Abstract
Replication forks are arrested at specific sequences to facilitate a variety of DNA transactions. Forks also stall at sites of DNA damage, and the regression of stalled forks without rescue can cause genetic instability. Therefore, unraveling the mechanisms of fork arrest and of rescue of stalled forks is of considerable general interest. In Schizosaccharomyces pombe, products of two mating-type switching genes, swi1 and swi3, participate in fork arrest at the mating-type switch locus. Here, we show that these proteins also act at three termini (Ter) also called replication fork barriers in the spacer regions of rDNA but not at a fourth site, RFP4, which is nonfunctional when present in a plasmid. Two of the Swi1p- and Swi3p-dependent sites were also dependent on the transcription terminator Reb1p. Furthermore, hydroxyurea-induced replication stress mimicked the effect of swi1 or swi3 mutations at these sites. A swi1 mutant that failed to arrest forks at the mating-type fork barrier RTS1 was functional at the rDNA Ter sites, suggesting some specificity of action. Both WT and mutant forms of Swi1p were physically localized at the Ter sites in vivo. The results support the notion that Swi1p and Swi3p act at several different protein–DNA complexes in the rDNA spacer regions to arrest replication but that not all fork barriers required their activity to arrest forks.
Keywords: mating-type switching genes, genome stability, replication termination
Stalled DNA replication forks are subject to collapse if not processed by replication restart and/or recombination (1, 2). Such collapsed forks potentiate major DNA damage and can lead to genomic instability (1, 3). Replication forks may stall or arrest at DNA lesions or breaks, and mutations in replication proteins and genotoxic stress due to substrate depletion can also cause fork arrest (4, 5). Although such types of arrest are generally random events, natural site-specific replication terminators (Ter sites) or replication fork barriers (RFB) have also been identified in both prokaryotes and eukaryotes (reviewed in ref. 6). Natural fork barriers cause polar fork arrest and are presumably required to ensure proper completion of DNA replication, chromosome segregation, or other physiological events such as promotion of mating-type switching in Schizosaccharomyces pombe (6–8). Interestingly in this regard, fork arrest within the intergenic spacers of ribosomal DNA (rDNA) has been conserved from yeast to humans (9–14), presumably playing a critical role in controlling replication, recombination, and transcription within rDNA (15–18). In Saccharomyces cerevisiae, in addition to promoting replication termination, rDNA recombination, and extrachromosomal rDNA circle (ERC) formation, the terminator protein Fob1p has been implicated in Sir2-mediated rDNA silencing by its association with the regulator of nucleolar silencing and telophase exit (RENT) complex (19). Accumulation of an excess of ERC has been implicated in cellular aging (20).
The mechanisms of eukaryotic replication fork arrest remain ill defined as compared with prokaryotic systems, in which the interaction of a terminator protein with Ter sites suffices to cause orientation-specific or polar fork arrest by inhibiting the unwinding activity of the replicative helicase (6, 21–23). However, recent work in both Sa. cerevisiae and Sc. pombe has begun to unravel the mechanisms of eukaryotic fork arrest. In Sa. cerevisiae, Fob1p has been shown to bind to the intergenic rDNA termination (Ter or RFB) sites to arrest forks in a polar manner (24, 25). Similarly, the RNA polymerase I (polI) transcription terminator Reb1p causes polar fork arrest at two of three Ter sites in the intergenic spacer region of fission yeast (26). At each of the rDNA loci from different systems, only single terminator proteins that bind to one or more Ter sites have been identified. On the other hand, at the Sc. pombe mating-type switch locus mat1, four genes have been implicated in causing polar fork arrest at a site called RTS1. Arrest at this site is required to ensure unidirectional replication of mat1, which is essential for the formation of a strand-specific and site-specific imprint at this locus that subsequently initiates mating-type switching (7, 8, 27). Two of these genes, swi1 and swi3, are also required to stall the fork at MPS1, the imprinting site, whereas the two genes rtf1 and rtf2 are presumably required for fork arrest only at RTS1 (7, 28). The mechanism(s) of action of swi1 and swi3 in causing site-specific arrest remain unknown.
Swi1p and its Sa. cerevisiae homolog Tof1p have been implicated in S-phase replication checkpoint control (29–31). Tof1p was first identified as a topoisomerase I-interacting protein (32) and was subsequently found to mediate a checkpoint response in a pathway parallel with the one controlled by Rad9p (29). Furthermore, Tof1p may travel with the replication fork along with another checkpoint mediator, Mrc1p, to prevent uncoupling of the replisomal proteins from the replicated regions during genotoxic stress (30). Swi1p has been implicated in replication checkpoint by activating the checkpoint kinase Cds1p in response to hydroxyurea (HU)-induced replication stress, which depletes deoxyribonucleotides (dNTPs) and, thereby, causes replication forks to stall randomly throughout the genome (31). Swi1p stabilizes these stalled forks and probably prevents their collapse (31). Interestingly, for reasons not yet known, forks stalled at the natural Ter sites apparently do not activate the replication checkpoint response, as evidenced by normal cell-cycle progression in WT cells. Therefore, understanding the mechanism of action of Swi1p both at stress-induced randomly stalled replication forks and at natural fork barriers and understanding the mechanism of Swi1p and other proteins in fork stabilization and checkpoint induction (or lack thereof) is of considerable interest.
