Arabidopsis Aurora kinases phosphorylate MAP65-1 at its unfolded tail domain and dynamic switching of its phosphorylation status throughout mitosis is required for proper cell cycle progression
Abstract
Aurora kinases are key effectors of mitosis. Plant Auroras are functionally divided into two clades. The alpha Auroras (Aurora1 and Aurora2) associate with the spindle and the cell plate and are implicated in controlling formative divisions throughout plant development. The beta Aurora (Aurora3) localizes to centromeres and likely functions in chromosome separation. In contrast to the wealth of data available on the role of Aurora in other kingdoms, knowledge on their function in plants is merely emerging. This is exemplified by the fact that only histone H3 and the plant homolog of TPX2 have been identified as Aurora substrates in plants. Here we provide biochemical, genetic, and cell biological evidence that the microtubule-bundling protein MAP65-1—a member of the MAP65/Ase1/PRC1 protein family, implicated in central spindle formation and cytokinesis in animals, yeasts, and plants—is a genuine substrate of alpha Aurora kinases. MAP65-1 interacts with Aurora1 in vivo and is phosphorylated on two residues at its unfolded tail domain. Its overexpression and down-regulation antagonistically affect the alpha Aurora double mutant phenotypes. Phospho-mutant analysis shows that Aurora contributes to the microtubule bundling capacity of MAP65-1 in concert with other mitotic kinases.
Aurora is a highly conserved family of Ser/Thr kinases, critical for cell division regulation in all eukaryotes. The aberrations of its activity are directly linked with the occurrence of multiple human cancers, resulting in high interest in the field of oncology. While yeasts possess only one Aurora representative [Ipl1 in Saccharomyces cerevisiae (Chan and Botstein, 1993) and Ark1 in Schizosaccharomyces pombe (Petersen et al., 2001)], higher eukaryotes have evolved two [e.g. Caenorhabditis elegans (Schumacher et al., 1998a; 1998b), Drosophila melanogaster (Glover et al., 1995), Xenopus laevis (Roghi et al., 1998; Adams et al., 2000), and Oryza sativa and Physcomitrella patens (Demidov et al., 2005; Guo et al., 2009)] or three homologs [e.g. mammals (Crane et al., 2004), Arabidopsis (Demidov et al., 2005; Van Damme et al., 2011)]. Mammalian Aurora kinases, designated A, B, and C, have very high sequence similarity, but different subcellular localization and substrate specificity (Vader and Lens, 2008; Carmena et al., 2009). Whereas both A- and B-type Auroras are expressed in proliferating cells, Aurora A is associated mainly with centrosomes and the spindle apparatus from prophase through telophase, and Aurora B is present at the kinetochores during spindle biorientation and at the midzone during anaphase and telophase. Aurora C, on the other hand, is highly expressed in testis and tumor cells, where it localizes to spindle poles from anaphase to cytokinesis (Kimura et al., 1999). Arabidopsis (Arabidopsis thaliana) Auroras have been divided into an α-group (comprising Aurora 1 and 2) and β-group (Aurora 3) based on their functional diversity and targeting patterns (Demidov et al., 2005; Kawabe et al., 2005; Van Damme et al., 2011). Aurora 1 and 2 associate with spindle microtubules (MTs) and the forming cell plate, whereas Aurora 3 is present at centromeres in metaphase (Van Damme et al., 2004a, 2011; Demidov et al., 2005).
In contrast to other kingdoms, the insight into Aurora activities in plants is still scarce. In yeasts and animals, Auroras exert their roles through phosphorylating multiple substrates at distinct stages of cell division, such as histone H3 (Goto et al., 2002), TPX2 (Eyers and Maller, 2004), Ajuba (Hirota et al., 2003), CDC25B (Dutertre et al., 2004), CENP-A (Zeitlin et al., 2001), the kinesin MKLP1 (Guse et al., 2005), Survivin (Wheatley et al., 2004), and many more. Besides histone H3 and TPX2, no Aurora substrates have been reported in plants so far. Down-regulation of alpha Auroras in Arabidopsis leads to multiple phenotypic aberrations, likely due to hypo-phosphorylation of their substrates. Namely, the double alpha Aurora knockdown mutant aur1/aur2 exhibits defects in orienting formative cell divisions (Van Damme et al., 2011), while RNAi silencing displays impaired cytokinesis with defective cell plate formation (Petrovská et al., 2012). The preferential phosphorylation of the microtubule-associated protein TPX2 by Arabidopsis Aurora 1 (Tomaštíková et al., 2015) and the colocalization of TPX2 and Aurora 1 on MTs (Petrovská et al., 2012) hint toward a role of alpha Aurora kinases in the regulation of cytoskeletal dynamics.
AtMAP65-1 (MAP65-1) belongs to the MAP65/Ase1/PRC1 protein family, which comprises evolutionarily conserved nonmotor MT-cross-linking proteins implicated in central spindle formation and cytokinesis in animals, yeasts, and plants (Chang-Jie and Sonobe, 1993; Jiang et al., 1998; Chan et al., 1999; Smertenko et al., 2000, 2004; Mollinari et al., 2002; Schuyler et al., 2003; Müller et al., 2004). MAP65/Ase1/PRC1 proteins form dimers and bundle predominantly antiparallel MTs (Gaillard et al., 2008; Subramanian et al., 2010). This MT bundling activity is controlled via phosphorylation by mitotic kinases, including Aurora. Cyclin-dependent kinase (CDK) phosphorylation of the human MAP65 homolog, PRC1 (protein required for cytokinesis) decreases its MT bundling activity (Jiang et al., 1998; Mollinari et al., 2002). PRC1 is required for bipolar spindle assembly and is an Aurora B binding partner in cytokinesis (Ozlü et al., 2010), but is not likely an Aurora substrate (Neef et al., 2007). On the other hand, Ipl1-dependent phosphorylation of the yeast MAP65 homolog Ase1 (anaphase spindle elongation 1), is required for spindle pole body separation, but not for its anaphase function (spindle elongation). Furthermore, both in vertebrates and in fission yeast, PRC1 and Ase1 are required for the proper localization of Aurora at the spindle midzone (Yamashita et al., 2005; Kurasawa et al., 2004). Finally, MAP65-1 was hypothesized to be a likely substrate for plant Aurora kinases (Smertenko et al., 2006). All these observations led us to the question whether the activity of Arabidopsis MAP65-1 could be regulated by plant alpha Aurora kinases.
The Arabidopsis genome encodes nine MAP65 genes (Hussey et al., 2002). All of them have a similar structural organization as PRC1/Ase1: an N-terminal region that comprises the dimerization domain required for the bundling activity, and a C-terminal domain required for MT binding (Gaillard et al., 2008; Smertenko et al., 2008). The MT-binding domain is composed of two distinct parts: the N-terminal part adopts a spectrin fold, which is responsible for the binding to MTs (Gaillard et al., 2008; Subramanian et al., 2010), and an unfolded C terminus, which is highly divergent between the different MAP65 isoforms and carries several phosphorylation sites (Smertenko et al., 2006, 2008; Gaillard et al., 2008). Although the role of the unfolded C terminus is still not well understood, several studies indicate that it could be involved in MT polymerization and bundling properties, and contribute to the differences in MAP65s intracellular targeting (Van Damme et al., 2004a; Mao et al., 2005b). The functionality of the phosphorylation sites at the C terminus was exemplified in several studies revealing that phosphorylation by mitotic kinases down-regulates the MT binding/bundling properties of MAP65-1 during metaphase, required for a timely cell cycle progression (Mao et al., 2005a; Sasabe et al., 2006; Smertenko et al., 2006).
Although Aurora-dependent C-terminal phosphorylation of MAP65-1 has been predicted, it was never demonstrated to be a plant Aurora substrate and it is unknown to what extent this phosphorylation controls its function on the cellular and developmental level. Here we establish MAP65-1 as a genuine substrate of alpha Aurora kinases. We provide in vivo evidence for their interaction and show biochemically that Arabidopsis alpha Auroras phosphorylate two predicted residues at its C terminus. Live imaging and phospho-mutant analysis demonstrate that Aurora phosphorylation affects the targeting of MAP65-1 during cell cycle progression. Furthermore, genetic analysis shows a functional interaction between Auroras and MAP65-1 required for plant development, but also indicates that Aurora-dependent phospho-regulation of MAP65-1 operates largely in concert with other kinases. Together, a complex regulatory network emerges where MAP65-1 is subject to multiple phosphorylation events by diverse kinases during consecutive stages of the cell cycle.
RESULTS
MAP65-1 Is a Substrate of Arabidopsis Alpha Auroras
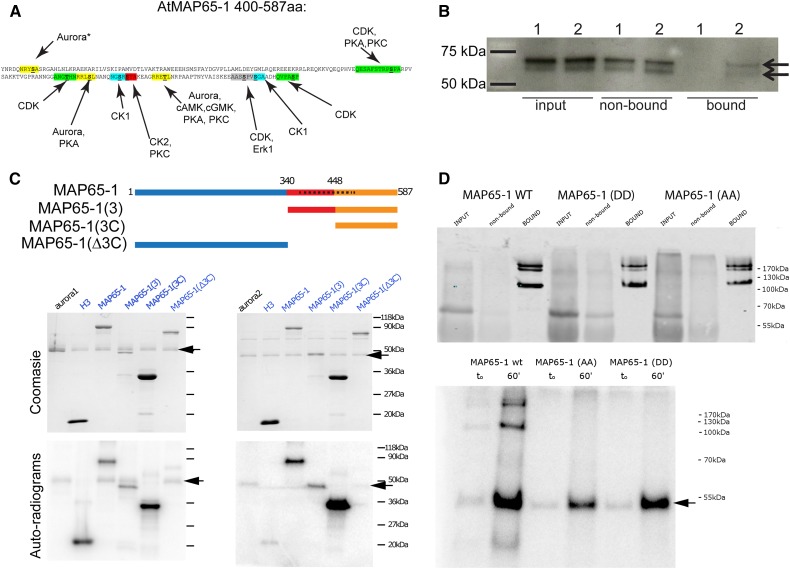
In plants, MAP65-1-dependent MT bundling was shown to be modulated by the phosphorylation status of nine sites at the C-terminal domain and two of these sites were predicted to be targets of Arabidopsis Aurora 1 (Sasabe et al., 2006; Smertenko et al., 2006; Fig. 1A). Analysis of all Arabidopsis MAP65 protein sequences using the Aurora consensus phosphorylation motif determined for budding yeast Ipl1 (Cheeseman et al., 2002) and human Aurora A (Ferrari et al., 2005) identified putative Aurora phosphorylation sites in all AtMAP65 proteins positioned downstream of their dimerization domain (Supplemental Fig. S1). This motif search identified also a third putative Aurora phosphorylation site in MAP65-1, which was not described previously (Supplemental Figs. S1 and S2; Smertenko et al., 2006). For MAP65-1, MAP65-2, MAP65-6, and MAP65-8, several of these putative sites were also reported to be phosphorylated in vivo in large-scale mass spectrometry experiments (PhosPhAt 4.0 Database). These include the three identified Aurora phospho-sites in MAP65-1 (Supplemental Fig. S2): Ser-532 (Umezawa et al., 2013), and Ser-408 and Thr-552 (Roitinger et al., 2015). Although the large-scale phospho-proteomics analyses did not specify the kinases responsible for the observed phosphorylation of these three residues, they do prove that they are genuine phosphorylation targets in planta.
Figure 1.
Arabidopsis MAP65-1 is a substrate of alpha Aurora kinases. A, The C-terminal domain of Arabidopsis MAP65-1 carries multiple putative phosphorylation sites (colored). Within each peptide sequence the phosphorylatable S/T residue is highlighted in bold and underlined. All putative kinase motifs (except for the first predicted Aurora site, marked with a star) were identified in Smertenko et al. (2006). The three putative Aurora phosphorylation sites (marked in yellow) were predicted based on the Aurora consensus phosphorylation motif identified in budding yeast Ipl1 (Cheeseman et al., 2002) and human Aurora A (Ferrari et al., 2005). B, Co-immunoprecipitation of MAP65-1 from control Col-0 plants (1) and complemented aur1/aur2 double knockdown mutant seedlings expressing genomic Aurora1-GFP (2). In contrast to the control, endogenous MAP65-1 (present as two bands and detected using a MAP65-1 antibody) is specifically pulled down with AUR1-GFP in the bound fraction (indicated with the arrows). C, Schematic representation of MAP65-1 [Projection domain (blue), C-terminal moiety (red and orange) with an unfolded divergent C terminus domain (orange), spectrin-like domain (dashed black bar)] and the truncations used for the in vitro phosphorylation assays using recombinant Arabidopsis Aurora1 (left) and Aurora2 (right). The alpha Auroras phosphorylate MAP65-1 at its C-terminal domain (domain 3C). Top gel pictures are Coomassie staining, bottom gel pictures are autoradiograms. Phosphorylation of histone H3 was included as a positive control for Aurora phosphorylation. Arrows mark the position of Aurora1 and Aurora2. D, GFP-fused MAP65-1 isoforms (wild type, AA, and DD), expressed in Nicotiana benthamiana leaves were immunoprecipitated and confirmed using Western blot (upper panel). The higher MW bands at 170 probably represent MAP65-1 dimers. In vitro Aurora1 kinase assays using the immunoprecipitated GFP-fused MAP65-1 isoforms as substrates (autoradiogram, lower panel, t0 and t60 min) show that wild-type MAP65-1 is phosphorylated by Aurora1, in contrast to the Aurora phosho-mimicking form [MAP65-1(DD) and the nonphosphorylatable form MAP65-1(AA)]. The arrow marks the autophosphorylation of Aurora1.
Alignment of MAP65-1 from 15 plant species [ClustalW (McWilliam et al., 2013)], yeast (Ase1) and vertebrates (PRC1), aimed to verify evolutionary conservation of these putative sites, revealed that the putative Aurora phosphorylation sites are not conserved between the kingdoms. The animal (Homo sapiens and X. laevis) PRC1 does not contain any Aurora consensus motif, in agreement with experimental evidence that X. laevis PRC1 is not phosphorylated by Aurora (Neef et al., 2007). Aurora phosphorylates five residues of S. cerevisiae Ase1, but these sites are scattered throughout the protein and completely divergent from the Arabidopsis ones. One of the three putative Aurora phosphorylation sites is very well conserved in plants (Ser-532), one is conserved between Arabidopsis and several monocot species but not in other dicots (Ser-408), and the third one (Thr-552) is not conserved among plant species (Supplemental Fig. S2). The presence of diverged phosphorylation sites hints toward a plant-specific MAP65-1 control mechanism by Aurora kinases, which might be linked to the functional expansion of this protein family in plants compared to the single PRC1 and Ase1 proteins in animals and yeasts (Supplemental Fig. S1).
