR3 is reminiscent of an earlier report that YFP-PEN1 and callose are delivered to the papilla via a BFA-sensitive pathway, which requires the ADP ribosylation factor-GTP exchange factor GNOM (Nielsen et al., 2012). Hence, it is likely that the same GNOM-dependent pathway is engaged for delivery of YFP-HR3 to the fungal penetration sites. Given that, unlike in animals, BFA mainly inhibits certain endosome recycling pathways and there exists BFA-insensitive exocytic trafficking in plants (Richter et al., 2007; Teh and Moore, 2007), it is reasonable to infer that EHM-targeting of YFP-HR3 (and RPW8.2-RFP) is via BFA-insensitive exocytosis.
Unlike the haustorial complex formed by powdery mildew and rust where a haustorial neckband has been well-documented (Heath, 1976; Chong and Harder, 1980), no apparent haustorial neckband is clearly observed in the haustorial complex formed by oomycete pathogens (Mims et al., 2004; Soylu, 2004). Whether the origin and biogenesis of the EHM induced by oomycete pathogens shares the same cell biological principles remains to be determined. In this regard, it is worth noting that several PM-proteins (PEN1, FLS2, and REM1.3, PDLP1, etc.), Golgi marker proteins (SYP32 and Got1p) and endosomal markers (RabC1 and Ara7) were found in the perihaustorial membrane encasing oomycete haustoria in tobacco or Arabidopsis cells (Lu et al., 2012; Bozkurt et al., 2014; Caillaud et al., 2014). However, based on our observations, PM-localized proteins such as GFP-PEN1 (Wang et al., 2009) and YFP-SYP131 (Fig. 5C) were apparently excluded from the EHM induced by powdery mildew even when stably expressed from the RPW8.2 promoter. This seemingly more relaxed selectivity for EHM-resident proteins in the case of oomycete haustorial complexes might reflect differential contribution of endosomal trafficking pathways to EHM biogenesis under different pathogen contexts and suggest that recruitment of EHM resident proteins may be more stringent in powdery mildew haustorium-invaded cells. More recently, Bozkurt et al. (2015) observed localization of RabG3c, a Rab7 GTPase to the EHM during haustorial biogenesis of Phytophthora infestans in N. benthamiana. Given that (1) RabG3c is localized to late endosomes and the tonoplast in uninfected cells, and (2) a tonoplast-localized Suc transporter is not found in the EHM, the authors suggested that there exists specific rerouting of vacuole-targeted late endosomes to the EHM during haustorial biogenesis (Bozkurt et al., 2015). It would be interesting to test if such traffic rerouting also occurs during haustorial biogenesis of powdery mildew in plant epidermal cells.
Apparently, more definitive cell biological and/or genetic evidence is needed to make a solid conclusion with regard to the origin and biogenesis of the EHM induced by fungal and oomycete pathogens. Conceivably, pharmacological inhibitors specifically inhibiting plant endocytosis or exocytosis and/or genetic mutants defective only in EHM-oriented trafficking would provide novel means and insight to allow more critical assessment of the contribution of different intracellular trafficking pathways to the formation of the EHM in haustorium-invaded host cells. Future research will be directed to these two aspects.
MATERIALS AND METHODS
Plant Materials and Cultivation
Arabidopsis (Arabidopsis thaliana) accessions Col-0 and/or Col-gl (Col-0 carrying the glabrous mutation) were used for generation of all transgenic lines. All genetic analyses including crossing, genotyping, and phenotyping were conducted in accordance with previous reports (Xiao et al., 2003; Xiao et al., 2005). Unless otherwise indicated, seeds were sown in Sunshine Mix no. 1 or Metro-Mix 360 soil (MD Plant & Suppliers) and cold-treated (4°C for 1 to 2 d). Seedlings were kept under 22°C, 75% RH, short-day (8 h light at approximately 125 μm·m−2·s−1, 16 h dark) conditions for 5 to 6 weeks before pathogen inoculation and/or other treatments.
Pathogens Strains, Inoculation, and Phenotyping
Powdery mildew isolate Golovinomyces cichoracearum UCSC1 (Gc-UCSC1 was maintained on live Col-0 or pad4-1 mutant plants for generation of fresh conidia for inoculation purposes. Inoculation, visual scoring, photographing, and quantification of disease susceptibility were done as described in Xiao et al. (2005). All pathogen infection experiments were repeated at least three times.
