Abstract
With current anti-HIV treatments targeting only 4 of the 15 HIV proteins, many potential viral vulnerabilities remain unexploited. We report small-molecule inhibitors of the HIV-1 protein Nef. In addition to expanding the anti-HIV arsenal, small-molecule inhibitors against untargeted HIV proteins could be used to dissect key events in the HIV lifecycle. Numerous incompletely characterized interactions between Nef and cellular ligands, for example, present a challenge to understanding molecular events during HIV progression to AIDS. Assays with phage-displayed Nef from HIVNL4-3 were used to identify a series of guanidine alkaloid-based inhibitors of Nef interactions with p53, actin, and p56lck. The guanidines, synthetic analogs of batzellidine and crambescidin natural products, inhibit the Nef–ligand interactions with IC50 values in the low micromolar range. In addition, sensitive in vivo assays for Nef inhibition are reported. Although compounds that are effective in vitro proved to be too cytotoxic for cellular assays, the reported Nef inhibitors provide proof-of-concept for disrupting a new HIV target and offer useful leads for drug development.
To counter the HIV pandemic, inhibitors targeting HIV protease, reverse transcriptase, and virus-cell fusion are currently available; integrase inhibitors are in clinical trials (1); and viral entry (2) and maturation inhibitors (3) are under development. However, current treatments do not target all HIV proteins, and expanding the anti-HIV repertoire may improve treatment options. For example, strains of HIV missing the gene encoding Nef can fail to progress to AIDS (4–6). Nef-deleted strains of HIV provide a genetic proof-of-concept for the potential efficacy of Nef inhibitors. Nef inhibition by the guanidine alkaloid congeners reported here could provide lead compounds for pharmaceutical development and tools for understanding the enigmatic roles of Nef in the HIV lifecycle.
The HIV-1 Nef protein is ≈200 aa in length, with the exact length varying by HIV isolate. The Nef C terminus and myristolyated N terminus are relatively flexible (7, 8). Although many cellular binding partners to Nef have been identified (9, 10), how Nef contributes to the viral lifecycle is not well understood; however, Nef expression is essential for HIV propagation and maintenance of viral loads (11, 12). Variations in Nef sequences isolated from HIV-infected individuals can also correlate with the rate of HIV progression (13). Nef interactions with CD4 (14–16) and major histocompatibility complex molecules (17, 18) are particularly important and can lead to down-regulation of key immune-system receptors.
Nef contributes to CD4 down-regulation by means of multiple interactions with cellular ligands. These interactions internalize CD4 and direct it to cellular-degradation pathways (19–21). For example, CD4 is usually protected from degradation by interaction with cellular p56lck (19). Nef binding to p56lck disrupts the CD4–p56lck complex, exposing a CD4 dileucine motif and directing the protein to endocytic pathways (22, 23). Nef can tether the cytoplasmic tail of CD4 to adaptor-protein complexes on the surface of clathrin-coated pits, also leading to CD4 endocytosis (16). Moreover, Nef can link CD4 to the endosomal β-COP protein (16, 24), directing CD4 to the lysosome for degradation. Loss of CD4 has been correlated directly with increased HIV replication in T lymphocytes (15).
The N terminus of Nef (residues 1 to ≈57) is essential to interactions with p56lck, p53, actin (25–28), and perhaps other cellular proteins. Nef binding to p53 can block p53-mediated apoptosis (26). The Nef interaction with actin could influence subcellular localizations of Nef (27, 28). Here, we demonstrate by ELISA that the N terminus of Nef from the NL4-3 infectious molecular clone of HIV is essential for Nef binding to p53, actin, and p56lck. Because the Nef N terminus binds multiple targets, an inhibitor directed against this region of the protein could potentially block multiple Nef–ligand interactions during viral replication.
Guanidine alkaloids of the batzelladine family, which were isolated from the sponge Crambe crambe, can inhibit gp120–CD4 binding and induce CD4–p56lck dissociation (29–31). Crambescidin guanidine alkaloids from the same sponge have also been shown to inhibit HIV-1 cell fusion (32, 33). The capability of guanidine-based natural products for disrupting these protein–protein interactions suggested that analogs of batzellidine and crambescidin alkaloids might inhibit other protein–protein interactions. In this article, selected batzelladine and crambescidin analogs are shown to disrupt Nef–p53, Nef–actin, and Nef–p56lck interactions.
Materials and Methods
Nef-Phagemid Construction. The NL4-3 nef gene was cloned from HIVNL4-3 (34) into a M13 phage display vector between the signal peptide and the p8 gene with the NsiI and AflII restriction sites.
Compounds. Batzelladine and crambescidin analogs were synthesized as described (33, 35–38). Each compound was stored at –20°C at an average concentration of 10 mg/ml in DMSO.