We report here that the mating-type switching proteins Swi1p and Swi3p caused site-specific replication fork arrest outside the mating-type switch locus mat1 in the intergenic spacer regions of rDNA. High-resolution mapping of this region by using 2D gel electrophoresis revealed the existence of four sites of fork arrest, Ter1–Ter3 and RFP4, three of which, Ter1–Ter3, depend on Swi1p and Swi3p for activity. Swi1p and Swi3p acted in concert with Reb1p at two of these sites, Ter2 and Ter3. RFP4, so named by convention (33) because it is not a genuine Ter site and may or may not depend on transcription, did not require Swi1p or Swi3p for activity. Interestingly, replication stress induced by HU treatment completely abolished fork arrest at Ter1–Ter3, thereby mimicking the effect of swi1 or swi3 mutants at these sites. Thus, Swi1p association with natural fork barriers may be dynamic, such that Swi1p could easily be recruited to stress-induced stalled forks. We also show that Swi1p demonstrates specificity at natural termini, because a mutant form of the protein (E662K) that was completely defective in fork arrest at RTS1 remained fully functional at all rDNA Ter sites. Thus, Swi1p appeared to act with some selectivity in promoting fork arrest at certain fork barriers.
Materials and Methods
Plasmids and Strains. SP976 (h90, ade6-M210, ura4-D18, leu1-32), SP785 (h90, his2, ade6-M216, swi1-111), SP918 (h90, ade6-M216, leu1-32, swi3-146), and JZ277 (h90, his2, ade6-M210, rtf3-1) were provided by A. J. S. Klar (National Cancer Institute, Frederick, MD). SP976 represents the WT strain used in these studies. JZ277 harbors the swi1 E662K mutation. SPGK100 (h90, ade6-M210, ura4-D18, leu1-32, reb1Δ::kanMX) and SPGK301C (h90, ade6-M210, ura4-D18, leu1-32, swi1Δ::ura4) were derived from SP976 by using standard gene-replacement techniques (34). pREP1-NTAP was a generous gift from K. L. Gould (Vanderbilt University, Nashville, TN), and pRL3 was kindly provided by R. Dhar (National Institutes of Health, Bethesda). DNA sequencing was performed by the Medical University of South Carolina Biotechnology Resource Laboratory. Details of strain and plasmid construction are described in detail in Supporting Text, which is published as supporting information on the PNAS web site. The sequences of primers and oligos used are available upon request.
2D Agarose Gel Electrophoresis. Replication intermediates were isolated for 2D gel analysis as described by Huberman et al. (35) with modifications. For details, see Supporting Text.
Chromatin Immunoprecipitation (ChIP) Assays. SPKG301C transformed with either pcGST-Swi1 or pcGST-Swi1 E662K was used for all ChIP assays. Protein–protein and protein–DNA cross-linking were performed as described by Kurdistani and Grunstein (36). ChIP assays were performed as described in ref. 37 and references therein. Anti-GST antibody and protein A-Sepharose are available commercially (Amersham Biosciences). Products were analyzed on 2.7% agarose gels, stained with ethidium bromide, and analyzed with a GelDoc 2000 ChemiDoc image analyzer (Bio-Rad).
Results
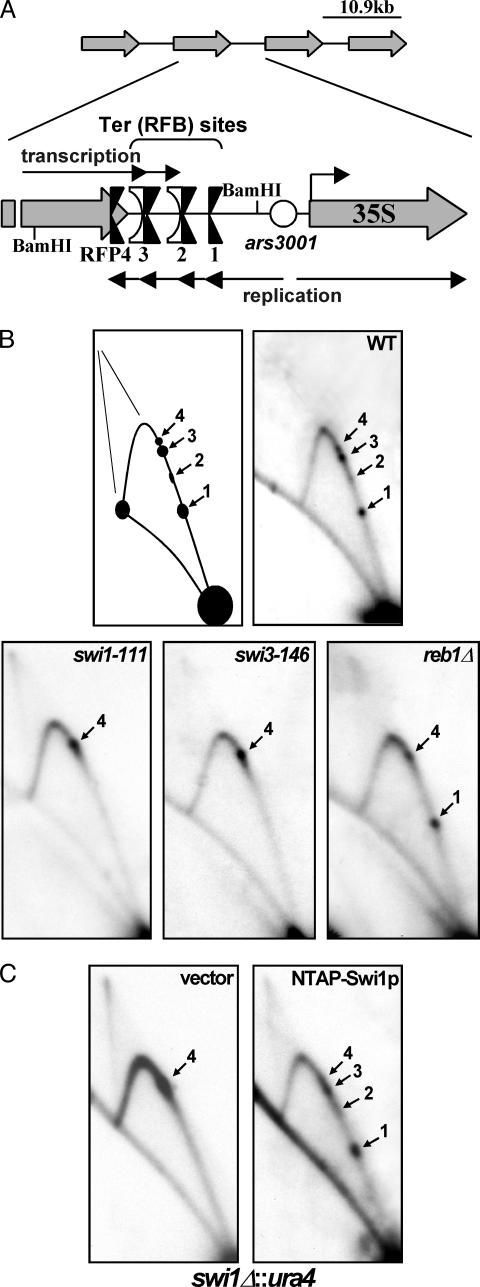
Four Sites of Fork Arrest Operated at the Intergenic Spacer Regions of S. pombe rDNA. Previous work demonstrated the presence of a fork-arresting region in the intergenic spacer of fission yeast rDNA (10) that was subsequently resolved as three replication fork barriers RFB1–RFB3 (26). To investigate and begin to identify the protein(s) needed for fork arrest in this region, we performed 2D gel analysis of the BamHI fragment, which is most of the intergenic spacer region upstream of ars3001 as well as the end of the region encoding the rRNA (Fig. 1A) from WT and selected mutant strains. Contrary to previous reports in ref. 26, we discovered the presence of four (not three) sites that impeded fork movement called Ter (RFB) 1–TER3 and RFP4 (Fig. 1 A and B) within this region. All four fork barriers arrested replication forks traveling counter to the direction of transcription (unpublished results). Ter2 and Ter3 were dependent on the RNA polI transcription terminator Reb1p (Fig. 1B) as reported in ref. 26. RFP4 had not been identified, probably because it is very close to Ter3 and, therefore, could not be resolved as a separate site in 2D gels. Also, RFP4 activity is sometimes weak in WT cells if fork arrest at Ter1–Ter3 is very efficient (unpublished results). Interestingly, because RFP4 lies upstream of the three RNA polI terminators mapped in vivo (38), this site is located within rather than downstream of the transcription unit (see below).