DNA Constructs and Generation of Transgenic Lines
All primer sequences and related information are summarized in Supplemental-Table 1. Overexpression constructs for HR1, HR2, HR3, and HR4 were created after amplification of the genomic sequence including approximately 1000 bp of the 5′ UTR/promoter region (496 bp in the case of AtHR3) from the T20E23 BAC clone obtained from the Arabidopsis Biological Resource Center into the binary vector pKMB downstream of the 35S promoter (Mylne and Botella, 1998).
The pCX-DG TA cloning vector described in Chen et al. (2009) was modified to include an approximately 1.8 kb RPW8.2 promoter region by releasing the 35S::GFP fragment upstream of the XcmI and plant NOS terminator using EcoRI digestion and replacement (pCX-DG-R82p). We used this modified TA cloning vector to express the full-length HR3 and HR3 variants, and HR3-RPW8.2 chimeric genes.
HR1, HR2, HR3, and HR4 genes with their NPs (approximately 1000 bp upstream of the ATG start codon) were translationally fused at the C terminus with YFP using the pPZPYFP23′ vector described in (Wang et al., 2007), a modified derivative of pPZP211 (Hajdukiewicz et al., 1994). HR1, HR2, HR3, and HR4 genes were also translationally fused to YFP at the N terminus under control of the 35S promoter using the pEarleyGate gateway compatible vector series (Earley et al., 2006). Recombination reactions were completed using pENTR/D-TOPO clones of the full genomic DNA sequences. Gateway LR Clonase II enzyme mix was used to complete the recombination reaction between the entry clones and destination vector (pEG104).
Chimeric domain swapping constructs between the N-domain (amino acid 1 to 147; HR3n) and the C-domain (amino acid 148 to 213; HR3c) of HR3 and the N-domain (amino acid 1 to 145; R82n) and the C-domain (amino acid 146 to 174; R82c) of RPW8.2 were created using NcoI adapted primers designed at the end of the second predicted coiled-coil domain of each gene. HR3p::HR3n-R82c and R82p::R82n-HR3c chimeric genes were translationally fused to YFP at the C terminus under control of either the HR3 or RPW8.2 promoter using the previously mentioned pPZPYFP23′ homemade vector (Wang et al., 2007). Additionally, HR3n-R82c-YFP was also cloned into the homemade pCX-DG-R82p.
TA cloning using the modified binary vector pCX-DG-R82p was also used for a variety of other constructs including: YFP-HR1, YFP-HR2, YFP-HR3, YFP-HR4 and RPW8.2-RFP. All generated constructs in binary vectors were introduced into Agrobacterium strain GV3101 and stable transgenic Arabidopsis plants were generated by floral dip (Clough et al., 2000).
Pharmacological Treatments and Gel Blotting
Fully expanded leaves of approximately 7 week-old RPW8.2-YFP or YFP-HR3/RPW8.2-RFP Col-gl background plants were detached from the base of the petioles and inserted into sterilized Murashige and Skoog agar medium in petri dishes. Detached leaves were inoculated evenly with Gc-UCSC1 and at different time points (0, 12, or 16 hpi) leaf sections (approximately 0.25 cm2) were examined using a model no. LSM710 confocal microscope (Zeiss) at 36 to 42 hpi after haustorial staining with 0.5% propidium iodide (PI). For FM4-64 dye uptake experiments, leaf sections were immersed in a fresh 20 µM FM4-64 solution containing 0.002% Silwet l-77 (Lehele Seeds) for facilitating dye uptake by leaf epidermal cells under nondisruptive conditions. Infiltration of FM4-64 into leaf tissues was done by applying vacuum to leaf samples immersed in an FM4-64 solution containing 0.002% Silwet l-77 in a container for 3 to 5 min followed by immediate confocal imaging. BFA treatments were conducted with a 300 μM solution of BFA dissolved in a 1% DMSO solution in a similar manner as reported in Nielsen et al. (2012).
For western blotting, Arabidopsis leaves infected with Gc UCSC1 was collected at 5 dpi for preparation of total microsomal proteins as described in Wang et al. (2007). An anti-GFP antibody (A290; Abcam) was used to detect YFP-fusion proteins from various Arabidopsis transgenic lines.
