Oryza sativa PHO2 interacts with two h-type thioredoxins, which links phosphate homeostasis with redox balance in rice.
Abstract
Phosphate overaccumulator2 (PHO2) encodes a ubiquitin-conjugating E2 enzyme that is a major negative regulator of the inorganic phosphate (Pi)-starvation response-signaling pathway. A yeast two-hybrid (Y2H) screen in rice (Oryza sativa; Os) using OsPHO2 as bait revealed an interaction between OsPHO2 and two h-type thioredoxins, OsTrxh1 and OsTrxh4. These interactions were confirmed in vivo using bimolecular fluorescence complementation (BiFC) of OsPHO2 and OsTrxh1/h4 in rice protoplasts and by in vitro pull-down assays with 6His-tagged OsTrxh1/h4 and GST-tagged OsPHO2. Y2H assays revealed that amino acid Cys-445 of OsPHO2 and an N-terminal Cys in the “WCGPC” motif of Trxhs were required for the interaction. Split-ubiquitin Y2H analyses and BiFC assays in rice protoplasts confirmed the interaction of OsPHO2 with PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 (OsPHF1), and PHOSPHATE1;2 (OsPHO1;2) in the endoplasmic reticulum and Golgi membrane system, where OsPHO2 mediates the degradation of OsPHF1 in both tobacco (Nicotiana benthamiana) leaves and rice seedlings. Characterization of rice pho2 complemented lines, transformed with an endogenous genomic OsPHO2 or OsPHO2C445S (a constitutively reduced form) fragment, indicated that OsPHO2C445S restored Pi concentration in rice to statistically significant lower levels compared to native OsPHO2. Moreover, the suppression of OsTrxh1 (knockdown and knockout) resulted in slightly higher Pi concentration than that of wild-type Nipponbare in leaves. These results demonstrate that OsPHO2 is under redox control by thioredoxins, which fine-tune its activity and link Pi homeostasis with redox balance in rice.
Phosphorus (P) is an essential macronutrient for growth and development of plants. Although abundant P may exist in soils, inorganic phosphate (Pi), the major available form of P for plants, is often limiting for plant growth (Raghothama, 2000; Vance et al., 2003). Thus, to maintain cellular Pi homeostasis, plants have evolved a broad array of sophisticated strategies to cope with Pi-limiting conditions in soil, from transcriptional and posttranscriptional to metabolic and morphological levels to coordinate Pi sensing, signaling, uptake, allocation, and recycling coupled with the regulation of growth and development (Raghothama, 1999; Ticconi and Abel, 2004; Misson et al., 2005; Chiou and Lin, 2011).
To date one clue of our understanding of Pi starvation signaling is via the PHOSPHATE STARVATION RESPONSE/microRNA399 (miR399)/Phosphate overaccumulator2 (PHO2) regulatory system. As orthologs have been identified in various plant species and the inverse relationship of their expression patterns in Pi starvation response, this regulatory pathway is thought to be of biological importance and conserved in several plants, including both Arabidopsis (Arabidopsis thaliana; At) and rice (Oryza sativa; Os; Bari et al., 2006; Valdés-López et al., 2008; Zhou et al., 2008; Hu et al., 2011; Kuo and Chiou, 2011; Wang et al., 2013). Arabidopsis pho2 mutants accumulate excessive Pi in shoot tissue and display Pi toxicity symptoms (Delhaize and Randall, 1995; Dong et al., 1998). PHO2 encodes a ubiquitin-conjugating (UBC) E2 enzyme UBC24 and functions as a repressor to prevent Pi overaccumulation by controlling Pi uptake and root-to-shoot translocation of Pi (Fujii et al., 2005; Aung et al., 2006; Bari et al., 2006). In both Arabidopsis and rice, loss of PHO2 activity mimics a Pi starvation response, including up-regulation at the level of transcript abundance of several Phosphate Starvation Induced genes, stimulation of phosphatase and RNase activities, as well as altered lipid composition, which results in increased Pi uptake, enhanced translocation from roots to shoots, and altered Pi remobilization in leaves (Aung et al., 2006; Bari et al., 2006; Wang et al., 2009; Hu et al., 2011).
Suppression of PHO2 by miR399 overexpression leads to Pi toxicity (Aung et al., 2006; Chiou et al., 2006). Upon Pi starvation, miR399 is up-regulated by PHOSPHATE STARVATION RESPONSE1, a key transcriptional regulator of Pi starvation responses (Rubio et al., 2001; Bustos et al., 2010), and the induced miR399 targets PHO2 transcripts for cleavage (Bari et al., 2006; Chiou et al., 2006). In addition, the miR399-PHO2 pathway is regulated by target mimicry, where a noncoding RNA INDUCED BY PHOSPHATE STARVATION1 has a complementary region to miR399, thus ultimately inhibiting the miR399-mediated silencing on PHO2 mRNA (Franco-Zorrilla et al., 2007). Results of reciprocal grafting between wild-type and miR399-overexpressing plants revealed that the shoot-derived mature miR399s move through the phloem and across graft junction to suppress PHO2 in the root, indicating that the shoot-to-root movement of miR399 likely serves as a long-distance systemic signal of Pi deficiency (Lin et al., 2008; Pant et al., 2008). In Arabidopsis, AtPHO2 has been shown to modulate the degradation of PHO1, which is involved in Pi loading to the xylem (Poirier et al., 1991; Stefanovic et al., 2007) and several other Pi transporters in the PHOSPHATE TRANSPORTER1 (PHT1) family, through direct physical interaction at the endomembranes, including the endoplasmic reticulum (ER) membrane system, under Pi-replete conditions (Liu et al., 2012; Huang et al., 2013; Park et al., 2014). These findings give a mechanistic basis of how PHO2 (or more correctly, lack of PHO2) enhances Pi uptake and facilitates root-to-shoot translocation of Pi.
An alternative mRNA isoform of OsPHO2, which is referred to as OsPHO2.2 and specifically expressed upon Pi starvation, has been detected in rice (Secco et al., 2013). Sequencing results of both OsPHO2 isoforms revealed that the difference is within their 5′ untranslated region (5′UTR), where the first exon of the 5′UTR of OsPHO2.1 is replaced by two upstream fragments in OsPHO2.2, but no alteration in the protein coding sequence is predicted. OsPHO2.2 has been shown to be more actively translated into protein than isoform OsPHO2.1, because OsPHO2.1 RNA was enriched in the nonpolysomal fraction, while OsPHO2.2 was preferentially associated with polysomes (Secco et al., 2013). The functional implications of Pi starvation-induced OsPHO2.2 are unknown but reveal another possible layer of regulation of this central mediator, i.e. OsPHO2, of Pi status in rice.
In this study, another level of regulation of OsPHO2 was uncovered. Using a yeast two-hybrid (Y2H) screen, two OsPHO2-interacting proteins were identified, namely thioredoxins (Trxs) OsTrxh1 and OsTrxh4, that was confirmed using biomolecular fluorescence complementation (BiFC) visualization in rice protoplasts and in vitro pull-down. Characterization of the nature of this interaction revealed that Cys residues in both OsPHO2 and OsTrxhs are required. In addition, it was shown that OsPHO2 interacts with OsPHO1;2 (Secco et al., 2010) and PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 (OsPHF1; Chen et al., 2011) and mediates the degradation of OsPHF1. The constitutively reduced form OsPHO2C445S complemented rice pho2 more effectively than native OsPHO2, and the suppression of OsTrxh1 led to increased Pi concentration of leaves, revealing a cross-talk between Pi homeostasis and redox status in rice.
RESULTS
OsPHO2 Interacts with OsTrxh1 and OsTrxh4
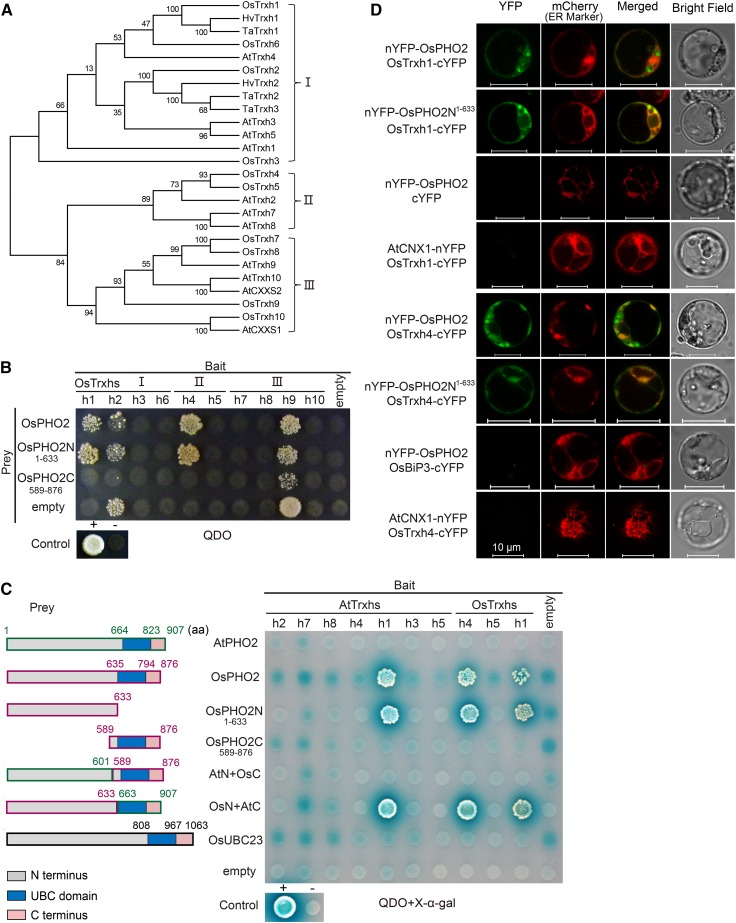
In order to identify interacting proteins of OsPHO2, Y2H assays were performed using OsPHO2 as bait to screen a rice cDNA library, resulting in the identification of several OsPHO2-interacting partners. Among these, two h-type thioredoxins, OsTrxh1 and OsTrxh4, were observed. Trxs are small thiol-disulfide oxidoreductase and ubiquitously distributed in prokaryotes and eukaryotes. Trxs contain a highly conserved redox-active domain (CXXC/S; Holmgren, 1989). Phylogenetic analyses of Trxhs from four plant organisms revealed the homologs of OsTrxh1 and OsTrxh4 in rice and Arabidopsis (Fig. 1A). To confirm whether OsPHO2 could interact with other Trxhs in rice, a direct Y2H assay was conducted using ten OsTrxh proteins as bait (Fig. 1B). Only the mated yeast cells with positive interaction of prey and bait proteins could survive on selective quadruple-dropout synthetically defined medium (QDO, SD/-Leu-Trp-His-Ade) plates. The Y2H results showed that OsPHO2 could bind to OsTrxh1 and OsTrxh4, but not bind to OsTrxh2/h3/h5/h6/h7/h8/h9/h10. OsTrxh2 and OsTrxh9 exhibited autoactivation of the selective markers, since the yeast cells harboring pGBKT7-OsTrxh2/h9 and pGADT7-Rec could grow on QDO medium (Fig. 1B).
