A complex consisting of an atypical HLH protein, OsBUL1, a protein containing KxDL-motif, LO9-177, and basic HLH protein, OsBC1, positively affects leaf bending and grain size in rice.
Abstract
Rice atypical HLH protein Oryza sativa BRASSINOSTEROID UPREGULATED 1-LIKE1 (OsBUL1) is preferentially expressed in the lamina joint where it controls cell elongation and positively affects leaf angles. OsBUL1 knockout mutant (osbul1) and transgenic rice for double-stranded RNA interference (dsRNAi) of OsBUL1 produced erect leaves with smaller grains, whereas OsBUL1 overexpressors and an activation tagging line of OsBUL1 exhibited increased lamina inclination and grain size. Moreover, OsBUL1 expression was induced by brassinolide (BL) and osbul1 did not respond to BL treatment. To understand the molecular network of OsBUL1 function in rice, we isolated a novel OsBUL1-interacting protein, LO9-177, an uncharacterized protein containing a KxDL motif, and functionally studied it with respect to the lamina inclination and grain size of rice. OsBUL1 COMPLEX1 (OsBC1) is a basic helix-loop-helix (bHLH) transcriptional activator that interacts with OsBUL1 only in the presence of LO9-177 forming a possible trimeric complex for cell elongation in the lamina joint of rice. Expression of OsBC1 is also upregulated by BL and has a similar pattern to that of OsBUL1. Transgenic rice plants expressing OsBC1 under the control of OsBUL1 promoter showed increased grain size as well as leaf bending, while transgenic lines for dsRNAi and/or expressing a dominant repressor form of OsBC1 displayed reduced plant height and grain size. Together, these results demonstrated that a novel protein complex consisting of OsBUL1, LO9-177, and OsBC1 is associated with the HLH-bHLH system, providing new insight into the molecular functional network based on HLH-bHLH proteins for cell elongation.
Plant architecture is an important factor for efficient photosynthesis and high yield (Sakamoto et al., 2006). In rice (Oryza sativa), the degree of lamina inclination is an important trait that determines architecture. The degree of lamina angles depends on cell division, expansion, and cell wall composition in the lamina joint (Nakamura et al., 2009; Zhang et al., 2009; Zhao et al., 2010; Ning et al., 2011). Erect leaf phenotypes are desired to avoid shade when plants are grown at high planting density (Van Camp, 2005).
Mutations in brassinosteroid (BR) biosynthesis genes (Hong et al., 2002, 2003; Sakamoto et al., 2006; Wu et al., 2008) and BR signaling genes (Bai et al., 2007) change lamina angle. Other phytohormones are also involved in controlling the lamina joint inclination. Ethylene participates in BR-induced lamina inclination. Indole-3-acetic acid influences lamina joint inclination at high concentrations and has a synergistic interaction with BR (Wada et al., 1981; Cao and Chen, 1995). A gain-of-function mutant for OsGH3-1 encoding an indole-3-acetic acid amido synthetase showed increased leaf angles due to stimulated cell elongation at the lamina joint (Zhao et al., 2013). Reduced expression of SPINDLY, a negative regulator of gibberellin signaling, also causes increased lamina inclination (Shimada et al., 2006).
Transcription factors that determine the angle have been identified. Mutations in OsLIGULELESS1, encoding a SQUAMOSA promoter binding domain protein, exhibit an erect leaf phenotype (Lee et al., 2007). Transgenic plants overexpressing rice ILI1-BINDING HLH1 (OsIBH1) also show erect leaves (Zhang et al., 2009). Inducing expression of genes encoding atypical helix-loop-helix (HLH) proteins such as BRASSINISTEROID UPREGULATED1 (BU1), INCREASED LAMININAR INCLINATION1 (ILI1), and POSITIVE REGULATOR OF GRAIN LENGTH1 (PGL1) conferred a higher lamina angle degree (Tanaka et al., 2009; Zhang et al., 2009; Heang and Sassa, 2012a).
Increased lamina angles are observed in transgenic rice plants overexpressing LAX PANICLE (Komatsu et al., 2003), a T-DNA insertion mutant of OsWRKY11 (Wang et al., 2005), and RNA interference transgenic lines for SHORT VEGETATIVE PHASE group MADS-box genes such as OsMADS22, OsMADS55, and OsMADS47 (Lee et al., 2008). Decreased expression of rice LEAF AND TILLER ANGLE INCREASED, encoding a CCCH-type zinc-finger protein, also results in increased lamina inclination through regulating BR signaling (Wang et al., 2008). LEAF INCLINATION2, encoding a VERNALIZATION INSENSITIVE3-like protein, acts as a repressor of cell division for regulation of collar development (Zhao et al., 2010). Enhancing expression of a gene encoding BAHD acyltransferase-like protein produces slender grains with enlarged leaf angles (Feng et al., 2016). Additionally, a gain-of-function epiallele of rice RELATED TO ABSCISIC ACID INSENSITIVE3/VIVIPAROUS1 (VP1) 6, encoding a B3 DNA-binding domain-containing protein, caused larger lamina inclination but smaller grain size by modulating BR homeostasis (Zhang et al., 2015).
Basic helix-loop-helix (bHLH) proteins are a group of important transcription factors that play diverse roles in both animals and plants. They occupy key positions in phytochrome signal transduction cascades, contributing to stomata differentiation, cell fate determination, and BR-response gene expression (Bernhardt et al., 2005; Duek and Fankhauser, 2005; Serna, 2007; Bhattacharya and Baker, 2011). In particular, several bHLH proteins including BRI1 ENHANCED EXPRESSION1 (BEE1) to BEE3 (Friedrichsen et al., 2002) and BES1-INTERACTING MYC-LIKE1 (BIM1) to BIM3 (Yin et al., 2005) are implicated in BR signaling in Arabidopsis (Arabidopsis thaliana). Recently, a group of typical bHLH proteins, ACTIVATORS OF CELL ELONGATIONs (ACEs), were also reported to act as positive regulators (Ikeda et al., 2012), whereas another bHLH protein, ANTAGONIST OF PGL1 (APG), was found to function as a negative regulator of cell elongation (Heang and Sassa, 2012a, 2012b, 2012c). Interestingly, some atypical HLH proteins, such as PACLOBUTRAZOL RESISTANCEs (PREs), BU1, ILI1, PGL1, and PGL2 (Hyun and Lee, 2006; Lee et al., 2006; Tanaka et al., 2009; Wang et al., 2009; Zhang et al., 2009; Heang and Sassa, 2012a, 2012b), also contribute to cell elongation, while others, including ATBS1-INTERACTING FACTORs (AIFs), IBH1, and OsIBH1, suppress cell elongation (Wang et al., 2009; Zhang et al., 2009; Ikeda et al., 2013). Recently, a triantagonistic bHLH system was reported to explain the relationship between bHLH and atypical HLH proteins in controlling cell elongation in Arabidopsis (Ikeda et al., 2012), although AIFs were missing in the model.
In this work, we functionally characterized OsBUL1 that encodes an atypical HLH protein belonging to the PRE group using gain-of-function and loss-of-function approaches in rice. To better understand the network of OsBUL1 function and to identify components of the network, we searched for OsBUL1 interacting proteins, and LO9-177, a KxDL motif-containing protein, was isolated. LO9-177 also showed cell elongation activity under the control of OsBUL1 promoter, and the protein was found to be a molecular mediator between OsBUL1 and OsBC1, a typical bHLH transcription factor that promotes increased lamina angles. Our findings reveal one more layer in the cell elongation machinery based on the atypical HLH-bHLH network by providing a novel complex positively acting on lamina inclination and/or grain size through cell elongation of rice.
RESULTS
OsBUL1 Knockout Rice Produces Erect Leaves with Small Grains
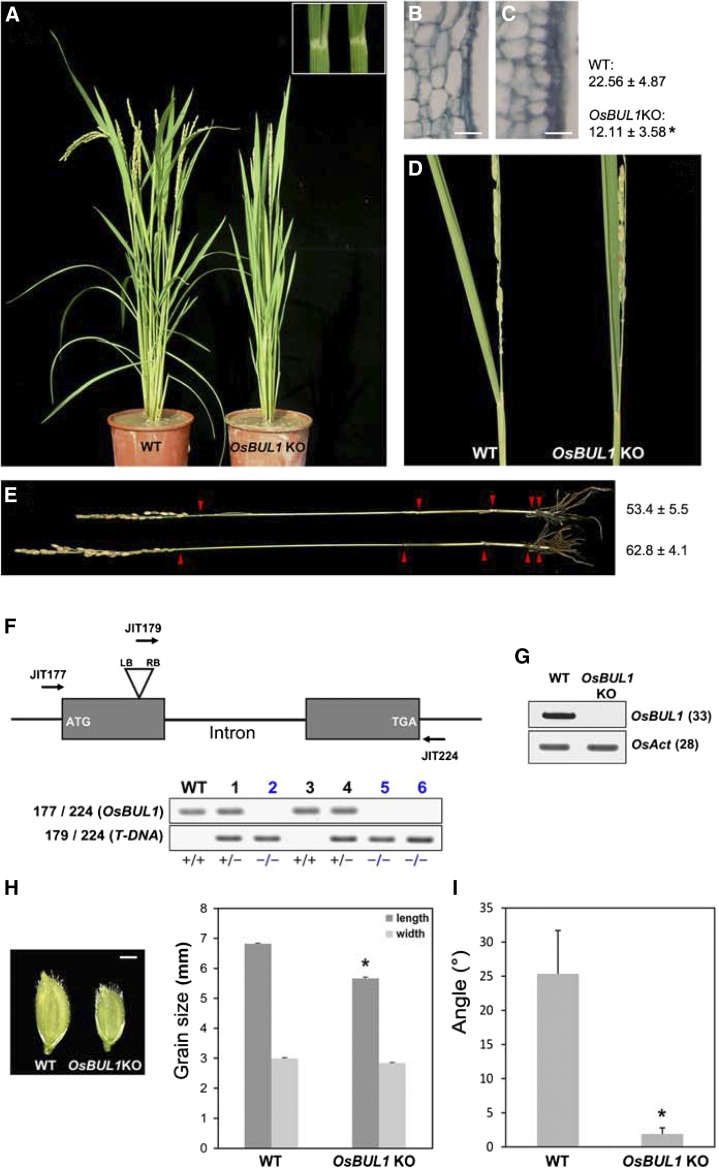
We identified an erect leaf mutant from a T-DNA insertion mutant population in rice (Fig. 1, A–E; Jeon et al., 2000; Jeong et al., 2002, 2006). Genotyping revealed that T-DNA was inserted in the first exon of Os02g51320 (Fig. 1F). The gene encodes an atypical HLH protein and is highly homologous to BU1 (Tanaka et al., 2009). We named the gene O. sativa BU1-like 1 (OsBUL1). Intact OsBUL1 mRNA was not detected in the mutant line, demonstrating that osbul1 is a null allele (Fig. 1G). The mutant exhibited reduced lamina angles (Fig. 1, A and D). Average length of cells in the lamina joint was reduced by 46.3% compared to wild-type control (Fig. 1, B and C). Plant height was also reduced due to reduced internode length (Fig. 1E). The mutant showed small spikelets and consequently produced small grains (Fig. 1H). Leaf angle was also reduced in the mutant (Fig. 1I). Transgenic rice plants generated by double-stranded RNA interference (dsRNAi) using OsBUL1 coding sequence showed similar phenotypes (Supplemental Fig. S1, A–C), indicating that the mutant phenotypes were due to mutations in OsBUL1. In addition to the OsBUL1 transcript, transcript levels of OsBUL1 homologous genes such as OsBU, OsBUL2, OsBUL3, and OsILI1 were also less abundant in the dsRNAi plants, indicating a multigene knockdown effect (Supplemental Fig. S1, D and E).