Isolation of Haustorial Complexes
This protocol is modified from the “Isolation of intracellular hyphae by isopycnic centrifugation technique” described in Pain et al. (1994). Extracted haustoria suspensions were obtained from pooled T2 lines of appropriate transgenic backgrounds between 7 and 8 dpi with Gc UCSC1. Approximately 1 to 2 total grams of infected leaf tissue was macerated 2× in a conventional kitchen blender in a minimum volume of 20 to 30 mL of fresh, chilled haustoria extraction buffer (3-[N-morpholino]-propane sulphonic acid 0.02 M, pH 7.2 containing 0.2 M Suc) for 45 to 60 s. After each 1× blending procedure, macerated plant material was filtered through a 40 µm nylon mesh material using vacuum filtration to remove large fragments of plant debris as most mature Gc UCSC1 haustoria are typically between 15 and 25 μm in size. Final filtrates were centrifuged at 1080g for 15 min at 4°C. Pelleted material containing haustoria was resuspended in 2 to 3 mL of haustoria extraction buffer and kept at 4°C before imaging or further analysis.
Confocal Microscopy
Laser scanning confocal microscopy was done using a model no. LSM710 confocal microscope (Zeiss) as previously described in Wang et al. (2009, 2013). All images and videos presented are single optical sections or Z-stack projected of 15 to 30 images unless otherwise indicated. For FM4-64 staining, detached leaf sections (approximately 0.25 cm2) were submerged in 20 μM FM4-64 (Molecular Probes) in water for 15 to 45 min. For PI staining, detached leaf sections (approximately 0.25 cm2) were submerged in 0.5% PI solution for 45 to 60 min then washed briefly (10 to 15 min) in water before imaging. Sirofluor staining of leaf sections for callose was conducted with a 0.1 mg mL−1 dissolved in a 1% DMSO solution (Biosupplies). Image data were processed using Zen 2009 Light Edition and Adobe Photoshop CS5.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis genome initiative or GenBank/EMBL database under accession numbers AT3G50450 (HR1), AT3G50460 (HR2), AT3G50470 (HR3), AT3G50480 (HR4), AY225587 (BoHRa), and AY225588 (BoHRb).
SUPPLEMENTAL DATA
The following supplemental materials are available.
Supplemental Figure S1. Schematic gene structures and the location and direction of the T-DNA insertions (arrowheads) in HR1, HR2, and HR3.
Supplemental Figure S2. Protein sequence alignment of the RPW8 family.
Supplemental Figure S3. Expression of HR1-YFP, HR2-YFP, and HR3-YFP is detectable in N. benthamiana when transiently coexpressed with a viral RNAi suppressor.
Supplemental Figure S4. HR3 is not required for RPW8.1 and RPW8.2-mediated resistance.
Supplemental Table S1. Information about the DNA constructs used in this study.
Supplementary Material
Acknowledgments
We thank Frank Coker for maintaining the growth facility, Dr. Ying Wu for critical reading of this manuscript, and two anonymous reviewers for their constructive comments, which helped us to improve the manuscript.
Footnotes
This project was supported by National Science Foundation grants no. IOS-1146589 and IOS-1457033 to S.X.
Articles can be viewed without a subscription.