Figure 1.
OsPHO2 interacts with OsTrxh1 and OsTrxh4. A, Phylogeny of h-type thioredoxin (Trxh) proteins from Arabidopsis (At), rice (Os), Hordeum vulgare (Hv), and Triticum aestivum (Ta). Phylogenetic analyses were conducted in MEGA6, and the tree was generated using the neighbor-joining method. Trxh proteins were divided into three subgroups as indicated. B, Identification of the interaction between OsPHO2 and rice Trxh family members by Y2H assays. For bait, OsTrxh1/h2/h3/h6 (subgroup I), OsTrxh4/h5 (II), and OsTrxh7/h8/h9/h10 (III) were used. For prey, full-length OsPHO2, OsPHO2N1–633 (N-terminal amino acids 1 to 633) and OsPHO2C589–876 (C-terminal amino acids 589 to 876) were used. Yeast transformants were cultured on selection medium QDO (SD/-Leu-Trp-His-Ade). Positive control (+), pGAD-SV40 mated with pGBK-53; negative control (−), pGAD-SV40 mated with pGBK-Lam; empty, pGBKT7 or pGADT7-Rec without any insert. C, Y2H assays to probe for interaction between PHO2 and Trxh proteins of rice and Arabidopsis. OsTrxh1, OsTrxh4, and their homologs in Arabidopsis were used as bait. For prey, full-length AtPHO2, full-length OsPHO2 or OsPHO2N1–633, OsPHO2C589–876, and chimeric PHO2 proteins, AtPHO2 N-terminal fused with OsPHO2 C-terminal (AtN+OsC), OsPHO2 N-terminal fused with AtPHO2 C-terminal (OsN+AtC), and the OsPHO2 homologous protein OsUBC23 were used. Yeast cells were cultured on selection medium QDO and using X-α-gal as substrate for colorimetric detection of α-galactosidase activity. Blue box indicates the UBC domain. D, BiFC visualization of the interaction between OsPHO2 and OsTrxh1/h4 in rice protoplasts. Images were captured 12 to 15 h after polyethylene glycol-mediated transformation under a ZEISS LSM710nlo confocal laser scanning microscope. The ER marker was AtWAK2 (Nelson et al., 2007). The combinations of nYFP-OsPHO2 with empty vector cYFP, AtCNX1-nYFP with OsTrxh1-cYFP, nYFP-OsPHO2 with OsBiP3-cYFP, and AtCNX1-nYFP with OsTrxh4-cYFP, respectively, served as negative controls. Bars = 10 μm.
To investigate whether the UBC domain that functions as an E2 enzyme is involved in interaction, OsPHO2 was truncated into OsPHO2N1–633 (N-terminal amino acids 1 to 633) and OsPHO2C589–876 (C-terminal amino acids 589 to 876) as prey. OsUBC23, the homologous protein of OsPHO2 in rice, and the chimeric PHO2 proteins, AtN+OsC (AtPHO2 N-terminal fused with OsPHO2 C-terminal) and OsN+AtC (OsPHO2 N-terminal fused with AtPHO2 C-terminal), were constructed and tested for interaction as well. The Y2H results showed that OsTrxh1 and OsTrxh4 only interacted with prey containing OsPHO2N1–633, but not OsPHO2C589–876, OsUBC23, or AtN+OsC (Fig. 1, B and C). This indicated that the binding motif was located in the N-terminal 1 to 633 amino acids of OsPHO2 and the UBC domain was not involved in the interaction. Noticeably, while PHO2 displays 45% identity and 62% similarity between rice and Arabidopsis, the highest levels of identity are observed in the UBC domain that is not involved in binding (Supplemental Fig. S1, A and B).
To confirm the OsPHO2-OsTrxhs protein interaction occurred in vivo, a BiFC assay was performed using rice protoplasts isolated from stems and leaves of etiolated seedlings (Fig. 1D). Meanwhile, the subcellular localization of OsPHO2 and two OsTrxhs was assessed using green or yellow fluorescence protein (GFP or YFP)-fused proteins, resulting in OsPHO2 being mainly located to the ER, OsTrxh1 widely distributed in the intracellular regions including the cytoplasm and nucleus, and OsTrxh4 mainly in the cytoplasm and ER (Supplemental Fig. S2). Moreover, OsTrxh1 could be secreted into the extracellular regions (Zhang et al., 2011), implying its origins in the rough ER. Using OsPHO2 and OsPHO2N1–633 fused to the N-terminal YFP fragment (nYFP-OsPHO2 and nYFP-OsPHO2N1–633) and OsTrxh1/h4 fused to the C-terminal YFP fragment (OsTrxh1-cYFP and OsTrxh4-cYFP), the YFP signal could be restored, mostly overlapping with the mCherry signal of the ER marker Arabidopsis wall-associated kinase (AtWAK2), confirming in vivo interaction between OsPHO2 and OsTrxh1/h4 (Fig. 1D). In contrast, the negative controls consisting of nYFP-OsPHO2 and the empty cYFP vector, as well as CALNEXIN1 (AtCNX1)-nYFP and OsTrxh1-cYFP, nYFP-OsPHO2 and Binding protein3 (OsBiP3)-cYFP, and AtCNX1-nYFP and OsTrxh4-cYFP, failed to produce any YFP signal (Fig. 1D). AtCNX1 is a known ER membrane protein (Huang et al., 1993; Helenius et al., 1997), and OsBiP3 is a known ER lumen chaperone protein (Park et al., 2010).
Taken together, these results showed that OsPHO2 interacts with OsTrxh1 and OsTrxh4 in vivo. Moreover, the protein levels of OsTrxh1 and OsTrxh4 remained relatively constant when they were coexpressed with OsPHO2 in tobacco (Nicotiana benthamiana) leaves (Supplemental Fig. S3), indicating that OsPHO2 did not mediate the degradation of OsTrxh1 or OsTrxh4.
Cys Residue 445 of OsPHO2 and the “WCGPC” Motif of Trxhs Are Required for Interaction of OsPHO2 with OsTrxh1 and OsTrxh4
Cys residues in the “WCGPC” motif of Trxhs are the active amino acids for redox regulation. The reduced form Trx-(SH)2, can reduce disulfide bonds of target proteins and convert into the oxidized form Trx-S2, which contains a disulfide linkage within the active site and is reduced to Trx-(SH)2 by NADPH and Trx reductases (Kallis and Holmgren, 1980; Carrie et al., 2008).
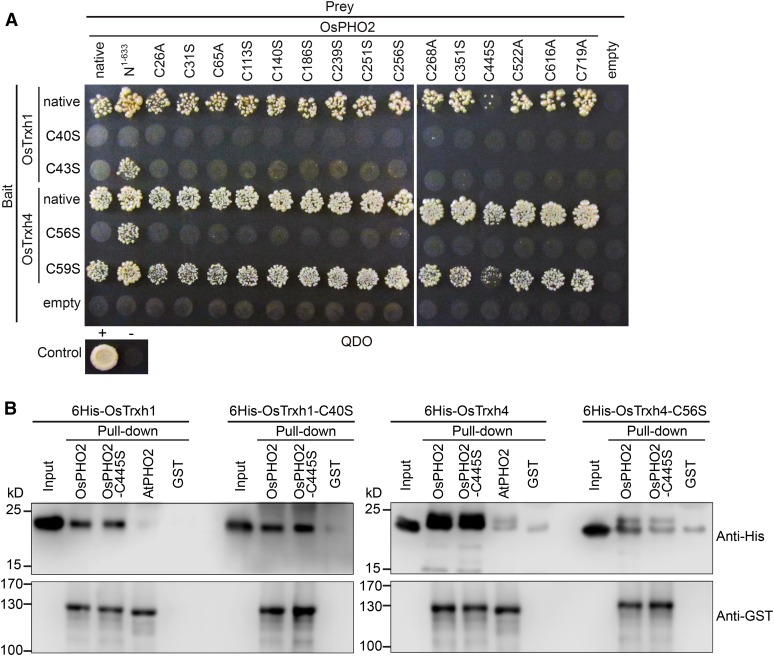
To investigate whether these two Cys residues in the “WCGPC” motif of OsTrxh1/h4 are involved in the interaction with OsPHO2 and if Cys residues of OsPHO2 play a role in binding to OsTrxh1/h4, Y2H assays were carried out using OsPHO2 and OsTrxh1/h4 mutants with site-directed mutations of Cys residues (Fig. 2A). Cys residues in the “WCGPC” motif of OsTrxh1/h4 (highlighted in Supplemental Fig. S1C) were changed to Ser and indicated as C40S, C43S, C56S, and C59S for bait, respectively. On the other hand, Cys residues in the N terminus of OsPHO2 (highlighted in Supplemental Fig. S1B), were changed to Ala or Ser, and indicated as C26A, C31S, C65A, C113S, C140S, C186S, C251S, C256S, C268A, C351S, C445S, C522A, C616A, and C719A for prey, respectively. The Y2H results (Fig. 2A) showed that for OsPHO2 mutations, only OsPHO2-C445S resulted in impaired interaction with OxTrxh1 and OsTrxh4. For the OsTrxh1 mutations, OsTrxh1-C40S could not interact with native OsPHO2, OsPHO2 mutants, or OsPHO2N1–663, while C43S failed to interact with OsPHO2 or OsPHO2 mutants, although was still able to interact with OsPHO2N1–663. For OsTrxh4, site mutation of C56S suppressed the interaction with OsPHO2 and OsPHO2 mutants except with OsPHO2N1–663. However, OsTrxh4-C59S could still interact with all OsPHO2 alleles, though the interaction with OsPHO2-C445S was impaired. These suggested that the N terminus Cys in the “WCGPC” motif of OsTrxh1/h4 and the Cys-445 of OsPHO2 are important for the interaction of OsPHO2 with OsTrxh1 and OsTrxh4.
Figure 2.