Figure 1.
OsBUL1 null mutants show erect leaves. A and D, OsBUL1 knockout mutant plants exhibit erect leaves. The lamina joint area of the second leaf from the wild type (left) and OsBUL1KO is shown in the box (A). B and C, Comparison of cells in the lamina joint area by longitudinal sections in wild-type (B) and OsBUL1KO (C) plants. Length of cells in the lamina joint of wild-type control and OsBUL1KO plants is presented. Values are given as means ± sd (µm; n > 12; *P < 0.01, Student’s t test). Bar = 20 µm in B and C. E, Mild reduction of plant height by retarded elongation of lower internodes is observed in OsBUL1KO plants (top). Arrowheads indicate each node of plants, and plant height of each genotype is presented as means ± sd (cm, n > 12). F, OsBUL1KO plant is a T-DNA insertion mutant, and #2, #5, and #6 plants are OsBUL1 KO homozygous plants. Genotyping primers were as follows: (JIT177) 5′-GGAATTCATGTCGAGCAGAAGGTCGTCGCGTG-3′, (JIT179) 5′-CCACAGTTTTCGCGATCCAGACTG-3′, and (JIT224) 5′-CGCTTGCTGCTGCTGCTTGCCGATC-3′. +/+ Means wild type/wild type segregant, and +/− and −/− indicate heterozygote and homozygote for T-DNA insertion, respectively. G, OsBUL1 transcripts are not detected in OsBUL1KO plants. The numbers in the parentheses indicate PCR cycles. H, The mutant plants produce smaller grains. Bar = 2 mm. Error bars indicate sd (n = 30; *P < 0.0001, Student’s t test). I, Leaf angles between wild-type and OsBUL1KO plants. The third leaf from the top of the main stem was used for measurement and values are given as means ± sd in the graph (n = 18; *P < 0.001, Student’s t test).
Isolation of OsBUL1D, a Gain-of-Function Mutant of OsBUL1
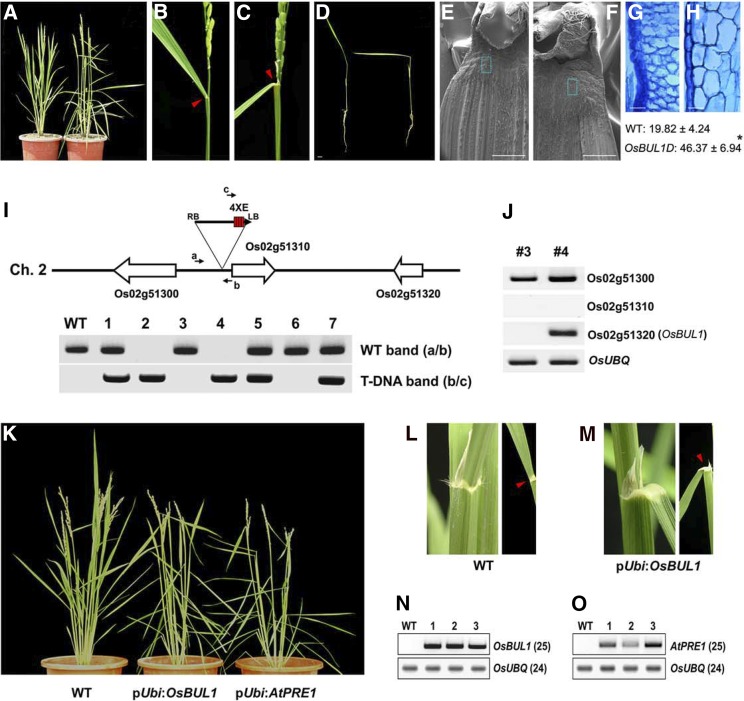
We isolated an activation tagging line in which OsBUL1 expression was significantly increased (Fig. 2). The mutant plants displayed increased lamina inclination. The leaf-bending phenotype was observed even at the seedling stage (Fig. 2D). Scanning electron microscopy and sectional anatomy revealed that the lamina joint area of the mutant was expanded and the cells in the region were enlarged (Fig. 2, F and H). Indeed, average length of cells in the lamina joint of OsBUL1D was more than 2-fold longer than that of wild-type control (Fig. 2, G and H). The heterozygous plants were selfed, and the progeny showed a 3:1 ratio of the mutant to the wild type, indicating that the mutant is dominant. T-DNA was inserted 8.1 kb downstream of OsBUL1 near Os02g51310 on chromosome 2 (Fig. 2I). While Os02g51320 transcript level was dramatically increased, expression of nearby genes was not significantly affected (Fig. 2J).
Figure 2.
Larger lamina angles caused by an OsBUL1 activation tagging line and recapitulation of the increased lamina inclination phenotype by OsBUL1 or its Arabidopsis homolog, PRE1 overexpression. A, A gain-of-function mutant of OsBUL1 (OsBUL1D; right) and the wild type. B to D, Compared to the wild type (B), OsBUL1D showed increased leaf angles (C) in a flag leaf and young leaves (right in D). Arrowheads indicate the lamina joint of flag leaf (B and C). Bar = 1 cm in D. E to H, Magnified images show that the lamina joint area is extended in OsBUL1D (F and H) compared to the wild type (E and G) due to enlarged cells. G and H, Longitudinal sections of the lamina joint area in the wild type (G) and OsBUL1D (H). Length of cells in the lamina joint of wild-type control and OsBUL1D plants is presented. Values are given as means ± sd (µm; n > 15; *P < 0.001, Student’s t test). Bars = 1 mm (E and F) and 25 µm (G and H). I, Genotyping of the OsBUL1D line. 4XE means four tandem copies of CaMV 35S enhancer. Primer sequences are (a) 5′-TGCCACCTCAGTAAAAACCGGACAC-3′, (b) 5′-CGATGACAAGTTGAGGGAGCTTTGG-3′, and (c) 5′-CGTCCGCAATGTGTTATTAAG-3′. J, Higher expression level of OsBUL1 (Os02g51320) in the OsBUL1D. K, Overexpression of OsBUL1 or its Arabidopsis homolog, PRE1 in wild-type rice phenocopies the gain-of-function mutant of OsBUL1. L and M, Compared with the wild type (L), OsBUL1 overexpressors show increased lamina inclination (M). Arrowheads indicate the lamina joint of flag leaf (L and M). N and O, Expression of OsBUL1 and AtPRE1 in transgenic rice plants shown in K. The numbers in the parentheses indicate PCR cycles.
The OsBUL1D mutant plants produced grains with increased size (Supplemental Fig. S2). Recapitulated phenotype of OsBUL1D was observed in transgenic rice overexpressing OsBUL1 by maize (Zea mays) Ubi1 promoter (Fig. 2, K to N; Supplemental Fig. S2). These results are consistent with a previous report stating that grain size was increased by cell elongation in transgenic rice plants expressing PGL2 (the same gene as OsBUL1) under the control of chitinase promoter (Heang and Sassa, 2012b).
To investigate whether OsBUL1 is functionally conserved in dicot plants, we produced rice plants overexpressing Arabidopsis PRE1 that are homologous to rice OsBUL1. These transgenic plants showed similar phenotypes to OsBUL1-overexpressing plants (Fig. 2, K and O; Supplemental Fig. S2). We expressed the OsBUL1 gene in Arabidopsis and tobacco (Nicotiana benthamiana) using CaMV 35S promoter. The transgenic plants showed phenotypes of elongated petioles due to cell elongation (Supplemental Fig. S3). Transgenic Arabidopsis with reduced expression of PRE1 showed short petioles with tiny cells (Supplemental Fig. S3, A and B).
OsBUL1 Is Induced by Exogenous Brassinolide and Is Involved in BR Signaling
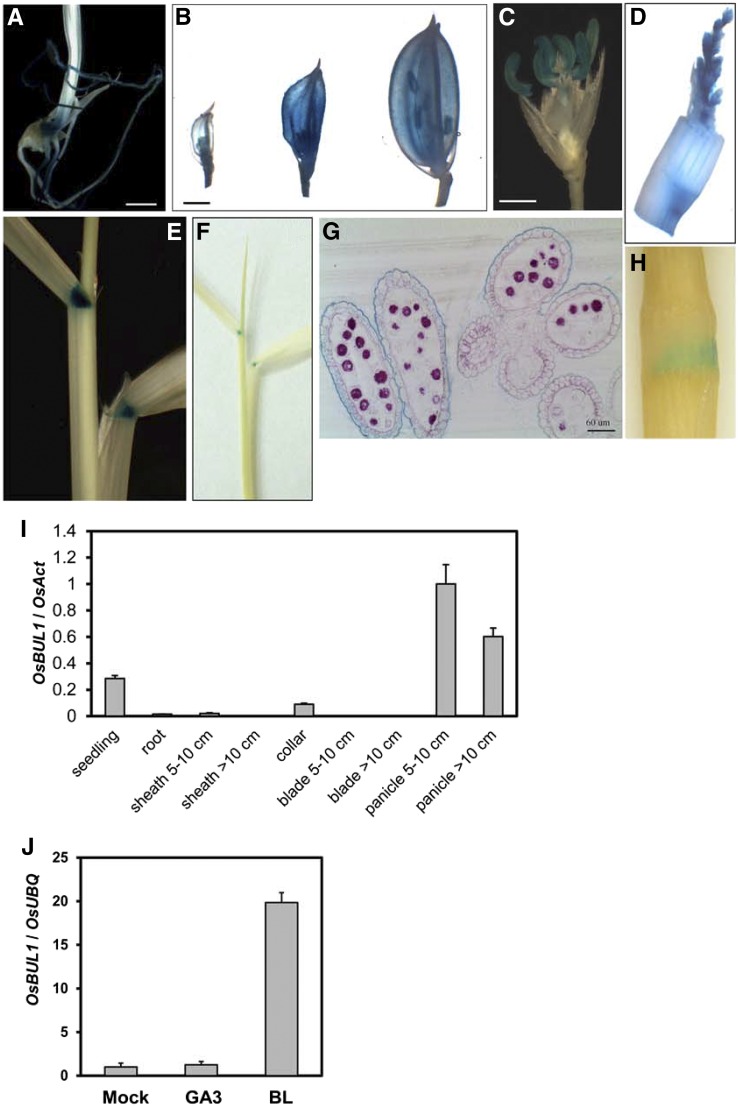
Transgenic rice plants carrying GUS driven by the 2.2-kb OsBUL1 promoter were generated and their GUS activity was examined. GUS expression was detected in seedlings, lamina joints, nodes, panicles, and floral organs including palea, lemma, and anthers (Fig. 3, A–H). Expression was not found in leaf sheath, leaf blade, and internode. The results coincided with the expression pattern of OsBUL1 transcripts analyzed by qRT-PCR (Fig. 3I).