To test whether MAP65-1 interacts with plant Aurora kinases, we performed an in vivo coimmunoprecipitation assay, where MAP65-1 was found to specifically copurify along with functional Aurora1-GFP in the complemented Aurora double knockdown background (aur1/aur2, Fig. 1B). In vitro phosphorylation assays using recombinant full-length MAP65-1 and its truncated forms established proof for Aurora phosphorylation of MAP65-1 and confirmed the in vivo interaction. Recombinant Arabidopsis Aurora 1 and 2 phosphorylated full-length MAP65-1 and the analysis of truncated forms of MAP65-1 showed that phosphorylation was restricted to its C-terminal domain (Fig. 1C). Mass spectrometry analysis of the in vitro phosphorylated peptides (Supplemental Table S1) revealed that the phosphorylated residues overlapped with those that were predicted, S532 and T552 (Smertenko et al., 2006). As no peptides were identified in the mass spectrometry analysis covering S408, we performed in vitro kinase assays using transiently expressed full-length MAP65-1-GFP, as well as two MAP65-1 phosphomutant forms, carrying either substitutions of S532 and T552 to Ala (constituting a double phosphorylation-dead “AA” form) or to Asp (a phospho-mimicking “DD” form), to determine whether MAP65-1 could be phosphorylated by Aurora 1 on other residues as well. In contrast to the wild-type MAP65-1-GFP, none of the phosphomutant forms [MAP65-1(AA) and MAP65-1(DD)] were phosphorylated by recombinant Aurora 1 in vitro (Fig. 1D). Therefore, although Ser-408 matches the consensus Aurora phosphorylation motif and was identified as an in vivo phospho-peptide (Roitinger et al., 2015), our analysis indicates specificity of Aurora 1 for S532 and T552.
Phosphorylation by Aurora Does Not Significantly Change the MT Bundling of MAP65-1 In Vitro
Because MT bundling is the canonical activity of MAP65s, we aimed to verify whether Aurora-dependent phosphorylation of MAP65-1 affected its bundling capacity. For this purpose, recombinant MAP65-1 and both phospho-mutant forms [MAP65-1(AA) and MAP65-1(DD)] were purified. Cosedimentation assays showed that the recombinant Aurora-phosphorylated MAP65-1 and the MAP65-1(DD) forms had a lower affinity for MTs than the wild-type and MAP65-1(AA) forms (Supplemental Fig. S3A). In vitro total internal reflection fluorescence (TIRF) experiments, aiming to determine the bundling of MTs by either the wild-type MAP65-1 or the phospho-mutant forms, showed that phospho-mimicking the alpha Aurora phosphorylation sites did not abolish MAP65-1 MT bundling in vitro (Supplemental Fig. S3, B and C) and in vitro MT dynamics were not significantly different between MAP65-1 and both phospho-mutant forms (Supplemental Fig. S3D). Thus, Aurora-dependent phosphorylation alone does not strongly affect the in vitro bundling capacity of MAP65-1.
Aurora-dependent Phosphorylation Alters MAP65-1 MT Binding Properties In Vivo
As the next functional assay of these phospho-sites, the subcellular localization of MAP65-1 relative to Aurora 1, as well as the localization of the phospho-mutants, was analyzed in vivo during cell division. For this purpose, additional phospho-mutant forms were created: a phospho-mimicking form in all nine putative phospho-sites [called thereafter 9D, reported previously (Smertenko et al., 2006)] and a phospho-mimicking form in seven putative phospho-sites (all non-Aurora sites, called “7D”). Tobacco BY-2 cells have been used extensively to study the dynamic localization of MAP65 proteins (Van Damme et al., 2004b; Smertenko et al., 2006; Gaillard et al., 2008; Fache et al., 2010; Sasabe et al., 2011). We therefore used this system to analyze the colocalization between MAP65-1 and Aurora 1, as well as the effect of Aurora-phospho-mutations on the localization of MAP65-1 throughout mitosis.
In vivo imaging of BY-2 cells demonstrated that the microtubule bundling regulation (decrease in metaphase and increase in anaphase) of MAP65-1 was comparable between C-terminal and N-terminal GFP fusions (Supplemental Fig. S4A; Supplemental Movies S1 and S2). Also, the timing of cell division duration did not differ between the two forms (Supplemental Fig. S4B). However, in cells expressing the C-terminal fusion there were some cytoplasmic aggregates visible and the internal MTs were more prominently labeled (Supplemental Fig. S4A preprophase). Based on our previous work and these observations, we continued our analyses with the C-terminal fusion.
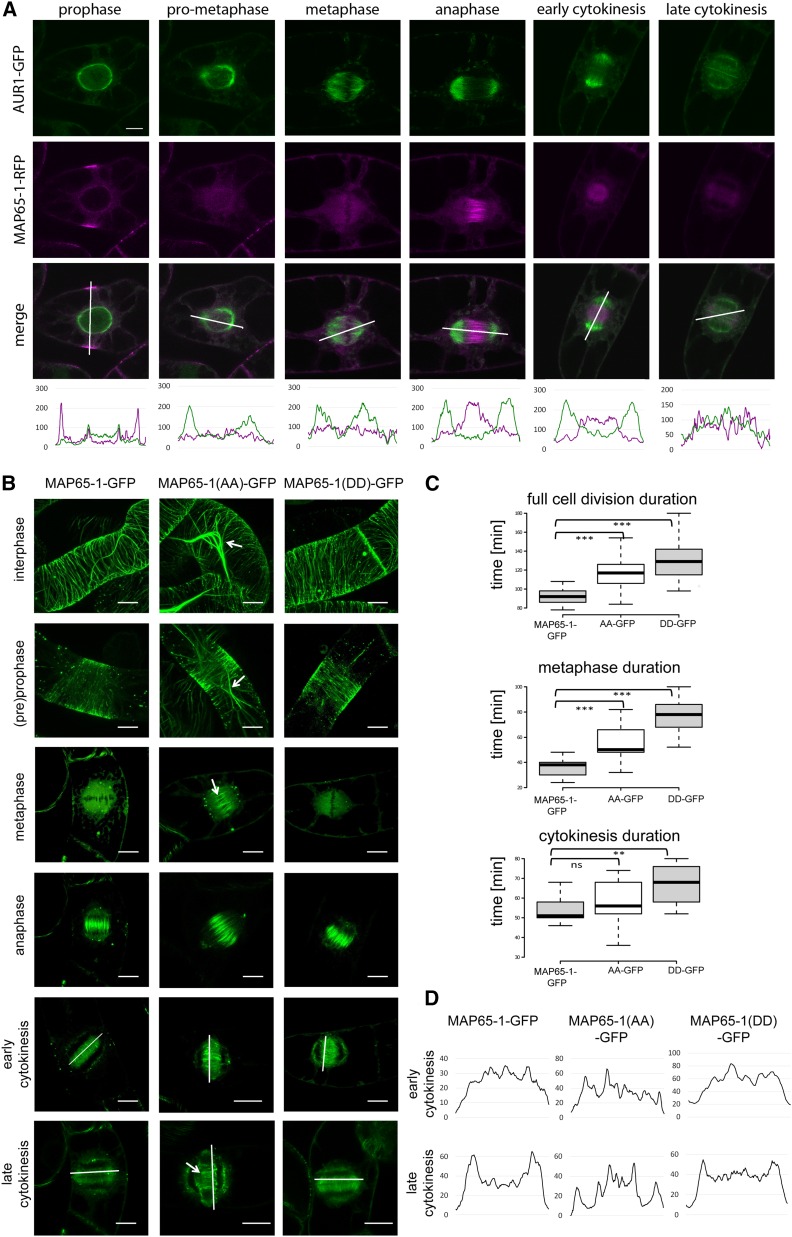
Colocalization analysis between Aurora1-GFP and MAP65-1-TagRFP throughout mitosis revealed that both proteins colocalized at various cell cycle phases such as the preprophase band (PPB) stage, where both proteins accumulated around the nuclear envelope before its disassembly or at the phragmoplast during cytokinesis (visualized by the fluorescent plots below the images in Fig. 2A). A clear negative correlation was observed between the presence of Aurora 1 and the MT bundling of MAP65-1 (Fig. 2A), which would be in agreement with Aurora-dependent phosphorylation decreasing the MT bundling capacity of MAP65-1. The lack of localization overlap of the kinase and its substrate, therefore, correlated with the deactivation of MAP65-1 MT bundling.
Figure 2.
Phospho-mutations at S532 and T552 alter MAP65-1 properties in vivo. A, Colocalization between Aurora1-GFP and MAP65-1-RFP throughout mitosis. The panels are taken from three different time-lapse movies. The fluorescence intensity plots (shown below the merged images) demonstrate colocalization of Aurora1-GFP and MAP65-1-RFP e.g. at the nuclear envelope during the PPB stage and during cytokinesis as well as a negative correlation between Aurora1 localization and MAP65-1 MT bundling from prophase to cytokinesis. Scale bar = 10 μm. B, Representative images (n > 10) of Arabidopsis MAP65-1-GFP, MAP65-1(AA)-GFP, and MAP65-1(DD)-GFP expressed in tobacco BY-2 cells during consecutive stages of cell division. Arrows indicate excessive microtubule bundling observed in prophase, metaphase, and late cytokinesis in cells expressing MAP65-1(AA)-GFP. Images of cells in interphase and preprophase are projections, while the other phases are represented by single images. Scale bar = 10 μm. C, Box plot representation of full cell division (top), metaphase (middle), and cytokinesis (bottom) duration in BY-2 cells expressing MAP65-1-GFP (n = 13), MAP65-1(AA)-GFP (n = 14), and MAP65-1(DD)-GFP (n = 9). Overexpression of MAP65-1(AA), but also MAP65-1(DD) prolongs cell division duration compared to the wild-type MAP65-1 (t test; triple asterisk: P < 0.0001; double asterisk: P < 0.001; ns (not significant): P > 0.01). Center lines show the medians; box limits indicate the 25th and 75th percentiles as determined by R software; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles. D, Fluorescence intensity plots (taken along the white lines in B) across the phragmoplasts in early or late cytokinesis in BY-2 cells. MAP65-1(AA)-expressing cells show delayed depolymerization of central phragmoplast microtubules in late cytokinesis.
When the different forms of MAP65-1 were compared, MAP65-1(AA) ectopically bundled prophase, metaphase, and phragmoplast MTs compared to the wild-type MAP65-1 (Fig. 2B, Supplemental Movies S5 and S6). MAP65-1(DD), on the other hand, localized similarly to the wild-type MAP65-1. Quantification of the timing of each cell division phase revealed that MAP65-1(AA) delayed the anaphase onset (prolonged metaphase, defined from the PPB disappearance until just before chromosome separation), in agreement with previous data (Smertenko et al., 2006). Surprisingly, MAP65-1(DD) delayed this phase even more, although the protein was not visibly associated with the spindle MTs (Fig. 2C). It is thus likely that the delay caused by MAP65-1(DD) expression is MT-independent.
In late cytokinesis, BY-2 cells expressing MAP65-1(AA) showed remnant MT bundling in the center of the phragmoplast, in contrast to cells expressing wild-type MAP65-1 and MAP65-1(DD), where the highest fluorescence signal was associated with the outer rim of the expanding cell plate (Fig. 2D). These observations are indicative of a function of Aurora-controlled MAP65-1 inactivation during the transition from cross-linking anaphase spindle MTs to cell plate formation, which likely coincides with the translocation of Aurora 1 to the cell plate in early cytokinesis (Fig. 2A).
To distinguish between the effect of Aurora phosphorylation and the phosphorylation on the other seven putative MAP65-1 phosphorylation sites, MAP65-1(7D) and MAP65-1(9D) were localized during cell division in BY-2 cells. Phospho-mimicries in all nine and seven phospho-sites did not significantly affect the timing of cell division, although MAP65-1(7D)-GFP did prolong metaphase duration (Supplemental Fig. S4B and Supplemental Movies S5 and S6). However, their binding to MTs was altered. MAP65-1(7D) only faintly decorated the cortical MTs, was clearly present at the PPB, and failed to reappear on the anaphase MTs. MAP65-1(9D) lost all visible MT association capacity with the exception of the PPB. The recruitment to the cortical MTs of MAP65-1(7D), in contrast to MAP65-1(9D), showed that both phospho-mimicking Aurora-dependent residues have an additional negative effect on the MT binding capacity of MAP65-1, while the absence of recruitment to metaphase and anaphase spindle and phragmoplast MTs suggests that MAP65-1(7D) is phosphorylated on its Aurora residues during these cell cycle stages. MAP65-1 association with the PPB, independent of its phosphorylation status, points to additional factors controlling its recruitment compared to cortical or other mitotic MT arrays.
To corroborate the specificity of the in vivo function of MAP65-1 phosphorylation by Aurora, we treated BY-2 cells with a specific Aurora inhibitor, MLN8237. MLN8237 (Alisertib) is known to target A- and B-type Auroras in a dose-dependent manner (Sells et al., 2015; Nair and Schwartz, 2016). Protein sequence alignment revealed that all key interacting residues between MLN8237 and Aurora kinases are conserved between human and plant Auroras (Supplemental Fig. S5A), indicating that plant kinases have all the required features to be inhibited by the drug. Moreover, hypersensitivity of the Aurora double mutant compared to wild type as well as the absence of Histone H3 phosphorylation in the presence of the drug demonstrated that MLN8237 inhibits Aurora kinase activity in plants (Supplemental Fig. S5, B and E). We specifically monitored the localization of MAP65-1(7D)-GFP in the presence or absence of MLN8237. This MAP65-1 form has both Aurora sites active, while all the other seven phosphorylatable targets at its C-terminal domain are phospho-mimicked. Consequently, any observed change in localization is unlikely to originate from blocking off-target kinases by the inhibitor and, therefore, reflects the reduced Aurora-dependent phosphorylation of MAP65-1. In BY-2 cells treated with MLN8237, and in contrast to the control situation, MT bundling of MAP65-1(7D)-GFP around the interphase and prophase nuclei was observed in roughly 70% of the cells (Supplemental Fig. S5F). This localization pattern was also detected when MAP65-1(AA)-GFP (a non-Aurora phosphorylatable form) was imaged (Supplemental Fig. S5G; Fig. 2B). The altered localization in the presence of the Aurora inhibitor provides additional in vivo evidence for a function of plant Aurora kinases in controlling the MT bundling capacity of MAP65-1.