Fig. 1.
2D gel autoradiograms of replication intermediates of genomic rDNA showing the topology of fork movement in the region. (A) Diagram depicting the array of tandemly repeated rDNA units located on both arms of chromosome III. A closer view of the intergenic region shows the presence of the four rDNA fork barriers Ter (RFB) 1–3 and RFP4 near the 3′ end of the 35S transcription unit. Note the presence of RFP4 upstream of Reb1p binding sites Ter2 and Ter3, which represent two of three in vivo transcription terminators. (B) Autoradiograms of 2D gel analyses of the replication intermediates of the BamHI fragment depicted in A. Ter1–Ter3 and RFP4 localize to the ascending Y-arc of this fragment in WT cells (black arrows). Ter1–Ter3 are absent in swi1-111 and swi3-146 cells, whereas only Ter2 and Ter3 are absent in reb1Δ::kanMX cells. Note that forks accumulate at RFP4 in the swi1 and swi3 (and reb1Δ) mutants. (C) TAP-tagged Swi1p fully complements swi1Δ cells at Ter1–Ter3. Two-dimensional gel analysis of a swi1Δ::ura4 strain (SPGK301C) expressing only the TAP epitope (vector) or N-terminally tagged Swi1p (NTAP-Swi1p).
Swi1p and Swi3p Were Needed for Fork Arrest at Ter1–Ter3 but Not at RFP4. Swi1p (31) and its Sa. cerevisiae homolog Tof1p (30) stabilize stalled forks during replication stress. Swi1p and Swi3p are also needed for site-specific fork arrest at RTS1 and at the imprinting site MPS1 of the mat1 locus (7). Therefore, we investigated by 2D gel electrophoresis whether Swi1p and/or Swi3p are needed for stable fork arrest at natural fork barriers other than those at mat1. We discovered that Swi1p and Swi3p were needed for stable fork arrest at Ter1–Ter3 of rDNA. The mutant strains swi1-111 and swi3-146, which are defective in fork arrest at RTS1 and also in mating-type switching (7), lacked detectable fork-arresting activity at Ter1–Ter3 (Fig. 1B). As expected, we also observed a lack of fork arrest at the same three Ter sites in a swi1Δ strain (SPGK301C). To ensure that the effect was due directly to swi1 and not to a cryptic mutation in some other gene, we expressed Swi1p under control of the thiamine-inducible nmt promoter in the swi1Δ strain SPGK301C and thereby were able to restore fork arrest at Ter1–Ter3 (Fig. 1C).
We also examined replication intermediates of the same fragment from a reb1Δ strain by 2D gel electrophoresis and confirmed that Reb1p was required for fork arrest at Ter2 and Ter3 but not at Ter1 or RFP4 (Fig. 1B). Therefore, Swi1p and Swi3p along with Reb1p were needed to promote fork arrest at Ter2 and Ter3.
As contrasted with Ter1–Ter3, RFP4 did not require Swi1p or Swi3p for fork arrest. In fact, the spot at RFP4 increased in intensity in the absence of the other three barriers, probably because all forks accumulated at RFP4 when Ter1–Ter3 were rendered nonfunctional (Fig. 1B). The independence of RFP4 on Swi1p and Swi3p is particularly intriguing, because it represents the only known natural fork barrier (of six, including this report) in Sc. pombe of its kind. Because RFP4 does not function extrachromosomally and, therefore, may depend on rRNA transcription or on interaction with sites that may be located outside the region (see below), we conclude that Swi1p and Swi3p are needed for stable fork arrest at terminator protein-mediated natural Ter sites.