The Cys-445 of OsPHO2 and the N terminus Cys in “WCGPC” motif of Trxhs are important for the interaction of OsPHO2 with OsTrxh1 and OsTrxh4. A, Y2H analyses of the Cys site-mutated OsPHO2 with OsTrxh1/h4. For prey, the Cys residues were changed to Ala or Ser and indicated as C26A, C31S, C65A, C113S, C140S, C186S, C251S, C256S, C268A, C351S, C445S, C522A, C616A, and C719A, respectively. For bait, the Cys residues in the “WCGPC” motif of OsTrxh1/h4 were changed to Ser and indicated as C40S, C43S, C56S, and C59S, respectively. The OsPHO2, OsTrxh1, and OsTrxh4 without mutation were indicated as “native,” respectively. B, In vitro interaction of 6His-tagged OsTrxh1, OsTrxh1-C40S, OsTrxh4, and OsTrxh4-C56S with GST-tagged OsPHO2, OsPHO2-C445S, and AtPHO2. GST alone was used as a control for nonspecific binding. Anti-His and anti-GST antibodies were used to detect OsTrxh1/h4 and PHO2 proteins, respectively.
In vitro pull-down assays were conducted, using GST-tagged OsPHO2/OsPHO2-C445S, and 6His-tagged OsTrxh1/OsTrxh1-C40S/OsTrxh4/OsTrxh4-C56S. The results confirmed the interaction between OsPHO2 and OsTrxh1/h4 and, furthermore, confirmed that the mutation of C56S in OsTrxh4 impaired direct protein-protein interaction with OsPHO2 or OsPHO2-C445S (Fig. 2B). Surprisingly, the interaction of OsTrxh1-C40S with OsPHO2 was not suppressed, and OsPHO2-C445S could still bind to OsTrxh1 and OsTrxh4.
Arabidopsis PHO2 Does Not Interact with Trxs
The interaction of AtPHO2 was tested for interactions with Trxhs using three subgroup I members AtTrxh2/h7/h8, and four subgroup II members, AtTrxh1/h3/h4/h5, as well as OsTrxh1/h4, as bait in the Y2H assays (Fig. 1C). No detectable interactions between AtPHO2 and OsTrxh1, OsTrxh4, or AtTrxhs were observed, implying that the interaction of OsPHO2 with OsTrxh1/h4 is unique in rice. This is supported by in vitro pull-down assays demonstrating that OsTrxh1 and OsTrxh4 could bind to OsPHO2, but not to AtPHO2 (Fig. 2B). Interestingly, OsPHO2 also interacted with one Arabidopsis Trx protein AtTrxh1 (Fig. 1C).
Complementation of Rice pho2 Mutant
In order to complement rice pho2 mutant (Tos17 insertion line NE6022; Supplemental Fig. S4), a 9.8-kb genomic region encompassing OsPHO2 or OsPHO2C445S (a C445S mutant) was transformed into rice pho2 background. Fifteen OsPHO2/pho2 and fourteen OsPHO2C445S/pho2 independent complementation lines were used for further analyses (Fig. 3).
Figure 3.
Expression of OsPHO2 and OsPHO2C445S under the native OsPHO2 promoter could complement the rice pho2 mutant. A, The Pi concentration in leaves of rice pho2 recovery lines, transformed with endogenous genomic OsPHO2 sequence with or without C445S site mutation, under control of endogenous OsPHO2 promoter, respectively. Rice seedlings were grown in a Pi-sufficient solution (0.32 mm Pi) for 24 d. Nip, Wild-type Nipponbare. Fifteen OsPHO2/pho2 and fourteen OsPHO2C445S/pho2 complemented lines were analyzed. For each line, the average fourth leaf Pi concentration of three seedlings were used. The Pi concentration are presented as box plots (n = 9, 9, 15, and 14, respectively). The median is marked by a black line within the box. The boundaries of the boxes indicate the 25th and 75th percentiles. Errors bars indicate the 10th and 90th percentiles. Each data of individual complemented line outside the 10th and 90th percentiles is presented as a single dot. Data significantly different from the pho2 and between indicated pairs are marked by asterisks (Tukey’s ANOVA test; *P < 0.05, **P < 0.01, ***P < 0.001). No significant difference between OsPHO2C445S/pho2 lines and Nip. FW, Fresh weight. B, Growth performance of 24-d-old rice pho2 recovery lines with the similar expression level of OsPHO2 as Nip. Four OsPHO2/pho2 and four OsPHO2C445S/pho2 recovery lines were taken pictures. Bars = 4 cm.
The Pi concentration was assessed in leaves of wild type Nipponbare (Nip), pho2 mutant, and complemented lines. It was observed that the leaf Pi concentrations in all OsPHO2/pho2 and OsPHO2C445S/pho2 plants were significantly decreased, compared with that in rice pho2 (Fig. 3A). Also, the growth performance of transgenic lines with similar OsPHO2 expression level compared to Nip showed that both OsPHO2 isoforms could complement rice pho2, based on reversion of Pi toxicity symptoms (Fig. 3B). Notably, the leaf Pi concentration of OsPHO2C445S complemented lines was similar to wild-type Nip (no significant difference); however, in OsPHO2/pho2 plants, the average Pi levels in leaves were still significantly higher than that of Nip or OsPHO2C445S/pho2 plants (Fig. 3A). These results indicated that the mutated OsPHO2C445S, the constitutively reduced form, was more effective at restoring Pi levels than native OsPHO2, when it was directed by endogenous OsPHO2 promoter.
OsPHO2 Interacts with OsPHO1;2 and OsPHF1
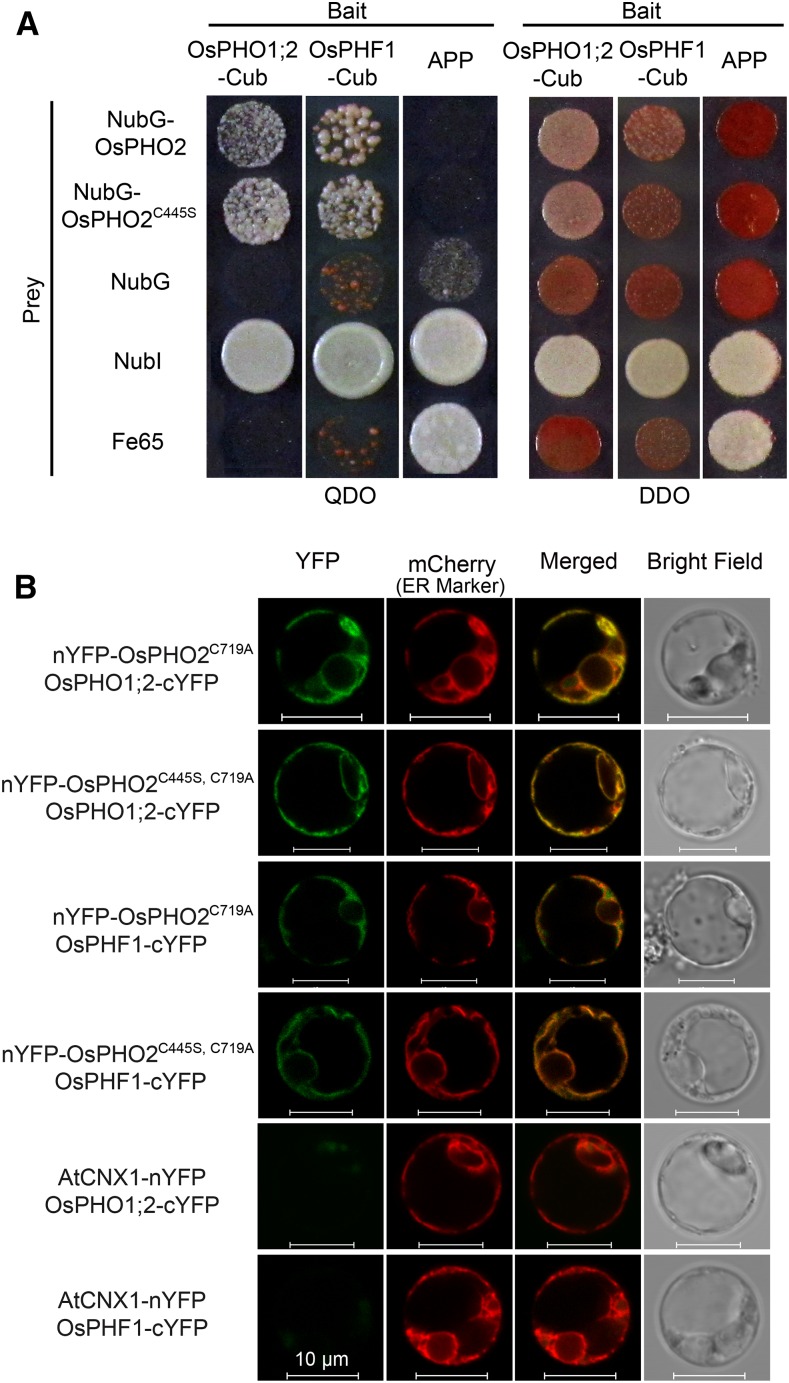
In Arabidopsis, AtPHO2 has been shown to modulate the degradation of AtPHO1 and AtPHT1;1/4, through direct interaction at the endomembranes, including the ER membrane system (Liu et al., 2012; Huang et al., 2013; Park et al., 2014). Also, the protein abundance of AtPHF1, which regulates the localization of PHT1 from ER to the plasma membrane (Bayle et al., 2011), was increased in pho2 mutants (Huang et al., 2013), indicating that PHF1 might be the target of PHO2. To determine whether OsPHO2 directly interacts with OsPHF1 (Chen et al., 2011), OsPHO1;2 (Secco et al., 2010), and Pi transporters OsPTs, BiFC assays in rice protoplasts and split-ubiquitin Y2H assays were conducted. In the BiFC assays, the conserved catalytic active residue Cys-719 in the UBC domain of OsPHO2 was changed to Ala (OsPHO2C719A) to disrupt the UBC activity. The pho2 seedlings were used to produce the rice protoplasts to avoid the effect of endogenous OsPHO2 on the interaction with potential targets.
For the split-ubiquitin Y2H assays between OsPHO2 and OsPHF1/OsPHO1;2 (Fig. 4A), OsPHF1 and OsPHO1;2 were fused with C-terminal ubiquitin (Cub) as bait, and OsPHO2 fused with N-terminal of mutated ubiquitin with Gly-3 (NubG) as prey. Coexpression of N-terminal of wild-type ubiquitin with Ile-3 (NubI) or NubG with baits was used as positive and negative controls, respectively. Also, the bait amyloid A4 precursor protein (APP) could interact with NubI and amyloid β-A4 precursor protein-binding family B member 1 (Fe65) but not with NubG and NubG-OsPHO2. It was shown that OsPHO2 could interact with OsPHF1 and OsPHO1;2 to activate the expression of reporter genes, while the negative controls did not survive on the selective medium QDO (Fig. 4A).
Figure 4.