Figure 3.
Spatiotemporal expression of OsBUL1. A to H, GUS staining of various tissues from pOsBUL1:GUS transgenic rice plants: seedlings (A), spikelets with different developmental stages (B), a spikelet after removing palea and lemma (C), young panicle (D), lamina joint (E and F), anthers (G), and node (H). Bar = 1 mm in A to C. I, Relative expression level of OsBUL1 in various organs at different developmental stages. J, Induction of OsBUL1 expression by GA3 and BL at the 24-h time point after treatment. Error bars indicate sd of three biological replicates.
We examined whether OsBUL1 expression is regulated by phytohormones that play roles in cell elongation. The level of OsBUL1 transcripts was increased 19.5-fold by 24-h brassinolide (BL) treatment. However, gibberellin (GA3) did not influence the gene expression (Fig. 3J). Whereas dark-grown wild-type seedlings had longer shoots, roots, and coleoptiles compared to light-grown seedlings, osbul1 seedlings had shorter shoots when they were grown under dark conditions compared to light conditions (Supplemental Fig. S4). OsBUL1 transcripts are more abundant in plants grown in the dark (Supplemental Fig. S4C). Lamina inclination assay revealed that osbul1 did not respond to various concentrations of BL (Supplemental Fig. S5, A and B). These observations indicate that OsBUL1 is linked to BR signaling (Supplemental Fig. S4A)
OsBUL1 Interacts with LO9-177
To understand functional roles of OsBUL1 at the molecular level, we performed yeast two-hybrid screening of a rice cDNA library prepared from aboveground parts of rice plants. The screening resulted in identification of LO9-177 (Fig. 4A), which is an uncharacterized conserved protein with a predicted monomeric mass of 13.7 kD, containing a characteristic KxDL motif toward its C terminus (Hayes et al., 2011). The spatiotemporal expression pattern of LO9-177 overlapped with that of OsBUL1 (Fig. 3I; Supplemental Figs. S11B and S12, A–D). They were colocalized in the nucleus as well as the cytoplasm (Supplemental Figs. S9A).
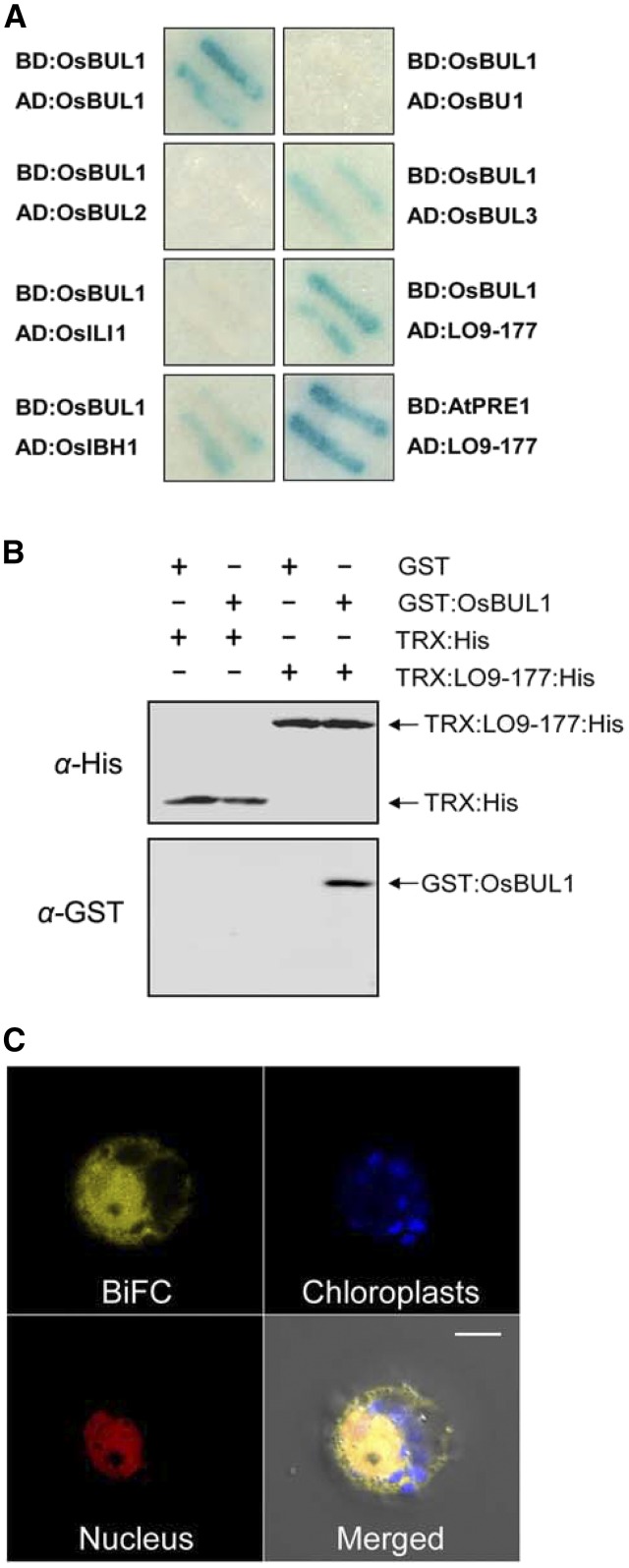
Figure 4.
OsBUL1 interacts with LO9-177. A, Yeast two-hybrid systems demonstrated that OsBUL1 interacts with LO9-177, a small protein containing the KxDL motif. Also, OsBUL1 forms a homodimer and interacts with OsBUL3 and OsIBH1 but not with OsBU1, OsBUL2, and ILI1 (OsILI1). B, His pull-down assays with bacterial recombinant proteins.OsBUL1-GST fusion proteins were pulled down only with LO-177:His (6x His residues) fusion proteins. C, BiFC assays showed yellow florescent signals from reconstructed YFP in both the cytoplasm and nucleus of rice protoplasts containing YFPn:OsBUL1 and YFPc:LO9-177. The blue color is autofluorescence from chloroplasts, and nuclear localization signal (NLS)-RFP marker was used for nuclear labeling. Merged fluorescence signals include reconstructed YFP by BiFC, RFP by NLS-RFP, and autofluorescence in chloroplasts. The negative controls that were conducted by cotransfecting unfused YFP fragments together with a single complementary BiFC interactor-protein resulted in no detectable signals (Supplemental Fig. S9C). Bar = 5 µm.
OsBUL1 formed homodimers and selectively interacted with OsBUL3 among OsBUL1 homologs including OsBU1, OsBUL2, OsBUL3, and OsILI1 (the same as ILI1). OsBUL1 also interacted with OsIBH1, an OsILI1-interacting protein (Zhang et al., 2009; Figure 4A; Supplemental Fig. S7A). Of note, OsIBH1 interacted with OsBUL1 and its homologs but could not form a homodimer (Supplemental Fig. S7A).
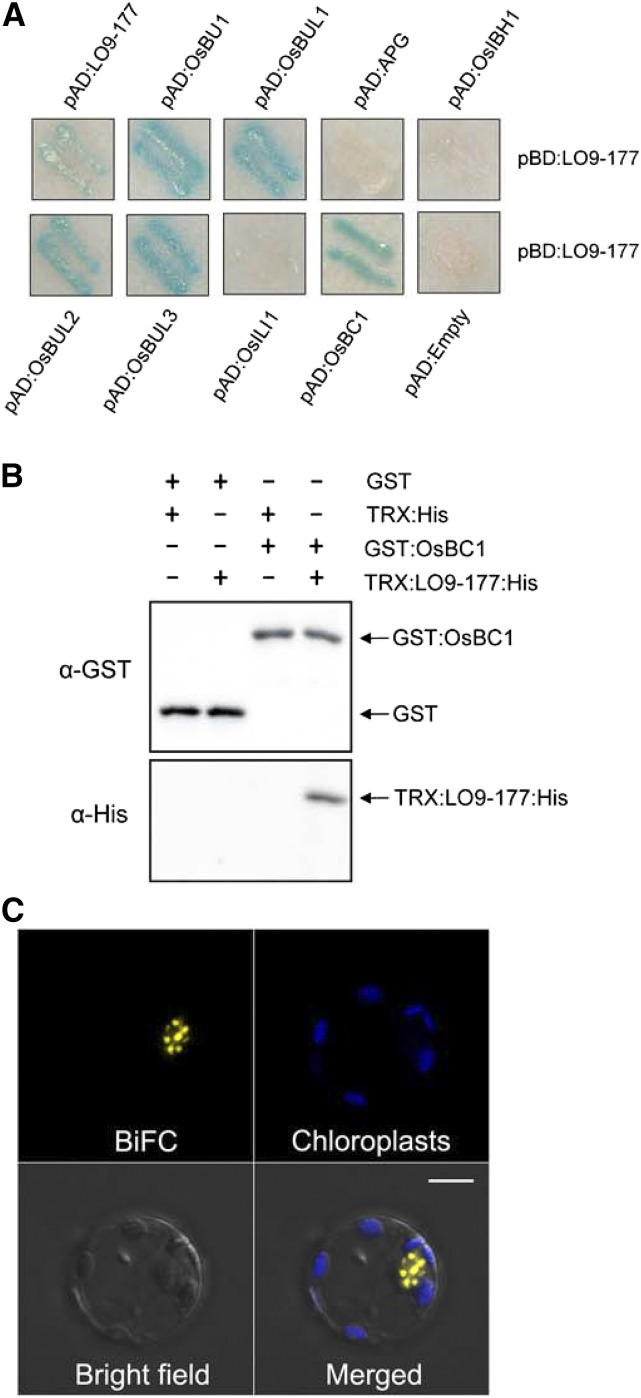
In vitro pull-down assays using Escherichia coli-expressed recombinant proteins, GST-tagged OsBUL1 (GST:OsBUL1) interacted with His-tagged LO9-177 (TRX:LO9-177:His), but not with TRX:His (Fig. 4B). Interaction between OsBUL1 and LO9-177 was also verified in rice cells by bimolecular fluorescence complementation (BiFC) assays using nYFP:LO9-177 and cYFP:OsBUL1 (Fig. 4C).