MAP65-1 Phosphorylation by Aurora Affects the MAP65-1 Localization in Metaphase and during Cytokinesis in Arabidopsis
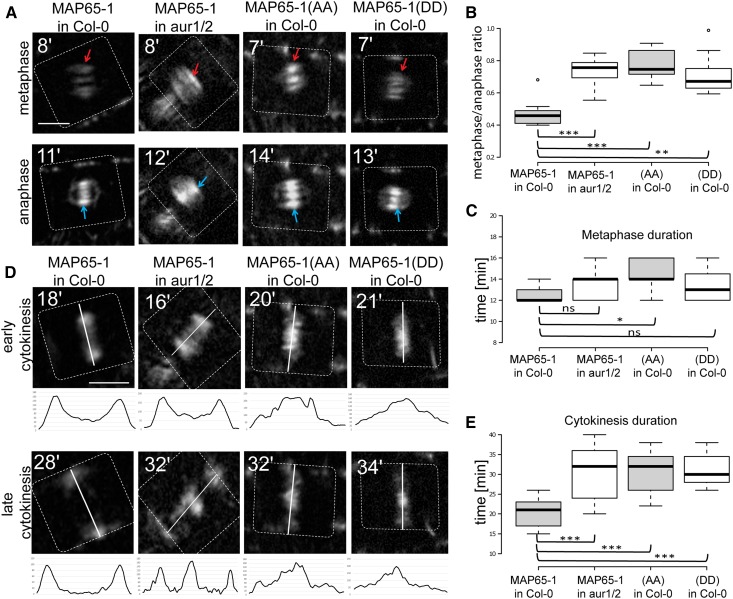
To confirm the effect of the Aurora-dependent phosphorylation of MAP65-1 in Arabidopsis cells, MAP65-1 as well as MAP65-1(AA) were localized in vivo in wild-type (Col-0) and aur1/aur2 double knockdown mutant seedlings throughout mitosis. While expression of MAP65-1(AA) under the AUR1 promoter did reveal enhanced recruitment to the nuclear surface during the PPB stage in Arabidopsis wild-type root cells (in 76% cells for the mutant (n = 42) vs. 18% for the wild-type (n = 34) MAP65-1 form; Supplemental Fig. S6A), 4D spinning disc imaging revealed no significant change in MAP65-1 prophase spindle intensity or metaphase duration (wild-type Col-0 vs aur1/aur2 double mutant) when MAP65-1 was either driven by the MAP65-1 endogenous promoter or when ectopically expressed under the strong mitotic AUR1 promoter (Supplemental Fig. S7), indicating that the Aurora double mutant, likely due to remnant alpha Aurora expression, does not mirror the effect of the MAP65-1(AA) form. However, when metaphase and anaphase localization patterns of wild-type MAP65-1, MAP65-1(AA), and MAP65-1(DD) were compared in Arabidopsis, the ratio of MAP65-1 metaphase to anaphase spindle signal increased in the aur1/aur2 mutant background, due to a stronger metaphase recruitment (Fig. 3, A and B). This ratio pattern was similar to the effect of MAP65-1(AA) in Col-0 when compared to the wild-type MAP65-1 in the Col-0 background (Fig. 3, A and B) and to the observations in BY-2. A ratio increase was also observed for MAP65-1(DD), although statistically less significant. Dynamic Aurora-dependent phosphorylation and dephosphorylation of MAP65-1 is therefore required for the differential accumulation of this protein between the metaphase and anaphase spindle in Arabidopsis. The labeling of metaphase spindles by MAP65-1 observed here is likely due to mitotic overproduction of the fluorescent fusion as DAPI staining of these lines linked the spindle fluorescence signals with metaphase (Supplemental Fig. S6B).
Figure 3.
Localization of MAP65-1 and its phospho-mutants MAP65-1(AA) and MAP65-1(DD) during cell division in Arabidopsis seedlings. A, Representative metaphase and anaphase localization images of prAUR1:MAP65-1-TagRFP in Col-0 versus the aur1/aur2 double mutant (aur1/2), and MAP65-1(AA) versus MAP65-1(DD) in Col-0 root meristem cells. Red arrows indicate metaphase spindles; blue ones indicate anaphase midzone. The timing of cell division (in min, calculated from the disappearance of the PPB as time point zero) is given in the top-left corner. Dotted lines indicate cell borders. The observed metaphase MAP65-1 signal is likely a consequence of overexpression (see Supplemental Fig. S6). Scale bar = 5 μm. B, Fluorescence intensity ratios (metaphase over anaphase spindle signal) comparing prAUR1:MAP65-1-TagRFP expressed in wild-type Col-0 (n = 9) versus the aur1/aur2 double mutant background (aur1/2; n = 10), and prAUR1:MAP65-1(AA)-TagRFP (n = 10) or prAUR1:MAP65-1(DD)-TagRFP (n = 11) in Col-0. Altered Aurora phosphorylation capacity of MAP65-1 significantly increases the metaphase to anaphase ratio (t test; triple asterisk: P < 0.0001, double asterisk P = 0.00012). Center lines show the medians; box limits indicate the 25th and 75th percentiles as determined by R software; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles, outliers are represented by dots. C, Metaphase duration (calculated from the PPB disappearance to anaphase midzone appearance) in Arabidopsis root meristem cells expressing prAUR1:MAP65-1-TagRFP in Col-0 (n = 9) and in the aur1/aur2 double mutant (aur1/2; n = 10), as well as prAUR1:MAP65-1(AA)-TagRFP (n = 10) and prAUR1:MAP65-1(DD)-TagRFP (n = 11) in Col-0. The asterisk indicates a statistically significant difference between lines expressing prAUR1:MAP65-1-TagRFP and prAUR1:MAP65-1(AA)-TagRFP in the Col-0 background (t test P = 0.00974). D, Localization of prAUR1:MAP65-1-TagRFP in Col-0 and aur1/aur2 double mutant (aur1/2), and prAUR1:MAP65-1(AA)-TagRFP and prAUR1:MAP65-1(DD)-TagRFP in Col-0 in early and late cytokinesis. The lines indicate the division planes. The fluorescence intensity plots measured across the phragmoplasts (along the white lines) are shown under the respective images. The timing of cell division (in min, calculated from the disappearance of the PPB as time point zero) is given in the top-left corner. Dotted lines indicate cell borders. Altered Aurora phosphorylation capacity of MAP65-1 results in delayed MT turnover at the central region of the phragmoplast. Scale bar = 5 μm. E, Cytokinesis duration (calculated from the beginning of anaphase spindle expansion to the disappearance of the MTs at the phragmoplast rim) in Arabidopsis root meristem cells expressing prAUR1:MAP65-1-TagRFP in Col-0 and in the aur1/aur2 double mutant (aur1/2), as well as prAUR1:MAP65-1(AA)-TagRFP and prAUR1:MAP65-1(DD)-TagRFP in Col-0 (n = 18, 17, 11, 15 sample points, respectively). Altered Aurora phosphorylation capacity of MAP65-1 results in a statistically significant prolongation of cytokinesis compared to the control (triple asterisks; t test P < 1E-05).
The calculation of the timing of the cell division phases disclosed that metaphase was slightly longer in Col-0 cells expressing MAP65-1(AA) than MAP65-1 wild-type or MAP65-1(DD) (t test P = 0.00974 for MAP65-1(AA) versus wild-type MAP65-1 and P = 0.174 for MAP65-1(DD) versus wild-type MAP65-1; Fig. 3C). Thus, the inability to phosphorylate MAP65-1 by Aurora might delay the anaphase onset also in Arabidopsis cells due to ectopic spindle MT bundling. Remnant Aurora expression in the aur1/aur2 double knockdown likely alleviates this effect compared to the MAP65-1(AA) expression.
Similar to the observations in BY-2, cytokinesis in Col-0 cells expressing MAP65-1(AA) or wild-type MAP65-1 in aur1/aur2 was affected: MTs in the center of the phragmoplast were not depolymerized as efficiently as in the control situation (Fig. 3D). This was also the case for MAP65-1(DD) in Col-0, although the latter was not observed in BY-2. Consequently, the duration of cytokinesis (measured from the beginning of the phragmoplast expansion through the disappearance of the MTs as the phragmoplast rim reached the plasma membrane) was significantly (approximately 50%) longer in cells expressing MAP65-1 in aur1/aur2, MAP65-1(AA) in Col-0, and MAP65-1(DD) in Col-0 when compared to the control (Fig. 3E). Hence, similar to the metaphase to anaphase transition, MAP65-1 might need to undergo dynamic Aurora-dependent phosphorylation changes in cytokinesis to allow efficient phragmoplast MT dynamics.
Mitotic Overexpression and Knockout of MAP65-1 Antagonistically Affect the Mutant Phenotype of the aur1/aur2 Double Knockdown Mutant
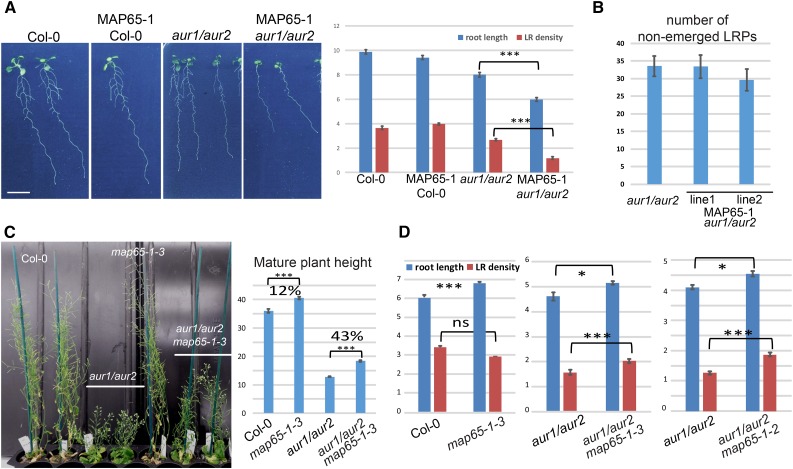
If Aurora-dependent phosphorylation restricts MAP65-1 MT bundling capacity throughout the cell cycle, then ectopic (mitotic) expression of MAP65-1 in the aur1/aur2 double knockdown mutant background should aggravate the aur1/aur2 developmental phenotype due to its reduced alpha Aurora activity. To verify this hypothesis, MAP65-1 was expressed under a strong mitotic promoter (prAUR1) in wild-type Col-0 and in the aur1/aur2 mutant. In contrast to Col-0, MAP65-1 mitotic overexpression aggravated the primary root length, lateral root (LR) density, rosette size, and bushy shoot phenotypes of the aur1/aur2 mutant (Fig. 4A and Supplemental Fig. S8, A and B). Mitotic overexpression of MAP65-1 did not significantly reduce the number of nonemerged primordia up to the first emerged primordium, indicating that the LR density reduction is likely caused by LR outgrowth perturbations rather than initiation defects (Fig. 4B and Supplemental Fig. S9). To rule out the possibility that the aur1/aur2 mutant is simply hypersensitive to alterations in expression levels of proteins that bind the cytoskeleton, we expressed the MT binding domain (MBD) of MAP4 as a TagRFP fusion and driven by the Aurora 1 promoter in both Col-0 wild-type and the aur1/aur2 mutant backgrounds. In contrast to the overexpression of MAP65-1, mitotic overexpression of MBD did not aggravate the Aurora double mutant phenotype (Supplemental Fig. S10). Thus, the aggravation of the aur1/aur2 mutant phenotype was not caused by overproduction of a MT binding protein, but was linked specifically to MAP65-1.
Figure 4.
Deregulating MAP65-1 in the Aurora double mutant background genetically confirms interaction between MAP65-1 and alpha Auroras. A, Representative 13 d-old seedlings (n > 30) of Col-0, Col-0 expressing prAUR1:MAP65-1-TagRFP (MAP65-1 Col-0), aur1/aur2, and aur1/aur2 expressing prAUR1:MAP65-1-TagRFP (MAP65-1 aur1/aur2), and their root length and LR density quantifications. Ectopic overexpression of MAP65-1 in the aur1/aur2 double mutant (t test; triple asterisk; P < 0.0001) reduces main root length as well as severely reduces lateral root outgrowth. Scale bar = 1 cm. B, Calculation of the nonemerged lateral root primordia (LRPs) in the aur1/aur2 double mutant and two independent lines of aur1/aur2 double mutant expressing prAUR1:MAP65-1-TagRFP. LRPs were counted from the root meristem up to the first emerged LRP. The total number of nonemerged LRPs between the analyzed lines is not statistically different (t test P = 0.98 for line 1 and P = 0.38 for line 2). Root length is depicted in cm; LR density is the number of LRs/cm main root. Images of the LRP are shown in Supplemental Fig. S9. C, Representative images of mature Col-0 (wild type), aur1/aur2 double mutant, map65-1-3 single mutant and aur1/aur2 map65-1-3 triple mutant plants grown in soil and quantification of the stem length (n ≥ 22); triple asterisk, t test P < 0.0001. Values (in %) indicate the increase in stem length between the respective lines and show that the effect of knocking out MAP65-1 in the Aurora double mutant is not merely additive. D, Quantification of the primary root length and lateral root density of 10 d-old seedlings (n > 25) grown in vitro in continuous light conditions of Col-0 (wild type), map65-1-3, aur1/aur2, and both triple mutant combinations (aur1/aur2 map65-1-3 and aur1/aur2 map65-1-2; t test; triple asterisk: P < 0.0001; single asterisk: P < 0.01; ns: P > 0.01). Knocking out MAP65-1 in the Aurora double mutant background partially rescues the LR density phenotype, while there is no difference (t test P = 0.16) in LR density between wild type (Col-0) and map65-1-3 KO plants.
As the overexpression of MAP65-1 aggravated the aur1/aur2 double knockdown mutant phenotype, we anticipated that removing this substrate might lead to a partial rescue. To test this, we crossed a map65-1 mutant (GABI-Kat_198A01; map65-1-3) into the aur1/aur2 mutant background. Western blotting using a MAP65-1 antibody (Smertenko et al., 2004) confirmed the strong down-regulation of MAP65-1 in map65-1-2 and map65-1-3 (Supplemental Fig. S8C), and the remaining signal likely represents a cross reaction with other MAP65 isoforms (Lucas et al., 2011).