ELISA of Phage-Displayed Nef. Cultures of Escherichia coli harboring individual phagemids were grown for 7 h at 37°C in 1 ml of 2YT medium (16 g of tryptone/10 g of yeast extract/5 g of NaCl per liter of water) and 50 μg/ml carbenicillin. M13-K07 helper phage (1010 phage per ml) was added, and cultures were transferred to 2YT medium (30 ml), supplemented with carbenicillin (50 μg/ml) and kanamycin (25 μg/ml), before incubation overnight at 37°C. Cells were removed by centrifugation (10 min at 8,000 × g), and the phage was precipitated from the supernatant by addition of 8 ml of 20% polyethylene glycol/2.5M NaCl. Phage were isolated by centrifugation (10 min at 8,000 × g and 2 min at 2,000 × g), resuspended in 1 ml of PBS (0.14 M NaCl/2.7 mM KCl/8 mM Na2HPO4/2 mM KH2PO4, pH 7.2, in 1 liter of water) and isolated by centrifugation (10 min at 8,000 × g). Phage concentration was determined by UV–Vis absorbance (OD268 = 1, 8.31 × 10–9 M, 5 × 1012 phage per ml). Phages were diluted to 5 × 1012 phage per ml in PBS. Specific wells of a 96-well Maxisorp immunoplate were coated with p53 (Santa Cruz Biotechnology), actin (Sigma), p56lck (Upstate Biotechnology, Charlottesvile, VA), or anti-Nef antibody AE6 ascites (100 μl, 5 μg/ml in 50 mM carbonate buffer, pH 9.6) for 2 h at room temperature. The plate was then blocked for 30 min with ovalbumin (0.2%) in PBS and washed five times with PT buffer (0.05% Tween 20 in PBS). Separately, phage displaying HIV Nef were serially diluted with dilution buffer (0.2% ovalbumin/0.05% Tween 20/PBS). The phage solution was added directly to the corresponding wells of target-coated assay plates. The assay plates were shaken for 1 h at room temperature and washed five times with PT buffer. After washing, plates were incubated with anti-M13/horseradish peroxidase (HRP) conjugate (100 μl; 1:5,000 dilution, Amersham Biosciences) in dilution buffer for 30 min, and washed eight times with PT buffer and twice with PBS. Plates were developed with o-phenylenediamine dihydrochloride/H2O2 solution (100 μl; 1 mg/ml OPD/0.02% H2O2) in citric acid buffer (50 mM citric acid/50 mM Na2HPO4, pH 5.0). After 10 min, the absorbance was measured spectrophotometrically at 450 nm by using a 96-well microtiter-plate reader (μQuant, Bio-Tek, Burlington, VT). Phage-displayed Nef constructs were prepared from overnight cultures grown under identical conditions.
Small-Molecule Competition ELISA. Potential inhibitors were diluted 1:100 or 1:1,000 in deionized water. Dilution buffer (120 μl) was added to specific wells of a 96-well microtiter plate (Costar). For initial single-point screens of potential Nef inhibitors, compounds were added to specific wells for a final concentration of 5 μM. Phage displaying Nef (40 μl, to a final concentration of 2.8 nM) were added. The solutions of Nef and potential inhibitors were incubated for 1 h. The solutions (100 μl) from corresponding wells of the dilution plate were added to the blocked, ligand-bound assay plates, which were prepared as described above. The plate was developed as described. For dose–response assays of 14, 17, and 22, inhibitors were prepared to a final concentration of 30 μM in water and diluted in dilution buffer. Phage-displayed Nef was added to each well, and the assay was performed as described for the single-point screens.
Direct Assay of Small-Molecule Binding. Crambescidin analog 47 was tested for binding to nonphage-displayed Nef (US Biologicals, Swampscott, MA), actin, or ovalbumin blocking agent. Maxisorp plates were coated with Nef (100 μl; 5 μg/ml in 50 mM carbonate buffer, pH 9.6) or actin (100 μl, 5 μg/ml) for 2 h at room temperature. The plate was blocked and washed as described for the phage-displayed ELISA. Crambescidin analog 47, which was chosen for its availability, was diluted to a final concentration of 5.0 μM and incubated on the coated wells for 1 h (100 μl). The plate was washed seven times with PT buffer and three times with water, and 100 μl of 100 mM HCl was added to each well. We analyzed 80 μl of each well by reverse-phase HPLC–MS (model LCT, Waters). Crambescidin analog 47 was separated on a 2 mm × 15 cm C18 column by using a gradient from 98% solvent A (98% water/2% methanol/0.2% acetic acid) to 95% solvent B (methanol/0.2% acetic acid) over 20 min, followed by 7 min at solvent B before returning to solvent A over 3 min. With a flow rate of 0.2 ml/min, the retention time for crambescidin analog 47 was 23.2 min.
Assay for Compound Cytotoxicity and Anti-HIV Activity. We tested 18 compounds for cytotoxicity by the method described in ref. 39. Briefly, compounds were dissolved in ethanol, serially diluted, and incubated with MT-2 cells in triplicate. After cellular growth at 37°C for 72 h, cells were stained with Finter's neutral red dye, and the percentage of viable cells was quantified at 540 nm.