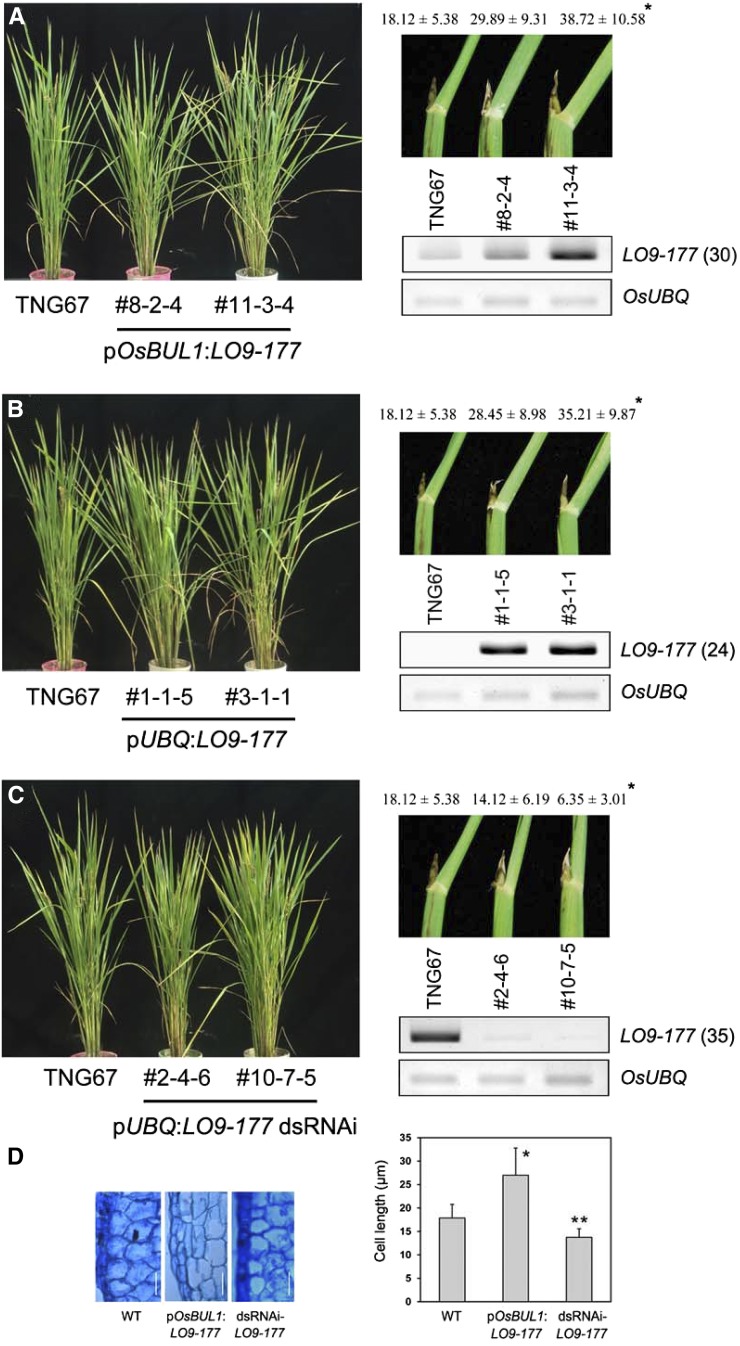
To study the functional roles of LO9-177, a plasmid for overexpression of LO9-177 by OsBUL1 promoter was constructed and introduced into rice plants. They showed phenotypes of increased lamina angles with elongated cells in the lamina joint and grain size (Fig. 5, A and D; Supplemental Fig. S6). Transgenic plants overexpressing LO9-177 by the maize ubi1 promoter displayed similar phenotypes (Fig. 5B). On the contrary, transgenic lines expressing LO9-177 dsRNAi showed reduced leaf angles with mildly reduced length of cells in the lamina joint (Fig. 5, C and D). These indicate that LO9-177 functions similar to OsBUL1.
Figure 5.
Transgenic rice plants for functional characterization of LO9-177. A to C, Transgenic rice plants expressing LO9-177 driven by OsBUL1 promoter (A) and constitutive ubiquitin promoter (B) caused increased leaf angles while dsRNAi lines displayed erect leaves (C). The full-length LO9-177 coding region (372 bp) was used for dsRNAi construction, and endogenous expression level of LO9-177 was examined by RT-PCR using primers, 5′-AGACTCGATAGGGGAGAGGA-3′ and 5′-TCGGCAAAAGTGCAGACAAAAC-3′. The third leaf from the top of the main stem was used for lamina angle measurement and values for leaf angles are given as means ± sd (n = 10 to 14; *P < 0.001, Student’s t test). RNA was extracted from collars for cDNA synthesis. The numbers in parentheses indicate PCR cycles. D, Longitudinal sections of the lamina joint area in the wild type, pOsBUL1:LO9-177, and dsRNAi of LO9-177 plants. Length of cells in the lamina joint of wild-type control, pOsBUL1:LO9-177, and dsRNAi of LO9-177 plants is also presented. Values are given as means ± sd (n > 15; *P < 0.01; **P < 0.05, Student’s t test). Bars = 20 µm.
LO9-177 Interacts with OsBC1, a bHLH Protein
We screened the rice library using the LO9-177 protein as a bait and isolated a novel bHLH protein (Os09g33580) that we named OsBUL1 COMPLEX1 (OsBC1; Fig. 6A). Expression patterns of OsBC1 and LO9-177 were similar to each other in several organs including collars and panicles (Supplemental Figs. S11B and S12). LO9-177 was able to form a homodimer and interact with OsBUL1 homologs such as OsBU1, OsBUL2, and OsBUL3, except OsILI1. Moreover, LO9-177 did not interact with OsIBH1 and APG, which was reported to interact with PGL2 as a bHLH protein (Fig. 6A; Heang and Sassa, 2012b). GST pull-down assays with GST:OsBC1 and TRX:LO9-177:His confirmed the interaction between them (Fig. 6B). OsBC1 was localized in the nucleus whereas LO9-177 was found in both the nucleus and cytoplasm (Supplemental Fig. S9, A and B). BiFC assays using rice protoplasts displayed florescent signals in the nucleus with speckles from reconstructed YFP by nYFP:OsBC1 and cYFP:LO9-177 (Fig. 6C), suggesting the interaction may occur near active transcription sites (Reddy et al., 2012). Interestingly, OsBC1 was able to interact with OsIBH1 although it could not interact with OsBUL1 or OsBUL1 homologs (Supplemental Fig. S7B).
Figure 6.
LO9-177 interacts with OsBC1. A, LO9-177 is able to make a homodimer. It interacts with OsBUL1 and its homologs, such as OsBU1, OsBUL2, and OsBUL3, except ILI1 (OsILI1), with different affinities and with a typical bHLH protein, OsBC1, but does not interact with OsIBH1 and APG. B, GST pull-down assays showed LO9-177:His fusion proteins were pulled down only together with GST:OsBC1 fusion proteins. C, Reconstructed yellow fluorescence from YFPn:OsBC1 and YFPc:LO9-177 was detected only in the nucleus with speckles of transfected rice protoplasts. The negative controls that were conducted by cotransfecting unfused YFP fragments together with a single complementary BiFC interactor protein resulted in no detectable signals (Supplemental Fig. S9C). Bar = 5 µm.
OsBUL1, LO9-177, and OsBC1 Form a Complex
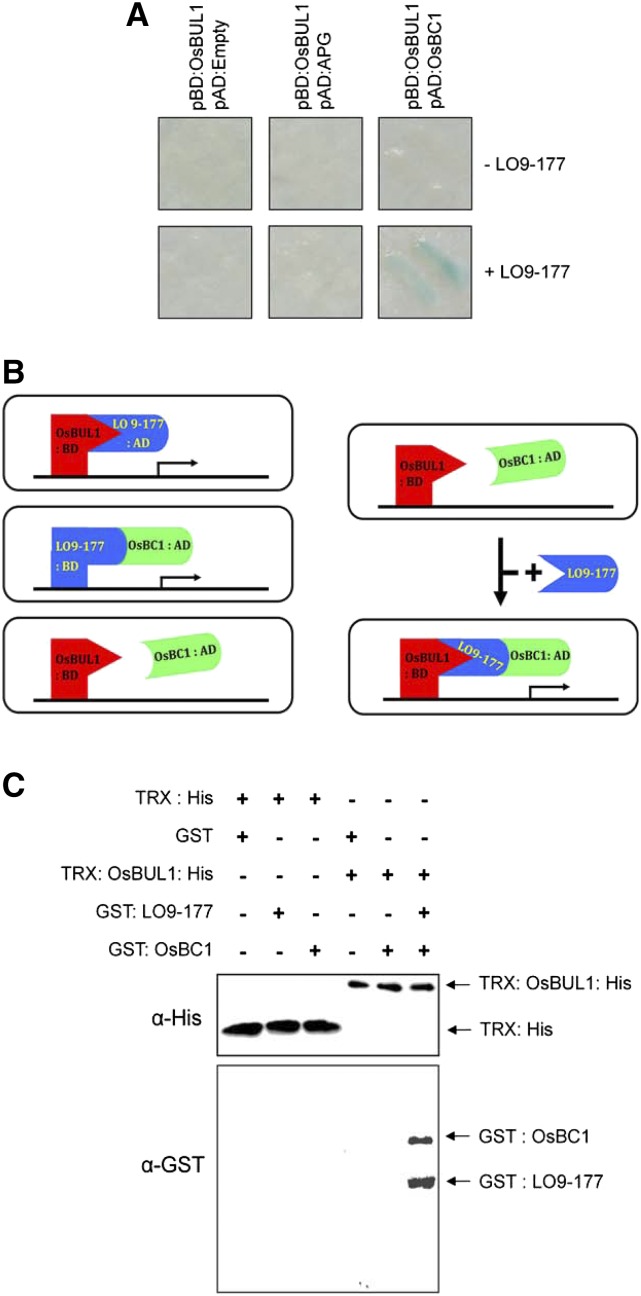
LO9-177 interacted with both OsBUL1 and OsBC1 while OsBUL1 did not directly interact with OsBC1. A yeast three-hybrid system was used to evaluate whether LO9-177 is a molecular mediator between OsBUL1 and OsBC1. The experiment revealed that OsBUL1 interacts with OsBC1 in the presence of LO9-177, indicating formation of a trimeric complex consisting of OsBUL1, LO9-177, and OsBC1 (Fig. 7, A–C). Interaction between OsBUL1 and APG was not enhanced by LO9-177 (Heang and Sassa, 2012b). We confirmed the formation of a complex by in vitro pull-down assays using recombinant proteins obtained from E. coli culture. GST:OsBC1 fusion proteins were pulled down together with TRX:OsBUL1:His fusion proteins only in the presence of LO9-177 (Fig. 7C). Notably, OsBC1 has transcriptional activation activity and forms a homodimer (Supplemental Fig. S8). GUS staining of rice plants harboring pOsBC1:GUS construct indicated that the gene is preferentially expressed in anthers and lamina joint. This expression pattern is similar to that of OsBUL1 (Fig. 8). In addition, we examined the expression of OsBUL1 and OsBC1 in oslg1-1 mutant that is defective in collar formation (Lee et al., 2007). In the mutant, both OsBUL1 and OsBC1 transcripts were rarely detectable in the lamina joint area, suggesting that both genes are preferentially expressed in the lamina joint rather than in the sheath or blade of mature leaves (Supplemental Fig. S2, G and H).
Figure 7.
OsBUL1 interacts with OsBC1 through LO9-177. A, Yeast three-hybrid systems demonstrated that OsBUL1 interacts with OsBC1 only in the presence of LO9-177. However, coexpression of LO9-177 did not affect the interaction between OsBUL1 and APG. B, Schema showing the positive interaction between OsBUL1 and OsBC1 mediated by LO9-177 in yeast cells. C, His pull-down assays demonstrated that OsBC1 can be pulled down with OsBUL1 only in the presence of LO9-177.
Figure 8.