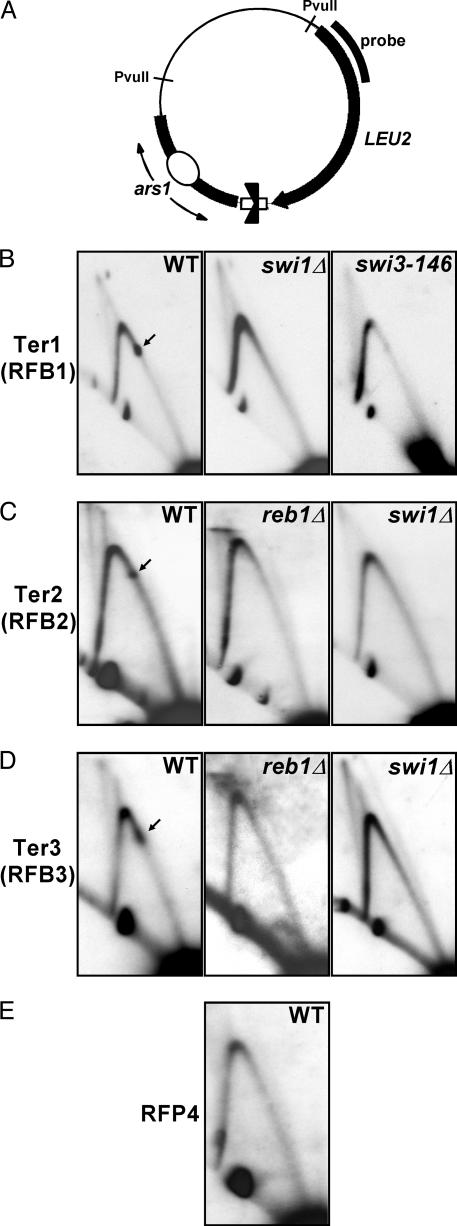
To study further the effects of Swi1p and Swi3p on the rDNA termini and to localize Ter1 more precisely, we cloned DNA fragments that included Ter1, Ter2, Ter3, or RFP4 into the shuttle vector pIRT2 at the BamHI site (Fig. 2A) and analyzed PvuII-digested replication intermediates by 2D gel electrophoresis. A 252-bp fragment upstream of the intergenic HindIII site (with respect to rDNA transcription) and corresponding to Ter1 exhibited efficient fork-arresting activity in its native orientation. Fork arrest at this site was completely abolished in swi1Δ and swi3-146 strains (Fig. 2B). Similarly, a 250-bp fragment encompassing the farthest downstream Reb1p binding site and corresponding to Ter2 caused stable fork arrest in its native orientation. Fork arrest was completely abolished in both reb1Δ and swi1Δ strains (Fig. 2C). Because Ter3 and RFP4 are difficult to resolve in 2D gels due to their close proximity to one another, further characterization of both sites was also carried out in plasmid replicons. 2D analysis of replication intermediates revealed that a 257-bp fragment encompassing the upstream Reb1p binding site and corresponding to Ter3 possessed efficient fork-arresting activity that depended on both Reb1p and Swi1p (Fig. 2D). These data thus confirmed results obtained from such analysis on whole chromosomes. The location of RFP4 within the rRNA transcription unit suggested that fork arrest at this site may result from collision of the transcription and replication machineries and, thus, differ mechanistically from Ter1–Ter3. We therefore cloned ≈400 bp of DNA sequence immediately upstream of Ter3 into the shuttle vector pIRT2, thereby separating this sequence from elements required to initiate rRNA transcription. Indeed, analysis of replication intermediates revealed that this sequence did not function as a replication fork barrier extrachromosomally (Fig. 2E). Identical results were obtained when analyzing ≈1 kb of upstream sequence (data not shown). Although not definitive, this experiment taken together with the location of RFP4 within the transcription unit strongly suggests that RFP4 may be transcription-dependent. Therefore, we referred to this site as replication fork pause 4 based on convention (33). It is mechanistically interesting to note that the stability of forks stalled by such unconventional means is Swi1p- and Swi3p-independent, as contrasted with forks stalled at terminator protein–DNA complexes such as Ter1–Ter3.
Fig. 2.
2D gel autoradiograms of Ter1–Ter3 and RFP4 cloned into plasmid replicons. Ter1–Ter3 are Swi1p- and Swi3p-dependent, whereas RFP4 is nonfunctional in plasmid context. (A) Diagram of pIS8B.IRT2, pTer2.IRT2, p1REB1.IRT2, and pRFP4b.IRT2, containing Ter1, Ter2, Ter3, and sequences corresponding to RFP4, respectively, cloned in native orientation with respect to fork movement. The region of probe hybridization is shown. PvuII-digested replication intermediates were analyzed in all plasmid 2D gels. (B) 2D gel analyses of pIS8B.IRT2 replication intermediates. Ter1 was narrowed down to 252 bp of intergenic sequence contained in pIS8B.IRT2. This sequence causes fork arrest in WT cells but not in swi1Δ::ura4 or swi3-146 cells. (C) 2D gel analysis of pTer2.IRT2 replication intermediates. Ter2, corresponding to the downstream Reb1p binding element (with respect to transcription) and ≈250 bp of surrounding sequence, functions as a fork barrier in WT but not in reb1Δ::kanMX or swi1Δ::ura4 cells. (D) 2D gel analysis of p1REB1.IRT2 replication intermediates. The farthest upstream Reb1p binding element (with respect to transcription) and ≈250 bp of surrounding sequence, corresponding to Ter3, cause efficient fork arrest in WT cells that depends on both reb1 and swi1 function. (E) 2D gel analysis of pRFP4b.IRT2 replication intermediates. Approximately 400 bp of DNA sequence immediately upstream of Ter3 (with respect to transcription) fails to arrest replication forks when cloned extrachromosomally, in contrast to Ter1–Ter3.