References
- Aist JR. (1976) Papillae and related wound plugs of plant cells. Annu Rev Phytopathol 14: 145–163 [Google Scholar]
- Assaad FF, Qiu J-L, Youngs H, Ehrhardt D, Zimmerli L, Kalde M, Wanner G, Peck SC, Edwards H, Ramonell K, Somerville CR, Thordal-Christensen H (2004) The PEN1 syntaxin defines a novel cellular compartment upon fungal attack and is required for the timely assembly of papillae. Mol Biol Cell 15: 5118–5129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S, Talbot C, Boutte Y, Catrice O, Read ND, Satiat-Jeunemaitre B (2004) FM-dyes as experimental probes for dissecting vesicle trafficking in living plant cells. J Microsc 214: 159–173 [DOI] [PubMed] [Google Scholar]
- Bozkurt TO, Belhaj K, Dagdas YF, Chaparro-Garcia A, Wu CH, Cano LM, Kamoun S (2015) Rerouting of plant late endocytic trafficking toward a pathogen interface. Traffic 16: 204–226 [DOI] [PubMed] [Google Scholar]
- Bozkurt TO, Richardson A, Dagdas YF, Mongrand S, Kamoun S, Raffaele S (2014) The plant membrane-associated REMORIN1.3 accumulates in discrete perihaustorial domains and enhances susceptibility to Phytophthora infestans. Plant Physiol 165: 1005–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushnell WR, Bergquist SE (1975) Aggregation of host cytoplasm and the formation of papillae and haustoria in powdery mildew of barley. Phytopathology 65: 310–318 [Google Scholar]
- Bushnell WR, Gay JL (1978) Accumulation of solutes in relation to the structure and function of haustoria in powdery mildews. In Spencer DM, ed, The Powdery Mildews. Academic Press, London, pp 183-235 [Google Scholar]
- Caillaud MC, Wirthmueller L, Sklenar J, Findlay K, Piquerez SJ, Jones AM, Robatzek S, Jones JD, Faulkner C (2014) The plasmodesmal protein PDLP1 localises to haustoria-associated membranes during downy mildew infection and regulates callose deposition. PLoS Pathog 10: e1004496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campe R, Langenbach C, Leissing F, Popescu GV, Popescu SC, Goellner K, Beckers GJ, Conrath U (2016) ABC transporter PEN3/PDR8/ABCG36 interacts with calmodulin that, like PEN3, is required for Arabidopsis nonhost resistance. New Phytol 209: 294–306 [DOI] [PubMed] [Google Scholar]
- Chandran D, Hather G, Wildermuth MC (2011) Global expression profiling of RNA from laser microdissected cells at fungal-plant interaction sites. Methods Mol Biol 712: 263–281 [DOI] [PubMed] [Google Scholar]
- Chen S, Songkumarn P, Liu J, Wang GL (2009) A versatile zero background T-vector system for gene cloning and functional genomics. Plant Physiol 150: 1111–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J, Harder DE (1980) Ultrastructure of haustorium development in Puccinia-Coronata-avenae. 1. Cyto-chemistry and electron-probe x-ray analysis of the haustorial neck ring. Can J Bot-Rev Can De Bot 58: 2496–2505 [Google Scholar]
- Clough SJ, Fengler KA, Yu IC, Lippok B, Smith RK Jr., Bent AF (2000) The Arabidopsis dnd1 “defense, no death” gene encodes a mutated cyclic nucleotide-gated ion channel. Proc Natl Acad Sci USA 97: 9323–9328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins NC, Thordal-Christensen H, Lipka V, Bau S, Kombrink E, Qiu JL, Hückelhoven R, Stein M, Freialdenhoven A, Somerville SC, Schulze-Lefert P (2003) SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425: 973–977 [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Ellinger D, Naumann M, Falter C, Zwikowics C, Jamrow T, Manisseri C, Somerville SC, Voigt CA (2013) Elevated early callose deposition results in complete penetration resistance to powdery mildew in Arabidopsis. Plant Physiol 161: 1433–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil F, Gay JL (1977) Ultrastructural and physiological properties of the host interfacial components of haustoria of Erysiphe pisi in vivo and in vitro. Physiol Plant Pathol 10: 1–12 [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25: 989–994 [DOI] [PubMed] [Google Scholar]
- Halterman D, Zhou F, Wei F, Wise RP, Schulze-Lefert P (2001) The MLA6 coiled-coil, NBS-LRR protein confers AvrMla6-dependent resistance specificity to Blumeria graminis f. sp. hordei in barley and wheat. Plant J 25: 335–348 [DOI] [PubMed] [Google Scholar]