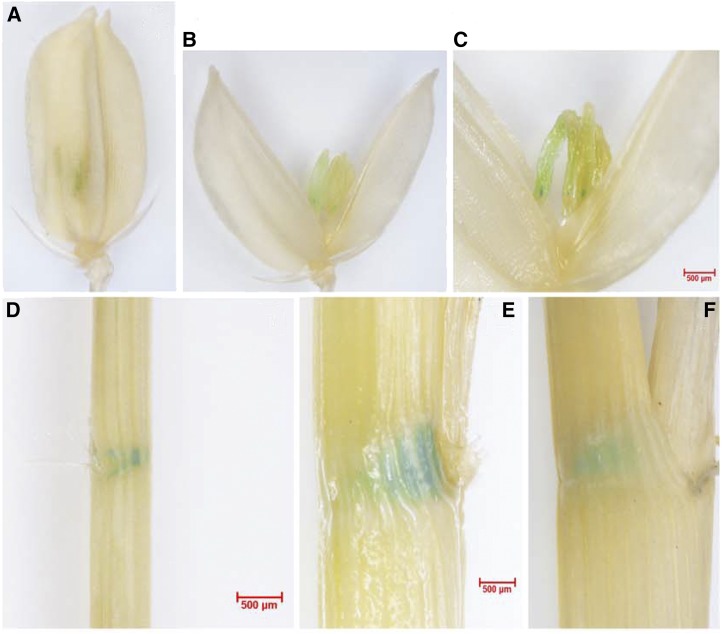
Histochemical GUS staining of rice plants harboring pOsBC1:GUS. A to C, Closed and open spikelets. D to F, Lamina joint parts of 10-d-old rice seedlings (D) and mature plants at heading stage (E and F).
Expression of OsBC1 Driven by OsBUL1 Promoter Increased Lamina Inclination and Grain Size
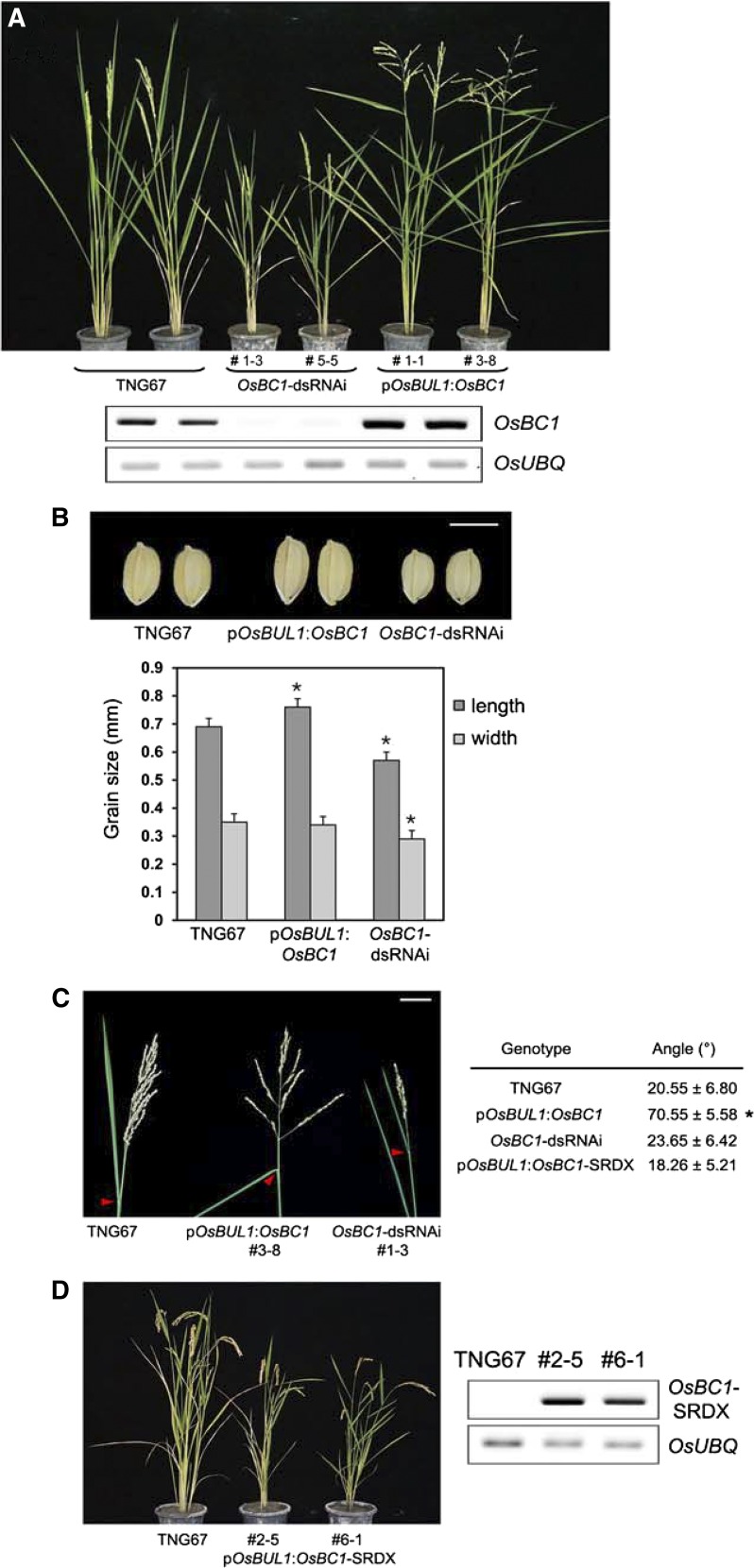
We examined the functional activity of OsBC1 by expressing the gene under the control of OsBUL1 promoter in rice. Transgenic plants containing pOsBUL1:OsBC1 showed phenotypes of increased lamina angles and grain size (Fig. 9, A–C). Epidermal cells of mature grains from the transgenic plants were enlarged, and the expression level of genes involved in cell elongation such as expansins was higher compared to the wild type (Supplemental Fig. S14). Conversely, OsBC1-dsRNAi transgenic rice lines exhibited semidwarf phenotype with small grains (Fig. 9, A–C) but did not show any obvious reduction in lamina inclination. Panicle branches were widely spread in the pOsBUL1:OsBC1 plants, whereas they were short and compact in the OsBC1-dsRNAi lines (Fig. 9C). Interestingly, transgenic Arabidopsis overexpressing OsBC1 under the control of the 35S promoter produced narrow and elongated leaves that were composed of elongated cells (Supplemental Fig. S10).
Figure 9.
Observation of functional activity of OsBC1 at locations where OsBUL1 is expressed. A, Transgenic rice plants containing pOsBUL1:OsBC1 had dramatically increased angles of leaves and panicle branches, whereas dsRNAi of OsBC1 lines were semidwarf without obvious changes of leaf angles. RNAs were extracted from young panicles for cDNA synthesis for RT-PCR. The 273-bp fragment of OsBC1 coding region amplified by primers, 5′-GACCACTCTCAGAAGATGGAAG-3′ and 5′-CTACTGGAAAGAGCACATG-3′, was used for dsRNAi construction. B, The grain size of transgenic plants with pOsBUL1:OsBC1 also increased, while dsRNAi lines of OsBC1 produced small grains (*P < 0.001, Student’s t test). Bar = 5 mm. C, Angles for panicle branches are also increased in transgenic plants with pOsBUL1:OsBC1, while dsRNAi lines of OsBC1 produced compact panicle architecture. Red arrowheads indicate the lamina joint of flag leaf in each genotype. Bar = 5 cm. The third leaf from the top of the main stem was used for lamina angle measurement, and values for leaf angles are given as means ± sd (n = 8 to 12; *P < 0.001, Student’s t test). D, Expression of OsBC1-SRDX, a repressor form of OsBC1 under the control of OsBUL1 promoter, rendered rice plants to exhibit similar phenotypes to OsBC1 dsRNAi lines. RNAs were isolated from collars for cDNA synthesis and RT-PCR using primers, 5′-CATCCCTGAAGATGCCTCAATG-3′ and 5′-TATGCGAATCCTAGTTCCAGTTCGAGATC-3′.
We also generated a construct for chimeric OsBC1 repressor by inserting oligomers coding for the SRDX domain (LDLDLELRLGFA; Hiratsu et al., 2003) in front of the stop codon of OsBC1. Fusion of the SRDX domain to a transcriptional activator can convert it into a repressor that overrides activation of endogenous transcription factors, resulting in a dominant-negative phenotype. On the contrary, the SRDX fusion to a native repressor enhances transcriptional repression, causing overexpression phenotypes (Ikeda and Ohme-Takagi, 2009). Transgenic rice plants expressing the transgene under the control of the OsBUL1 promoter showed phenotypes similar to those of OsBUL1 dsRNAi lines (Fig. 9D), indicating that OsBC1 may act as a transcriptional activator in lamina joints and panicles of rice.
DISCUSSION
In this study, we screened rice T-DNA mutant pools to identify plants showing abnormal lamina angles, and a T-DNA tagging line for OsBUL1 KO was isolated and characterized. Previously, atypical HLH proteins similar but different from OsBUL1 have been reported to play roles in cell elongation in an antagonistic manner with other atypical HLH and bHLH proteins in Arabidopsis and rice (Wang et al., 2009; Zhang et al., 2009; Ikeda et al., 2012). Arabidopsis AIFs, as atypical HLH proteins, act as negative regulators of BR signaling and interacting antagonists of PREs (Wang et al., 2009), another group of atypical HLH proteins including OsBUL1. Moreover, Arabidopsis PREs are able to interact with IBH1 as rice ILI1 does with OsIBH1 to have antagonistic effects on cell elongation (Zhang et al., 2009). Recently, a triantagonistic model has been suggested for PRE1, IBH1, and ACEs/CRYPTOCHROME INTERACTING BASIC-HELIX-LOOP-HELIX5 (CIB5) in Arabidopsis without AIFs (Ikeda et al., 2012). Based on this model, PRE1 positively regulates cell elongation by forming heterodimers with IBH1 that suppresses IBH1 inhibition of ACEs. AIFs, like IBH1, are also known to interact not only with PREs but also with ACE1, implying a complicated network of HLH-bHLH proteins in cell elongation (Ikeda et al., 2013; Supplemental Fig. S13).
OsBUL1 is upregulated by exogenous BL similar to OsBU1 (Tanaka et al., 2009), and the OsBUL1D and OsBUL1-overexpressing rice exhibited increased lamina inclination and grain size resulting from cell elongation. On the contrary, osbul1 did not respond to BL treatment in lamina bending assays and produced erect leaves with small spikelets. This phenotypic alteration is well matched with the spatial expression pattern of OsBUL1.