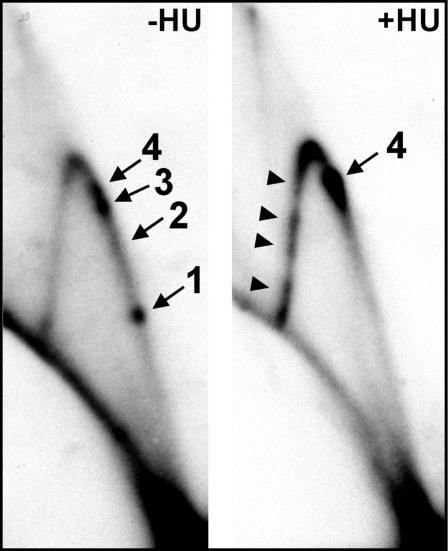
HU-Induced Replication Stress Mimicked swi1 and swi3 Deletion at Ter1–Ter3. Swi1p has been shown to stabilize stalled forks generated by HU-induced stress, at least within the rDNA, and to subsequently signal to the checkpoint kinase Cds1p (31). Therefore, we chose to analyze the Swi1p- and Swi3p-dependent Ter sites in the presence of HU during which Swi1p is presumably engaged at a multitude of unstable forks throughout the genome. 2D gel electrophoresis revealed that fork arrest at all three Swi1p- and Swi3p-dependent fork barriers (Ter1–Ter3) was abolished after growth of unsynchronized cultures for one cell cycle (≈3–3.5 h) in the presence of 12 mM HU (Fig. 3). It should be noted that these conditions are sufficient to cause fork destabilization that would predicate rescue by Swi1p and also Swi1p-dependent Cds1p activation (31). Thus, induction of replication stress by HU caused removal by competition of Swi1p and/or Swi3p at the natural rDNA fork barriers.
Fig. 3.
Autoradiogram of 2D gels of replication intermediates generated in the presence of HU. HU-induced replication stress eliminates fork arrest at the natural fork barriers Ter1–Ter3 but introduces numerous new pause sites. Shown is 2D gel analysis of the BamHI fragment of WT (SP976) cells, grown in the absence (–HU) and presence (+HU) of 12 mM HU for 3.5 h. Although the region is actively replicated, the Swi1p- and Swi3p-dependent Ter1–Ter3 are abolished in the presence of HU, whereas the Swi1p- and Swi3p-independent RFP4 is unaffected, accumulating an excess of forks. Note also the appearance of numerous pause sites on the descending arc (arrowheads).
The Swi1p- and Swi3p-independent fork arrest at RFP4 was not abolished by HU treatment. In fact, the intensity of the spot at RFP4 significantly increased in intensity (Fig. 3), resembling the effects of swi1 and swi3 mutations on RFP4 (Fig. 1B). These results would be expected if the induced genotoxic stress titrated Swi1p away from Ter1–Ter3 and onto stalled forks throughout the genome, rendering Ter1–Ter3 inactive. Therefore, Swi1p appeared to represent a novel dynamic link between natural replication fork barriers and stress-induced fork stalling.
In addition to its effects on the natural termini, HU treatment also caused the appearance of at least four other pause sites on the descending Y-arc that were absent in unstressed cells (Fig. 3, arrowheads). These sites are within the 3′ end of the rRNA transcription unit.
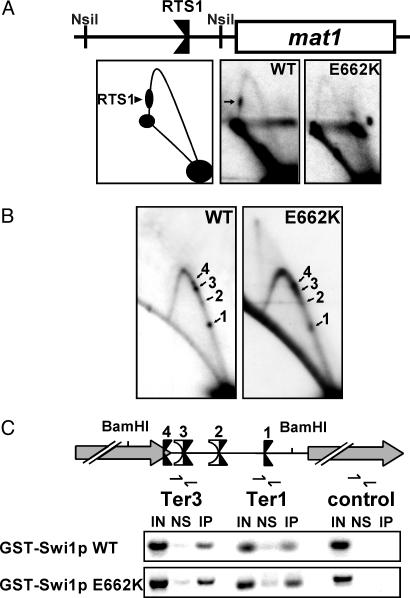
Specificity of Fork Arrest at Ter1–Ter3 Differs from That at RTS1 of mat1. Swi1p terminator activity at RTS1 and pausing at MPS1 have been separated by a point mutation in swi1 (Swi1p E662K). The mutant is completely defective in causing fork arrest at RTS1 but still functional in causing fork pausing at MPS1 (7). To examine further Swi1p function at the rDNA termini, we analyzed rDNA replication in this mutant strain. As shown in Fig. 4A, RTS1 activity was reproducibly absent in this strain. However, the E662K mutant form of Swi1p still promoted fork arrest at all three Swi1p-dependent rDNA Ter sites, Ter1–Ter3 (Fig. 4B). These results suggest some specificity of Swi1p action at natural fork barriers.
Fig. 4.