OsPHO2 interacts with OsPHO1;2 and OsPHF1. A, Split-ubiquitin Y2H analysis of the interaction between OsPHO2/OsPHO2C445S and OsPHF1/OsPHO1;2. Coexpression of NubI or NubG with OsPHO1;2 or OsPHF1 was used as positive and negative controls, respectively. Also, the bait APP served as a control that could interact with NubI and Fe65, but not with NubG and NubG-OsPHO2. QDO, SD/-Leu-Trp-His-Ade; DDO, SD/-Leu-Trp. B, BiFC visualization of the interaction between OsPHO2 and OsPHF1/OsPHO1;2 in rice pho2 protoplasts. Coexpression of AtCNX1-nYFP and OsPHF1-cYFP or OsPHO1;2-cYFP served as negative controls. The ER marker was AtWAK2-mCherry. Bars = 10 μm.
To assess the subcellular colocalization of OsPHO2 and OsPHF1/OsPHO1;2, the YFP- or mCherry-fused constructs were used and transformed into rice protoplasts. The ER and Golgi markers are AtWAK2 and Glycine max mannosyl-oligosaccharide 1,2-α-mannosidase (GmMan1), respectively (Nelson et al., 2007). It was seen that YFP signal of OsPHO2 was overlapping with mCherry signal of both ER and Golgi marker, meanwhile overlapping with the mCherry signal of OsPHO1;2 and OsPHF1 (Supplemental Fig. S5). For the BiFC assays, OsPHO2C719A or OsPHO2C445S, C719A fused to nYFP was colocalized with OsPHF1 and OsPHO1;2 fused to cYFP in rice protoplasts (Fig. 4B). The AtCNX1-nYFP was used as a negative control. The BiFC results further showed the interaction between OsPHO2 and OsPHF1/OsPHO1;2 in the ER and Golgi membrane system (Fig. 4B). Moreover, the C445S mutation in OsPHO2 did not affect the interaction between OsPHO2 and OsPHF1/OsPHO1;2 (Fig. 4).
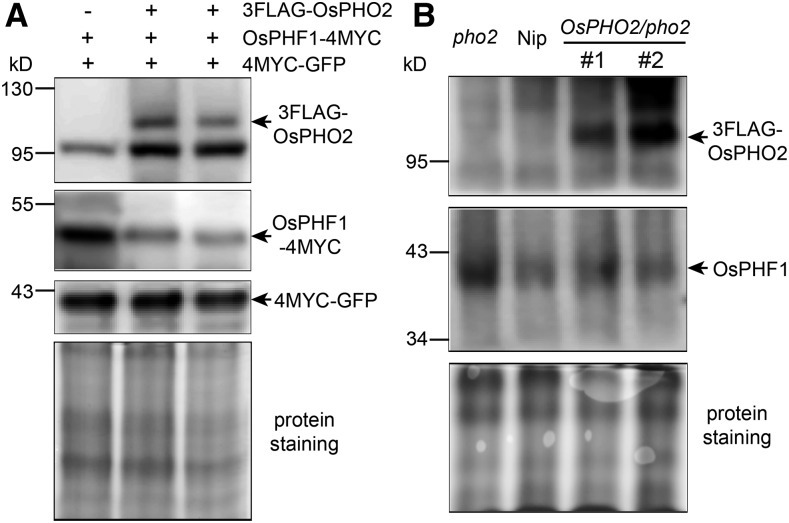
In order to investigate whether OsPHO2 is involved in the degradation of OsPHF1, OsPHO2 fused to 3FLAG was coexpressed with 4MYC-tagged OsPHF1 in tobacco leaves using Agrobacterium infiltration. Immunoblot analysis using tag-specific antibodies revealed that the protein level of OsPHF1-4MYC decreased when it was coexpressed with 3FLAG-tagged OsPHO2 (Fig. 5A). The role of OsPHO2 in mediating the degradation of OsPHF1 was also investigated in wild-type Nip, rice pho2 and OsPHO2/pho2 recovery plants. Using anti-OsPHF1 antibodies, it was seen that the OsPHF1 protein accumulated in pho2 mutant and declined to normal level in OsPHO2/pho2 recovery lines, compared with that in wild type (Fig. 5B).
Figure 5.
OsPHO2 modulates the degradation of OsPHF1. A, Protein levels of 3FLAG-tagged OsPHO2 with 4MYC-tagged OsPHF1 in transiently expressed tobacco (Nicotiana benthamiana) leaves. The tag-specific antibodies were used in the immunoblot analysis. The amount of OsPHF1-4MYC protein was reduced when it was coexpressed with 3FLAG-OsPHO2. Coinfiltration of the 4MYC-GFP construct was used as an internal control. B, Immunoblot analysis of OsPHF1 and 3FLAG-OsPHO2 proteins in seedlings of rice wild-type Nip, pho2 mutant and two OsPHO2/pho2 recovery lines. Anti-FLAG and anti-OsPHF1 antibodies were used. The protein staining (bottom) was used as a loading control.
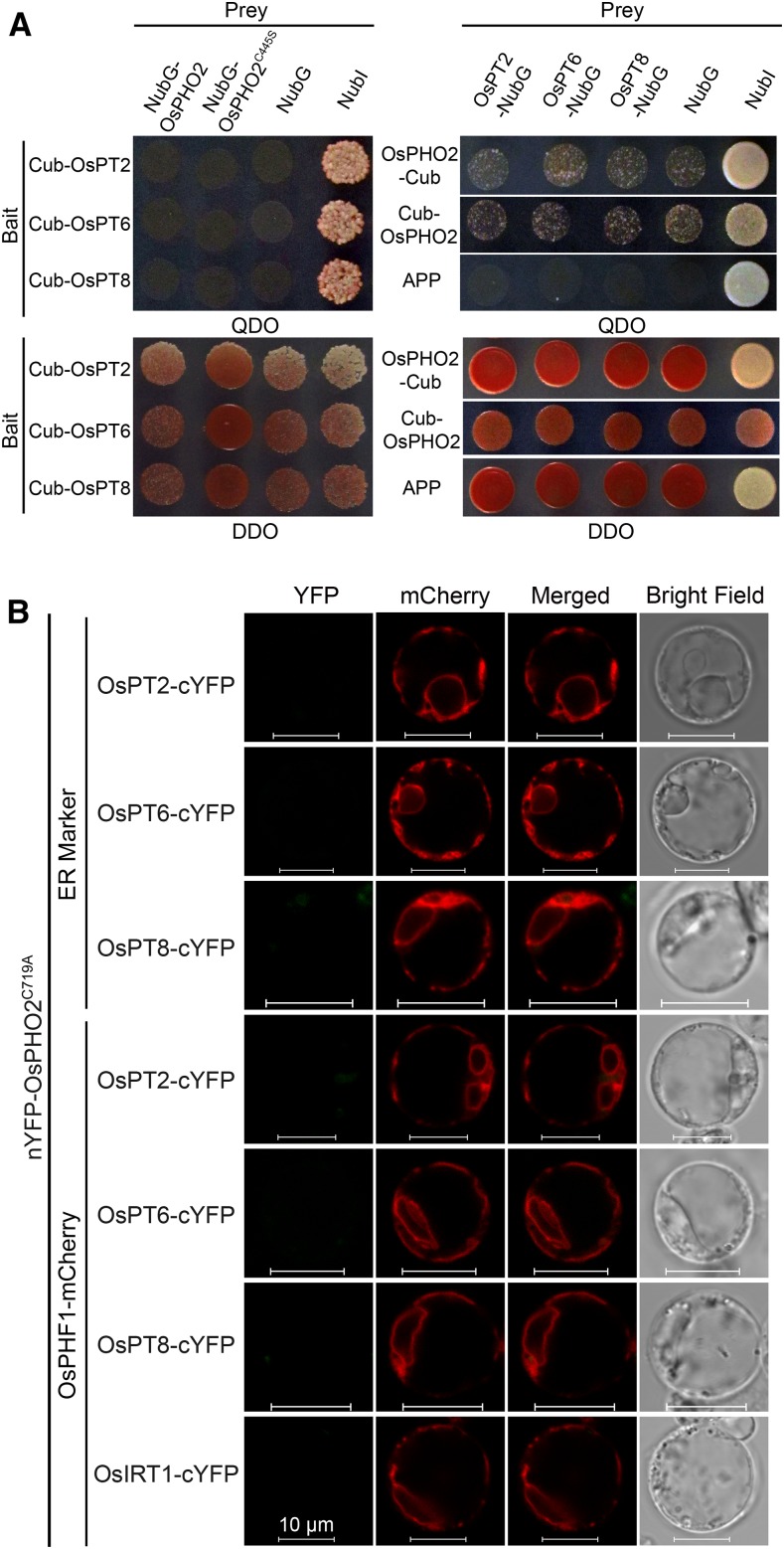
For OsPTs, OsPT2, OsPT6, and OsPT8 were used. In split-ubiquitin Y2H assays, it was seen that OsPT2/6/8-Cub exhibited strong autoactivation activities of reporter genes (Supplemental Fig. S6). Using OsPT2/6/8-NubG as prey or Cub-OsPT2/6/8 as bait, no interaction between OsPHO2 and OsPT2/6/8 was detected (Fig. 6A). The BiFC assays were carried out in rice protoplasts. IRON-REGULATED TRANSPORTER1 (OsIRT1) fused to cYFP was used as negative control. It was seen that the coexpression of nYFP-OsPHO2C719A and OsPT2/6/8-cYFP, with or without the OsPHF1-mCherry, did not produce any detectable YFP signal (Fig. 6B).
Figure 6.
Interaction assays between OsPHO2 and OsPT2/6/8. A, Split-ubiquitin Y2H analysis detected no interaction between OsPHO2 and OsPT2/6/8. Coexpression of baits with NubG or NubI were used as negative or positive controls, respectively. Also, coexpression of NubG preys with APP were used as negative controls. B, BiFC assays showed that OsPHO2 did not interact with OsPT2/6/8 in rice pho2 protoplasts. AtWAK2-mCherry (ER marker) or OsPHF1-mCherry was cotransformed. Coexpression of nYFP-OsPHO2C719A and OsIRT1-cYFP was used as a negative control. Bars = 10 μm.