To understand the molecular functional networks of OsBUL1 in rice, the OsBUL1 bait protein was screened against the rice cDNA library using yeast two-hybrid methods, and a small uncharacterized protein, LO9-177, was isolated as an OsBUL1-interacting protein. Although a bHLH protein, APG was reported to interact with OsBUL1 as a negative regulator of cell elongation (Heang and Sassa, 2012a, 2012b, 2012c), we could not detect a positive interaction between APG and OsBUL1 or its homologs including BU1 (OsBU1), OsBUL2, OsBUL3, and ILI1 in our yeast systems (Supplemental Fig. S7B). Phenotypic analyses of transgenic rice using gain-of-function and loss-of-function approaches demonstrated that LO9-177 is a genetic factor affecting lamina inclination as well as encoding an interacting protein of OsBUL1. Mild lamina inclination phenotype of LO9-177 overexpressing plants might be due to the amount of interacting proteins for functional complex formation in the lamina joint. Actually, the KxDL domain in LO9-177 is regarded as being critical in selective interaction with a protein or protein complex (Hayes et al., 2011). In mammals, the KXD protein containing a KxDL motif interacts with biogenesis of lysosome-related organelles complex-1 subunit 1 (BLOS1) whose mutation is responsible for Hermansky-Pudlak syndrome and transgenic KXD knockout mouse showed similar symptoms of Hermansky-Pudlak syndrome (Yang et al., 2012). Thus, LO9-177 was used as a bait for screening of interacting proteins and interestingly a bHLH protein, OsBC1 was isolated as a LO9-177 interactor. LO9-177 was able to interact with atypical HLH proteins such as OsBU1, OsBUL1, OsBUL2, and OsBUL3 (but not OsIBH1), but did not interact with APG showing interaction specificity and complexity among HLH/bHLH and LO9-177 proteins. Despite LO9-177 being localized both in the nucleus and the cytoplasm, the interaction with OsBC1 occurs only in the nucleus, which is likely due to subcellular localization of OsBC1, a nuclear protein. Next, we tested the possibility of formation of a complex consisting of OsBUL1, LO9-177, and OsBC1. Yeast two- and three-hybrid systems together with pull-down assays suggested that the three proteins form a complex.
In order to examine the functional activities of OsBC1, we generated transgenic rice containing pOsBUL1:OsBC1 and the transgenic lines exhibited a significant increase in lamina angles and grain size. In contrast, dsRNAi and pOsBUL1:OsBC1-SRDX lines showed reduced height and produced smaller grains, implying that OsBC1 is a transcriptional activator that plays a positive role in cell elongation and the transcriptional activity of OsBC1 was shown in yeast cells (Supplemental Fig. S8). However, the lamina angle of OsBC1 knockdown plants was not affected significantly, suggesting that the loss-of-function effect of OsBC1 may be compensated for by the function of other homologous genes. For example, Arabidopsis ACEs/CIBs, which are bHLH transcription factors, play a similar role in cell elongation (Ikeda et al., 2012). Of note, OsBC1 as well as OsBUL1 are upregulated by BL (Supplemental Fig. S11). Although it is not obvious yet whether OsBC1 is a homolog of Arabidopsis ACEs, it seems that the triantagonistic model suggested in Arabidopsis research (Ikeda et al., 2012) can be applied to rice in cell elongation (Fig. 10): Atypical HLH proteins including OsBUL1 interact with OsIBH1, OsIBH1-overexpressing transgenic rice produced small cells (Zhang et al., 2009), and we identified OsBC1, a transcriptional activator containing a bHLH domain for cell elongation that interacts with OsIBH1. In Arabidopsis, there is another group of atypical HLH proteins that includes AIF1, AIF2, AIF3, and AIF4 whose mRNAs are expressed ubiquitously. They act as negative regulators in cell elongation by interacting with both PREs and ACE1 (Ikeda et al., 2013). Identification and functional characterization of rice AIF homologs will provide more information about plant cell elongation through the HLH-bHLH regulation systems in plants.
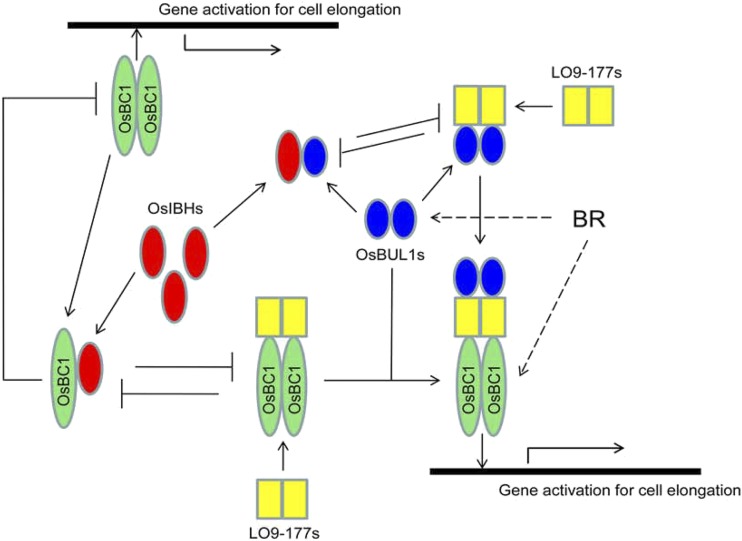
Figure 10.
Simplified working model for cell elongation through a network of HLH-bHLH proteins. Previous studies (Tanaka et al., 2009; Wang et al., 2009; Zhang et al., 2009; Ikeda et al., 2012) and data provided by this study were used together for the construction of the model. The formation of a hexameric complex was drawn based on yeast two- and three-hybrid results with pull-down assays in this study. The arrows and T-shaped lines signify positive and negative actions, respectively. The dashed arrows were drawn based on the up-regulation of OsBUL1 and OsBC1 by BL.
In addition, we provide evidence of a novel complex consisting of OsBUL1, LO9-177, and OsBC1 that has a positive effect on cell elongation. Moreover, each component in the complex is capable of making a homodimer implying that assembly of a hexameric complex might be possible (Fig. 10).
Regulation of plant cell elongation is governed by multiple signals and is also important for normal development and adaptation to various environmental conditions. The balance among atypical HLH and bHLH proteins seems to be critical for cell elongation, and the fine-tuning of the balance or stabilization of molecular activity required for cell elongation by OsBC1 is likely achieved by the formation of a complex with LO9-177 and atypical HLHs including OsBUL1 (Fig. 10). By better understanding the machineries controlling the balance of HLH-bHLH proteins, we may manipulate the lamina inclination of crop plants, which is an important agronomic trait for improved productivity.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
OsBUL1 activation tagging line, PFG_3A_01926, and OsBUL1null mutant line, PFG_2D_01383, are japonica rice cultivars Dongjin and Hwayoung, respectively. Transgenic rice plants were produced with Dongjin or Tainung67 (TNG67) japonica rice cultivars. Rice plants (Oryza sativa) were grown in the field under natural long days or in the greenhouse with 28°C day/25°C night cycles. Lamina angles were measured between a stem and a leaf blade at the second or the third leaf from 8 to 12 individual plants per line. To assess the leaf angles of mature rice plants in the paddy field, leaves were numbered from top to bottom. Thus, each flag leaf of the main tiller was regarded as the first leaf. Arabidopsis (Arabidopsis thaliana) strain, Columbia (Col), and tobacco (Nicotiana benthamiana) plants were used for transformation and grown in a growth chamber under long days with 23°C day/20°C night cycles.
Lamina Joint Inclination Bioassay
Sterilized seeds were germinated and grown for 8 d in a growth chamber. The lamina joint inclination assays were performed as previously described (Jeong et al., 2007). Seedlings were sampled by excising approximately 2-cm segments that contained lamina joints at the same position from each plant under dim light conditions. They were floated on distilled water containing various concentrations of BL. After incubation at 28°C, the angle induced between the lamina joint and the sheath was measured.
Vector Construction and Transformation
The whole/parts of open reading frames (ORFs) of OsBUL1, PRE1, LO9-177, OsBC1, and OsBC1-SRDX were cloned into pGA3426 or its derivatives for overexpression and/or dsRNAi purposes in rice. For expression by OsBUL1 promoter, the ubiquitin promoter of pGA3426 was replaced with the 2.2-kb OsBUL1 promoter. Vector pGA3383 was used for analyzing OsBUL1 promoter activity using the GUS reporter in rice (Kim et al., 2009). Constructed plasmids were individually transformed into embryonic calli of Dongjin or TNG67 rice cultivars by Agrobacterium tumefaciens-LBA4404 mediation as described previously (Jeon et al., 2000). pGA643 vector and pJawohl8-RNAi silencing vector (kindly provided by I.E. Somssich, Max Planck Institute for Plant Breeding Research) were used for Arabidopsis and tobacco transformation by floral dipping and tissue culture via A. tumefaciens-mediated DNA delivery (Clough and Bent, 1998; Jang et al., 2002), respectively.
Hormone Treatment
Eight-day-old rice (O. sativa cv TNG67) seedlings grown in the growth chamber were treated with brassinolide (1 µm, BL from Sigma-Aldrich) or gibberellin (100 µM GA3 from Sigma-Aldrich). Whole parts above roots were harvested for RNA extraction at the 24 h time point after treatment.
Total RNA Isolation and Quantitative RT-PCR Analysis
Total RNAs of all the materials harvested were isolated using RNeasy plant mini kit (Qiagen) or Trizol solution (Invitrogen) according to the manufacturer’s instructions. DNase-treated RNA was subjected to reverse transcriptase reactions using oligo(dT) primer and Superscript III reverse transcriptase (Invitrogen) according to the manufacturer’s protocol. Subsequent PCR was performed with the first-strand cDNA mixture and EX-Taq polymerase (Takara Bio). qPCR was performed on a CFX96TM real-time system (Bio-Rad) using Maxima SYBR Green qPCR Master Mix (Thermo). For PCR, each sample was analyzed in triplicate. The run protocol was as follows: denaturation at 95°C for 10 min and annealing/extension repeated 45 times (95°C for 15 s and 60°C for 30 s, data acquisition was performed). Housekeeping genes such as OsUBQ (Komiya et al., 2008), OsAct (Caldana et al., 2007), AtPEX4 (Gregis et al., 2013), and AtACT (Jang et al., 2009) were included in the reactions as internal controls for normalizing the variations in the amount of cDNA used (Guénin et al., 2009). The threshold cycle (CT) was automatically determined for each reaction by the system set with default parameters. The specificity of the qRT-PCR was determined by curve analysis of the amplified products using the standard method installed in the system. Information on primers used is presented in Supplemental Table S1.
GUS Staining
For promoter analysis, about 2.2 kb of OsBUL1 5′ region was amplified with primers 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTGGCGCGCGATGATTTCGTGACATG-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCGTCAACAGCTAGCCTCTTCTACCAAACAC-3′ and cloned into pDONR207 by BP reaction (Invitrogen). For OsBC1 promoter, an ∼3-kb fragment of OsBC1 5′ region was amplified with primers 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTACTTAATTTAGTGTCATGTAAG-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTGCCAATGCCCTTGGTGTCCTAGATG-3′ and cloned into pDONR207. The entry clones for OsBUL1 promoter and OsBC1 promoter were used for LR reaction with pGA3383-Gateway vector for GUS fusion, respectively. The resulting plasmids were transformed into rice, and GUS staining was performed according to the method described previously (Jefferson, 1989).
Histological Analyses
Lamina joint samples were fixed by 4% paraformaldehyde in 0.1 m sodium phosphate buffer, dehydrated through a graded ethanol series, replaced with xylene, and embedded in Paraplast plus (Sigma-Aldrich). Paraffin sections (12 μm) were cut and stained with filtered 1% toluidine blue. Anthers stained with GUS were embedded using Epon812 resin (Fluka) and polymerized at 60°C. Cross sections (3 μm) were cut with a rotary microtome (Leica). The sections were photographed under a light microscope (Olympus BX51).