Generation of nef-Deleted HIVNL4-3. A BamHI–XhoI fragment encoding nucleotides 8,465–8,892 of HIV-1NL4-3 (GenBank accession no. M19921), containing the first 105 nucleotides of nef, was subcloned into pGEX-4T (Pharmacia). QuikChange mutagenesis with Pfu DNA polymerase I (Stratagene) was used to introduce two stop codons (italicized) between nucleotides 8,847 and 8,852. The plus-strand primer (5′-GTAAGGGAAAGAATGTGATGAGCTGAGCCAGCAGC-3′; nucleotides 8,832–8,866) and the minus-strand primer (5′-GCTGCTGGCTCAGCTCATCACATTCTTTCCCTTAC-3′; nucleotides 8,866–8,832) were used for this reaction. The mutations deleted a BsmBI site, and the absence of this site was used to indicate successful mutagenesis. The DNA fragment containing the change was then ligated into HIVNL4-3 to generate HIVNL4-3Δnef.
Infectious-plasmid DNA was introduced into H9 cells by single-pulse electroporation at 200 mV for 50 msec. Cultures were grown and refed every other day in growth media (RPMI 1640 containing 25 mM Hepes/2mM l-glutamine supplemented with 11.5% heat-inactivated FBS). Cells were monitored for infection by indirect immunofluorescence assay (40). When the cultures were 100% positive for HIV antigens, supernatant fluids were clarified of cells by low-speed centrifugation, followed by filtration through 0.45-μm pore-size cellulose acetate filters. Aliquots of HIVNL4-3 and HIVNL4-3Δnef were frozen at –80°C until use.
Real-Time PCR. We inoculated 1 × 107 CD4+ lymphoblastoid H9 cells in triplicate flasks with either HIVNL4-3 or HIVNL4-3Δnef at ≈2.5 × 106 cpm of reverse-transcriptase activity, resulting in a final multiplicity of infection of <1.0. Cells were harvested and lysed for real-time PCR as described to measure minus-strand strong-stop DNA, completely synthesized HIV cDNA, or integrated HIV cDNA by using AA55/M667, M661/M667, and nested alu PCR primers, respectively (41, 42). Infections were monitored over the first 48 h, which includes the first round of replication (42).
HIV Spread. We inoculated ≈1.5 × 107 MT-2 CD4+ lymphoblastoid cells with either HIVNL4-3 or HIVNL4-3Δnef at ≈1.0 × 106 cpm of reverse-transcriptase activity, resulting in a final multiplicity of infection of <1.0. Cells were harvested at various times after infection and fixed by using 1:1 acetone/methanol. Cells were stained by using polyclonal human Ig containing antibodies to HIV and fluorescein-conjugated goat anti-human IgG. The percentage of HIV antigen-positive cells was determined by using an epifluorescent microscope (Nikon) as described (40, 42).
Results
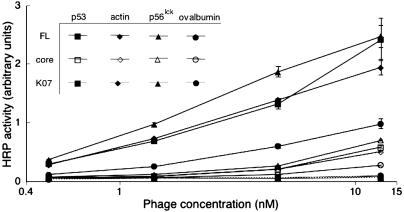
Phage Display and Assay of HIV-1 Nef. After subcloning the nef gene into an appropriate phagemid, display of Nef on the surface of M13 phage was verified by phage ELISA by using three antibodies specific to Nef (data not shown). Phage-displayed full-length Nef (from HIVNL4-3, residues 1–206) and core-domain Nef (residues 56–205) were examined by ELISA for binding to p53, actin, and p56lck (Fig. 1). Only full-length Nef bound to the three ligands, which confirms the requirement for the N terminus of Nef to mediate binding. By this assay, the three cellular ligands exhibited similar affinities for full-length Nef. Negative controls included binding of phage without Nef displayed (K07) to p53, actin, and p56lck and binding to the ovalbumin blocking agent by phage-displayed Nef; the negative controls exhibited greatly reduced binding (Fig. 1). Thus, the in vitro assays reported here examine interactions with full-length Nef. For a competition ELISA to identify potential small-molecule inhibitors of Nef, small molecules were added to a solution of phage-displayed full-length Nef and incubated for 1 h before binding to p53, actin, or p56lck.
Fig. 1.
Phage-displayed NL4-3 full-length (FL) or core domain (core) Nef binding to p53, actin, p56lck, or ovalbumin. The negative control represents phage (K07) without displayed Nef. Error bars for all phage-displayed Nef binding data indicate SE for the average of three independent assays.
Screening Guanidine Alkaloids to Identify Nef Inhibitors. A library containing the natural products ptilomycalin A, batzelladine F, and 46 batzelladine- and crambescidin-based analogs (33, 37, 38) were initially screened by competition ELISA (Fig. 2 and Figs. 6 and 7, which are published as supporting information on the PNAS web site). The control compounds arginine, guanidine, tetramethylguanidine, and tetramethylguanidine trifluoroacetic acid salt were also assayed. At compound concentrations of 5 μM, nine guanidines from this library inhibited the three Nef–ligand interactions at levels of >80% (Table 1). Three compounds (14, 17, and 22) exhibited >94% inhibition of the three Nef–ligand interactions.
Fig. 2.