Knockdown or Knockout of OsTrxh1 Leads to Higher Pi Concentration in Leaves
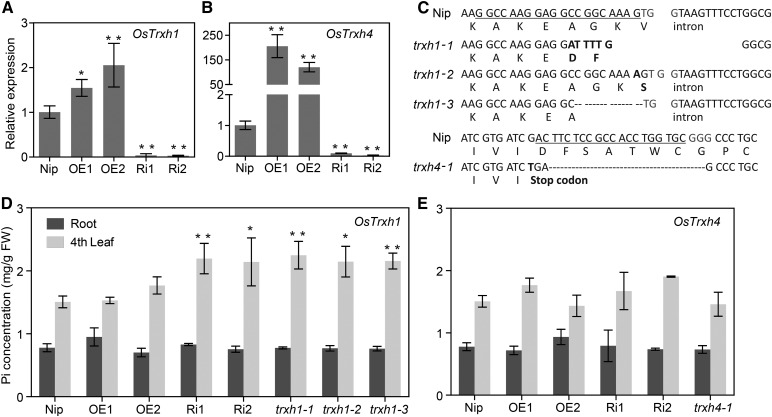
The effects of altering expression of OsTrxh1/h4 on Pi homeostasis were tested by generating the overexpression (OE), RNA interference (Ri), or mutant transgenic lines for OsTrxh1 and OsTrxh4, respectively (Fig. 7). Overexpression and knockdown efficiency of OsTrxh1 and OsTrxh4 were confirmed by quantitative real-time PCR (qRT-PCR) analyses (Fig. 7, A and B). Two independent transgenic lines of each OE/Ri construct transformation were selected for further experimental analysis. Under Pi-sufficient condition, OsTrxh1-OE plants showed only 1.5 and 2 times enhanced expression of OsTrxh1 due to the high abundance of OsTrxh1 transcripts in nontransgenic Nip (as shown by microarray data from the GENEVESTIGATOR; Supplemental Fig. S7), whereas the transcript abundance reduced significantly in OsTrxh1-Ri plants (Fig. 7A). On the other hand, transcript abundance of OsTrxh4 increased dramatically in leaves of OsTrxh4-OE lines and decreased significantly in OsTrxh4-Ri plants, compared to that in Nip (Fig. 7B). Mutant plants of OsTrxh1 and OsTrxh4 were obtained by Clustered Regularly Interspersed Short Palindromic Repeats (CRISPR)/Cas9 DNA editing system. Three trxh1 and one trxh4 homozygous mutant lines were identified by sequencing (Supplemental Fig. S8). The trxh1-1 has a very complicated mutation with total 16-bp deletion. The trxh1-2 has a 1-bp insertion, and trxh1-3 has an 8-bp deletion. The trxh4-1 has a 1-bp insertion and a 21-bp deletion, resulting in a stop codon (Fig. 7C). Morphological appearance of all these transgenic plants is presented in Supplemental Figure S9.
Figure 7.
Characterization OE, Ri, and CRISPR/Cas9 mutants of OsTrxh1 and OsTrxh4. Rice seedlings were grown in a Pi-sufficient solution (0.32 mm Pi) for 28 d. A and B, Identification of OE and Ri lines of OsTrxh1 and OsTrxh4. The expression of OsTrxh1/h4 in nontransgenic Nip and transgenic plants was analyzed by qRT-PCR. Expression of OsACTIN1 served as the internal control. C, Identification of trxh1 and trxh4 mutants. Four independent homozygous mutant lines were obtained. The trxh1-1 has a very complicated mutation with a 22-bp deletion and a 6-bp substitution, overall a 16-bp deletion. The trxh1-2 has a 1-bp insertion, and trxh1-3 has an 8-bp deletion. The trxh4-1 mutant has a 1-bp insertion and a 21-bp deletion, resulting in a stop codon. The underlined sequences are the target sites for CRISPR/Cas9 editing; the three bps in gray indicate the protospacer adjacent motif sequences. D and E, The roots and fourth leaf were sampled to determine Pi concentration. FW, Fresh weight. For A, B, D, and E, errors bars indicate sd and n = 3. Data significantly different from the Nip controls were indicated by asterisks (Dunnett’s ANOVA test; *P < 0.05, **P < 0.01).
Measurement of the Pi concentration in root of Nip and the OE, Ri, or mutant lines of OsTrxh1 and OsTrxh4 revealed no significant difference compared with that of Nip (Fig. 7, D and E). There was also no significant difference of the Pi levels in the fourth leaf between the Nip and OsTrxh1-OE, OsTrxh4-OE, or OsTrxh4-Ri plants (Fig. 7, D and E). However, the Pi concentration in the fourth leaf of two OsTrxh1-Ri lines and three trxh1 lines were significantly higher (1.4 to 1.5 times) than that of Nip (Fig. 7D), indicating that a functional OsTrxh1 is required to maintain normal Pi levels.
DISCUSSION
In this study, OsPHO2, an E2 enzyme of the protein ubiquitination system and a negative regulator of Pi uptake and translocation, was shown to interact with two Trxh proteins, OsTrxh1 and OsTrxh4, in rice. The Y2H and BiFC data indicated that the binding motif of OsPHO2 was located in the OsPHO2N1–663 (N-terminal 1 to 633 amino acids) and the UBC domain at the C terminus was not involved in the interaction (Fig. 1). Furthermore, OsPHO2 could not target OsTrxh1 and OsTrxh4 for degradation in tobacco leaves (Supplemental Fig. S3). The Trx proteins control the thiol-disulfide exchange of target proteins functional in various cellular processes. Taken together, it suggests that OsPHO2 is the downstream target of OsTrxh1/h4, but OsTrxh1/h4 are not the targets of OsPHO2.
In regard to the tissue-specific expression analysis, the transcriptional activities of OsPHO2 and OsTrxh1 have been investigated by qRT-PCR and promoter-reporter assays in the previous studies. Both OsPHO2 and OsTrxh1 are ubiquitously expressed in all tissues, such as roots, leaves, culms, and grain husks (Hu et al., 2011; Zhang et al., 2011). In particular, the OsPHO2 is preferentially expressed in the vascular systems of all tissues with highest level in roots (Hu et al., 2011). Coincidently, the expression of OsTrxh1 can be detected in the vascular tissues of roots, leaves, culms, and grain husks (Zhang et al., 2011). This implies the similar spatial-temporal expression pattern of OsPHO2 and OsTrxh1.
The protein-protein interaction assays clearly showed a difference between OsPHO2 and AtPHO2 in terms of interaction with Trxhs, in that no interaction of AtPHO2 could be detected with Trxhs (Figs. 1 and 2B), suggesting different regulatory factors affect OsPHO2 compared with AtPHO2. To test this hypothesis, cross-complementation of Atpho2 and Ospho2 mutants was carried out with two constructs expressing 35S::AtPHO2 and 35S::OsPHO2, respectively (Supplemental Figs. S10 and S11). The ability of the different constructs to complement the pho2 mutation in Arabidopsis and rice was tested using growth performance and measuring the shoot Pi concentration in complemented lines. The shoot Pi concentration and growth performance showed that the Pi toxicity phenotype could be rescued in Arabidopsis with overexpression of AtPHO2 (35S::AtPHO2/Atpho2), while overexpression of OsPHO2 (35S::OsPHO2/Atpho2) did not rescue the growth phenotype of Atpho2 mutant (Supplemental Fig. S10). Five of eight 35S::AtPHO2/Atpho2 lines showed significantly reduced shoot Pi concentration compared to Atpho2, and even lower Pi concentration than that of wild-type Col-0, as might be expected from overexpression of this negative regulator of Pi acquisition (Supplemental Fig. S10). In contrast, for the Ospho2 mutant, overexpression of AtPHO2 (35S::AtPHO2/Ospho2) was unable to complement the growth phenotype caused by shoot Pi accumulation in rice pho2 (Supplemental Fig. S11). This confirms a difference in function between AtPHO2 and OsPHO2 as hypothesized from the interaction assays. Considering the presence of OsPHO2.2, an alternative mRNA isoform of OsPHO2 with altered splicing at the 5′UTR (Secco et al., 2013), together with our results, it suggested that the regulation difference between AtPHO2 and OsPHO2 is not only at the transcriptional level but also at the posttranslational level.
In Arabidopsis, it has been shown that AtPHO2 mediates the degradation of AtPHT1;1/4 and AtPHO1 through direct interaction and ubiquitination, subsequently to suppress both Pi acquisition and xylem loading of Pi (Liu et al., 2012; Huang et al., 2013; Park et al., 2014). However, it has not been confirmed that AtPHO2 could interact with AtPHF1 (Huang et al., 2013), which is required for PHT1 proteins transiting through the ER to plasma membrane (Bayle et al., 2011). In this study, we found that OsPHO2 did interact with OsPHO1;2 (which is consistent with AtPHO2) and OsPHF1 (which has not been demonstrated for AtPHO2) in the ER and Golgi (Fig. 4; Supplemental Fig. S5) and modulated the degradation of OsPHF1 (Fig. 5). Another noticeable point is that no detectable interactions between OsPHO2 and OsPT2/6/8 were observed by both split-ubiquitin Y2H analyses and BiFC assays in rice protoplasts (Fig. 6), which is different from AtPHO2.
The N-terminal Cys of the Trx is essential for disulfide binding. In the reduced state, the N-terminal Cys of the Trx active site motif targets disulfide bonds in protein substrates with formation of an intermolecular disulfide intermediate. A subsequent attack by the C-terminal Cys of the active site releases the reduced target protein and the oxidized Trx (Kallis and Holmgren, 1980; Montrichard et al., 2009; Shahpiri et al., 2009). The Y2H and pull-down assays using site-mutated OsPHO2 and OsTrxh1/h4 suggest that the N terminus Cys in “WCGPC” motif of Trxhs and the Cys-445 of OsPHO2 are required for the interaction between OsPHO2 and OsTrxh1/h4 (Fig. 2). However, it was revealed by in vitro pull-down assays that the mutation of OsPHO2-C445S and OsTrxh1-C40S did not affect the interactions. We speculated that the discrepancy was due to the alteration of protein folding between yeast and Escherichia coli cells. The prokaryotic OsTrxh1-C40S protein might still bind to OsPHO2 since it contains another amino acid Cys-11 (Supplemental Fig. S1C). For the OsPHO2, the second Cys that is involved in disulfide binding was not identified by Y2H assays, whereas the presence of this Cys enabled the interaction of OsPHO2-C445S with OsTrxh1/h4 in the pull-down assays. Overall, the Y2H and pull-down results implied that OsPHO2 likely has reduced/oxidized forms and its redox status is regulated by OsTrxh1/h4. We hypothesized that OsPHO2C445S mimics the reduced OsPHO2, the more active form.