The rosette size of the triple aur1/aur2/map65-1-3 mutant did not statistically differ from the aur1/aur2 mutant and the single map65-1-3 mutant was morphologically indistinguishable from Col-0 at this developmental stage (25 d-old, Supplemental Fig. S8B). However, although mature map65-1-3 mutant plants were slightly taller than Col-0 wild-type plants, depletion of MAP65-1 in the aur1/aur2 double mutant background (aur1/aur2/map65-1-3 triple mutant) had a stronger and statistically significant effect on the overall plant height compared to the control (43% length increase in aur1/aur2/map65-1-3 versus aur1/aur2 compared to 12% between wild-type and the map65-1-3 single mutant; P = 3.7E-11). Furthermore, depletion of MAP65-1 also partially rescued the bushy aur1/aur2 mutant phenotype (Fig. 4C). Additionally, the aur1/aur2/map65-1-3 triple mutant substantially rescued the LR density phenotype of the aur1/aur2 mutant, which was also confirmed using the independent map65-1-2 mutant allele (SALK_118225; Lucas et al., 2011; Fig. 4D). The antagonistic effects of overexpression versus knockdown further indicate that MAP65-1 phosphorylation by alpha Aurora regulates its function. Taken together, our biochemical, imaging, and genetic interaction data reveal a regulatory system in which the activity of MAP65-1 is controlled by alpha Aurora phosphorylation in Arabidopsis.
Aurora-Dependent Control of MAP65-1 Is Required for Proper Meristematic Activity
To test whether the enhanced LR mutant phenotypes were Aurora phosphorylation-dependent, MAP65-1(AA) and MAP65-1(DD) forms were introduced into the Col-0 and aur1/aur2 mutant backgrounds. If MAP65-1 hypo-phosphorylation (due to diminished levels of Aurora) is the underlying cause of the phenotype enhancement in the aur1/aur2 background, the overexpression of the phospho-mimicking MAP65-1(DD) form should not aggravate the mutant phenotype. However, primary root length and LR density analyses showed that both MAP65-1(AA) and MAP65-1(DD) fusions aggravated the LR density and main root length phenotypes of the aur1/aur2 mutant. For the main root length, the effect was even more severe than the original wild-type MAP65-1 overexpression (Supplemental Fig. S11A).
However, phospho-mimicry of all nine phospho-sites in MAP65-1 (9D) as well as in the two putative CDK sites [MAP65-1(2D)] did not aggravate the mutant phenotypes (Supplemental Fig. S11A). Immunoblot analyses demonstrated that the differential effect between the wild-type and Aurora double knockdown mutant backgrounds could not be simply attributed to differential overexpression levels (Supplemental Fig. S11B). The aggravation of the LR phenotype in the aur1/aur2 mutant is, therefore, dependent on the phosphorylation status of MAP65-1, rather than on its mere overexpression, but not causal to direct Aurora-dependent phosphorylation of MAP65-1. Hence, the phenotype is probably due to indirect phosphorylation events, which differ between the wild-type and the aur1/aur2 mutant backgrounds.
As the Aurora double mutant is mostly affected in LR outgrowth by altered orientation of the first divisions, and knowing that mitotic overexpression of MAP65-1 aggravates the LR phenotype even more, this represents the most likely system to identify the causal effect of the aggravated mutant phenotype. The severe alterations in LR primordia patterning in the aur1/aur2 double knockdown mutant, however, hinder proper characterization of the effect of mitotic overexpression of MAP65-1 (Van Damme et al., 2011; Supplemental Fig. S9). During lateral root primordia divisions, mitotically expressed MAP65-1-TagRFP did reveal stronger metaphase spindle signals in the aur1/aur2 double mutant and ectopic bundling in the center of the expanding phragmoplast compared to the wild-type control (Supplemental Fig. S12), similar to the effects observed in the primary root epidermal cells.
We therefore examined the effect on cell divisions in the main root apical meristem (RAM). Quantification of the number of meristematic cells in the primary roots revealed that mitotic overexpression of MAP65-1 reduced the amount of meristematic cells in the aur1/aur2 double mutant (Supplemental Fig. 11, C and D). Moreover, this RAM phenotype enhancement in aur1/aur2 is dependent on the phosphorylation status of MAP65-1, but not directly controlled by Aurora kinase, as this decrease of the number of cells in the RAM is reverted to aur1/aur2-like values by the expression of the phospho-mimicry of all MAP65-1 9 phospho-sites (9D), as well as the putative CDK sites (2D; Supplemental Fig. 11, C and D).
It is therefore likely that the enhanced lateral root outgrowth defects observed in the aur1/aur2 mutant upon mitotic overexpression of MAP65-1 are similarly caused by a decreased number of divisions.
DISCUSSION
Aurora kinases exert multiple roles in diverse cell cycle-related processes by phosphorylating a wide variety of substrates (Biggins and Murray, 2001; Carmena et al., 2009; Fu et al., 2009; Hans et al., 2009; Van Damme et al., 2011; Petrovská et al., 2012; Hochegger et al., 2013; Demidov et al., 2014; Goldenson and Crispino, 2015; Weimer et al., 2015). However, to date, only histone H3 (Hsu et al., 2000) and TPX2 have been reported as alpha Aurora substrates in plants (Demidov et al., 2005; Tomaštíková et al., 2015).
Here, we show that the microtubule cross-linking protein MAP65-1 is a genuine substrate of Arabidopsis alpha Aurora kinases. The in vitro phosphorylation data confirmed the biochemical interaction in vivo. The interaction was also corroborated genetically, as mitotic MAP65-1 overexpression and knock-out of MAP65-1 antagonistically affected the phenotypes of the Aurora double knockdown mutant, likely by affecting meristematic activity. Furthermore, we showed that the phosphorylation status of the Aurora-dependent residues of MAP65-1 alters its MT-binding properties at specific cell cycle phases.
MAP65-1 Is a Substrate of the Arabidopsis Alpha Auroras
We identified S532 and T552, located at the C-terminal part of MAP65-1, to be alpha Aurora kinase targets. Aurora-dependent phosphorylation of MAP65-1 at its C terminus probably evolved specifically in plants, because PRC1 (the animal homolog of MAP65-1) is an Aurora binding partner (Ozlü et al., 2010), but likely not a substrate (Neef et al., 2007), while Ase1 (the MAP65-1 counterpart in yeast) is phosphorylated in vitro by Ipl1 (the Aurora homolog in S. cerevisiae) on completely different residues (Kotwaliwale et al., 2007). The Aurora phosphorylation consensus motif (Ferrari et al., 2005; Sardon et al., 2010) predicted three putative phosphorylation sites in Arabidopsis MAP65-1 and we could find experimental evidence for two of those, pointing out that the consensus motif is largely conserved between eukaryotic kingdoms, but also that it is only a strong indication rather than a fixed rule.
Functionally significant phosphorylation motifs tend to be better conserved during evolution than random motifs (Malik et al., 2008). One of the detected Arabidopsis Aurora sites in MAP65-1 (Ser-532) is perfectly conserved across plant species, the Ser-408 one is shared between Arabidopsis and some monocots but other dicots lack the essential Ser residue at this position, and the Thr-552 one is hardly conserved. While we show S532 and T552 to be genuine Aurora targets, we could not find evidence for Aurora-dependent phosphorylation of S408. The fact that the Arabidopsis MAP65-1 peptide with Ser-408 was found to be phosphorylated in a high-throughput phospho-proteomics screen indicates that it is truly an in vivo kinase residue, but not necessarily a target for Aurora. Other candidates might be the PKA-, PKC-type, or PAK1 isoforms of AGC kinases, as this motif matches their consensus sequence as well (Xue et al., 2005; 2011). It is plausible that the Ser-532 and Thr-552 sites together modulate MAP65-1 activity, necessary to distinguish it from the other eight MAP65s in Arabidopsis but it remains to be verified whether the nonconserved Thr-552 is equally functional to the conserved Ser-532. Nevertheless, the high conservation of the S532 phosphorylation motif, together with the high number of analogous motifs located at the C-terminal domain of other MAP65 isoforms, strongly suggests that MAP65-1 is not the sole family member controlled by the Aurora kinases.
Several Mitotic Kinases Differentially Control MAP65-1
The C-terminal region of MAP65-1 is predicted to be intrinsically disordered, which is in line with the dominant structural disorder of microtubule-associated proteins (MAPs), such as MAP2 or tau protein (Guharoy et al., 2013). As is the case for MAP65-1, such intrinsically disordered regions often harbor multiple regulatory posttranslational modification (primarily, phosphorylation) sites (Iakoucheva et al., 2004), the synergy of which results in complex regulatory phenomena.
The key question underlying this work was to elucidate the role of MAP65-1 phosphorylation by plant alpha Aurora kinases. Our in vitro experiments argue against Aurora-dependent phosphorylation of MAP65-1 directly controlling its MT bundling or dimerization capacity. The observed slight reduction in MT binding and bundling properties of the phospho-mimicry version (MAP65-1-DD) compared to the wild-type form rather fits with a combined role of Aurora and other mitotic kinases controlling the MT binding capacities of MAP65-1, in agreement with previous data (Smertenko et al., 2006).
In vivo, localized Aurora 1 accumulation negatively correlated with MAP65-1 MT bundling and phospho-mutant forms of MAP65-1, including forced absence of Aurora-dependent phosphorylation induced by pharmacological inhibition of Aurora activity, which altered its subcellular localization pattern throughout cell division. Phosphorylation by Aurora, therefore, likely controls the accumulation of MAP65-1 at MTs at the nuclear surface but not at the PPB, between metaphase and anaphase and between anaphase and cell plate formation, allowing timely transitions of these phases. In light of these observations, it was striking that the movement of Aurora1 from the anaphase spindle poles to the cell plate (during the anaphase spindle to phragmoplast transition) correlated with reduced MT-bundling of MAP65-1.
Our observations also resemble the effect of MAP65-1 phosphorylation by MAPK. Namely, both Aurora- and MAPK-dependent phosphorylations delay cell cycle progression and affect phragmoplast expansion in vivo (Sasabe et al., 2006; this work). However, MAPK-dependent phosphorylation of MAP65-1 was shown to substantially influence its MT bundling properties (Sasabe et al., 2006), while Aurora has only a mild effect. A nonphosphorylatable MAP65-1 version in its predicted CDK phosphorylation site (S503A; Mao et al., 2005a) also prematurely labels the metaphase spindle MTs. However, in contrast to this CDK phospho-null mutant form, Aurora phospho-mutants affected the timing of cell division in Arabidopsis and BY-2 cells. Thus, those and our results highlight differential strengths and functions for MAP65-1 phosphorylation by MAPK, CDK, and Aurora, although their interdependency cannot be ruled out for now (Fig. 5).
Figure 5.
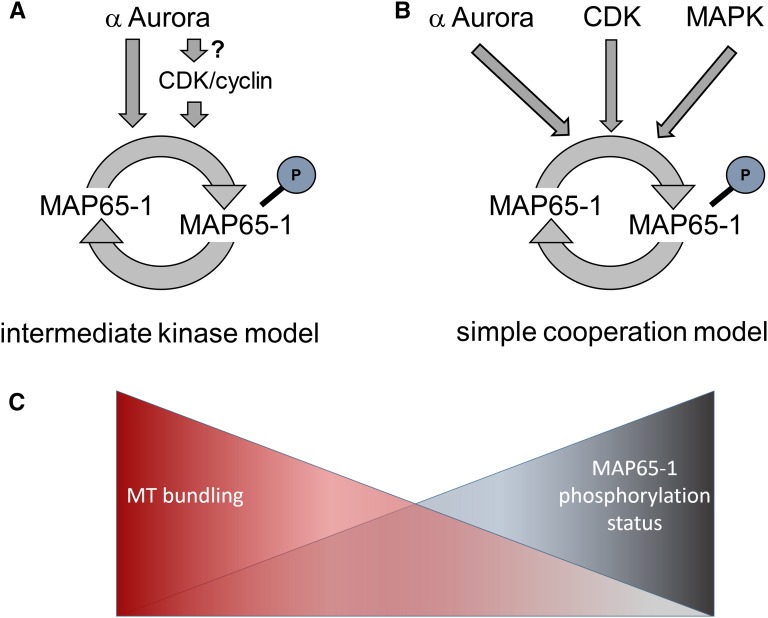
Proposed models of the role of Aurora phosphorylation on MAP65-1 in Arabidopsis. The intermediate kinase model (A) hypothesizes that MAP65-1 phosphorylation by CDK/cyclin complexes (or another kinase) depends on CDK activation by alpha Aurora kinases and is consistent with the observations that mitotic overexpression of MAP65-1, phospho-mimicked in its CDK sites (2D) does not aggravate the Aurora double mutant phenotype while overexpression of Auroras’ phospho-mimicry or nonphosphorylatable forms of MAP65-1 do. The simple cooperation model (B) argues for different mitotic kinases to independently contribute (with differential strengths) to the regulation of the MAP65-1 C-terminal unfolded domain charge and/or conformation to control its MT bundling activity. This model is consistent with the small reduction in MT bundling capacity of the MAP65-1(DD) in vitro compared to the nonphosphorylated forms and with the partial recovery of cortical MT bundling of the MAP65-1(7D) compared to MAP65-1(9D) in tobacco BY-2 cells. Whether the kinases perform the phosphorylations independently or whether they rely on each other’s activity remains to be addressed. C, The proposed models imply that phosphorylating kinases can control distinct properties of MAP65-1 MT bundling. A gradient of MAP65-1 phosphorylation might thus exist, where intermediate phosphorylation (e.g. by alpha Auroras) could change the specificity of MT bundling, without drastic alternations in the overall MT bundling capacity. Further phosphorylation by additional kinases is likely to affect MT bundling/binding properties of MAP65-1.
Mitotic Overexpression of MAP65-1 Reduces the Number of Divisions in the Aurora Double Mutant
The aggravated Aurora double mutant phenotypes (smaller rosette size, shorter roots, and fewer emerged lateral roots) by mitotic overexpression of MAP65-1 are likely caused by a reduced number of meristematic divisions. Indeed, as overexpression of MAP65-1 did not alter lateral root primordia initiation, the LR density reduction plausibly originates from fewer divisions and/or aborted primordia. Unfortunately, the severity of the alterations in lateral root patterning, already present in the Aurora double mutant, prevented quantification of the increase in this aggravation by mitotic overexpression of MAP65-1. However, the lateral root primordia effect is likely mirrored in the main root where a clear reduction in the number of mitotic RAM cells could be observed. In fact, both in the main root and in lateral root primordia cells, excessive MT bundling in metaphase and delayed depolymerization of phragmoplast MTs could be observed, which behaviors are probably causal to the reduced cell divisions.