2D gel analysis of fork movement at the mating-switch locus and ChIP analyses of the rDNA Ter sites. The E662K mutant form of Swi1p, which is completely defective in fork arrest at RTS1, is fully functional at the rDNA fork barriers and localizes to this region as effectively as WT Swi1p. (A) 2D gel analysis of the centromere-proximal NsiI fragment of mat1 containing RTS1, confirming results in ref. 7 that Swi1p E662K fails to arrest forks at this site. RTS1 (arrow) can be seen on the descending Y-arc in WT cells but not in cells harboring the swi1 E662K mutation (E662K). A diagram of the analyzed region is shown. (B) Ter1–Ter3 are functional in swi1 E662K mutant cells. 2D gel analysis of the intergenic BamHI fragment of WT and swi1 E662K (E662K) cells. (C) ChIP. WT GST-Swi1p localizes to the rDNA Ter sites as shown by enrichment of this region as compared with a region within the rDNA transcription unit located ≈5 kb from the fork barriers. Consistent with 2D replication analysis (see B), GST-Swi1p E662K localizes to the Ter sites at least as efficiently as the WT protein. Note that this assay cannot clearly resolve the closely associated Ter sites. A diagram of the analyzed region is shown.
Because both WT Swi1p and the E662K mutant arrested forks at rDNA Ter1–Ter3, we expected that both the WT and the mutant form of Swi1p would be preferentially localized near the Ter sites in comparison with other regions of the repeated rDNA units. To test this prediction, we expressed GST-tagged WT and E662K Swi1p under the control of the constitutively active Prae promoter (of medium strength) in swi1Δ cells (SPGK301C) and performed ChIP assays. We found that WT GST-Swi1p preferentially associated with the Ter sites in vivo but not with a region located ≈5 kb away within the rDNA transcription unit (Fig. 4C Middle). Of note, formaldehyde cross-linking (which mainly promotes protein–DNA cross-linking but much less efficient protein–protein cross-links) alone was insufficient to immunoprecipitate the Swi1p-associated Ter sites under the conditions used. Instead, use of the protein–protein cross-linker dimethyl adipimidate (DMA) before formaldehyde fixation was necessary. These data suggest that Swi1p association with the Ter sites was probably not due to direct binding to DNA. As expected, the E662K mutant form of Swi1p also associated with the rDNA termini in vivo at least as efficiently as WT Swi1p (Fig. 4C Bottom). The results obtained from ChIP analysis, therefore, were consistent with the 2D gel results in demonstrating that, in addition to the WT protein, Swi1p E662K remains functional at the rDNA fork barriers. Taken together, these results suggest that Swi1p mediates fork arrest at different types of site-specific replication termini, probably by specific interactions.
Discussion
We report here that Swi1p and Swi3p act with some site specificity outside the mating-type locus mat1 to arrest DNA replication at three of four natural sites of fork arrest in fission yeast rDNA. Two of the sites, Ter2 and Ter3, were also dependent on Reb1p (26), implicating multiple proteins in rDNA fork arrest. Although Swi1p (and Swi3p) act at different Ter sites, a Swi1p point mutant E662K was defective in fork arrest at RTS1 but not at the rDNA Ter sites, suggesting specificity at different terminator complexes. Genome-wide replication stress induced by HU also abolished arrest at the Swi1p- and Swi3p-dependent Ter1–Ter3. Importantly, replication forks stalled at the fourth barrier RFP4, which is nonfunctional in an extrachromosomal replicon, did not require Swi1p or Swi3p for stable arrest. RFP4 is expectedly unaffected by HU, which engages Swi1p in genome maintenance (31). The results point toward an expanded function for Swi1p and Swi3p in mediating site-specific replication fork arrest and begin to reconcile Swi1p's function at natural fork barriers with its function in maintaining genomic stability through its fork-stabilizing and S-phase checkpoint-inducing activity.
Swi1 is conserved among eukaryotes, and homologs include TOF1 of Sa. cerevisiae and the Tim family of genes in higher eukaryotes (7). These genes appear to share conserved function, at least among yeasts, in maintenance of genomic integrity (29, 31). The results presented here, taken together with reports in refs. 30 and 31, suggest that Swi1p and Tof1p may act as generalized terminator proteins to arrest the replisome at natural fork barriers and at other areas of fork stalling. After HU-mediated genotoxic stress, Tof1p prevents replisomal proteins present at stalled forks from uncoupling and drifting past the forks to associate instead with chromosomal regions farther downstream (30). Similarly, Swi1p appears to stabilize forks stalled by HU-induced stress and prevent their collapse (31). Like Tof1p, it may pause forks to prevent uncoupling of the replisome from replicated areas. At natural fork barriers, Swi1p acts (with Swi3p) to ensure fork pausing. This function may resemble Swi1p-mediated prevention of replisomal uncoupling at HU-stalled forks, suggesting that a randomly stalled replisome may share some similarity with a replisome stalled at Ter sites.
Before this study, terminator proteins were believed to act alone at specific rDNA fork barriers to mediate fork arrest. However, our results demonstrate that multiple proteins are needed for this function. For example, Reb1p acts with Swi1p and Swi3p to mediate arrest at Ter2 and Ter3. Reb1p binds to its two cognate binding elements in this region (39), which presumably represents the initial step in fork arrest.