Structures of batzelladine and crambescidin analogs. Depicted compounds inhibited at least one Nef–ligand interaction >80% in the initial competition ELISA. Compounds 14, 17, 22, 28, 42, and 46 were identified by the initial screen as the most potent inhibitors of the three Nef–ligand interactions.
Table 1. Percentage of inhibition for the most effective Nef–ligand inhibitors at a compound concentration of 5 μM.
| Compound | % Inhibition |
|---|---|
| Nef—p53 | |
| 17 | 101.1 ± 2.3 |
| 22 | 96.8 ± 0.9 |
| 14 | 94.7 ± 1.0 |
| 42 | 93.9 ± 1.0 |
| 46 | 93.3 ± 0.6 |
| 28 | 87.2 ± 0.6 |
| 34 | 84.5 ± 1.8 |
| 45 | 82.9 ± 1.0 |
| 47 | 81.2 ± 0.4 |
| 44 | 81.0 ± 0.6 |
| Nef—actin | |
| 22 | 102.9 ± 0.4 |
| 17 | 102.4 ± 2.0 |
| 14 | 102.2 ± 0.4 |
| 42 | 89.5 ± 0.8 |
| 28 | 85.5 ± 1.1 |
| 18 | 85.4 ± 3.6 |
| 10 | 84.2 ± 0.5 |
| 46 | 83.9 ± 2.0 |
| 45 | 80.6 ± 1.0 |
| 47 | 78.1 ± 1.8 |
| Nef—p56lck | |
| 17 | 101.0 ± 0.4 |
| 22 | 96.5 ± 0.4 |
| 14 | 95.8 ± 0.8 |
| 42 | 93.6 ± 1.4 |
| 46 | 89.3 ± 1.9 |
| 28 | 86.8 ± 0.6 |
| 18 | 82.7 ± 4.3 |
| 10 | 82.5 ± 1.0 |
| 40 | 80.6 ± 0.3 |
| 45 | 79.8 ± 1.7 |
Data are an average of three assays with the indicated SE.
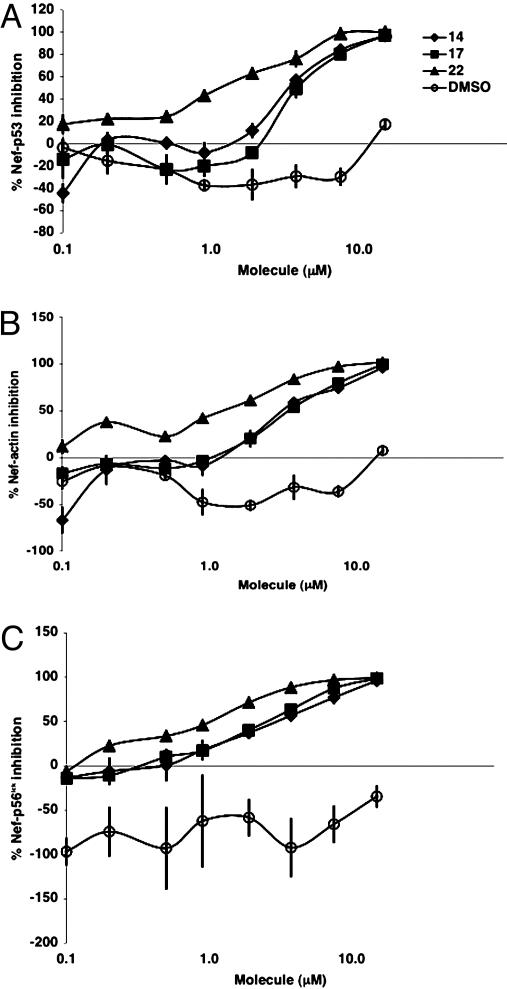
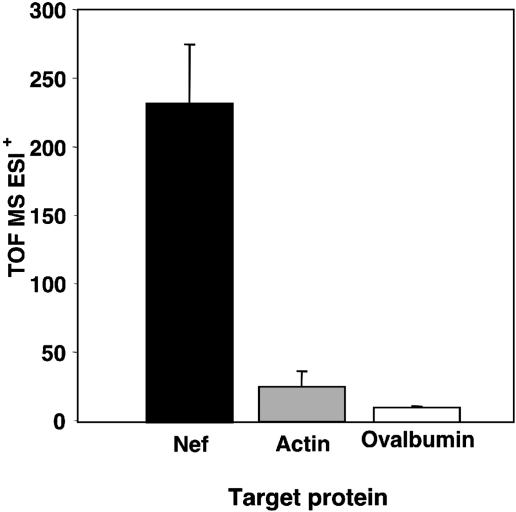
Inhibition of Nef Interactions with p53, Actin, and p56lck. As shown by competition ELISA, addition of specific batzelladine and guanidine compounds at low micromolar concentrations decreased phage-displayed Nef affinity for p53, actin, and p56lck to background levels. Interestingly, each compound exhibits similar relative efficacies for the different Nef–ligand interactions (Fig. 3). For example, the best inhibitors across each assay were 14, 17, and 22 (Table 1 and Fig. 2). Conversely, poor inhibitors failed to inhibit the three Nef–ligand interactions. In summary, compounds either inhibited all three or no Nef–ligand interactions. Additionally, overexpressed Nef, which was not displayed on the phage surface, was used to evaluate binding to crambescidin analog 47. The compound bound to Nef but not to actin (Fig. 4).