The similarity of the defects observed using both Aurora phospho-mimicry and Aurora nonphosphorylatable forms of MAP65-1 regarding increased metaphase spindle recruitment, as well as in delaying phragmoplast MT depolymerization, hints toward the necessity for a swift conversion between the phosphorylation states. In yeast cells, phospho-mutations in the Ipl1 substrate Dam1p (the outer kinetochore microtubule-associated protein) to either a phospho-mimicry or a nonphosphorylatable form were equally deleterious, demonstrating the requirement for the dynamic nature of phospho-regulation by Aurora (Cheeseman et al., 2002). Alternatively, the fact that MAP65-1(DD) delays the metaphase to anaphase transition similarly to MAP65-1(AA) might indicate that this Aurora-phospho-mimicry of MAP65-1 acts as a dominant negative form, plausibly through scavenging an interactor. This could be due to insufficient phospho-mimetic properties of MAP65-1(DD), which might interfere with a critical function at the phosphorylation sites. Even though both Asp (D) and Glu (E) are commonly used to mimic phosphorylation of a Ser or a Thr, they do not always fulfill the same role in vivo. For example, in HeLa cells, expression of a double CDK phospho-mimic PRC1(EE) mutant excessively bundled spindle MTs similarly to a nonphosphorylatable PRC1(AA) mutant, even though the EE mutant had no effect on MT binding or bundling of PRC1 in vitro (Mollinari et al., 2002).
Direct and Indirect Control of MAP65-1 Activity by Alpha Auroras
Genetic interactions established a causative relationship between MAP65-1 and the alpha Auroras. Mitotic overproduction of MAP65-1 in the aur1/aur2 mutant enhanced, while depletion of MAP65-1 in this background partially rescued, the mutant phenotypes. Moreover, we could show that the aggravation of the Aurora double mutant phenotype is not a consequence of a disturbed cytoskeleton in a sensitized background, caused by overexpression of a MT-targeted protein, but it is instead specific to MAP65-1. Moreover, the results with MAP65-1(9D) corroborate that the aggravated Aurora double mutant phenotypes are not caused by mere mitotic overexpression of MAP65-1, but instead are a consequence of altered phosphorylation control in this background. However, this phosphorylation control of MAP65-1 activity by Aurora is to some extent indirect, because a phospho-mimicking form MAP65-1(DD) equally aggravated the Aurora mutant phenotype similarly to the MAP65-1(AA) form, in contrast to two other phospho-mimicries (9D and 2D). The observation that a phospho-mimicking form of two CDK sites, MAP65-1(2D), did not aggravate the mutant phenotypes suggests altered CDK activity toward MAP65-1 in the Aurora double mutant background.
In the light of these results, an attractive hypothesis arises, in which Aurora controls the activity of a yet to be identified kinase (e.g. a CDK or its cyclin partners), which together phosphorylate MAP65-1 (Fig. 5A). Accordingly, when mitotically overexpressed, MAP65-1 would be hypo-phosphorylated in the aur1/aur2 double knockdown mutant on both the Aurora sites as well as other ones, leading to the aggravated phenotypic defects observed. This intermediate kinase hypothesis (Fig. 5A) is also based on the evidence that MAP65-1, Ase1, and PRC1 are CDK substrates (Jiang et al., 1998; Ubersax et al., 2003; Mao et al., 2005a). Moreover, several Arabidopsis cyclin-dependent kinases (e.g. CDKA;1, CDKB2s) and their regulatory cyclins (e.g. A- and B-type cyclins) have motifs matching the Aurora phosphorylation consensus sequence, making them candidates to be under control of Aurora. Nevertheless, we cannot rule out the possibility of Aurora controlling a different kinase (e.g. MAPK) as well.
Because it is widely accepted that phosphorylation alters the charge of a protein, leading to a switch of electrostatic interactions, an alternative and nonexclusive hypothesis is that Aurora phosphorylation of MAP65-1 predominantly functions in cooperation with other mitotic kinases to increase the negative charge at the C terminus of MAP65-1 (Fig. 5B). In this scenario, the charge modification, which is under Aurora control, might not be enough to drastically alter the function of MAP65-1 and no profound changes in MAP65-1 bundling activity would be observed for its phospho-mutants. This fits with our in vitro data, which showed only a slight reduction in MT binding and bundling capacity of the DD-form of MAP65-1. Likewise, a CDK-phospho-mimic AtMAP65-1 could still cosediment with MTs (Smertenko et al., 2004), similar to a phosphorylation-mimic of PRC1, which binds and bundles MTs equally to the nonphosphorylatable protein (Mollinari et al., 2002). However, an accumulation of negative charges due to phosphorylation by multiple kinases leads to functional changes in MAP65-1 (Fig. 5C). This is substantiated by the fact that functions of many eukaryotic proteins are regulated by multisite phosphorylations to maximize activation (McCoy et al., 2007). This is, for instance, the case for the retinoblastoma protein and its several CDK activators, where complete inactivation occurs only after sequential action of at least two distinct CDK complexes (Zarkowska and Mittnacht, 1997; Lundberg and Weinberg, 1998). Also, the phosphorylation of glycogen synthase kinase on certain sites by glycogen synthase kinase 3 requires prior phosphorylation on other sites by casein kinase II (Roach, 1990). This phenomenon might increase the strength and specificity of protein-protein (or protein-DNA) interactions and/or also induce conformational changes (Burke et al., 2010).
One version of this scenario could be that one protein kinase primes phosphorylation at a nearby site by another protein kinase (Picton et al., 1982; Flotow et al., 1990; Lundberg and Weinberg, 1998). Because all nine putative phosphorylation sites at the C terminus of MAP65-1 are positioned in close proximity, it is conceivable that Aurora could prime this domain for further phosphorylation steps by other kinases and/or Aurora might require prior MAP65-1 phosphorylation by other kinases. However, the aggravation of the Aurora mutant phenotypes by MAP65-1(DD) argues against sequential phosphorylation starting with Aurora. Whether this favors the cooperative model rather than the sequential model, or can be explained by the DD-form not acting as a true phospho-mimic variant, as shown before for CDKA;1 (Dissmeyer et al., 2007, 2009), needs to be determined.
Also, the affinity of MAP65-1 for MTs is unlikely solely controlled by mere protein phosphorylation, because a complete phospho-mimicry in all nine MAP65-1 sites at its C terminus still retained partial MT association in vitro (Smertenko et al., 2006) and associated with the PPB MTs in vivo in BY-2 cells (our results). Therefore, it is reasonable to envisage additional, plausibly cell cycle-dependent mechanisms regulating MAP65-1 properties and activity. Future work, leading to a spatio-temporal resolution of Aurora activity in plant cells with respect to the other mitotic kinases, will allow us to test this model of progressive MAP65-1 phosphorylation.
MATERIALS AND METHODS
Cloning
All full-length open reading frames of the proteins of interest were recombined into the pDONR221 or pDONR207 entry vector (Invitrogen) by a BP reaction. MAP65-1 ORF (without the stop codon) in pDONR207 vector was described in Van Damme et al. (2004a). MAP65-1 ORF containing the stop codon was created by amplifying the clone without stop with STOP codon in the reverse primer (Supplemental Table S2). MAP65-1(AA) and MAP65-1(DD) clones were generated via a site-directed mutagenesis using the wild-type MAP65-1 clone in pDONR207 as a template. MAP65-1(9D) and MAP65-1(2D) clones were created by amplifying the previously published sequences (Smertenko et al., 2006) and cloning them into the Gateway entry vector pDONR207. The MAP65-1(7D) clone was created by reverting two Aurora phospho-mimicking sites of the MAP65-1(9D) clone into the wild-type sites via a site-directed mutagenesis. The multisite LR Gateway reaction resulted in translational fusions between the MAP65-1 (and its phospho-mutants) and GFP or RFP (for expression in BY-2 cells) or TagRFP (Mylle et al., 2013; for expression in Arabidopsis plants), driven by the pr35S or prAUR1, respectively. Pr35S::RFP-MBD was created in the same manner. Pr35S::MAP65-1-GFP and the MBD entry clone were described in Van Damme et al. (2004b). The endogenous prAUR1 and the complementing prAUR1:AUR1-GFP construct used for the coimmunoprecipitation were described in Van Damme et al. (2011). The expression clones were generated in the pB7m34GW (expression in Arabidopsis) or pK7FWG2 and pH7RWG2 (expression in BY-2 cells of GFP and RFP fusions) destination vectors (Karimi et al., 2007). For protein purification, MAP65-1(AA) and MAP65-1(DD) were subcloned in pENTR/D-TOPO plasmid (Invitrogen) using the primers ForM65.1 and RevM65.1 (Supplemental Table S2) and then introduced into an homemade vector (Gaillard et al., 2008) by LR Gateway reaction. Recombinant proteins were expressed as GFP-tagged at their N terminus end and His-tagged at their C terminus.
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 (Col-0) plants and all mutant lines were grown under standard growth conditions in continuous light on vertical plates containing half-strength Murashige and Skoog (MS) medium supplemented with 8 g/L plant tissue culture agar and 1% Suc. Wild-type Col-0 and aur1/aur2 double mutant plants were transformed using the floral dip protocol (Clough and Bent, 1998). The aur1/aur2 double mutant and the complemented double mutant line were described in Van Damme et al. (2011). For the leaf series (Supplemental Fig. S8B), twelve 25-d-old plants grown in soil were analyzed. The Arabidopsis mutant line map65-1-3 Gabi-KAT_198A01 was acquired from the Gabi kat collection (Kleinboelting et al., 2012). The aur1/aur2 map65-1-3 triple mutant was created by crossing the aur1/aur2 double mutant with the map65-1-3 single mutant. The aur1/aur2 map65-1-2 triple mutant was created by crossing the aur1/aur2 double mutant with the complemented map65-1-2 single mutant (SALK_118225) expressing prMAP65-1:GFP-MAP65-1 [the line was a gift from Professor Sidney Shaw (Lucas et al., 2011)] and then selecting the progeny without the complementing transgene by fluorescence microscopy and PCR against GFP. Lateral root density measurements were performed as described in Van Damme et al. (2011).
Wild-type tobacco (Nicotiana benthamiana) plants were grown under a normal light regime (14 h of light, 10 h of darkness) at 25°C and 70% relative humidity. Tobacco infiltration with Agrobacterium tumefaciens strain C58 was performed as described in Boruc et al. (2010). However, in addition, coexpression of p19 protein from tomato bushy stunt virus was used for suppression of transgene silencing (Voinnet et al., 2000).
Stable tobacco BY-2 cell (N. tabacum “Bright Yellow-2”) transformation was carried out as described in Geelen and Inzé (2001). For the AUR1-GFP and MAP65-1-RFP colocalization analysis, BY-2 cells were cotransformed with both constructs, selected on kanamycin only, and screened for the presence of both the AUR1 and MAP65-1 by fluorescence microscopy on an Axiovert epifluorescence microscope (Zeiss). Several independently transformed calli were imaged for each construct.
Immunoblot Analysis
For the detection of MAP65-1-TagRFP protein, 4 to 5 d-old plants were harvested, frozen in liquid N2, and ground. A quantity of 500 µL of protein extraction buffer [50 mM Tris-Cl, pH 7.6, NaCl 150 mM, EDTA 5 mM, SDS 0,1% (w/v), β-mercaptoethanol 0,1% (w/v), EDTA-free Complete Protease Inhibitor Cocktail (1 tablet/10 mL, Roche Diagnostics), PhosSTOP Inhibitor Cocktail (1 tablet/10 mL; Roche Diagnostics)] was added to the ground plant powder on ice. Samples were vortexed and incubated on ice for 10 min. Protein crude extract was spun down at 14,000 rpm for 30 min at 4°C. Protein concentration (determined by Bradford method with BioRad Quick Start) was adjusted to 50 µg/mL for equal loading. A quantity of 5 μL 5× SDS loading dye was added to all samples and incubated for 5 min at 95°C. Samples were run on precast gels (7%; NuPAGE 7% Tris-Acetate Protein Gels; Novex Life Technologies) in MES-SDS buffer (Amresco) for approximately 45 min at 150 V. After blotting (iBlot System; Novex Life Technologies), membranes were transferred to TBS-T buffer (50 mM Tris, 150 mM NaCl, pH 7.6, 0.05% Tween) + 5% skim milk powder (Difco) and incubated at 4°C overnight on a rocking platform. Membranes were incubated with the MAP65-1 primary antibody (Smertenko et al., 2004) in TBS-T + 5% milk for 3 h at room temperature (1:1000 dilution). Membranes were incubated with the secondary antibody (a horseradish-conjugated anti-rabbit sera in TBS-T + 1% milk, 1:10,000 dilution) for 2 h at RT. Membranes were developed (Western Lightning Plus-ECL solutions; Perkin-Elmer) on a Molecular Imager, Chemidoc XRS+ (BioRad).
Proteolysis and Mass Spectrometry
In-solution digestion was performed using Trypsin (Promega) at a protein/enzyme ratio of 1:30 (w/w) in 1 M urea and 50 mM NH4HCO3 at 37°C. After 3 h of incubation, samples were diluted (4×) and incubated overnight at 37°C. Digestion was stopped by adding formic acid (0.1% final concentration). Samples were purified using C18 Macro SpinColumns (Harvard Apparatus) and dried using vacuum centrifugation. Digests were resolubilized in 50 μL of 98/2 (v/v) water/acetonitrile solution containing 0.1% formic acid before being analyzed.
For nano-LC-MS/MS analyses, 6 μL of the peptides mixture was analyzed by on-line nano-LC-MS/MS (Ultimate 3000; Dionex and LTQ-Orbitrap Velos; Thermo Fisher Scientific). Peptides were sampled on a 300 μm × 5 mm PepMap C18 precolumn, and separated on a 75 μm × 250 mm C18 column (PepMap; Dionex). The nano-LC method consisted of a 60 min gradient at a flow rate of 300 nL/min, ranging from 5 to 45% acetronitrile in 0.1% formic acid for 54 min before reaching 90% for the last 6 min. MS and MS/MS data were acquired using Xcalibur (Thermo Fisher Scientific). Spray voltage and heated capillary were, respectively, set at 1.4 kV and 200°C. Survey full-scan MS spectra (m/z = 400 to 1600) were acquired in the Orbitrap with a resolution of 30,000. The 20 most intense ions from the preview survey scan delivered by the Orbitrap were fragmented by collision-induced dissociation (collision energy, 35%) in the LTQ. Data were processed automatically using Mascot Daemon software (Ver. 2.3.2; Matrix Science). Searches against the UniProt database (August 2012 Ver.; Arabidopsis thaliana taxonomy). ESI-TRAP was chosen as the instrument and trypsin/P as the enzyme, and two missed cleavages were allowed. Precursor and fragment mass error tolerances were set, respectively, at 10 ppm and 0.6 Da. Peptide modifications allowed during the search were oxidation (M, variable), phosphorylation (S, T, Y as variable), and DeStreak (C, fixed).