Aside from the function of Reb1p in termination of transcription, mechanisms of replication fork arrest similar to those occurring at Ter2 and Ter3 are likely to occur at Ter1, except that a cognate DNA-binding protein that is yet to be identified is predicted to act at Ter1. Fork arrest at RTS1 is likely to occur by analogous means. It has been suggested that the Reb1p-like Rtf1p probably acts at RTS1 by binding to tandemly repeated binding elements (28). A second protein, Rtf2p, has been suggested to act just upstream of this region (28). Precisely how multiple proteins cooperate at different Swi1p- and Swi3p-dependent Ter sites remains unknown. Three mechanistic possibilities come to mind: (i) a multiprotein complex involving a Ter-binding protein as well as Swi1p and Swi3p may associate with the site to cause fork arrest, (ii) the Ter-binding protein may interact with replisome-associated Swi1p or Swi3p to halt the replication machinery, or (iii) fork pausing by a Ter-binding protein may be followed by stabilization of the stalled structure by Swi1p and Swi3p. It is interesting to note in this regard that the spot at RFP4 increased in intensity in the absence of Ter1–Ter3, suggesting that the stalled forks were able to replicate through Ter1–Ter3 in the absence of Swi1p or Swi3p to accumulate at RFP4.
Swi1p is shown to arrest forks at five unrelated site-specific natural fork barriers (ref. 7 and this article) as well as to stabilize randomly stalled forks (31). Thus, its fork-arresting activity appears to be somewhat nonspecific. However, the Swi1p point mutant E662K is specifically defective in fork arrest at RTS1 and fully functional at Ter1–Ter3 [as well as MPS1 (7)]. It is difficult to reconcile this result with a nonspecific fork-protecting function for Swi1p. Rather, the protein appears to demonstrate some selectivity for certain barriers. On the other hand, abolition of the natural fork barriers Ter1–Ter3 by HU-induced replication stress is interesting with respect to Swi1p specificity. Swi1p specificity for natural fork barriers appears to be sufficiently low such that stalled forks due to HU treatment are able to titrate this pool of Swi1p away from the natural fork barriers and onto the larger number of randomly stalled forks. Indeed, Swi1p levels at natural fork barriers may be limiting as cis mutations that affect localization of the protein at MPS1 increase its presence at RTS1 (40). This dynamic nature of Swi1p action at natural Ter sites versus stress-induced stall sites demonstrates for the first time that site-specific fork arrest can be modulated in vivo.
Six natural fork barriers have now been identified in fission yeast: RTS1 and MPS1 at the mat1 locus (7) and Ter1–Ter3 and RFP4 within the rDNA spacer region as reported here. RFP4 is the only natural fork barrier that acts independently of Swi1p and Swi3p. RFP4 is also unique because it appears to be located within the rDNA transcription unit (38, 39). RFP4 is nonfunctional in an extrachromosomal context, as contrasted with Ter1–Ter3. Two scenarios could account for this observation. Perhaps more than one unit sequence is required for RFP4 function, which may require looping between two sites. It is more likely, however, that RFP4 is rRNA-transcription-dependent and that replication forks accumulate at this site because of the collision of the RNA polI and replication machineries. It is interesting in this respect that forks stalled at RFP4 do not depend on Swi1p or Swi3p for stabilization as do forks stalled at natural replication fork barriers or HU-induced pause sites.
An apparent puzzle regarding natural replication fork barriers is that forks stalled at these sites presumably do not activate the replication checkpoint efficiently, whereas forks stalled because of genotoxic stress do so robustly (4, 5). The proteins recruited to forks stalled at natural Ter sites may differ from those recruited to forks stalled because of lesions or genotoxic stress. These differences could result from differences in fork structure or in replisome composition. Swi1p is associated with both types of arrested forks and is necessary for checkpoint activation at HU-stalled forks (31). Why, then, does Swi1p not activate cell-cycle arrest in WT cells by its association with stalled forks at the natural fork barriers?
Supplementary Material
Acknowledgments
We thank Drs. R. Dhar and K. L. Gould for providing strains and plasmids. This work was supported by grants from the National Institute of Allergy and Infectious Diseases and the National Institute of General Medical Sciences.
Abbreviations: ChIP, chromatin immunoprecipitation; HU, hydroxyurea; RFB, replication fork barriers; rDNA, ribosomal DNA; Ter sites, site-specific replication terminators.
References
- 1.McGlynn, P. & Lloyd, R. G. (2002) Nat. Rev. Mol. Cell Biol. 3, 859–870. [DOI] [PubMed] [Google Scholar]
- 2.Cox, M. M., Goodman, M. F., Kreuzer, K. N., Sherratt, D. J., Sandler, S. J. & Marians, K. J. (2000) Nature 404, 37–41. [DOI] [PubMed] [Google Scholar]
- 3.Kolodner, R. D., Putnam, C. D. & Myung, K. (2002) Science 297, 552–557. [DOI] [PubMed] [Google Scholar]
- 4.Boddy, M. N. & Russell, P. (2001) Curr. Biol. 11, R953–R956. [DOI] [PubMed] [Google Scholar]
- 5.Osborn, A. J., Elledge, S. J. & Zou, L. (2002) Trends Cell Biol. 12, 509–516. [DOI] [PubMed] [Google Scholar]
- 6.Bastia, D. & Mohanty, B. K. (1996) in DNA Replication in Eukaryotic Cells, ed. DePamphilis, M. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 177–215.