- Heath MC. (1976) Ultrastructural and functional similarity of haustorial neckband of rust fungi and Casparian strip of vascular plants. Can J Bot-Rev Can Bot 54: 2484–2489 [Google Scholar]
- Jordan T, Seeholzer S, Schwizer S, Töller A, Somssich IE, Keller B (2011) The wheat Mla homologue TmMla1 exhibits an evolutionarily conserved function against powdery mildew in both wheat and barley. Plant J 65: 610–621 [DOI] [PubMed] [Google Scholar]
- Kim H, O’Connell R, Maekawa-Yoshikawa M, Uemura T, Neumann U, Schulze-Lefert P (2014) The powdery mildew resistance protein RPW8.2 is carried on VAMP721/722 vesicles to the extrahaustorial membrane of haustorial complexes. Plant J 79: 835–847 [DOI] [PubMed] [Google Scholar]
- Koh S, André A, Edwards H, Ehrhardt D, Somerville S (2005) Arabidopsis thaliana subcellular responses to compatible Erysiphe cichoracearum infections. Plant J 44: 516–529 [DOI] [PubMed] [Google Scholar]
- Lawton K, Weymann K, Friedrich L, Vernooij B, Uknes S, Ryals J (1995) Systemic acquired resistance in Arabidopsis requires salicylic acid but not ethylene. Mol Plant Microbe Interact 8: 863–870 [DOI] [PubMed] [Google Scholar]
- Lipka V, Dittgen J, Bednarek P, Bhat R, Wiermer M, Stein M, Landtag J, Brandt W, Rosahl S, Scheel D, Llorente F, Molina A, et al. (2005) Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 310: 1180–1183 [DOI] [PubMed] [Google Scholar]
- Lu YJ, Schornack S, Spallek T, Geldner N, Chory J, Schellmann S, Schumacher K, Kamoun S, Robatzek S (2012) Patterns of plant subcellular responses to successful oomycete infections reveal differences in host cell reprogramming and endocytic trafficking. Cell Microbiol 14: 682–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XF, Li Y, Sun JL, Wang TT, Fan J, Lei Y, Huang YY, Xu YJ, Zhao JQ, Xiao S, Wang WM (2014) Ectopic expression of RESISTANCE TO POWDERY MILDEW8.1 confers resistance to fungal and oomycete pathogens in Arabidopsis. Plant Cell Physiol 55: 1484–1496 [DOI] [PubMed] [Google Scholar]
- Meyer D, Pajonk S, Micali C, O’Connell R, Schulze-Lefert P (2009) Extracellular transport and integration of plant secretory proteins into pathogen-induced cell wall compartments. Plant J 57: 986–999 [DOI] [PubMed] [Google Scholar]
- Mims CW, Richardson EA, Holt Iii BF, Dangl JL (2004) Ultrastructure of the host-pathogen interface in Arabidopsis thaliana leaves infected by the downy mildew Hyaloperonospora parasitica (vol 82, pg 1001, 2004). Can J Bot-Rev Can Bot 82: 1545 [Google Scholar]
- Mylne J, Botella JR (1998) Binary vectors for sense and antisense expression of Arabidopsis ESTs. Plant Mol Biol Report 16: 257–262 [Google Scholar]
- Nelson BK, Cai X, Nebenführ A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Nielsen ME, Feechan A, Böhlenius H, Ueda T, Thordal-Christensen H (2012) Arabidopsis ARF-GTP exchange factor, GNOM, mediates transport required for innate immunity and focal accumulation of syntaxin PEN1. Proc Natl Acad Sci USA 109: 11443–11448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orgil U, Araki H, Tangchaiburana S, Berkey R, Xiao S (2007) Intraspecific genetic variations, fitness cost and benefit of RPW8, a disease resistance locus in Arabidopsis thaliana. Genetics 176: 2317–2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pain NA, Green JR, Gammie F, O’Connell RJ (1994) Immunomagnetic isolation of viable intracellular hyphae of Colletotrichum lindemuthianum (Sacc. & Magn.) Briosi & Cav. from infected bean leaves using a monoclonal antibody. New Phytol 127: 223–232 [DOI] [PubMed] [Google Scholar]
- Qu F, Ren T, Morris TJ (2003) The coat protein of turnip crinkle virus suppresses posttranscriptional gene silencing at an early initiation step. J Virol 77: 511–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter S, Geldner N, Schrader J, Wolters H, Stierhof YD, Rios G, Koncz C, Robinson DG, Jürgens G (2007) Functional diversification of closely related ARF-GEFs in protein secretion and recycling. Nature 448: 488–492 [DOI] [PubMed] [Google Scholar]
- Roberts AM, Mackie AJ, Hathaway V, Callow JA, Green JR (1993) Molecular differentiation in the extrahaustorial membrane of pea powdery mildew haustoria at early and late stages of development. Physiol Mol Plant Pathol 43: 147–160 [Google Scholar]