Fig. 3.
Dose–response inhibition of phage-displayed Nef (2.8 nM) binding to p53 (A), actin (B), and p56lck (C). Percentage of inhibition is relative to ligand binding by phage-displayed Nef without the addition of inhibitor or DMSO.
Fig. 4.
Crambescidin analog 47 binding to Nef, actin, or ovalbumin negative control. Time-of-flight MS ESI+ (TOF MS ESI+) indicates the intensities of peaks at the expected retention time and molecular weight (520) of crambescidin analog 47. Error bars indicate the SE for the average of three independent assays.
The control compounds arginine, guanidine, tetramethylguanidine, and tetramethylguanidine trifluoroacetic acid salt can decrease Nef–ligand binding by up to 40%, but other guanidine alkaloid-based compounds are significantly more active. At low concentrations, DMSO, which was used here as a compound vehicle, slightly agonizes Nef interactions with p53, actin, and p56lck (Fig. 3). The batzellidine and crambescidin inhibitors reported here do not indiscriminately inhibit other protein–protein interactions, as demonstrated by a competition assay with a control interaction between phage-displayed HIV-1 Vif and p55 Gag (data not shown and S. Arrantinis, A.O., and G.A.W., unpublished data).
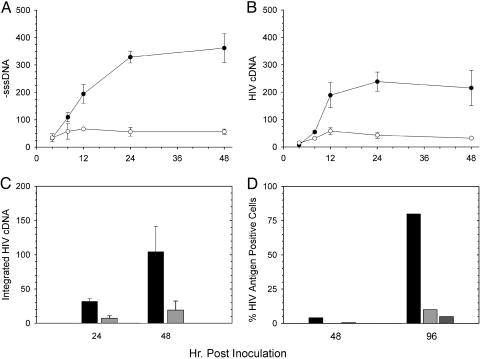
In Vivo Assays for Anti-Nef Activity. Sensitive assays were developed to quantitate the effects of Nef inhibitors on viral spread for cell culture-propagated HIV. First, stop-codon mutations were introduced into the nef gene in the HIVNL4-3 background (HIVNL4-3Δnef). Such mutations are expected to prevent Nef translation. A decrease in viral spread for HIVNL4-3Δnef was observed (Fig. 5). These results were confirmed with analysis by real-time PCR. Measurement of minus-strand strong-stop DNA, the first product of reverse transcription, and completely synthesized HIV cDNA, the final product of reverse transcription, demonstrated that HIVNL4-3Δnef was defective for spread after an inefficient initial infection (Fig. 5 A and B). This experiment also resulted in little integrated HIV cDNA (Fig. 5C). Finally, when measuring HIV-antigen synthesis, HIV lacking Nef fails to spread with the same efficiency as virus containing an intact nef gene (Fig. 5D). Results were the same when using HIV with the nef gene deleted (data not shown). HIVNL4-3Δnef demonstrated similar replication defects in both H9 (Fig. 5 A–C) and MT-2 cells (Fig. 5D). These experiments demonstrate that compounds capable of disrupting Nef activity could have dramatic effects on HIV replication in cell culture. Such activities can be readily detected by either immunofluorescence or real-time PCR assays.
Fig. 5.
Replication of HIVNL4-3Δnef in CD4+ lymphoblastoid cell lines. The first round of replication for HIVNL4-3Δnef (○) in H9 cells was compared with HIVNL4-3 containing an intact nef gene (•) by real-time PCR to measure minus-strand strong-stop DNA synthesis (A) and completely synthesized HIV cDNA (B). (C) Integrated HIV cDNA (provirus) was also quantified for both HIVNL4-3Δnef (light-gray bars) and HIVNL4-3 (dark-gray bars). (D) Replication of two distinct clones of HIVNL4-3Δnef (light-gray and medium-gray bars) as measured by an immunofluorescence assay was also decreased compared with HIVNL4-3 (black bars) in MT-2 cells. In A–C, data are the average from three infections; error bars are one SD. Values are in equivalent numbers of chronically HIV-infected H9 cells (41, 42). Cells were infected with equal amounts of each clone based on input reverse-transcriptase activity.
We chose 18 compounds, including 14, 17, and 22, for assays to quantitate anti-HIV activity. However, at submicromolar concentrations, all compounds were cytotoxic to MT-2 cells (data not shown). Unfortunately, this cellular toxicity prevents experiments with the 18 compounds to assess anti-HIV effects in human cell cultures.