- 7.Dalgaard, J. Z. & Klar, A. J. (2000) Cell 102, 745–751. [DOI] [PubMed] [Google Scholar]
- 8.Dalgaard, J. Z. & Klar, A. J. (2001) Genes Dev. 15, 2060–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brewer, B. J., Lockshon, D. & Fangman, W. (1992) Cell 71, 267–271. [DOI] [PubMed] [Google Scholar]
- 10.Sanchez, J. A., Kim, S. M. & Huberman, J. A. (1998) Exp. Cell Res. 238, 220–230. [DOI] [PubMed] [Google Scholar]
- 11.Hernandez, P., Martin-Parras, L., Martinez-Robles, M. L. & Schvartzman, J. B. (1993) EMBO J. 12, 1475–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lopez-Estrano, C., Schvartzman, J. B., Krimer, D. B. & Hernandez, P. (1999) Plant Mol. Biol. 40, 99–110. [DOI] [PubMed] [Google Scholar]
- 13.Little, R. D., Platt, T. H. & Schildkraut, C. L. (1993) Mol. Cell. Biol. 13, 6600–6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiesendanger, B., Lucchini, R., Koller, T. & Sogo, J. M. (1994) Nucleic Acids Res. 22, 5038–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayashi, T. & Horiuchi, T. (1996) Genes Cells 1, 465–474. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi, T., Heck, D. J., Nomura, M. & Horiuchi, T. (1998) Genes Dev. 12, 3821–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johzuka, K. & Horiuchi, T. (2002) Genes Cells 7, 99–113. [DOI] [PubMed] [Google Scholar]
- 18.Takeuchi, Y., Horiuchi, T. & Kobayashi, T. (2003) Genes Dev. 17, 1497–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang, J. & Moazed, D. (2003) Genes Dev. 17, 2162–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Defossez, P. A., Prusty, R., Kaeberlein, M., Lin, S. J., Ferrigno, P., Silver, P. A., Keil, R. L. & Guarente, L. (1999) Mol. Cell 3, 447–455. [DOI] [PubMed] [Google Scholar]
- 21.Khatri, G. S., MacAllister, T., Sista, P. R. & Bastia, D. (1989) Cell 59, 667–674. [DOI] [PubMed] [Google Scholar]
- 22.Bussiere, D. E. & Bastia, D. (1999) Mol. Microbiol. 31, 1611–1618. [DOI] [PubMed] [Google Scholar]
- 23.Lee, E. H., Kornberg, A., Hidaka, M., Kobayashi, T. & Horiuchi, T. (1989) Proc. Natl. Acad. Sci. USA 86, 9104–9108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohanty, B. K. & Bastia, D. (2004) J. Biol. Chem. 279, 1932–1941. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi, T. (2003) Mol. Cell. Biol. 23, 9178–9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanchez-Gorostiaga, A., Lopez-Estrano, C., Krimer, D. B., Schvartzman, J. B. & Hernandez, P. (2004) Mol. Cell. Biol. 24, 398–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dalgaard, J. Z. & Klar, A. J. (1999) Nature 400, 181–184. [DOI] [PubMed] [Google Scholar]
- 28.Codlin, S. & Dalgaard, J. Z. (2003) EMBO J. 22, 3431–3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foss, E. J. (2001) Genetics 157, 567–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katou, Y., Kanoh, Y., Bando, M., Noguchi, H., Tanaka, H., Ashikari, T., Sugimoto, K. & Shirahige, K. (2003) Nature 424, 1078–1083. [DOI] [PubMed] [Google Scholar]
- 31.Noguchi, E., Noguchi, C., Du, L. L. & Russell, P. (2003) Mol. Cell. Biol. 23, 7861–7874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park, H. & Sternglanz, R. (1999) Yeast 15, 35–41. [DOI] [PubMed] [Google Scholar]
- 33.Deshpande, A. M. & Newlon, C. S. (1996) Science 272, 1030–1033. [DOI] [PubMed] [Google Scholar]
- 34.Moreno, S., Klar, A. & Nurse, P. (1991) Methods Enzymol. 194, 795–823. [DOI] [PubMed] [Google Scholar]
- 35.Huberman, J. A., Spotila, L. D., Nawotka, K. A., el-Assouli, S. M. & Davis, L. R. (1987) Cell 51, 473–481. [DOI] [PubMed] [Google Scholar]
- 36.Kurdistani, S. K. & Grunstein, M. (2003) Methods 31, 90–95. [DOI] [PubMed] [Google Scholar]
- 37.Liu, Y., Kung, C., Fishburn, J., Ansari, A. Z., Shokat, K. M. & Hahn, S. (2004) Mol. Cell. Biol. 24, 1721–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Melekhovets, Y. F., Shwed, P. S. & Nazar, R. N. (1997) Nucleic Acids Res. 25, 5103–5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao, A., Guo, A., Liu, Z. & Pape, L. (1997) Nucleic Acids Res. 25, 904–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaykov, A., Holmes, A. M. & Arcangioli, B. (2004) EMBO J. 23, 930–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.