The characterization of rice pho2 complementation lines demonstrated that the Pi concentration in OsPHO2C445S/pho2 lines was lower than that of OsPHO2/pho2 lines (Fig. 3A), suggesting that the OsPHO2C445S is more effective at restoring Pi levels than native OsPHO2. One might notice that some pho2 complementation lines transformed by native OsPHO2 still displayed relatively higher leave Pi concentration than wild-type Nip. We assumed that the expression level of OsPHO2 was not in the balance with the activity of OsTrxh1/h4 in these lines, and then the decrement of reduced OsPHO2 resulted in Pi accumulation in leaves. The protein-protein interaction assays showed that the C445S mutation did not affect the interaction between OsPHO2 and OsPHF1/OsPHO1;2 (Fig. 4). In addition, the reduced expression of OsTrxh1 in rice led to increased Pi accumulation in leaves under Pi-sufficient conditions (Fig. 7D). This is proposed to be due to the fact that the lack of OsTrxh1 results in an oxidized OsPHO2 that is not as active, in the degradation of OsPHF1/OsPHO1;2. Noticeably, the leaf Pi concentration did not change in OsTrxh4 suppression lines compared with Nip (Fig. 7E). The microarray data collected from the GENEVESTIGATOR showed that the expression of OsTrxh4 is much lower than that of OsTrxh1 in different tissues and development stages in rice (Supplemental Fig. S7). We concluded that the high and ubiquitous expression of OsTrxh1 could complement the decreased expression of OsTrxh4; however, when the OsTrxh1 is down-expressed, the OsTrxh4 could not complement for its function due to the more restricted expression pattern of OsTrxh4.
Analysis of the corresponding Cys-445 amino acid of OsPHO2 among multiple monocot and dicot plant species revealed that most plants contain Cys at the corresponding site of PHO2 proteins, whereas it is Ser in Arabidopsis, Capsella rubella (a very close relative of Arabidopsis), and Physcomitrella patens (Supplemental Fig. S12). This suggested that the Cys of PHO2 at this site is conserved in the monocots and most of the dicots, however, the AtPHO2 is different. Nevertheless, whether PHO2 protein interacts with Trxhs in other plants remains to be determined.
CONCLUSION
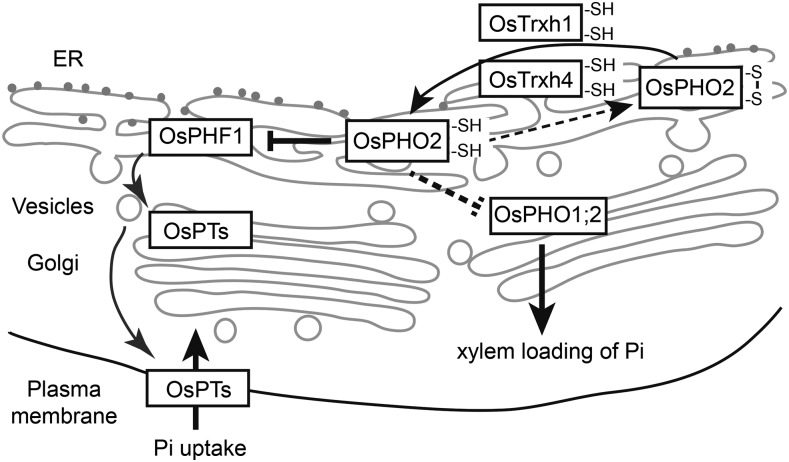
In conclusion, we propose a molecular mechanism of how OsPHO2 is regulated by OsTrxh1 and OsTrxh4 at the posttranslational level to modulate the Pi homeostasis in rice (Fig. 8). Under Pi-sufficient conditions, OsPHO2 is localized to the ER and Golgi, where it interacts with OsPHF1 and OsPHO1;2 and mediates the protein degradation of OsPHF1 and in turn regulates Pi homeostasis. We hypothesize that the reduced form of OsPHO2 (with two -SH) is active, and the redox status of OsPHO2 is modulated by two Trx proteins, OsTrxh1 and OsTrxh4. This mechanism is not observed in Arabidopsis, as AtPHO2 does not interact with Trxhs. It has been shown that AtPHO2 modulates the ubiquitination of AtPHT1;1/4 through direct interaction, and there is no evidence demonstrating the interaction between AtPHO2 and AtPHF1. Nevertheless, our study showed that OsPT2/6/8 of PHT1 families are not the direct downstream targets of OsPHO2, whereas OsPHO2 mediates the degradation of OsPHF1 to suppress the ER to plasma membrane localization of Pi transporters.
Figure 8.
Mechanism model of OsTrxh1/h4 interaction with OsPHO2 to regulate Pi homeostasis. OsPHO2 localizes to the ER and Golgi, interacts with OsPHO1;2 and OsPHF1, and mediates the protein degradation of OsPHF1. OsPHO1;2 might be the downstream target of OsPHO2 as well. The redox status of OsPHO2 is regulated by OsTrxh1 and OsTrxh4. See text for more details.
MATERIALS AND METHODS
Plant Materials and Growth Condition
The rice (Oryza sativa L. japonica) cv Nipponbare and pho2 mutant were used for physiological experiments and rice transformations. Hydroponic experiments were performed using a modified culture solution as described (Yoshida et al., 1976), containing 1.425 mm NH4NO3, 0.323 mm NaH2PO4, 0.513 mm K2SO4, 0.998 mm CaCl2, 1.643 mm MgSO4, 9 μm MnCl2, 0.155 μm CuSO4, 0.152 μm ZnSO4, 0.075 μm (NH4)6Mo7O24, 0.019 μm H3BO3,125 μm EDTA-Fe, and 0.25 mm NaSiO3, with pH 5.5. Rice seeds were germinated in tap water for 2 d and then transferred into hydroponic culture. Rice seedlings were grown in a growth room with a day/night temperature of 30°C/22°C and a 12 h photoperiod (200 µmol photons m−2 s−1).
For Arabidopsis (Arabidopsis thaliana), seeds of ecotype Columbia-0 (Col-0), pho2 mutant, and transgenic lines were surface-sterilized and sown on Murashige and Skoog (MS) medium plates supplemented with 2% (w/v) Suc, 0.7% (w/v) agar, pH 5.8. To select pho2 complementation lines, 18 mg/L Hygromycin B was supplied in MS plates. After vernalization at 4°C in the dark for 2 d, plates were incubated in a growth chamber with a 16 h light period (120 µmol photons m−2 s−1) and 23°C/18°C (day/night) temperature. Nine-day-old seedlings were transferred onto new MS plates and grown for another 9 d. Then, shoots were collected for Pi concentration determination.
Y2H Assay
Y2H assays were performed using the GAL4-based two-hybrid system (Clontech). The coding sequences (CDS) of OsPHO2, OsPHO2N (amino acids 1–633), OsPHO2C (amino acids 589–876), and OsUBC23 were subcloned in frame into pGADT7-Rec to generate pGAD-Preys. For chimeric PHO2, namely AtN+OsC and OsN+AtC, the coding regions of AtPHO2N (amino acids 1–601) and AtPHO2C (amino acids 663–907), were inserted into pGAD-OsPHO2C and pGAD-OsPHO2N, respectively. The coding regions of OsTrxh1/h2/h3/h4/h5/h6/h7/h8/h9/h10 and AtTrxh1/h2/h3/h4/h5/h7/h8 were cloned in frame into pGBKT7 to generate pGBK-Baits. For site-directed mutations of Cys to Ala or Ser in OsTrxh1, OsTrxh4 and OsPHO2, two fragments harboring desired change and 16 to 18 bp overlapping, were amplified, assembled, and subcloned into pGBKT7 or pGADT7-Rec, using GBclonart Seamless Cloning Kit (GBI) to generate pGBK-Baits and pGAD-Preys, respectively. The corresponding primers are listed in Supplemental Table S1.
The pGBK-Baits constructs were transformed into yeast strain Y187 and pGAD-Preys into Y2HGold (Clontech). Transformed cells were grown on synthetically defined (SD) medium lacking Leu or Trp for 4 d, respectively. Afterward, positive transformants were mated overnight and spotted onto double-dropout SD medium DDO (SD/-Leu-Trp) and QDO plates. X-α-gal was used as substrate for colorimetric detection of α-galactosidase activity. Plates were incubated at 28°C for 6 to 12 d. Positive control mating was as follows: pGAD-SV40 in Y2HGold and pGBK-53 in Y187. Negative control mating was as follows: pGAD-SV40 in Y2HGold and pGBK-Lam in Y187.
Subcellular Localization and BiFC
For the constructs transiently expressing in rice protoplasts, the coding regions of OsPHO2, OsPHO2N, OsTrxh1, and OsTrxh4 were amplified using attB-containing primers and cloned into pDEST-CGFP (Carrie et al., 2008) and BiFC vectors (http://www.bio.purdue.edu/people/faculty/gelvin/nsf/protocols_vectors.htm) pSAT4-DEST-nEYFP-C1 and pSAT5(A)-DEST-cEYFP-N1 using Gateway recombination technology (Invitrogen) to generate OsTrxh1-GFP, OsTrxh4-GFP, nYFP-OsPHO2, nYFP-OsPHO2N1–663, OsTrxh1-cYFP, and OsTrxh4-cYFP. The OsPHO2 CDS was subcloned into vector pSAT6-EYFP-C1 to obtain YFP-OsPHO2. The coding regions of OsBiP3 and AtCNX1 were ligated with the vectors pSAT6-EYFP-N1, pSAT1-cEYFP-C1-B, and pSAT1-nEYFP-C1 to generate OsBiP3-YFP, AtCNX1-YFP, OsBiP3-cYFP, and AtCNX1-nYFP, respectively. The coding regions of OsPHF1/OsPHO1;2/OsPT2/OsPT6/OsPT8/OsIRT1 and OsPHO2C719A/OsPHO2C445S, C719A were subcloned into the vectors pSAT1A-cEYFP-N1 and pSAT1-nEYFP-C1, respectively. The coding regions of OsPHF1/OsPHO1;2 were subcloned into the vector pSAT4A-mCherry-N1. The primers are given in Supplemental Table S1.
Rice protoplast isolation and polyethylene glycol-mediated transformation were performed as described previously (Chen et al., 2011). In brief, 10 μg plasmid DNA of each construct was transformed into 0.2 mL protoplast suspension. The ER marker was AtWAK2 and Golgi marker was GmMan1 as described previously (Nelson et al., 2007). After incubation at 30°C dark for 12 to 15 h, fluorescence signals in rice protoplasts were detected. Confocal microscopy images were taken using a LSM710nlo confocal laser scanning microscope (Zeiss). Excitation/emission wavelengths were 488 nm/506 to 538 nm for YFP and GFP, and 561 nm/575 to 630 nm for mCherry.
In Vitro Pull-Down
The attB-containing coding regions of OsPHO2/OsPHO2-C445S and AtPHO2 were subcloned into pDEST15 (Invitrogen) using Gateway system to generate GST-tagged prokaryotic expression vectors. The coding regions of OsTrxh1/OsTrxh1-C40S and OsTrxh4/OsTrxh4-C56S were subcloned into pET30a to generate corresponding 6His-tagged vectors. The primers are listed in Supplemental Table S1. Constructs were transformed into Escherichia coli TransB (DE3) (Transgen) and then treated with 0.5 mm isopropyl-β-d-thiogalactoside for 14 h under 18°C to induce expression of fusion proteins. GST-tagged proteins were purified using Glutathione Agarose (Pierce). For in vitro binding, 50 μL of glutathione-agarose beads with bound GST or GST-PHO2 proteins was added to 1 mL cell lysate of 6His-OsTrxh1/h4 in binding buffer (20 mm Tris-HCl, pH 8.0, 300 mm NaCl, 1% Triton X-100, and Complete EDTA-free Protease Inhibitor Cocktail [Roche]), and were incubated at 4°C for 3 h. The beads were washed three times with buffer (20 mm Tris-HCl, pH 8.0, 500 mm NaCl, and 0.5% NP-40). Bound proteins were eluted with 5×SDS loading buffer, analyzed by SDS-PAGE, and detected by immunoblotting using anti-His (Abbkine,1:5000) and anti-GST (EARTHOX,1:5000) antibodies.