Yeast Assays and BiFC Assays
Rice (O. sativa cv TNG67) cDNA library was constructed using poly(A)+ mRNAs extracted from whole aboveground parts including leaves, culms, and panicles at different developmental stages. The HybriZAP-2.1 XR cDNA library construction kit was used, and the initial plaque-forming units (pfu) of the constructed library was 1.8 × 106. Screening of the library with baits such as pBD:OsBUL1 and pBD:LO9-177 was performed as described previously (Jang et al., 2002). For yeast three-hybrid assays, we used an adaptor vector, pBridge (Clontech), together with prey vectors. EcoRI/XhoI fragment of amplified OsBUL1 full ORF (using 5′-GGAATTCATGTCGAGCAGAAGGTCGTCGCGTG-3′ and 5′-GCCTCGAGTCAGGAGCGGAGGATGCTGCGGATG-3′; restriction enzyme sites are underlined) was cloned into EcoRI/SalI sites fused to the binding domain (BD) and PspOMI/BglII fragment of amplified LO9-177 ORF (using 5′-GAGGGGCCCTCATGGAGAAGTCGCCGCCGGAG-3′ and 5′-GCCAGATCTTTAATCAAGCGGACTTTCAAG-3′; restriction enzyme sites are underlined) was inserted into NotI/BglII sites of the pBridge vector for independent expression in yeast cells. Verification of interactions with X-Gal filter assays were also conducted as reported by Jang et al. (2015). For BiFC assays in rice, each cDNA of OsBUL1, LO9-177, and OsBC1 was cloned into pVYCE vector or pVYNE vectors (Citovsky et al., 2006; Tzfira et al., 2005) for addition of half YFP to the each cDNA. YFP and CFP fusion for cellular localization of each protein was also conducted as previously described (Jang et al., 2008). Isolation and transfection of Arabidopsis and rice protoplasts were as described by Wu et al. (2009) and Zhang et al. (2011). Images of cells with fluorescence were taken by confocal microscopy (LSM 510 META NLO DuoScan; Carl Zeiss).
Protein Pull-Down Assays
The cDNAs encoding OsBUL1, LO9-177, and OsBC1 were cloned into the EcoRI/XhoI sites of the pGEX6P-1 for GST fusion and amplified OsBUL1 ORF with EagI/XhoI ends and LO9-177 ORF with BamHI/XhoI ends were also inserted into NotI/XhoI and BamHI/XhoI sites of pET201 vector (Bhalerao et al., 1999), respectively, for His fusion. The nucleotide sequences for the fusion proteins were confirmed by sequencing (Sequencing Core Facility, Scientific Instrument Center, Academia Sinica, Taiwan). Constructed plasmids were introduced into Escherichia coli BL21 (DE3), and cells transformed by construct of GST fusion were cultured to an OD600 followed by induction with 1 mm isopropyl-β-d-thiogalactopyranoside. After overnight induction at 16°C, cells were collected and homogenized with lysis buffer (10 mm Tris, pH 8.0, 5 mm DTT, 1% Triton X-100, and 150 mm NaCl). The soluble GST fusion proteins were extracted and immobilized on glutathione-MagBeads (GenScript) for subsequent pull-down assays. For His pull-down assays, cells transformed by His-fusion construct were cultured until the OD600 reached 0.6, and isopropyl-β-d-thiogalactopyranoside was added to a final concentration of 1 mm to start induction. After overnight induction at 16°C, cells were harvested and homogenized with lysis buffer (10 mm Na2HPO4, 10 mm NaH2PO4, 500 mm NaCl, and 20 mm imidazole). The soluble thioredoxin (TRX)-tagged His fusion proteins were extracted and immobilized on Ni-charged MagBeads (GenScript), and the recombinant proteins were purified according to the manufacturer’s instructions. In vitro pull-down assays were performed by incubation with a combined mixture of proteins in a binding buffer (50 mm Tris-HCl at pH 7.5, 100 mm NaCl, 0.25% Triton X-100, and 35 mm β-mercaptoethanol) for 4 to 6 h at 4°C with rotation. Collection of the beads was achieved using a MagRack 6 (GE Healthcare) followed by washing six times with binding buffer. The pulled-down proteins were separated on a 12% SDS-polyacrylamide gel and detected by western blotting using anti-GST or anti-His antibody.
In Situ Hybridization
Young spikelets of rice (cv TNG67) were collected and fixed, dehydrated, embedded, and sliced (10 µm thickness), and hybridization was performed as previously described (Ko et al., 2014). For preparation of digoxigenin-labeled RNA probes, DNA fragments containing the end part of the coding region to 3′ untranslated region or only 3′ untranslated region from each gene, LO9-177 (352 bp) and OsBC1 (326 bp), were amplified using following primers: LO9-177-For, 5′-GAACAATGCTTTGCGGAGGTGTC-3′ and LO9-177-Rev, 5′-CATAGGACTACAAGGTTACACAAC-3′; OsBC1-For, 5′-CAATCTCATGCCATCATGGAC-3′; and OsBC1-Rev, 5′-CAGACAAGGGGATGGACTCG-3′. Each amplified DNA fragment was cloned into the pGEM-T vector (Promega), and each sense and antisense probe was synthesized by T7 and SP6 RNA polymerases, respectively. Hybridization was performed at 62 or 63°C with 20 ng of digoxigenin-labeled RNA probes.
Accession Numbers
Genes in this article can be found in the GenBank/EMBL or RiceGE databases under the following accession numbers: ACE1 (At1g68920), ACE2 (At1g10120), ACE3 (At3g23690), AIF1 (At3g05800), AIF2 (At3g06590), AIF3 (At3g17100), AIF4 (At1g09250), APG (Os05g04740.1), CIB1 (At4g34530), CIB5 (At1g26260), PRE1 (At5g39860), PRE2 (At5g15160), PRE3 (At1g74500), PRE4 (At3g47710), PRE5 (At3g28857), PRE6 (At1g26945), IBH1 (At2g43060), LO9-177 (Os03g43910), OsBC1 (Os09g33580), OsBU1 (BU1, Os06g12210), OsBUL1 (Os02g51320), OsBUL2 (Os03g07540), OsBUL3 (Os10g26410), OsBUL4 (Os10g26460), OsBUL5 (Os11g39000.1), OsBUL6 (Os03g07510.1), OsEXPA1 (Os04g15840), OsEXPA2 (Os01g60770), OsEXPA3 (Os05g19570), OsEXPA4 (Os05g39990), OsILI1 (ILI1, Os04g54900), OsIBH1 (Os04g56500), OsXTH1 (Os04g51460), and OsXTR1 (Os11g33270).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. OsBUL1 dsRNAi lines phenocopy OsBUL1 null mutation.
Supplemental Figure S2. Increased OsBUL1 expression confers larger grains and leaf angles in rice.
Supplemental Figure S3. Introduction of rice OsBUL1 into dicot plants Arabidopsis and tobacco.
Supplemental Figure S4. Morphological changes of OsBUL1KO seedlings under light or dark conditions.
Supplemental Figure S5. Brassinolide response on wild-type and OsBUL1KO rice.
Supplemental Figure S6. Expression level of LO9-177 is positively linked to grain size.
Supplemental Figure S7. Protein interactions using yeast two-hybrid systems.
Supplemental Figure S8. OsBC1 has autotranscriptional activation activity.
Supplemental Figure S9. Subcellular localization of proteins.
Supplemental Figure S10. Transgenic Arabidopsis overexpressing OsBC1.
Supplemental Figure S11. Expression analyses of LO9-177 and OsBC1.
Supplemental Figure S12. In situ hybridization of genes in spikelets.
Supplemental Figure S13. A phylogenetic tree showing the relationships among atypical HLH and typical bHLH proteins.
Supplemental Figure S14. OsBC1 affects cell size in plants.
Supplemental Table S1. A list of primers used in this study.
Supplementary Material
Acknowledgments
We thank Pei-Chun Liao and Wei-Chih Lin for rice transformation, and Hsing-Hui Lee and Ya-Chen Liu for assistance with pull-down assays. We also thank members of core facility laboratories of Academia Sinica for microscopy, DNA sequencing, and in situ hybridization and Miranda Loney for help with English editing.