Discussion
In Vitro Screens for Nef Inhibition. Phage display has been used to explore complex protein–protein interactions, such as mapping functional epitopes and uncovering binding partners (43, 44). The technique also offers a sensitive format for high-throughput screening of inhibitors to the phage-displayed protein. For example, enzyme inhibitors have been identified from phage-displayed peptide libraries, and the resultant peptides have been used for high-throughput, small-molecule screening (45). The in vitro Nef–ligand screens described here represent what is, to our knowledge, the first application of phage display applied to the identification of protein–protein inhibitors. The technique offers some potential advantages over conventional protein–protein interaction screens, which often require cumbersome additional steps, such as biotinylation or fluorescent labeling of one target protein. Advantages of the phage-display technique include sensitive, robust binding assays and rapid adaptability of the target protein. Altering the phage-displayed protein to target a different protein variant requires a simple one-step mutagenesis. Other ELISAs to examine Nef–ligand interactions (including to p53, actin, and p56lck) have been reported (26, 27, 46).
To demonstrate the adaptability of phage display-based assays, we examined the importance of the N terminus of Nef to interactions with the cellular ligands p53, actin, and p56lck. Binding to full-length and core HIVNL4-3 Nef was compared. Anti-Nef antibodies and a similar ELISA format were used to verify the approximately equivalent display levels for the two constructs (data not shown). Essentially, no binding to the three ligands was observed for phage-displayed core Nef (Fig. 1). The requirement for the N terminus of Nef to mediate binding to p53, actin, and p56lck confirms known binding preferences (25–27) and demonstrates the efficacy of the assay. The reported phage-displayed binding assay could also be adapted for Nef library selections and screens.
Small-Molecule Nef Inhibitors. Next, we sought to determine whether the in vitro Nef assays reported here could be used to identify small molecules capable of disrupting Nef binding activities. Batzellidine and crambescidin alkaloids quickly became the focus of our efforts because of the known abilities for some batzellidine and crambescidin derivatives to disrupt other protein–protein interactions, including CD4 binding to p56lck (29, 30, 32). Competition-binding data demonstrate inhibition of three Nef–ligand interactions by batzellidine and crambescidin analogs (Fig. 3). As described above, the three cellular ligands, p53, actin, and p56lck, all bind to the N terminus of Nef. The ranking of best to worst inhibitor is essentially conserved across the three Nef–ligand assays (see Table 1 and Figs. 6 and 7). Additionally, overexpressed Nef, which is not displayed on the phage surface, binds crambescidin analog 47. However, crambescidin analog 47 fails to bind actin or the ovalbumin negative control (Fig. 4). Together, the data conclusively demonstrate that the identified compounds inhibit Nef–ligand interactions primarily by binding to Nef and not the cellular ligand.
Guanidines 14, 17, and 22 were the most potent inhibitors of the three Nef–ligand interactions (Table 1 and Fig. 2). The crambescidin alkaloid ptilomycalin A (44) was a slightly weaker inhibitor, whereas batzelladine F (26) showed low activity. In general, members of the library that contained two guanidine subunits were poor inhibitors. The aliphatic side chains of the best inhibitors are highly flexible, which could permit these molecules to assume various binding conformations. The conserved bicyclic or tricyclic guanidine motif of the best inhibitors suggests that this functionality is essential for modulating Nef inhibition. These guanidine units could form bidentate hydrogen bonds to an appropriate Nef functionality, possibly a carboxylate side chain.
The Nef-inhibitory activity reported here provides another example of protein–protein inhibition by guanidine alkaloids. Various batzellidine and crambescidin alkaloids have been reported to inhibit gp120–CD4, CD4–p56lck, and HIV-1 cell-fusion protein–protein interactions (29–33). With the planar guanidine functionality surrounded by hydrophobic rings, molecules of this class are ideally suited to disrupt the large, relatively flat surfaces common to protein–protein interactions. Interest in this class of molecules has been partially motivated by the cytotoxic properties of the natural variants. Having identified a large number of active compounds, tissue culture-based assays were developed to examine the potential for inhibiting Nef in vivo.
In Vivo Assay of Nef Inhibitors. Because Nef is an acronym for “negative effector of HIV function,” we were pleased to observe clearly distinguishable effects for growth of HIV constructs with a deleted or stop codon-terminated nef gene (Fig. 5). Cytotoxicity assays applied the methods reported in ref. 39. Unfortunately, high cytotoxicity for 18 guanidine alkaloids with in vitro Nef inhibitory activities precluded determination of in vivo anti-HIV activities.
In summary, we report small-molecule HIV-1 Nef inhibitors and analysis of HIVNL4-3Δnef by real-time PCR. Although the inhibitors proved to be too cytotoxic for anti-HIV assays, the assays and structure–activity relationship data reported here can guide development of Nef inhibitors. Such inhibitors could provide insight into the cellular consequence of Nef binding to p53, actin, and p56lck. Other Nef–ligand interactions could require the Nef N terminus, and the reported compounds could also inhibit such interactions.