Co-Immunoprecipitation
Arabidopsis seedlings were grown in liquid 1/2 MS medium for 3 d, harvested (approximately 150 μL), and immediately frozen in liquid N2. Seedlings were ground and 1.5 mL extraction buffer was added [150 mm NaCl, 50 mm Tris-HCl (pH 7.5), 2% Triton X-100, Protease Inhibitor and Phosphatase Inhibitor tablet (Roche)]. Samples were incubated on ice for 30 min, vortexed every 5 min, and spun down for 30 min at 14,000 rpm at 4°C. The supernatant was transferred to a new precooled tube and spun down for 20 min, 14,000 rpm, 4°C. Input sample was taken from the supernatant. GFP-Trap_A beads (equilibrated twice 50 μL beads/sample in 500 μL cold extraction buffer with 0.1% TritonX-100; Chromotek) were mixed with the protein extract and incubated at 4°C on a turning wheel for 3 h. Next, the supernatant was taken as the nonbound fraction. Magnetic beads were washed five times with cold extraction buffer with 0.1% TritonX-100 and resuspended in 100 μL extraction buffer with 0.1% Triton X-100 (the bound fraction). A quantity of 5 μL 5× SDS loading dye was added to all samples (input, nonbound, bound) and incubated for 5 min at 95°C. Afterward, samples were spun down for 30 s and run in two replicates on precast gels (7%; NuPAGE 7% Tris-Acetate Protein Gels; Novex Life Technologies) in MES-SDS buffer (Amresco) for approximately 5 min at 150 V. After blotting (the iBlot System; Novex Life Technologies), the membranes were blocked [in TBS-T buffer (50 mm Tris, 150 mm NaCl, pH 7.6, 0.05% Tween) + 5% bovine serum albumin] at room temperature overnight on a rocking platform. One of the duplicate membranes was incubated with the MAP65-1 primary antibody (Smertenko et al., 2004) for 3 h at room temperature followed by the secondary antibody (HRP-conjugated anti-rabbit sera in TBS-T + 1% bovine serum albumin in a 1:10.000 dilution for 2 h at RT). The second membrane was incubated with the GFP-HRP antibody (1:5000; Macs Miltenyi Biotec) for 3 h at RT. Membranes were developed (Western Lightning Plus-ECL solutions; Perkin-Elmer) on a Molecular Imager, Chemidoc XRS+ (BioRad).
Immunoprecipitation of MAP65-1 Isoforms from N. benthamiana
GFP-MAP65-1, GFP-MAP65-1(AA), and GFP-MAP65-1(DD) proteins were produced by agrobacterium infiltration of N. benthamiana leaves as described in Boruc et al. ( 2010). Tobacco transformation efficiency was checked by confocal microscopy. Leaves expressing MAP65-1 isoforms were frozen in liquid nitrogen and stored until lysis in extraction buffer (150 mM TRIS-HCl pH 7.5, 150 mM NaCl, 10% glycerol, 10 mM EDTA, 1 mM NaMoO4, 1 mM NaF freshly supplemented with 10 mM DTT, 0.5% PVPP PhosSTOP, and EDTA-free Complete tablets; Roche). Efficient extraction was performed using 1 mL of extraction buffer per gram of frozen material. Lysed material was stored on ice for 10 min and subsequently centrifuged at 4°C (16,000g). Supernatant was recovered and incubated for 2 h at 4°C with GFP-Trap_A beads (50 μL of dry beads previously equilibrated in extraction buffer; Chromotek). Upon completion of incubation the material was centrifuged at 0.5g and the supernatant was recovered. Beads were washed three times using 50 µL of kinase buffer and pelleted down at 0.5g each time. The presence of MAP65-1 isoforms for each pulldown experiment was confirmed by immunoblotting using monoclonal 1:1000 anti-GFP primary antibody (Living Colors JL-8; Clontech) and anti-Mouse (IRDye 800CW; LiCOR). Detection was done on a CLx system at 800 nm (Odyssey).
Recombinant Protein Purification
Aurora1 (At4g32830) and Aurora2 (At2g25880) cDNAs were obtained from Riken (PDA15493 for Aurora1 and PDA13561 for Aurora2). Both Auroras were purified as tagged proteins with His tag at N- and C termini using a home-made vector (Gaillard et al., 2008). Primers used to amplify cDNAs of AURORA1 and AURORA2 for recombinant kinase production can be found in Supplemental Table S2. His-AURORA-His proteins were expressed for 20 h at 12°C in Rosetta (DE3) pLysS bacterial strain (Novagen-Merck Biosciences) by induction with 1 mm IPTG. After induction, cells were suspended in cold lysis buffer (100 mm Tris-HCl pH 8.0, 0.1 M NaCl, 10% Glycerol, 2.5 mm β-mercaptoethanol, and 10 mm Imidazole), containing protease inhibitors (complete, EDTA free; Roche) and then sonicated. The cell lysate was centrifuged at 50,000g for 30 min and applied on Ni Sepharose columns (Amersham Bioscience) for 1 h at 4°C. Columns were washed with protein wash buffer 1 (50 mm Tris-HCl pH 8, 1 m NaCl, 20 mm Imidazole, and 10% Glycerol), and protein wash buffer 2 (50 mm Tris-HCl pH 8.0, 0.25 m NaCl, 20 mm Imidazole, and 10% Glycerol). Proteins were eluted with elution buffer (50 mm Tris-HCl pH 8.0, 0.25 m NaCl, and 0.25 m Imidazole), and complemented with 0.5 mm EDTA and 10 mm DTT before being applied to gel filtration chromatography in 20 mm Tris HCl pH 8, 150 m NaCl, and 5 mm DTT. Purified Auroras were frozen in liquid nitrogen in the presence of 10% of glycerol and stored at −80°C.
The different His-tagged MAP65-1 recombinant proteins were purified as described in Gaillard et al. (2008) and stored at −80°C in MAP buffer [10% (vol/vol) glycerol, 50 mm NaPi, 0.1 m NaCl, and 0.5 mm DTT, pH 7.9].
Bovine brain tubulin was purified according to Vantard et al. (1994) in BRB80 buffer (80 mm PIPES, pH 6.8, 1 mm EGTA, and 1 mm MgCl2). Fluorescent tubulin (Alexa488-labeled tubulin and Alexa568-labeled tubulin) and biotinylated tubulin were prepared according to Hyman et al. (1991).
Determination of the Apparent Steady-State Kd of MAP65-1
The apparent Kd value of MAP65-1(AA), MAP65-1(DD) and in vitro phosphorylated MAP65-1 by recombinant Aurora kinase was determined according to Portran et al. (2013). In brief, 2 μM of taxol-stabilized MTs were incubated with various concentrations of GFP-MAP65-1 proteins for 30 min at room temperature. After centrifugation (20 min, 100,000g), the amount of GFP-MAP65-1 proteins in the supernatant and the pellet was measured by spectrofluorimetry (excitation, 490 nm; emission, 512 nm). For all proteins, the intensity of the signal was converted into a concentration value using a linear calibration curve. The Kd value was calculated by fitting the binding data by Equation 1 according to Pollard (2010):
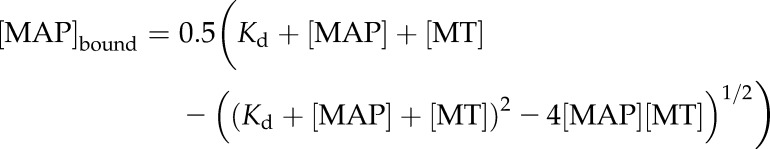 |
where [MAP]bound is the concentration of pelleted MAP65-1 bound to MT, [MAP] is the concentration of free MAP65-1, [MT] is the fixed concentration of MT, and Kd is the equilibrium dissociation constant.
In Vitro Kinase Assays Using Recombinant MAP65-1 Proteins and Their Truncations
Kinase assays were performed using a modified protocol from Hans et al. (2009). In brief, 1 μM of recombinant His-Aurora kinase was added to 30 μL of kinase buffer (50 mm Tris, pH 7.4, 10 mm MgCl2, 1 mm EGTA, 1 mm DTT, 5 mm NaF, 50 μM Na3VO4, 0.1 mm ATP, and 5 mm beta glycerophosphate) containing 0.75 μM of recombinant substrates. After addition of 0.15 μCi of [γ-32P]ATP, the reaction was allowed to proceed for 30 min at 30°C, and then stopped by addition of 20% TCA. After 1 h at 4°C, proteins were pelleted and separated on SDS polyacrylamide gel. The gel was dried and processed for autoradiography. Identification of phosphorylated residues of MAP65-1 by mass spectroscopy analysis was done by Etude de la Dynamique des Protéomes-Service (Commissariat à l’Énergie Atomique et aux Énergies Alternatives-Institut de Recherche en Technologies et Sciences pour le Vivant-Laboratoire d’Etude de la Dynamique des Protéomes, Grenoble, France) as described above.
In Vitro Kinase Assays Using GFP-Trap-purified MAP65-1, MAP65-1(AA), and MAP65-1(DD)
The kinase Master mix (10 μL per reaction) consisted of 0.05 μL (10 mm stock) ATP; 1 μL of 10 μCi/μL of 32P-γPO4-ATP; 1 μL of 100 μM recombinant AURORA1 kinase; 2 μL of 10× kinase buffer (10× buffer: 500 mM TRIS pH 7.5, 100 mM MgCl2, 10 mM EGTA, and 10 mM DTT); and 5.95 μL of distilled water.
Kinase assays were carried out by mixing 10 μL of GFP-beads (from pull down) with 10 μL of kinase master mix in duplicate 1.5 mL microcentrifuge tubes. The first tube (to) contains a reaction mix, which was immediately quenched by addition of 5 μL of SDS-PAGE loading buffer and boiled in a heating block. The second tube was incubated at 30°C for 1 h. After the incubation time, the reaction is quenched as described above.
Turbidimetry Bundling Assays
Taxol-stabilized MTs (500 nM) were incubated with 75 nm of MAP65-1 at room temperature in MAP buffer. The turbidity of the reaction was recorded at 350 nm.
MT Dynamics by TIRF Microscopy
Cleaning of slides and coverslips and preparation of perfusion chambers were performed according to Portran et al. (2013). MT seed bundles containing 2 to 3 MTs were obtained by incubating 0.3 μM of GMPCPP-stabilized MT seeds with 3 nM of MAP65-1 for 10 min at room temperature, and then used immediately. For TIRF assays, MT seeds were laid on neutravidin-coated slides for 5 min in the perfusion chamber. They were then elongated by gently flowing an elongation mix containing 18 μM tubulin (30% Alexa 568-labeled tubulin and 70% unlabeled tubulin), 1 mM GTP, an oxygen scavenger cocktail (1 mg/mL Glc, 81 μg/mL catalase, and 0.67 mg/mL Glc oxidase), 20 μM DTT, 1% BSA, and 0.025% methyl cellulose (1500 CP) in mix buffer (1 × BRB80 / 0.5 × MAP buffer) in the presence or the absence of 50 nM recombinant MAP65-1 proteins. MT dynamics were visualized using an objective-based azimuthal Ilas2 TIRF microscope (Nikon TE-2000-E, modified by Roper Scientific). The microscope stage was kept at 32°C using a warm stage controller (MC60; Linkam Tadworth). Excitation was achieved using 491 nm and 561 nm lasers (Roper Scientific). Time-lapse recordings (one frame every 2 s) were performed for 30 min using MetaMorph software (Ver. 7.7.5; Molecular Devices). Movies were processed to improve signal/noise ratio (equalize light, low-pass, and flatten background filters of MetaMorph software were used). MT bundle elongation and dynamics were analyzed using kymographs generated by MetaMorph and analyzed with ImageJ (National Institutes of Health). Parameters of MT dynamics (rate of elongation and shortening, frequency of catastrophe and rescue) were calculated for each MT in bundles.
Confocal and Spinning Disc Imaging
Fluorescence was analyzed with an inverted confocal microscope FluoView FV1000 (Olympus), equipped with a 63× water-corrected objective (NA 1.2). Fluorescence was imaged in a multichannel setting with 488-nm and 543-nm excitation light for GFP and RFP or TagRFP excitation, respectively. Emission fluorescence was captured in the frame-scanning mode via 500 nm to 550 nm and 560 nm to 660 nm bandpass emission windows for GFP and RFP/TagRFP fluorescence, respectively. Propidium iodide was visualized using 514-nm laser excitation and a 566 nm to 649 nm emission window for detection.
Time series of root epidermal cells were captured using an Ultra View Vox Spinning Disc Imaging System (Perkin-Elmer) with an inverted Eclipse Ti microscope (Nikon) equipped with a perfect focus system (PFSIII; Nikon) for Z-drift compensation. Movies were taken using a Plan Apo 60× water (NA 1.20) lens, a 488 nm laser or 543 nm laser, and Hamamatsu 512 × 512 BI (16 μm × 16 μm pixel size) CCD camera. Two- to three-h-long movies were acquired with 30 time points per h interval, including a Z-stack of 1 μm step size and a stitched region of approximately nine images (to capture the growing root), controlled by the Volocity software (Perkin-Elmer). The region of interest around the cells of interest was afterward corrected for movement and processed with the Volocity software.
Immunolocalization of Phosphorylated Histone3 Ser-10 in BY-2 Cells
Four-d-old BY-2 cells were treated with 2 μM MLN8237 (1000× stock diluted in DMSO; Selleck) for 1 h and fixed with 4% paraformaldehyde in PBS for 1 h. Cell walls were digested with 2% driselase for 15 min. The cells were incubated with 1:500 dilution of anti-H3S10ph antibody (Rabbit polyclonal anti-body; Merck Millipore) in PBS overnight at 4°C. For secondary antibodies reaction, the cells were incubated with 1:600 dilution of Alexa 488-conjugated goat anti-Rabbit IgG antibody (Invitrogen) for 3 h at room temperature. DNA was stained with 0.2 μg/mL DAPI solution.
DAPI Staining of Arabidopsis Roots
A quantity of 5-to-7-d old Arabidopsis seedlings was fixed with 4% paraformaldehyde in PBS for 5 to 10 min, then incubated with 1 μg/mL DAPI and imaged.