- Sáenz-Mata J, Jiménez-Bremont JF (2012) HR4 gene is induced in the Arabidopsis-Trichoderma atroviride beneficial interaction. Int J Mol Sci 13: 9110–9128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soylu S. (2004) Ultrastructural characterisation of the host-pathogen interface in white blister-infected Arabidopsis leaves. Mycopathologia 158: 457–464 [DOI] [PubMed] [Google Scholar]
- Stein M, Dittgen J, Sánchez-Rodríguez C, Hou BH, Molina A, Schulze-Lefert P, Lipka V, Somerville S (2006) Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell 18: 731–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh OK, Moore I (2007) An ARF-GEF acting at the Golgi and in selective endocytosis in polarized plant cells. Nature 448: 493–496 [DOI] [PubMed] [Google Scholar]
- Uemura T, Ueda T, Ohniwa RL, Nakano A, Takeyasu K, Sato MH (2004) Systematic analysis of SNARE molecules in Arabidopsis: dissection of the post-Golgi network in plant cells. Cell Struct Funct 29: 49–65 [DOI] [PubMed] [Google Scholar]
- Wang W, Berkey R, Wen Y, Xiao S (2010) Accurate and adequate spatiotemporal expression and localization of RPW8.2 is key to activation of resistance at the host-pathogen interface. Plant Signal Behav 5: 1002–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Devoto A, Turner JG, Xiao S (2007) Expression of the membrane-associated resistance protein RPW8 enhances basal defense against biotrophic pathogens. Mol Plant Microbe Interact 20: 966–976 [DOI] [PubMed] [Google Scholar]
- Wang W, Wen Y, Berkey R, Xiao S (2009) Specific targeting of the Arabidopsis resistance protein RPW8.2 to the interfacial membrane encasing the fungal Haustorium renders broad-spectrum resistance to powdery mildew. Plant Cell 21: 2898–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Zhang Y, Wen Y, Berkey R, Ma X, Pan Z, Bendigeri D, King H, Zhang Q, Xiao S (2013) A comprehensive mutational analysis of the Arabidopsis resistance protein RPW8.2 reveals key amino acids for defense activation and protein targeting. Plant Cell 25: 4242–4261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y, Wang W, Feng J, Luo MC, Tsuda K, Katagiri F, Bauchan G, Xiao S (2011) Identification and utilization of a sow thistle powdery mildew as a poorly adapted pathogen to dissect post-invasion non-host resistance mechanisms in Arabidopsis. J Exp Bot 62: 2117–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Brown S, Patrick E, Brearley C, Turner JG (2003) Enhanced transcription of the Arabidopsis disease resistance genes RPW8.1 and RPW8.2 via a salicylic acid-dependent amplification circuit is required for hypersensitive cell death. Plant Cell 15: 33–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Calis O, Patrick E, Zhang G, Charoenwattana P, Muskett P, Parker JE, Turner JG (2005) The atypical resistance gene, RPW8, recruits components of basal defence for powdery mildew resistance in Arabidopsis. Plant J 42: 95–110 [DOI] [PubMed] [Google Scholar]
- Xiao S, Ellwood S, Calis O, Patrick E, Li T, Coleman M, Turner JG (2001) Broad-spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science 291: 118–120 [DOI] [PubMed] [Google Scholar]
- Xiao S, Emerson B, Ratanasut K, Patrick E, O’Neill C, Bancroft I, Turner JG (2004) Origin and maintenance of a broad-spectrum disease resistance locus in Arabidopsis. Mol Biol Evol 21: 1661–1672 [DOI] [PubMed] [Google Scholar]
- Yahiaoui N, Srichumpa P, Dudler R, Keller B (2004) Genome analysis at different ploidy levels allows cloning of the powdery mildew resistance gene Pm3b from hexaploid wheat. Plant J 37: 528–538 [DOI] [PubMed] [Google Scholar]
- Yang X, Wang W, Coleman M, Orgil U, Feng J, Ma X, Ferl R, Turner JG, Xiao S (2009) Arabidopsis 14-3-3 lambda is a positive regulator of RPW8-mediated disease resistance. Plant J 60: 539–550 [DOI] [PubMed] [Google Scholar]
- Zhou F, Kurth J, Wei F, Elliott C, Valè G, Yahiaoui N, Keller B, Somerville S, Wise R, Schulze-Lefert P (2001) Cell-autonomous expression of barley Mla1 confers race-specific resistance to the powdery mildew fungus via a Rar1-independent signaling pathway. Plant Cell 13: 337–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.