Supplementary Material
Acknowledgments
We thank Dr. John Greaves (University of California, Irvine) for expert MS advice and Dr. John Guatelli (University of California at San Diego) for gifts of nef-deleted HIVNL4-3 and HIVNL4-3 containing a double-stop mutation that were used to ensure that the phenotype of the engineered nef double-stop reported here was consistent with other nef-deleted HIV clones. We obtained the pNL4-3 (Dr. Malcolm Martin, National Institute of Allergy and Infectious Diseases, Bethesda) and AE6 anti-Nef antibody reagents from the AIDS Research and Reference Reagent Program (Division of AIDS, National Insti-tute of Allergy and Infectious Diseases, Bethesda). This work was supported by a Young Investigator Award from the Arnold and Mabel Beckman Foundation (to G.A.W.), Burroughs Wellcome Fund Award 99-2609 (to W.E.R.), and National Institutes of Health Heart, Lung, and Blood Institute Grant HL-25854 (to L.E.O.). W.E.R. is a Burroughs Wellcome Clinical Scientist in Translational Research.
References
- 1.Gulick, R. M. (2003) Clin. Microbiol. Infect. 9, 186–193. [DOI] [PubMed] [Google Scholar]
- 2.Lin, P.-F., Blair, W., Wang, T., Spicer, T., Guo, Q., Zhou, N., Gong, Y.-F., Wang, H.-G. H., Rose, R., Yamanaka, G., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 11013–11018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li, F., Goila-Gaur, R., Salzwedel, K., Kilgore, N. R., Reddick, M., Matallana, C., Castillo, A., Zoumplis, D., Martin, D. E., Orenstein, J. M., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 13555–13560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salvi, R., Garbuglia, A. R., Di Caro, A., Pulciani, S., Montella, F. & Benedetto, A. (1998) J. Virol. 72, 3646–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kirchhoff, F., Greenough, T. C., Brettler, D. B., Sullivan, J. L. & Desrosiers, R. C. (1995) N. Engl. J. Med. 332, 228–232. [DOI] [PubMed] [Google Scholar]
- 6.Deacon, N. J., Tsykin, A., Solomon, A., Smith, K., Ludford-Menting, M., Hooker, D. J., McPhee, D. A., Greenway, A. L., Ellett, A., Chatfield, C., et al. (1995) Science 270, 988–991. [DOI] [PubMed] [Google Scholar]
- 7.Geyer, M. & Peterlin, B. M. (2001) FEBS Lett. 496, 91–95. [DOI] [PubMed] [Google Scholar]
- 8.Geyer, M., Munte, C. E., Schorr, J., Kellner, R. & Kalbitzer, H. R. (1999) J. Mol. Biol. 289, 123–138. [DOI] [PubMed] [Google Scholar]
- 9.Arold, S. T. & Baur, A. S. (2001) Trends Biochem. Sci. 26, 356–363. [DOI] [PubMed] [Google Scholar]
- 10.Geyer, M., Fackler, O. T. & Peterlin, B. M. (2001) EMBO Rep. 2, 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kestler, H. W., III, Ringler, D. J., Mori, K., Panicali, D. L., Sehgal, P. K., Daniel, M. D. & Desrosiers, R. C. (1991) Cell 65, 651–662. [DOI] [PubMed] [Google Scholar]
- 12.Greenway, A. L., Holloway, G. & McPhee, D. A. (2000) Adv. Pharmacol. 48, 299–343. [DOI] [PubMed] [Google Scholar]
- 13.Kirchhoff, F., Easterbrook, P. J., Douglas, N., Troop, M., Greenough, T. C., Weber, J., Carl, S., Sullivan, J. L. & Daniels, R. S. (1999) J. Virol. 73, 5497–5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stoddart, C. A., Geleziunas, R., Ferrell, S., Linquist-Stepps, V., Moreno, M. E., Bare, C., Xu, W., Yonemoto, W., Bresnahan, P. A., McCune, J. M. & Greene, W. C. (2003) J. Virol. 77, 2124–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lundquist, C. A., Tobiume, M., Zhou, J., Unutmaz, D. & Aiken, C. (2002) J. Virol. 76, 4625–4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piguet, V., Gu, F., Foti, M., Demaurex, N., Gruenberg, J., Carpentier, J.-L. & Trono, D. (1999) Cell 97, 63–73. [DOI] [PubMed] [Google Scholar]
- 17.Schindler, M., Würfl, S., Benaroch, P., Greenough, T. C., Daniels, R., Easterbrook, P., Brenner, M., Münch, J. & Kirchhoff, F. (2003) J. Virol. 77, 10548–10556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen, G. B., Gandhi, R. T., Davis, D. M., Mandelboim, O., Chen, B. K., Strominger, J. L. & Baltimore, D. (1999) Immunity 10, 661–671. [DOI] [PubMed] [Google Scholar]