MNL8237 Treatment
BY-2 cells were treated with 4 μM MLN8237 for 1 h and imaged. Arabidopsis seedlings were grown vertically on varying concentrations of MLN8237 (0, 0.1, 0.5, and 1 μM) to score the root growth inhibition.
Root Clearing
For the analysis of nonemerged lateral root primordia, roots were cleared according to the Malamy and Benfey clearing method (Malamy and Benfey, 1997).
RAM Size Analysis
RAM size was calculated as the distance between the quiescent center and the transition zone (indicating the position of the first elongating cortical cell). To calculate the RAM, measurements were performed on 7-d-old seedlings stained with propidium iodide and imaged by confocal microscopy.
Primers Used
All primers used in this study can be found in Supplemental Table S2.
Quantification
Box plot graphs were generated using the BoxplotR web tool (Spitzer et al., 2014). The statistical differences for Figure 4C were estimated with a Wald test. Assumptions were assessed by residual analysis. The analysis was performed with the glm procedure of SAS (Ver. 9.4; SAS Institute). Fluorescence intensities in confocal images were calculated as the average of the top 100 pixels within the region of interest using the histogram tool from the ImageJ software package, combined with a custom-made Excel spreadsheet calculation table.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AUR1, At4g32830; AUR2, At2g25880; The following reference sequences of proteins were used for the alignment in Supplemental Figure 1: At5g55230 (AtMAP65-1), At4g26760 (AtMAP65-2), At5g51600 (AtMAP65-3), At3g60840 (AtMAP65-4), At2g38720 (AtMAP65-5), At2g01910 (AtMAP65-6), At1g14690 (AtMAP65-7), At1g27920 (AtMAP65-8), At5g62250 (AtMAP65-9). Sequences used for the alignment in Supplemental Figure 2: NP_116582.1 (Saccharomyces cerevisiae), NP_003972.1 (Homo sapiens), NP_001080868.1 (Xenopus laevis), AT5G55230 (Arabidopsis thaliana), XP_003544755.1 (Glycine max), XP_010913972.1 (Elaeis guineensis), XP_008364470.1 (Malus domestica), XP_003617643.1 (Medicago truncatula), XP_009394695.1 (Musa acuminata subsp. malaccensis), XP_002277391.2 (Vitis vinifera), XP_006346185.1 (Solanum tuberosum), GenBank: CAC17794.1 (Nicotiana tabacum), XP_002298820.1 (Populus trichocarpa), XP_010227508.1 (Brachypodium distachyon), NP_001058056.1 (Oryza sativa Japonica Group), GenBank: AFW76336.1 (Zea mays), GenBank: BAJ86160.1 (Hordeum vulgare subsp. vulgare), GenBank: EMS44988.1 (Triticum urartu).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Protein sequence alignment of all nine Arabidopsis MAP65 proteins.
Supplemental Figure S2. Protein sequence alignment comparing yeast (S. cerevisiae) Ase1, vertebrate (H. sapiens and X. laevis) PRC1, A. thaliana MAP65-1, and 14 other plant species.
Supplemental Figure S3. Aurora phosphorylation of MAP65-1 has only a subtle effect on its bundling capacity in vitro.
Supplemental Figure S4. S532 and T552 contribute to MT binding of MAP65-1 in vivo.
Supplemental Figure S5. The Aurora kinase inhibitor MLN8237 affects the subcellular localization of MAP65-1 in vivo.
Supplemental Figure S6. Subcellular localization of MAP65-1 and MAP65-1(AA) in Arabidopsis root meristem.
Supplemental Figure S7. Localization of MAP65-1 in (pre)prophase, and metaphase timing in wild-type Col-0 compared to the aur1/aur2 double mutant seedlings.
Supplemental Figure S8. Overexpression of MAP65-1 aggravates the phenotype of aur1/aur2 double mutant plants.
Supplemental Figure S9. Nonemerged lateral root primordia patterning in wild type, aur1/aur2, and aur1/aur2 with mitotic overexpression of MAP65-1.
Supplemental Figure S10. Ectopic expression of the MBD of MAP4 does not aggravate the phenotype of the Aurora double mutant.
Supplemental Figure S11. Ectopic overexpression of MAP65-1 and its Aurora-dependent phospho-mutant forms aggravate the root phenotypes of the aur1/aur2 double mutant.
Supplemental Figure S12. Subcellular localization of prAUR1:MAP65-1-TagRFP during LR primordia development.
Supplemental Table S1. Mass spectrometry data obtained from recombinant MAP65-1 phosphorylated in vitro by recombinant Aurora1.
Supplemental Table S2. List of primers used in this study.
Supplemental Movie S1. Subcellular localization of pr35S:MAP65-1-GFP in BY-2 cells during cell division.
Supplemental Movie S2. Subcellular localization of pr35S:GFP-MAP65-1 in BY-2 cells during cell division.
Supplemental Movie S3. Subcellular localization of pr35S:MAP65-1(AA)-GFP in BY-2 cells during cell division.
Supplemental Movie S4. Subcellular localization of pr35S:MAP65-1(DD)-GFP in BY-2 cells during cell division.
Supplemental Movie S5. Subcellular localization of pr35S:MAP65-1(7D)-GFP in BY-2 cells during cell division.
Supplemental Movie S6. Subcellular localization of pr35S:MAP65-1(9D)-GFP in BY-2 cells during cell divisioN.
Acknowledgments
We thank Veronique Storme (Plant Systems Biology, VIB) for help with the statistical analysis of the differential stem elongation phenotypes; Jacobus (Co) Knetsch for cloning and Arabidopsis transformation; Jeremie Gaillard (le Laboratoire Physiologie Cellulaire & Végétale, Grenoble) for MAP65-1 purification; and Professor Gustavo Gutierrez (Vrije Universiteit Brussels, Laboratory of Pathophysiological Cell Signaling) for advice and infrastructure access for the in vitro kinase assays using immunoprecipitated MAP65-1 forms.
Footnotes
This work was supported by a grant from the Research Foundation of Flanders (G029013N to D.V.D. and T.B.); a Belgian Science Policy (BELSPO) Back to Belgium postdoctoral fellowship grant (selection year 2012 to J.B.); a grant from the French Research Agency, ANR (MITOTUBE to V.S.-M. and M.V.); and an Odysseus grant from the Research Foundation Flanders (G.0029.12 to P.T.). K.K. is supported by a Japan Society for the Promotion of Science postdoctoral fellowship for Research Abroad and the Advancing Strategic International Networks to Accelerate the Circulation of Talented Researchers.
Articles can be viewed without a subscription.
References
- Adams RR, Wheatley SP, Gouldsworthy AM, Kandels-Lewis SE, Carmena M, Smythe C, Gerloff DL, Earnshaw WC (2000) INCENP binds the Aurora-related kinase AIRK2 and is required to target it to chromosomes, the central spindle and cleavage furrow. Curr Biol 10: 1075–1078 [DOI] [PubMed] [Google Scholar]
- Biggins S, Murray AW (2001) The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev 15: 3118–3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boruc J, Van den Daele H, Hollunder J, Rombauts S, Mylle E, Hilson P, Inzé D, De Veylder L, Russinova E (2010) Functional modules in the Arabidopsis core cell cycle binary protein-protein interaction network. Plant Cell 22: 1264–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke JR, Deshong AJ, Pelton JG, Rubin SM (2010) Phosphorylation-induced conformational changes in the retinoblastoma protein inhibit E2F transactivation domain binding. J Biol Chem 285: 16286–16293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena M, Ruchaud S, Earnshaw WC (2009) Making the Auroras glow: regulation of Aurora A and B kinase function by interacting proteins. Curr Opin Cell Biol 21: 796–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CS, Botstein D (1993) Isolation and characterization of chromosome-gain and increase-in-ploidy mutants in yeast. Genetics 135: 677–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J, Jensen CG, Jensen LC, Bush M, Lloyd CW (1999) The 65-kDa carrot microtubule-associated protein forms regularly arranged filamentous cross-bridges between microtubules. Proc Natl Acad Sci USA 96: 14931–14936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang-Jie J, Sonobe S (1993) Identification and preliminary characterization of a 65 kDa higher-plant microtubule-associated protein. J Cell Sci 105: 891–901 [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Anderson S, Jwa M, Green EM, Kang JS, Yates JR III, Chan CS, Drubin DG, Barnes G (2002) Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell 111: 163–172 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant Cell 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Crane R, Gadea B, Littlepage L, Wu H, Ruderman JV (2004) Aurora A, meiosis and mitosis. Biol Cell 96: 215–229 [DOI] [PubMed] [Google Scholar]
- Demidov D, Lermontova I, Weiss O, Fuchs J, Rutten T, Kumke K, Sharbel TF, Van Damme D, De Storme N, Geelen D, Houben A (2014) Altered expression of Aurora kinases in Arabidopsis results in aneu- and polyploidization. Plant J 80: 449–461 [DOI] [PubMed] [Google Scholar]
- Demidov D, Van Damme D, Geelen D, Blattner FR, Houben A (2005) Identification and dynamics of two classes of Aurora-like kinases in Arabidopsis and other plants. Plant Cell 17: 836–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dissmeyer N, Nowack MK, Pusch S, Stals H, Inzé D, Grini PE, Schnittger A (2007) T-loop phosphorylation of Arabidopsis CDKA;1 is required for its function and can be partially substituted by an aspartate residue. Plant Cell 19: 972–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dissmeyer N, Weimer AK, Pusch S, De Schutter K, Alvim Kamei CL, Nowack MK, Novak B, Duan GL, Zhu YG, De Veylder L, Schnittger A (2009) Control of cell proliferation, organ growth, and DNA damage response operate independently of dephosphorylation of the Arabidopsis Cdk1 homolog CDKA;1. Plant Cell 21: 3641–3654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutertre S, Cazales M, Quaranta M, Froment C, Trabut V, Dozier C, Mirey G, Bouché JP, Theis-Febvre N, Schmitt E, Monsarrat B, Prigent C, et al. (2004) Phosphorylation of CDC25B by Aurora-A at the centrosome contributes to the G2-M transition. J Cell Sci 117: 2523–2531 [DOI] [PubMed] [Google Scholar]
- Eyers PA, Maller JL (2004) Regulation of Xenopus Aurora A activation by TPX2. J Biol Chem 279: 9008–9015 [DOI] [PubMed] [Google Scholar]
- Fache V, Gaillard J, Van Damme D, Geelen D, Neumann E, Stoppin-Mellet V, Vantard M (2010) Arabidopsis kinetochore fiber-associated MAP65-4 cross-links microtubules and promotes microtubule bundle elongation. Plant Cell 22: 3804–3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S, Marin O, Pagano MA, Meggio F, Hess D, El-Shemerly M, Krystyniak A, Pinna LA (2005) Aurora-A site specificity: a study with synthetic peptide substrates. Biochem J 390: 293–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flotow H, Graves PR, Wang AQ, Fiol CJ, Roeske RW, Roach PJ (1990) Phosphate groups as substrate determinants for casein kinase I action. J Biol Chem 265: 14264–14269 [PubMed] [Google Scholar]
- Fu J, Bian M, Liu J, Jiang Q, Zhang C (2009) A single amino acid change converts Aurora-A into Aurora-B-like kinase in terms of partner specificity and cellular function. Proc Natl Acad Sci USA 106: 6939–6944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard J, Neumann E, Van Damme D, Stoppin-Mellet V, Ebel C, Barbier E, Geelen D, Vantard M (2008) Two microtubule-associated proteins of Arabidopsis MAP65s promote antiparallel microtubule bundling. Mol Biol Cell 19: 4534–4544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geelen DN, Inzé DG (2001) A bright future for the bright yellow-2 cell culture. Plant Physiol 127: 1375–1379 [PMC free article] [PubMed] [Google Scholar]
- Glover DM, Leibowitz MH, McLean DA, Parry H (1995) Mutations in Aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell 81: 95–105 [DOI] [PubMed] [Google Scholar]
- Goldenson B, Crispino JD (2015) The Aurora kinases in cell cycle and leukemia. Oncogene 34: 537–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto H, Yasui Y, Nigg EA, Inagaki M (2002). Aurora-B phosphorylates Histone H3 at serine 28 with regard to the mitotic chromosome condensation. Genes to Cells 7: 11–17. [DOI] [PubMed] [Google Scholar]
- Guharoy M, Szabo B, Contreras Martos S, Kosol S, Tompa P (2013) Intrinsic structural disorder in cytoskeletal proteins. Cytoskeleton 70: 550–571 [DOI] [PubMed] [Google Scholar]
- Guo L, Ho CM, Kong Z, Lee YR, Qian Q, Liu B (2009) Evaluating the microtubule cytoskeleton and its interacting proteins in monocots by mining the rice genome. Ann Bot (Lond) 103: 387–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guse A, Mishima M, Glotzer M (2005) Phosphorylation of ZEN-4/MKLP1 by Aurora B regulates completion of cytokinesis. Curr Biol 15: 778–786 [DOI] [PubMed] [Google Scholar]
- Hans F, Skoufias DA, Dimitrov S, Margolis RL (2009) Molecular distinctions between Aurora A and B: a single residue change transforms Aurora A into correctly localized and functional Aurora B. Mol Biol Cell 20: 3491–3502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T, Kunitoku N, Sasayama T, Marumoto T, Zhang D, Nitta M, Hatakeyama K, Saya H (2003) Aurora-A and an interacting activator, the LIM protein Ajuba, are required for mitotic commitment in human cells. Cell 114: 585–598 [DOI] [PubMed] [Google Scholar]
- Hochegger H, Hégarat N, Pereira-Leal JB (2013) Aurora at the pole and equator: overlapping functions of Aurora kinases in the mitotic spindle. Open Biol 3: 120185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu JY, Sun ZW, Li X, Reuben M, Tatchell K, Bishop DK, Grushcow JM, Brame CJ, Caldwell JA, Hunt DF, Lin R, Smith MM, et al. (2000) Mitotic phosphorylation of histone H3 is governed by Ipl1/Aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102: 279–291 [DOI] [PubMed] [Google Scholar]
- Hussey PJ, Hawkins TJ, Igarashi H, Kaloriti D, Smertenko A (2002) The plant cytoskeleton: recent advances in the study of the plant microtubule-associated proteins MAP-65, MAP-190 and the Xenopus MAP215-like protein, MOR1. Plant Mol Biol 50: 915–924 [DOI] [PubMed] [Google Scholar]