Split-Ubiquitin Y2H Assay
Split-ubiquitin Y2H assays were performed according to the manufacturer’s instructions provided with the DUAL membrane pairwise interaction kit (Dualsystems Biotech). The coding regions of OsPHO2/OsPHF1/OsPHO1;2 and OsPHO2/PT2/PT6/PT8 were cloned in frame into the vectors pBT3-STE and pBT3-N to generate Baits-Cub and Cub-Baits, respectively. For prey, the coding regions of OsPHO2/PT2/PT6/PT8 and OsPHO2/OsPHO2C445S were cloned in frame into the vectors pPR3-STE and pPR3-N to generate Preys-NubG and NubG-Preys, respectively. The primers are listed in Supplemental Table S1.
Yeast strain NMY51 cells were cotransformed with a pair of bait and prey constructs and plated onto DDO. The protein-protein interactions were assessed by the growth of yeast colonies on QDO. The control prey pOstI-NubI expressing the wild-type NubI could interact with Cub, but mutated NubG displays almost no affinity for Cub. The prey pNubG-Fe65 expressing the cytosolic protein Fe65 and the bait pTSU2-APP expressing the type I integral membrane protein APP were used as the positive controls.
Identification of Rice and Arabidopsis pho2 Mutants
The rice Tos17 insertion mutant of OsPHO2, NE6022, was obtained from the Rice Genome Resource Center (Japan). The homozygous rice pho2 plants were identified by PCR using primers listed in Supplemental Table S2. The Arabidopsis pho2 (Delhaize and Randall, 1995) mutant CS8508 was obtained from the Arabidopsis Biological Resource Center. CS8508 is an ethyl methanesulfonate mutant, in which mutation in exon 5 changes a Trp-671 to a stop codon.
Constructs for Expression in Plants and Plant Transformation
For complementation of Atpho2 and Ospho2, the attB-containing CDS of AtPHO2 and OsPHO2 were constructed into pH2GW7(0) (Karimi et al., 2002) using Gateway system to obtain 35S::AtPHO2/OsPHO2. The 9.8-kb DNA fragment containing the endogenous genomic OsPHO2 sequence with 3×FLAG-tag was obtained by assembling two DNA fragments from PCR amplifications with overlapping sequences and inserted into modified pCAMBIA130 (Wang et al., 2009) using seamless cloning.
For overexpression constructs, the cDNA fragments of OsTrxh1 (519 bp) and OsTrxh4 (470 bp) were cloned into modified pTF101.1-ubi (Zheng et al., 2010). The RNA interference construct of OsTrxh1 and OsTrxh4 were obtained by subcloning 310-bp or 281-bp fragments into pH7GWIWG2(I) (Karimi et al., 2002) at both sense and antisense orientations, respectively. For CRISPR/Cas9 vectors of OsTrxh1 and OsTrxh4, the target sequences are 5′-GGCCAAGGAGGCCGGCAAAG-3′ and 5′-ACTTCTCCGCCACCTGGTGC-3′, respectively. The synthetic 20bp DNA oligos were inserted into the vector pRGEB31 according to the instruction as described (http://www.bio-protocol.org/e1225). All the corresponding primers are listed in Supplemental Table S2.
For Arabidopsis transformation, constructs were transformed into atpho2 mutant, using an Agrobacterium tumefaciens floral-dipping method, as described previously (Clough and Bent, 1998). Agrobacterium strain GV3101 was applied. For rice transformation, constructs were transformed into callus derived from mature embryo of Nip and pho2 via Agrobacterium strains EHA101 and EHA105.
Agrobacterium-Mediated Infiltration of Tobacco Leaves
The coding sequences of 3FLAG and omega sequences were subcloned into modified pCAMBIA1300 (Wang et al., 2009) driven by the CaMV 35S promoter to generate 1300-3F. The OsPHO2 CDS was subcloned in frame into 1300-3F to generate 3F-OsPHO2. The coding sequences of GFP, OsTrxh1/h4, and OsPHF1 were subcloned in frame into pBS-4MYC, and then the corresponding fragments with omega sequences and 4MYC coding region were subcloned into modified pCAMBIA1300 to generate 4M-GFP, 4M-OsTrxh1/OsTrxh4, and OsPHF1-4M, respectively. The primers are given in Supplemental Table S2. The resulting constructs were transiently expressed in tobacco (Nicotiana benthamiana) leaves by Agrobacterium infiltration. The Agrobacterium strain C58C1 harboring p19 was used to prevent the onset of posttranscriptional gene silencing in the infiltrated leaves. 4M-GFP served as internal control for infiltration and expression. Infiltrated tobacco plants were grown for another 3 d before sample collection for protein extraction.
Protein Isolation and Immunoblot Analysis
For total protein extraction, plant tissues were ground in liquid nitrogen and dissolved in SDS extraction buffer containing 2% (w/v) SDS, 60 mm Tris-HCl, pH 8.5, 2.5% (v/v) glycerol, 0.13 mm EDTA, 1 mm phenylmethylsulfonyl fluoride and protease inhibitor cocktail (Sigma). For membrane protein isolation, plant tissues were ground in liquid nitrogen and followed by the manufacturer’s instructions provided with the Plant Fractionated Protein Extraction Kit (PE0240, Sigma) to isolate hydrophobic proteins. Protein samples (30 to 50 μg) were loaded on SDS-PAGE gels, transferred to polyvinylidene fluoride membranes, and analyzed by immunoblot using anti-FLAG M2 (1:3000, Sigma-Aldrich), anti-MYC (1:2000, Merck), and anti-OsPHF1 (1:500; Chen et al., 2015) antibodies.
Measurement of Pi Concentration
Leaves and roots of the wild-type and transgenic seedlings were sampled separately. The Pi concentration was measured using the procedure modified as described previously (Wang et al., 2009). Briefly, 30 to 60 mg of fresh tissue samples were homogenized with 50 µL of 5 m H2SO4 by FastPrep-24 (MP Biomedical) and followed by adding 1 mL H2O in a 2 mL tube. The homogenate was centrifuged at 12,000 rpm for 10 min at 4°C, and the supernatant was collected. Then the supernatant was diluted to an appropriate concentration and mixed with a malachite green reagent in 3:1 ratio for 30 min. After that, the absorption values were determined at A650 by a POLARstar OPTIMA (BMG LABTECH). Pi concentration was calculated from a standard curve generated using varying concentrations of KH2PO4.
RNA Preparation and qRT-PCR
Total RNA was extracted from plant samples using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. RNA samples were collected with three biological replications. First-strand cDNAs were synthesized from total RNA using M-MLV reverse transcriptase (Promega). qRT-PCR were performed using LightCycler 480 SYBR Green I Master Kit on a LightCycler480 machine (Roche Diagnostics) according to the manufacturer’s instructions. The relative level of expression was calculated by the equation 2−ΔΔCt using housekeeping gene OsACTIN1 as an internal reference. The primers for qRT-PCR analysis are given in Supplemental Table S3.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL database under the following accession numbers: Oryza sativa japonica OsPHO2 (Os05g0557700), OsUBC23 (Os01g0125900), OsTrxh1 (Os07g0186000), OsTrxh2 (Os05g0508500), OsTrxh3 (Os09g0401200), OsTrxh4 (Os03g0800700), OsTrxh5 (Os07g0190800), OsTrxh6 (Os12g0281300), OsTrxh7 (Os01g0168200), OsTrxh8 (Os05g0169000), OsTrxh9 (Os05g0480200), OsTrxh10 (Os04g0629500), OsPHO1;2 (Os02g0809800), OsPHF1 (Os07g0187700), OsPT2 (Os03g0150800), OsPT6 (Os08g0564000), OsPT8 (Os10g0444700), OsACTIN1 (Os03g0718100), and OsBiP3 (Os02g0115900); Arabidopsis thaliana AtPHO2 (At2g33770), AtTrxh1 (At3g51030), AtTrxh2 (At5g39950), AtTrxh3 (At5g42980), AtTrxh4 (At1g19730), AtTrxh5 (At1g45145), AtTrxh7 (At1g59730), AtTrxh8 (At1g69880), AtTrxh9 (At3g08710), AtTrxh10 (At3g56420), ATCXXS1 (At1g11530), ATCXXS2 (At2g40790), AtWAK2 (At1g21270), AtCNX1 (At5g61790), and AtEF1b (At5g19510); Glycine max GmMan1 (NM_001251128.1); Hordeum vulgare HvPHO2 (GQ861514.1), HvTrxh1 (AY245454.1), and HvTrxh2 (AY245455.1); Triticum aestivum TaTrxh1 (AY072771.1), TaTrxh2 (AF286593.2), and TaTrxh3 (AF420472.2); Zea mays ZmPHO2 (XM_008661357.1); and Nicotiana benthamiana NbPHO2 (EU375892.1).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Alignments of amino acid sequences.
Supplemental Figure S2. Subcellular localization of OsPHO2, OsTrxh1, and OsTrxh4-FP fusion proteins in rice protoplasts.
Supplemental Figure S3. OsPHO2 could not target OsTrxh1 and OsTrxh4 to degradation in tobacco leaves.
Supplemental Figure S4. Identification of rice Tos17 insertion mutant pho2 (NE6022).
Supplemental Figure S5. Cosubcellular localization of YFP-OsPHO2 with ER/Golgi markers and OsPHF1/OsPHO1;2-mCherry fusion proteins in rice protoplasts.
Supplemental Figure S6. Validation of autoactivation activities of bait vectors in split-ubiquitin Y2H assays.
Supplemental Figure S7. Expression levels of OsTrxh1, OsTrxh4, and OsPHO2 in different tissues and different development stages based on microarray data retrieved from GENEVESTIGATOR (https://genevestigator.com/gv/).
Supplemental Figure S8. Sequencing results of the CRISPR/Cas9 mutants of OsTrxh1 and OsTrxh4.
Supplemental Figure S9. Morphological appearance of Nip, OE, Ri, and CRISPR/Cas9 mutants of OsTrxh1 and OsTrxh4.