Glossary
- BR
brassinosteroid
- HLH
helix-loop-helix
- bHLH
basic helix-loop-helix
- dsRNAi
double-stranded RNA interference
- BL
brassinolide
- BiFC
bimolecular florescence complementation
- ORF
open reading frame
References
- Bai M-Y, Zhang L-Y, Gampala SS, Zhu S-W, Song W-Y, Chong K, Wang Z-Y (2007) Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc Natl Acad Sci USA 104: 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt C, Zhao M, Gonzalez A, Lloyd A, Schiefelbein J (2005) The bHLH genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis. Development 132: 291–298 [DOI] [PubMed] [Google Scholar]
- Bhalerao RP, Salchert K, Bakó L, Okrész L, Szabados L, Muranaka T, Machida Y, Schell J, Koncz C (1999) Regulatory interaction of PRL1 WD protein with Arabidopsis SNF1-like protein kinases. Proc Natl Acad Sci USA 96: 5322–5327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, Baker NE (2011) A network of broadly expressed HLH genes regulates tissue-specific cell fates. Cell 147: 881–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldana C, Scheible W-R, Mueller-Roeber B, Ruzicic S (2007) A quantitative RT-PCR platform for high-throughput expression profiling of 2500 rice transcription factors. Plant Methods 3: 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Chen S (1995) Brassinosteroid-induced rice lamina joint inclination and its relation to indole-3-acetic acid and ethylene. Plant Growth Regul 16: 189–196 [Google Scholar]
- Citovsky V, Lee L-Y, Vyas S, Glick E, Chen M-H, Vainstein A, Gafni Y, Gelvin SB, Tzfira T (2006) Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta. J Mol Biol 362: 1120–1131 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Duek PD, Fankhauser C (2005) bHLH class transcription factors take centre stage in phytochrome signalling. Trends Plant Sci 10: 51–54 [DOI] [PubMed] [Google Scholar]
- Feng Z, Wu C, Wang C, Roh J, Zhang L, Chen J, Zhang S, Zhang H, Yang C, Hu J, et al. (2016) SLG controls grain size and leaf angle by modulating brassinosteroid homeostasis in rice. J Exp Bot 67: 4241–4253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrichsen DM, Nemhauser J, Muramitsu T, Maloof JN, Alonso J, Ecker JR, Furuya M, Chory J (2002) Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics 162: 1445–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregis V, Andrés F, Sessa A, Guerra RF, Simonini S, Mateos JL, Torti S, Zambelli F, Prazzoli GM, Bjerkan KN, et al. (2013) Identification of pathways directly regulated by SHORT VEGETATIVE PHASE during vegetative and reproductive development in Arabidopsis. Genome Biol 14: R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guénin S, Mauriat M, Pelloux J, Van Wuytswinkel O, Bellini C, Gutierrez L (2009) Normalization of qRT-PCR data: the necessity of adopting a systematic, experimental conditions-specific, validation of references. J Exp Bot 60: 487–493 [DOI] [PubMed] [Google Scholar]
- Hayes MJ, Bryon K, Satkurunathan J, Levine TP (2011) Yeast homologues of three BLOC-1 subunits highlight KxDL proteins as conserved interactors of BLOC-1. Traffic 12: 260–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heang D, Sassa H (2012a) Antagonistic actions of HLH/bHLH proteins are involved in grain length and weight in rice. PLoS One 7: e31325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heang D, Sassa H (2012b) An atypical bHLH protein encoded by POSITIVE REGULATOR OF GRAIN LENGTH 2 is involved in controlling grain length and weight of rice through interaction with a typical bHLH protein APG. Breed Sci 62: 133–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heang D, Sassa H (2012c) Overexpression of a basic helix-loop-helix gene Antagonist of PGL1 (APG) decreases grain length of rice. Plant Biotechnol 29: 65–69 [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M (2003) Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J 34: 733–739 [DOI] [PubMed] [Google Scholar]
- Hong Z, Ueguchi-Tanaka M, Shimizu-Sato S, Inukai Y, Fujioka S, Shimada Y, Takatsuto S, Agetsuma M, Yoshida S, Watanabe Y, et al. (2002) Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J 32: 495–508 [DOI] [PubMed] [Google Scholar]
- Hong Z, Ueguchi-Tanaka M, Umemura K, Uozu S, Fujioka S, Takatsuto S, Yoshida S, Ashikari M, Kitano H, Matsuoka M (2003) A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 15: 2900–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun Y, Lee I (2006) KIDARI, encoding a non-DNA Binding bHLH protein, represses light signal transduction in Arabidopsis thaliana. Plant Mol Biol 61: 283–296 [DOI] [PubMed] [Google Scholar]
- Ikeda M, Fujiwara S, Mitsuda N, Ohme-Takagi M (2012) A triantagonistic basic helix-loop-helix system regulates cell elongation in Arabidopsis. Plant Cell 24: 4483–4497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Mitsuda N, Ohme-Takagi M (2013) ATBS1 INTERACTING FACTORs negatively regulate Arabidopsis cell elongation in the triantagonistic bHLH system. Plant Signal Behav 8: e23448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Ohme-Takagi M (2009) A novel group of transcriptional repressors in Arabidopsis. Plant Cell Physiol 50: 970–975 [DOI] [PubMed] [Google Scholar]
- Jang S, An K, Lee S, An G (2002) Characterization of tobacco MADS-box genes involved in floral initiation. Plant Cell Physiol 43: 230–238 [DOI] [PubMed] [Google Scholar]
- Jang S, Marchal V, Panigrahi KCS, Wenkel S, Soppe W, Deng X-W, Valverde F, Coupland G (2008) Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J 27: 1277–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S, Torti S, Coupland G (2009) Genetic and spatial interactions between FT, TSF and SVP during the early stages of floral induction in Arabidopsis. Plant J 60: 614–625 [DOI] [PubMed] [Google Scholar]
- Jang S, Choi S-C, Li H-Y, An G, Schmelzer E (2015) Functional characterization of Phalaenopsis aphrodite flowering genes PaFT1 and PaFD. PLoS One 10: e0134987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA. (1989) The GUS reporter gene system. Nature 342: 837–838 [DOI] [PubMed] [Google Scholar]
- Jeon J-S, Lee S, Jung K-H, Jun S-H, Jeong D-H, Lee J, Kim C, Jang S, Yang K, Nam J, et al. (2000) T-DNA insertional mutagenesis for functional genomics in rice. Plant J 22: 561–570 [DOI] [PubMed] [Google Scholar]
- Jeong D-H, An S, Kang H-G, Moon S, Han J-J, Park S, Lee HS, An K, An G (2002) T-DNA insertional mutagenesis for activation tagging in rice. Plant Physiol 130: 1636–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong D-H, An S, Park S, Kang H-G, Park G-G, Kim S-R, Sim J, Kim Y-O, Kim M-K, Kim S-R, et al. (2006) Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J 45: 123–132 [DOI] [PubMed] [Google Scholar]
- Jeong D-H, Lee S, Kim SL, Hwang I, An G (2007) Regulation of brassinosteroid responses by phytochrome B in rice. Plant Cell Environ 30: 590–599 [DOI] [PubMed] [Google Scholar]
- Kim S-R, Lee D-Y, Yang J-I, Moon S, An G (2009) Cloning vectors for rice. J Plant Biol 52: 73–78 [Google Scholar]
- Ko S-S, Li M-J, Sun-Ben Ku M, Ho Y-C, Lin Y-J, Chuang M-H, Hsing H-X, Lien Y-C, Yang H-T, Chang H-C, Chan M-T (2014) The bHLH142 transcription factor coordinates with TDR1 to modulate the expression of EAT1 and regulate pollen development in rice. Plant Cell 26: 2486–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu K, Maekawa M, Ujiie S, Satake Y, Furutani I, Okamoto H, Shimamoto K, Kyozuka J (2003) LAX and SPA: major regulators of shoot branching in rice. Proc Natl Acad Sci USA 100: 11765–11770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya R, Ikegami A, Tamaki S, Yokoi S, Shimamoto K (2008) Hd3a and RFT1 are essential for flowering in rice. Development 135: 767–774 [DOI] [PubMed] [Google Scholar]
- Lee J, Park J-J, Kim SL, Yim J, An G (2007) Mutations in the rice liguleless gene result in a complete loss of the auricle, ligule, and laminar joint. Plant Mol Biol 65: 487–499 [DOI] [PubMed] [Google Scholar]
- Lee S, Choi SC, An G (2008) Rice SVP-group MADS-box proteins, OsMADS22 and OsMADS55, are negative regulators of brassinosteroid responses. Plant J 54: 93–105 [DOI] [PubMed] [Google Scholar]
- Lee S, Lee S, Yang K-Y, Kim Y-M, Park S-Y, Kim SY, Soh M-S (2006) Overexpression of PRE1 and its homologous genes activates gibberellin-dependent responses in Arabidopsis thaliana. Plant Cell Physiol 47: 591–600 [DOI] [PubMed] [Google Scholar]
- Nakamura A, Fujioka S, Takatsuto S, Tsujimoto M, Kitano H, Yoshida S, Asami T, Nakano T (2009) Involvement of C-22-hydroxylated brassinosteroids in auxin-induced lamina joint bending in rice. Plant Cell Physiol 50: 1627–1635 [DOI] [PubMed] [Google Scholar]
- Ning J, Zhang B, Wang N, Zhou Y, Xiong L (2011) Increased leaf angle1, a Raf-like MAPKKK that interacts with a nuclear protein family, regulates mechanical tissue formation in the lamina joint of rice. Plant Cell 23: 4334–4347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AS, Day IS, Göhring J, Barta A (2012) Localization and dynamics of nuclear speckles in plants. Plant Physiol 158: 67–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Morinaka Y, Ohnishi T, Sunohara H, Fujioka S, Ueguchi-Tanaka M, Mizutani M, Sakata K, Takatsuto S, Yoshida S, et al. (2006) Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat Biotechnol 24: 105–109 [DOI] [PubMed] [Google Scholar]
- Serna L. (2007) bHLH proteins know when to make a stoma. Trends Plant Sci 12: 483–485 [DOI] [PubMed] [Google Scholar]
- Shimada A, Ueguchi-Tanaka M, Sakamoto T, Fujioka S, Takatsuto S, Yoshida S, Sazuka T, Ashikari M, Matsuoka M (2006) The rice SPINDLY gene functions as a negative regulator of gibberellin signaling by controlling the suppressive function of the DELLA protein, SLR1, and modulating brassinosteroid synthesis. Plant J 48: 390–402 [DOI] [PubMed] [Google Scholar]
- Tanaka A, Nakagawa H, Tomita C, Shimatani Z, Ohtake M, Nomura T, Jiang C-J, Dubouzet JG, Kikuchi S, Sekimoto H, et al. (2009) BRASSINOSTEROID UPREGULATED1, encoding a helix-loop-helix protein, is a novel gene involved in brassinosteroid signaling and controls bending of the lamina joint in rice. Plant Physiol 151: 669–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T, Tian G-W, Lacroix B, Vyas S, Li J, Leitner-Dagan Y, Krichevsky A, Taylor T, Vainstein A, Citovsky V (2005) pSAT vectors: a modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol Biol 57: 503–516 [DOI] [PubMed] [Google Scholar]
- Van Camp W. (2005) Yield enhancement genes: seeds for growth. Curr Opin Biotechnol 16: 147–153 [DOI] [PubMed] [Google Scholar]
- Wada K, Marumo S, Ikekawa N, Morisaki M, Mori K (1981) Brassinolide and homobrassinolide promotion of lamina inclination of rice seedlings. Plant Cell Physiol 22: 323–325 [Google Scholar]
- Wang D, Zhang H, Hu G, Fu Y, Si H, Sun Z (2005) Genetic analysis and identification of alarge leaf angles (lla) mutant in rice. Chin Sci Bull 50: 492–494 [Google Scholar]
- Wang H, Zhu Y, Fujioka S, Asami T, Li J, Li J (2009) Regulation of Arabidopsis brassinosteroid signaling by atypical basic helix-loop-helix proteins. Plant Cell 21: 3781–3791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Xu Y, Zhang C, Ma Q, Joo S-H, Kim S-K, Xu Z, Chong K (2008) OsLIC, a novel CCCH-type zinc finger protein with transcription activation, mediates rice architecture via brassinosteroid signaling. PLoS One 3: e3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CY, Trieu A, Radhakrishnan P, Kwok SF, Harris S, Zhang K, Wang J, Wan J, Zhai H, Takatsuto S, et al. (2008) Brassinosteroids regulate grain filling in rice. Plant Cell 20: 2130–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F-H, Shen S-C, Lee L-Y, Lee S-H, Chan M-T, Lin C-S (2009) Tape-Arabidopsis Sandwich - a simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, He X, Yang L, Zhou Z, Cullinane AR, Wei A, Zhang Z, Hao Z, Zhang A, He M, et al. (2012) The BLOS1-interacting protein KXD1 is involved in the biogenesis of lysosome-related organelles. Traffic 13: 1160–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J (2005) A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120: 249–259 [DOI] [PubMed] [Google Scholar]
- Zhang L-Y, Bai M-Y, Wu J, Zhu J-Y, Wang H, Zhang Z, Wang W, Sun Y, Zhao J, Sun X, et al. (2009) Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 21: 3767–3780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Sun J, Cao X, Song X (2015) Epigenetic mutation of RAV6 affects leaf angle and seed size in rice. Plant Physiol 169: 2118–2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Su J, Duan S, Ao Y, Dai J, Liu J, Wang P, Li Y, Liu B, Feng D, Wang J, Wang H (2011) A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7: 30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S-Q, Hu J, Guo L-B, Qian Q, Xue H-W (2010) Rice leaf inclination2, a VIN3-like protein, regulates leaf angle through modulating cell division of the collar. Cell Res 20: 935–947 [DOI] [PubMed] [Google Scholar]
- Zhao S-Q, Xiang J-J, Xue H-W (2013) Studies on the rice LEAF INCLINATION1 (LC1), an IAA-amido synthetase, reveal the effects of auxin in leaf inclination control. Mol Plant 6: 174–187 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.