- 19.Kim, Y.-H., Chang, S. H., Kwon, J. H. & Rhee, S. S. (1999) Virology 257, 208–219. [DOI] [PubMed] [Google Scholar]
- 20.Bandres, J. C., Shaw, A. S. & Ratner, L. (1995) Virology 207, 338–341. [DOI] [PubMed] [Google Scholar]
- 21.Cheng, H., Hoxie, J. & Parks, W. P. (1999) Virology 264, 5–15. [DOI] [PubMed] [Google Scholar]
- 22.Aiken, C., Konner, J., Landau, N. R., Lenburg, M. E. & Trono, D. (1994) Cell 76, 853–864. [DOI] [PubMed] [Google Scholar]
- 23.Grzesiek, S., Stahl, S. J., Wingfield, P. T. & Bax, A. (1996) Biochemistry 35, 10256–10261. [DOI] [PubMed] [Google Scholar]
- 24.Benichou, S., Bomsel, M., Bodéus, M., Durand, H., Douté, M., Letourneur, F., Camonis, J. & Benarous, R. (1994) J. Biol. Chem. 269, 30073–30076. [PubMed] [Google Scholar]
- 25.Baur, A. S., Sass, G., Laffert, B., Willbold, D., Cheng-Mayer, C. & Peterlin, B. M. (1997) Immunity 6, 283–291. [DOI] [PubMed] [Google Scholar]
- 26.Greenway, A. L., McPhee, D. A., Allen, K., Johnstone, R., Holloway, G., Mills, J., Azad, A., Sankovich, S. & Lambert, P. (2002) J. Virol. 76, 2692–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fackler, O. T., Kienzle, N., Kremmer, E., Boese, A., Schramm, B., Klimkait, T., Kücherer, C. & Mueller-Lantzch, N. (1997) Eur. J. Biochem. 247, 843–851. [DOI] [PubMed] [Google Scholar]
- 28.Campbell, E. M., Nunez, R. & Hope, T. J. (2004) J. Virol. 78, 5745–5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patil, A. D., Kumar, N. V., Kokke, W. C., Bean, M. F., Freyer, A. J., De Brosse, C., Mai, S., Truneh, A., Faulkner, D. J., Carte, B., et al. (1995) J. Org. Chem. 60, 1182–1188. [Google Scholar]
- 30.Patil, A. D., Freyer, A. J., Taylor, P. B., Carté, B., Zuber, G., Johnson, R. K. & Faulkner, D. J. (1997) J. Org. Chem. 62, 1814–1819. [Google Scholar]
- 31.Patil, A. D., Freyer, A. J., Offen, P., Bean, M. F. & Johnson, R. K. (1997) J. Nat. Prod. 60, 704–707. [DOI] [PubMed] [Google Scholar]
- 32.Chang, L., Whittaker, N. F. & Bewley, C. A. (2003) J. Nat. Prod. 66, 1490–1494. [DOI] [PubMed] [Google Scholar]
- 33.Bewley, C. A., Ray, S., Cohen, F., Collins, S. K. & Overman, L. E. (July 8, 2004) J. Nat. Prod., 10.1021/np049958o. [DOI] [PubMed]
- 34.Adachi, A., Gendelman, H. E., Koenig, S., Folks, T., Willey, R., Rabson, A. & Martin, M. A. (1986) J. Virol. 59, 284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cohen, F. & Overman, L. E. (2001) J. Amer. Chem. Soc. 123, 10782–10783. [DOI] [PubMed] [Google Scholar]
- 36.Cohen, F., Overman, L. E. & Ly Sakata, S. K. (1999) Org. Lett. 1, 2169–2172. [DOI] [PubMed] [Google Scholar]
- 37.Aron, Z. D., Pietraszkiewicz, H., Overman, L. E., Valeriote, F. & Cuevas, C. (2004) Bioorg. Med. Chem. Lett. 14, 3445–3449. [DOI] [PubMed] [Google Scholar]
- 38.Coffey, D. S., McDonald, A. I., Overman, L. E., Rabinowitz, M. H. & Renhowe, P. A. (2000) J. Amer. Chem. Soc. 122, 4893–4903. [Google Scholar]
- 39.Montefiori, D. C., Robinson, W. E., Jr., Schuffman, S. S. & Mitchell, W. M. (1988) J. Clin. Microbiol. 26, 231–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson, W. E., Jr., Montefiori, D. C., Gillespie, D. H. & Mitchell, W. M. (1989) J. Acquired Immune Defic. Syndr. 2, 33–42. [PubMed] [Google Scholar]
- 41.Victoria, J. G., Lee, D. J., McDougall, B. R. & Robinson, W. E., Jr. (2003) AIDS Res. Hum. Retroviruses 19, 865–874. [DOI] [PubMed] [Google Scholar]
- 42.Lee, D. J. & Robinson, W. E., Jr. (2004) J. Virol. 78, 5835–5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sidhu, S. S., Fairbrother, W. J. & Deshayes, K. (2003) ChemBioChem 4, 14–25. [DOI] [PubMed] [Google Scholar]
- 44.Diaz, J. E., Howard, B. E., Neubauer, M. S., Olszewski, A. & Weiss, G. A. (2003) Curr. Issues Mol. Biol. 5, 129–146. [PubMed] [Google Scholar]
- 45.Hyde-DeRuyscher, R., Paige, L. A., Christensen, D. J., Hyde-DeRuyscher, N., Lim, A., Fredericks, Z. L., Kranz, J., Gallant, P., Zhang, J., Rocklage, S. M., et al. (2000) Chem. Biol. 7, 17–25. [DOI] [PubMed] [Google Scholar]
- 46.Greenway, A., Azad, A., Mills, J. & McPhee, D. A. (1996) J. Virol. 70, 6701–6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.