Supplemental Figure S10. Overexpression of AtPHO2 could restore Arabidopsis pho2 mutant, whereas overexpression of OsPHO2 could not.
Supplemental Figure S11. Rice pho2 mutants could not be recovered by expressing 35S::AtPHO2.
Supplemental Figure S12. Amino acid alignment of PHO2 and PHO2-like proteins in multiple plant species.
Supplemental Table S1. Primers used for the plasmid constructs of Y2H, BiFC, and pull-down.
Supplemental Table S2. Primers used for the plasmid constructs of transient expression in tobacco leaves, pho2 recovery, and OsTrxh1/h4 transgenic plants.
Supplemental Table S3. Primers used for RT and qRT-PCR analysis.
Supplementary Material
Acknowledgments
We thank Dr. Juan Du for the suggestion on membrane protein extraction and Shelong Zhang for the technical support on confocal microscope manipulation.
Glossary
- UBC
ubiquitin-conjugating
- ER
endoplasmic reticulum
- Y2H
yeast two-hybrid
- BiFC
biomolecular fluorescence complementation
Footnotes
This work was supported by the Ministry of Science and Technology of China (grant nos. 2016YFD0100703, 2016ZX08003005-001, and 2014ZX08009328-002), the National Natural Science Foundation of China (grant nos. 31572189, 31401934, and 31471929), and the Australian Research Council Centre of Excellence Program (grant no. CE140100008).
References
- Aung K, Lin SI, Wu CC, Huang YT, Su CL, Chiou TJ (2006) pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol 141: 1000–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R, Datt Pant B, Stitt M, Scheible WR (2006) PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol 141: 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayle V, Arrighi J-F, Creff A, Nespoulous C, Vialaret J, Rossignol M, Gonzalez E, Paz-Ares J, Nussaume L (2011) Arabidopsis thaliana high-affinity phosphate transporters exhibit multiple levels of posttranslational regulation. Plant Cell 23: 1523–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos R, Castrillo G, Linhares F, Puga MI, Rubio V, Pérez-Pérez J, Solano R, Leyva A, Paz-Ares J (2010) A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet 6: e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrie C, Murcha MW, Kuehn K, Duncan O, Barthet M, Smith PM, Eubel H, Meyer E, Day DA, Millar AH, et al. (2008) Type II NAD(P)H dehydrogenases are targeted to mitochondria and chloroplasts or peroxisomes in Arabidopsis thaliana. FEBS Lett 582: 3073–3079 [DOI] [PubMed] [Google Scholar]
- Chen J, Liu Y, Ni J, Wang Y, Bai Y, Shi J, Gan J, Wu Z, Wu P (2011) OsPHF1 regulates the plasma membrane localization of low- and high-affinity inorganic phosphate transporters and determines inorganic phosphate uptake and translocation in rice. Plant Physiol 157: 269–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Wang Y, Wang F, Yang J, Gao M, Li C, Liu Y, Liu Y, Yamaji N, Ma JF, et al. (2015) The rice CK2 kinase regulates trafficking of phosphate transporters in response to phosphate levels. Plant Cell 27: 711–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou T-J, Aung K, Lin S-I, Wu C-C, Chiang S-F, Su CL (2006) Regulation of phosphate homeostasis by MicroRNA in Arabidopsis. Plant Cell 18: 412–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou T-J, Lin S-I (2011) Signaling network in sensing phosphate availability in plants. Annu Rev Plant Biol 62: 185–206 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Delhaize E, Randall PJ (1995) Characterization of a phosphate-accumulator mutant of Arabidopsis thaliana. Plant Physiol 107: 207–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B, Rengel Z, Delhaize E (1998) Uptake and translocation of phosphate by pho2 mutant and wild-type seedlings of Arabidopsis thaliana. Planta 205: 251–256 [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA, Paz-Ares J (2007) Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 39: 1033–1037 [DOI] [PubMed] [Google Scholar]
- Fujii H, Chiou TJ, Lin SI, Aung K, Zhu JK (2005) A miRNA involved in phosphate-starvation response in Arabidopsis. Curr Biol 15: 2038–2043 [DOI] [PubMed] [Google Scholar]
- Helenius A, Trombetta ES, Hebert DN (1997) Calnexin, calreticulin and the folding of glycoproteins. Trends Cell Biol 7: 193–200 [DOI] [PubMed] [Google Scholar]
- Holmgren A. (1989) Thioredoxin and glutaredoxin systems. J Biol Chem 264: 13963–13966 [PubMed] [Google Scholar]
- Hu B, Zhu C, Li F, Tang J, Wang Y, Lin A, Liu L, Che R, Chu C (2011) LEAF TIP NECROSIS1 plays a pivotal role in the regulation of multiple phosphate starvation responses in rice. Plant Physiol 156: 1101–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Franklin AE, Hoffman NE (1993) Primary structure and characterization of an Arabidopsis thaliana calnexin-like protein. J Biol Chem 268: 6560–6566 [PubMed] [Google Scholar]
- Huang T-K, Han C-L, Lin S-I, Chen Y-J, Tsai Y-C, Chen Y-R, Chen J-W, Lin W-Y, Chen P-M, Liu T-Y, et al. (2013) Identification of downstream components of ubiquitin-conjugating enzyme PHOSPHATE2 by quantitative membrane proteomics in Arabidopsis roots. Plant Cell 25: 4044–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallis GB, Holmgren A (1980) Differential reactivity of the functional sulfhydryl groups of cysteine-32 and cysteine-35 present in the reduced form of thioredoxin from Escherichia coli. J Biol Chem 255: 10261–10265 [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kuo H-F, Chiou T-J (2011) The role of microRNAs in phosphorus deficiency signaling. Plant Physiol 156: 1016–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SI, Chiang SF, Lin WY, Chen JW, Tseng CY, Wu PC, Chiou TJ (2008) Regulatory network of microRNA399 and PHO2 by systemic signaling. Plant Physiol 147: 732–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TY, Huang TK, Tseng CY, Lai YS, Lin SI, Lin WY, Chen JW, Chiou TJ (2012) PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis. Plant Cell 24: 2168–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misson J, Raghothama KG, Jain A, Jouhet J, Block MA, Bligny R, Ortet P, Creff A, Somerville S, Rolland N, et al. (2005) A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci USA 102: 11934–11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montrichard F, Alkhalfioui F, Yano H, Vensel WH, Hurkman WJ, Buchanan BB (2009) Thioredoxin targets in plants: The first 30 years. J Proteomics 72: 452–474 [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Pant BD, Buhtz A, Kehr J, Scheible WR (2008) MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J 53: 731–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BS, Seo JS, Chua N-H (2014) NITROGEN LIMITATION ADAPTATION recruits PHOSPHATE2 to target the phosphate transporter PT2 for degradation during the regulation of Arabidopsis phosphate homeostasis. Plant Cell 26: 454–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C-J, Bart R, Chern M, Canlas PE, Bai W, Ronald PC (2010) Overexpression of the endoplasmic reticulum chaperone BiP3 regulates XA21-mediated innate immunity in rice. PLoS One 5: e9262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier Y, Thoma S, Somerville C, Schiefelbein J (1991) Mutant of Arabidopsis deficient in xylem loading of phosphate. Plant Physiol 97: 1087–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghothama KG. (1999) PHOSPHATE ACQUISITION. Annu Rev Plant Physiol Plant Mol Biol 50: 665–693 [DOI] [PubMed] [Google Scholar]
- Raghothama KG. (2000) Phosphate transport and signaling. Curr Opin Plant Biol 3: 182–187 [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15: 2122–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secco D, Baumann A, Poirier Y (2010) Characterization of the rice PHO1 gene family reveals a key role for OsPHO1;2 in phosphate homeostasis and the evolution of a distinct clade in dicotyledons. Plant Physiol 152: 1693–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secco D, Jabnoune M, Walker H, Shou H, Wu P, Poirier Y, Whelan J (2013) Spatio-temporal transcript profiling of rice roots and shoots in response to phosphate starvation and recovery. Plant Cell 25: 4285–4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahpiri A, Svensson B, Finnie C (2009) From proteomics to structural studies of cytosolic/mitochondrial-type thioredoxin systems in barley seeds. Mol Plant 2: 378–389 [DOI] [PubMed] [Google Scholar]
- Stefanovic A, Ribot C, Rouached H, Wang Y, Chong J, Belbahri L, Delessert S, Poirier Y (2007) Members of the PHO1 gene family show limited functional redundancy in phosphate transfer to the shoot, and are regulated by phosphate deficiency via distinct pathways. Plant J 50: 982–994 [DOI] [PubMed] [Google Scholar]
- Ticconi CA, Abel S (2004) Short on phosphate: Plant surveillance and countermeasures. Trends Plant Sci 9: 548–555 [DOI] [PubMed] [Google Scholar]
- Valdés-López O, Arenas-Huertero C, Ramírez M, Girard L, Sánchez F, Vance CP, Luis Reyes J, Hernández G (2008) Essential role of MYB transcription factor: PvPHR1 and microRNA: PvmiR399 in phosphorus-deficiency signalling in common bean roots. Plant Cell Environ 31: 1834–1843 [DOI] [PubMed] [Google Scholar]
- Vance CP, Uhde-Stone C, Allan DL (2003) Phosphorus acquisition and use: Critical adaptations by plants for securing a nonrenewable resource. New Phytol 157: 423–447 [DOI] [PubMed] [Google Scholar]
- Wang C, Ying S, Huang H, Li K, Wu P, Shou H (2009) Involvement of OsSPX1 in phosphate homeostasis in rice. Plant J 57: 895–904 [DOI] [PubMed] [Google Scholar]
- Wang J, Sun J, Miao J, Guo J, Shi Z, He M, Chen Y, Zhao X, Li B, Han F, et al. (2013) A phosphate starvation response regulator Ta-PHR1 is involved in phosphate signalling and increases grain yield in wheat. Ann Bot (Lond) 111: 1139–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Forno D, Cock J, Gomez K (1976) Laboratory Manual for Physiological Studies of Rice, Ed 3 The International Rice Research Institute, Los Baños, Philippines [Google Scholar]
- Zhang C-J, Zhao B-C, Ge W-N, Zhang Y-F, Song Y, Sun D-Y, Guo Y (2011) An apoplastic h-type thioredoxin is involved in the stress response through regulation of the apoplastic reactive oxygen species in rice. Plant Physiol 157: 1884–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Cheng Z, Ai C, Jiang X, Bei X, Zheng Y, Glahn RP, Welch RM, Miller DD, Lei XG, et al. (2010) Nicotianamine, a novel enhancer of rice iron bioavailability to humans. PLoS One 5: e10190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Jiao F, Wu Z, Li Y, Wang X, He X, Zhong W, Wu P (2008) OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol 146: 1673–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.