- Hyman A, Drechsel D, Kellogg D, Salser S, Sawin K, Steffen P, Wordeman L, Mitchison T (1991) Preparation of modified tubulins. Methods Enzymol 196: 478–485 [DOI] [PubMed] [Google Scholar]
- Iakoucheva LM, Radivojac P, Brown CJ, O’Connor TR, Sikes JG, Obradovic Z, Dunker AK (2004) The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res 32: 1037–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Jimenez G, Wells NJ, Hope TJ, Wahl GM, Hunter T, Fukunaga R (1998) PRC1: a human mitotic spindle-associated CDK substrate protein required for cytokinesis. Mol Cell 2: 877–885 [DOI] [PubMed] [Google Scholar]
- Karimi M, Depicker A, Hilson P (2007) Recombinational cloning with plant gateway vectors. Plant Physiol 145: 1144–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe A, Matsunaga S, Nakagawa K, Kurihara D, Yoneda A, Hasezawa S, Uchiyama S, Fukui K (2005) Characterization of plant Aurora kinases during mitosis. Plant Mol Biol 58: 1–13 [DOI] [PubMed] [Google Scholar]
- Kimura M, Matsuda Y, Yoshioka T, Okano Y (1999) Cell cycle-dependent expression and centrosome localization of a third human aurora/Ipl1-related protein kinase, AIK3. J Biol Chem 274: 7334–7340 [DOI] [PubMed] [Google Scholar]
- Kleinboelting N, Huep G, Kloetgen A, Viehoever P, Weisshaar B (2012) GABI-Kat SimpleSearch: new features of the Arabidopsis thaliana T-DNA mutant database. Nucleic Acids Res 40: D1211–D1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotwaliwale CV, Frei SB, Stern BM, Biggins S (2007) A pathway containing the Ipl1/Aurora protein kinase and the spindle midzone protein Ase1 regulates yeast spindle assembly. Dev Cell 13: 433–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurasawa Y, Earnshaw WC, Mochizuki Y, Dohmae N, Todokoro K (2004) Essential roles of KIF4 and its binding partner PRC1 in organized central spindle midzone formation. EMBO J 23: 3237–3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas JR, Courtney S, Hassfurder M, Dhingra S, Bryant A, Shaw SL (2011) Microtubule-associated proteins MAP65-1 and MAP65-2 positively regulate axial cell growth in etiolated Arabidopsis hypocotyls. Plant Cell 23: 1889–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg AS, Weinberg RA (1998) Functional inactivation of the retinoblastoma protein requires sequential modification by at least two distinct cyclin-Cdk complexes. Mol Cell Biol 18: 753–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN (1997) Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124: 33–44 [DOI] [PubMed] [Google Scholar]
- Malik R, Nigg EA, Körner R (2008) Comparative conservation analysis of the human mitotic phosphoproteome. Bioinformatics 24: 1426–1432 [DOI] [PubMed] [Google Scholar]
- Mao G, Chan J, Calder G, Doonan JH Lloyd CW (2005a) Modulated targeting of GFP-AtMAP65-1 to central spindle microtubules during division. Plant J 43: 469–478. [DOI] [PubMed] [Google Scholar]
- Mao T, Jin L, Li H, Liu B, Yuan M (2005b) Two microtubule-associated proteins of the Arabidopsis MAP65 family function differently on microtubules. Plant Physiol 138: 654–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy CE, MacDonald A, Morrice NA, Campbell DG, Deak M, Toth R, McIlrath J, Arthur JS (2007) Identification of novel phosphorylation sites in MSK1 by precursor ion scanning MS. Biochem J 402: 491–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliam H, Li W, Uludag M, Squizzato S, Park YM, Buso N, Cowley AP, Lopez R (2013) Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Res 41: W597–W600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollinari C, Kleman JP, Jiang W, Schoehn G, Hunter T, Margolis RL (2002) PRC1 is a microtubule binding and bundling protein essential to maintain the mitotic spindle midzone. J Cell Biol 157: 1175–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller S, Smertenko A, Wagner V, Heinrich M, Hussey PJ, Hauser MT (2004) The plant microtubule-associated protein AtMAP65-3/PLE is essential for cytokinetic phragmoplast function. Curr Biol 14: 412–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylle E, Codreanu MC, Boruc J, Russinova E (2013) Emission spectra profiling of fluorescent proteins in living plant cells. Plant Methods 9: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair JS, Schwartz GK (2016) MLN-8237: A dual inhibitor of Aurora A and B in soft tissue sarcomas. Oncotarget 7: 12893–12903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef R, Gruneberg U, Kopajtich R, Li X, Nigg EA, Sillje H, Barr FA (2007) Choice of Plk1 docking partners during mitosis and cytokinesis is controlled by the activation state of Cdk1. Nat Cell Biol 9: 436–444 [DOI] [PubMed] [Google Scholar]
- Ozlü N, Monigatti F, Renard BY, Field CM, Steen H, Mitchison TJ, Steen JJ (2010) Binding partner switching on microtubules and Aurora-B in the mitosis to cytokinesis transition. Mol Cell Proteomics 9: 336–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen J, Paris J, Willer M, Philippe M, Hagan IM (2001) The S. pombe Aurora-related kinase Ark1 associates with mitotic structures in a stage dependent manner and is required for chromosome segregation. J Cell Sci 114: 4371–4384 [DOI] [PubMed] [Google Scholar]
- Petrovská B, Cenklová V, Pochylová Z, Kourová H, Doskočilová A, Plíhal O, Binarová L, Binarová P (2012) Plant Aurora kinases play a role in maintenance of primary meristems and control of endoreduplication. New Phytol 193: 590–604 [DOI] [PubMed] [Google Scholar]
- Picton C, Woodgett J, Hemmings B, Cohen P (1982) Multisite phosphorylation of glycogen synthase from rabbit skeletal muscle. Phosphorylation of site 5 by glycogen synthase kinase-5 (casein kinase-II) is a prerequisite for phosphorylation of sites 3 by glycogen synthase kinase-3. FEBS Lett 150: 191–196 [DOI] [PubMed] [Google Scholar]
- Pollard TD. (2010) A guide to simple and informative binding assays. Mol Biol Cell 21: 4061–4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portran D, Zoccoler M, Gaillard J, Stoppin-Mellet V, Neumann E, Arnal I, Martiel JL, Vantard M (2013) MAP65/Ase1 promote microtubule flexibility. Mol Biol Cell 24: 1964–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach PJ. (1990) Control of glycogen synthase by hierarchal protein phosphorylation. FASEB J 4: 2961–2968 [PubMed] [Google Scholar]
- Roghi C, Giet R, Uzbekov R, Morin N, Chartrain I, Le Guellec R, Couturier A, Dorée M, Philippe M, Prigent C (1998) The Xenopus protein kinase pEg2 associates with the centrosome in a cell cycle-dependent manner, binds to the spindle microtubules and is involved in bipolar mitotic spindle assembly. J Cell Sci 111: 557–572 [DOI] [PubMed] [Google Scholar]
- Roitinger E, Hofer M, Köcher T, Pichler P, Novatchkova M, Yang J, Schlögelhofer P, Mechtler K (2015) Quantitative phosphoproteomics of the ataxia telangiectasia-mutated (ATM) and ataxia telangiectasia-mutated and rad3-related (ATR) dependent DNA damage response in Arabidopsis thaliana. Mol Cell Proteomics 14: 556–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardon T, Pache RA, Stein A, Molina H, Vernos I, Aloy P (2010) Uncovering new substrates for Aurora A kinase. EMBO Rep 11: 977–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasabe M, Kosetsu K, Hidaka M, Murase A, Machida Y (2011) Arabidopsis thaliana MAP65-1 and MAP65-2 function redundantly with MAP65-3/PLEIADE in cytokinesis downstream of MPK4. Plant Signal Behav 6: 743–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasabe M, Soyano T, Takahashi Y, Sonobe S, Igarashi H, Itoh TJ, Hidaka M, Machida Y (2006) Phosphorylation of NtMAP65-1 by a MAP kinase down-regulates its activity of microtubule bundling and stimulates progression of cytokinesis of tobacco cells. Genes Dev 20: 1004–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher JM, Ashcroft N, Donovan PJ, Golden A (1998a) A highly conserved centrosomal kinase, AIR-1, is required for accurate cell cycle progression and segregation of developmental factors in Caenorhabditis elegans embryos. Development 125: 4391–4402 [DOI] [PubMed] [Google Scholar]
- Schumacher JM, Golden A, Donovan PJ (1998b) AIR-2: an Aurora/Ipl1-related protein kinase associated with chromosomes and midbody microtubules is required for polar body extrusion and cytokinesis in Caenorhabditis elegans embryos. J Cell Biol 143: 1635–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuyler SC, Liu JY, Pellman D (2003) The molecular function of Ase1p: evidence for a MAP-dependent midzone-specific spindle matrix. Microtubule-associated proteins. J Cell Biol 160: 517–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sells TB, Chau R, Ecsedy JA, Gershman RE, Hoar K, Huck J, Janowick DA, Kadambi VJ, LeRoy PJ, Stirling M, Stroud SG, Vos TJ, et al. (2015) MLN8054 and Alisertib (MLN8237): discovery of selective oral Aurora A inhibitors. ACS Med Chem Lett 6: 630–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smertenko AP, Chang HY, Sonobe S, Fenyk SI, Weingartner M, Bögre L, Hussey PJ (2006) Control of the AtMAP65-1 interaction with microtubules through the cell cycle. J Cell Sci 119: 3227–3237 [DOI] [PubMed] [Google Scholar]
- Smertenko AP, Chang HY, Wagner V, Kaloriti D, Fenyk S, Sonobe S, Lloyd C, Hauser MT, Hussey PJ (2004) The Arabidopsis microtubule-associated protein AtMAP65-1: molecular analysis of its microtubule bundling activity. Plant Cell 16: 2035–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smertenko AP, Kaloriti D, Chang HY, Fiserova J, Opatrny Z, Hussey PJ (2008) The C-terminal variable region specifies the dynamic properties of Arabidopsis microtubule-associated protein MAP65 isotypes. Plant Cell 20: 3346–3358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smertenko A, Saleh N, Igarashi H, Mori H, Hauser-Hahn I, Jiang CJ, Sonobe S, Lloyd CW, Hussey PJ (2000) A new class of microtubule-associated proteins in plants. Nat Cell Biol 2: 750–753 [DOI] [PubMed] [Google Scholar]
- Spitzer M, Wildenhain J, Rappsilber J, Tyers M (2014) BoxPlotR: a web tool for generation of box plots. Nat Methods 11: 121–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian R, Wilson-Kubalek EM, Arthur CP, Bick MJ, Campbell EA, Darst SA, Milligan RA, Kapoor TM (2010) Insights into antiparallel microtubule crosslinking by PRC1, a conserved nonmotor microtubule binding protein. Cell 142: 433–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaštíková E, Dmitri Demidov D, Jeřábková H, Binarová P, Houben A, Doležel J, Petrovská B (2015) TPX2 protein of Arabidopsis activates Aurora kinase 1, but not Aurora kinase 3 in vitro. Plant Mol Biol Report 33: 1988–1995 [Google Scholar]
- Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, Shokat KM, Morgan DO (2003) Targets of the cyclin-dependent kinase Cdk1. Nature 425: 859–864 [DOI] [PubMed] [Google Scholar]
- Umezawa T, Sugiyama N, Takahashi F, Anderson JC, Ishihama Y, Peck SC, Shinozaki K (2013) Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci Signal 6: rs8. [DOI] [PubMed] [Google Scholar]
- Vader G, Lens SM (2008) The Aurora kinase family in cell division and cancer. Biochim Biophys Acta 1786: 60–72 [DOI] [PubMed] [Google Scholar]
- Vantard M, Peter C, Fellous A, Schellenbaum P, Lambert AM (1994) Characterization of a 100-kDa heat-stable microtubule-associated protein from higher plants. Eur J Biochem 220: 847–853 [DOI] [PubMed] [Google Scholar]
- Van Damme D, Bouget FY, Van Poucke K, Inzé D, Geelen D (2004a) Molecular dissection of plant cytokinesis and phragmoplast structure: a survey of GFP-tagged proteins. Plant J 40: 386–398 [DOI] [PubMed] [Google Scholar]
- Van Damme D, De Rybel B, Gudesblat G, Demidov D, Grunewald W, De Smet I, Houben A, Beeckman T, Russinova E (2011) Arabidopsis α Aurora kinases function in formative cell division plane orientation. Plant Cell 23: 4013–4024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme D, Van Poucke K, Boutant E, Ritzenthaler C, Inzé D, Geelen D (2004b) In vivo dynamics and differential microtubule-binding activities of MAP65 proteins. Plant Physiol 136: 3956–3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O, Lederer C, Baulcombe DC (2000) A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103: 157–167 [DOI] [PubMed] [Google Scholar]
- Weimer AK, Demidov D, Lermontova I, Beeckman T, Van Damme D (2015) Aurora kinases throughout plant development. Trends Plant Sci 21: 69–79 [DOI] [PubMed] [Google Scholar]
- Wheatley SP, Henzing AJ, Dodson H, Khaled W, Earnshaw WC (2004) Aurora-B phosphorylation in vitro identifies a residue of survivin that is essential for its localization and binding to inner centromere protein (INCENP) in vivo. J Biol Chem 279: 5655–5660 [DOI] [PubMed] [Google Scholar]
- Xue Y, Liu Z, Cao J, Ma Q, Gao X, Wang Q, Jin C, Zhou Y, Wen L, Ren J (2011) GPS 2.1: enhanced prediction of kinase-specific phosphorylation sites with an algorithm of motif length selection. Protein Eng Des Sel 24: 255–260 [DOI] [PubMed] [Google Scholar]
- Xue Y, Zhou F, Zhu M, Ahmed K, Chen G, Yao X (2005) GPS: a comprehensive www server for phosphorylation sites prediction. Nucleic Acids Res 33: W184–W187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Sato M, Fujita A, Yamamoto M, Toda T (2005) The roles of fission yeast ase1 in mitotic cell division, meiotic nuclear oscillation, and cytokinesis checkpoint signaling. Mol Biol Cell 16: 1378–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarkowska T, Mittnacht S (1997) Differential phosphorylation of the retinoblastoma protein by G1/S cyclin-dependent kinases. J Biol Chem 272: 12738–12746 [DOI] [PubMed] [Google Scholar]
- Zeitlin SG, Shelby RD, Sullivan KF (2001) CENP-A is phosphorylated by Aurora B kinase and plays an unexpected role in completion of cytokinesis. J Cell Biol 155: 1147–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]