Abstract
Genomic rearrangements are a frequent source of instability, but the mechanisms involved are poorly understood. A 2.5-kbp poly(purine·pyrimidine) sequence from the human PKD1 gene, known to form non-B DNA structures, induced long deletions and other instabilities in plasmids that were mediated by mismatch repair and, in some cases, transcription. The breakpoints occurred at predicted non-B DNA structures. Distance measurements also indicated a significant proximity of alternating purine-pyrimidine and oligo(purine·pyrimidine) tracts to breakpoint junctions in 222 gross deletions and translocations, respectively, involved in human diseases. In 11 deletions analyzed, breakpoints were explicable by non-B DNA structure formation. We conclude that alternative DNA conformations trigger genomic rearrangements through recombination-repair activities.
Gross chromosomal rearrangements are a common source of genetic instability (1). Thus, characterization of the underlying molecular mechanisms of mutagenesis is fundamental for our understanding of human disease. A hallmark of gross deletions is the presence of short homologous tracts (typically 2–8 bp) at the breakpoints (2), a finding that has prompted speculation as to the two distinct mechanisms postulated to be responsible for their formation. The slipped mispairing model (3) envisages that during lagging strand DNA synthesis, distantly located repeats are brought into close proximity by the looping out of the single-stranded region, thereby enabling the replication complex to “jump” from the proximal to the distal repeat and hence bypass the looped structure. Alternative models propose that various types of repetitive sequence elements may serve as substrates for intra- or intermolecular recombination (2, 4). Neither model is satisfactory; slipped mispairing is inconsistent with deletions greater than ≈500 bp and deletions manifesting <4-bp homologies (5–9), whereas the recombination model does not provide a rationale for the initiation of the process.
Specific sequence motifs such as direct and inverted repeats, (RY·RY)n and (R·Y)n, in which R represents purine and Y represents pyrimidine, and four closely spaced G-rich direct repeats [i.e., (G·C)3] undergo structural transitions from the orthodox right-handed B-helical duplex to higher energy state non-B structures (slipped hairpin/loops, cruciforms, left-handed Z-helices, triplexes, and tetraplexes, respectively) (10–12) under torsional stress (negative supercoiling) in vivo.
Early articles in bacteria and hamster cells reported isolated cases in which deletions could occur by a recombination-repair reaction mediated by cruciform structures forming at each breakpoint (13, 14). Recently, the breakpoint junctions of the human constitutional translocations t(1;22), t(4;22), t(11;22), and t(17;22), which involve a common locus on chromosome 22q11.2, were found to coincide with large (>95 bp) cruciform structures (15–18), suggesting that this conformation may predispose specific loci to genomic rearrangements.
The polycystic kidney disease 1 (PKD1) gene is mutated in ≈85% of patients with autosomal dominant polycystic kidney disease (ADPKD) (19, 20). We found (21) that a 2.5-kbp poly(R·Y) sequence from intron 21, known to form various types of non-B DNA structures (refs. 21 and 22 and references therein), conferred plasmid instability and led to bacterial cell death in a supercoil-dependent fashion. Because negative supercoiling stabilizes non-B DNA structural transitions (10–12, 23–27), a mutagenic role for non-B conformations was established.
Here, we show that the PKD1 tract increased the frequency of plasmid alterations, including long deletions, in conjunction with DNA repair processes and transcription. In all 16 deletions (0.4–4.3 kbp) examined, the breakpoint junctions occurred at predicted non-B DNA structures, both in the PKD1 tract and in the vector, and included all known DNA conformations. Furthermore, analyses of the DNA sequences at 222 characterized human rearrangement breakpoint junctions revealed a significant association with non-B DNA-forming elements; in 11 randomly selected cases, the breakpoint locations were invariably explicable in terms of non-B DNA structure formation.
Thus, we conclude that genomic rearrangements occur at alternative DNA conformations.
Materials and Methods
A transcription terminator cassette was cloned in pGFPuv (Clontech) to give pRW3619, and the poly(R·Y) tracts from the human PKD1 gene were cloned downstream of the GFP gene in pRW3619 to obtain pRW3620 and pRW3621. The plasmids were transformed in Escherichia coli strains, and the concentration of GFP was measured by fluorescence spectroscopy after a 6-h induction of transcription [in the presence or absence of isopropyl β-d-thiogalactoside (IPTG)] in lysates. Plasmids were harvested and used to transform nucleotide excision repair-deficient cells (to avoid bacterial cell death) (21) to monitor the PKD1 tract- and transcription-dependent loss of fluorescence. Loss of fluorescence was also determined in selected strains after recultivations. Mutations responsible for fluorescence loss were determined by DNA sequencing. Complexity analysis (4, 28) was used to analyze the DNA sequence motifs at rearrangement breakpoints, both in E. coli and in the human Gross Rearrangement Breakpoint Database (GRaBD; www.uwcm.ac.uk/uwcm/mg/grabd). Details on E. coli strains used, plasmid construction, fluorometric GFP measurements, IPTG induction of transcription, determination of frequency of white colony-forming units (CFUs), primer sequences, statistical analysis, and DNA breakage and complexity analysis are reported in Supporting Methods in Supporting Text, which is published as supporting information on the PNAS web site.
Results
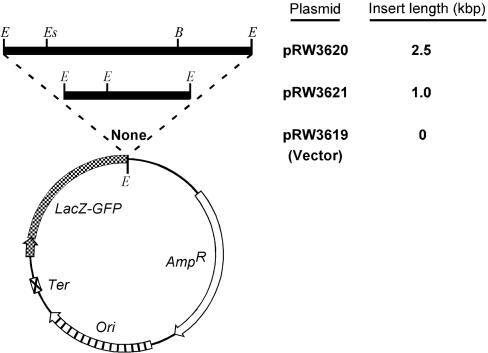
The 2.5-kbp Poly(R·Y) Tract Promotes Instability. To test the hypothesis that non-B DNA structures are associated with rearrangement mutagenesis, we constructed three plasmids (Fig. 1) containing GFP as a reporter gene (29). The 2.5-kbp poly(R·Y) sequence was cloned downstream of the GFP gene (pRW3620) in an orientation such that the purine-rich strand provided the template for transcription, as in the human PKD1 gene (30). pRW3621 lacked ≈1.5 kbp of the poly(R·Y) tract toward its 3′ end.
Fig. 1.
Plasmids used in the study. Striped arrow, ColE1 origin of replication; crossed box, transcriptional terminator; cross-hatched arrow and segment, lacZ promoter-operator and lacZ-GFPuv fusion gene, respectively (cross-hatched arrow shows the direction of transcription); open arrow, ampicillin-resistance gene; filled horizontal segments, PKD1 inserts; E, EcoRI recognition site; Es, EspI recognition site; and B, BbsI recognition site.
We cotransformed E. coli strains [two wild-type (wt), ΔUvrB, ΔMutL and ΔRecA; see part A of Table 4, which is published as supporting information on the PNAS web site] with both pIQ-kan (to repress GFP transcription) and pRW3619, pRW3620, or pRW3621 and grew the cells for 9 h in the absence of IPTG or in the presence of 2 mM IPTG for the last 6 h to induce GFP gene transcription (Fig. 5, which is published as supporting information on the PNAS web site). To identify single mutant plasmids from the intracellular pool, we isolated and retransformed plasmid DNA in ΔUvrB cells, which were then spread on IPTG-containing plates. Considering the composite results, we observed 2,027 (0.29%) white CFUs [372 (0.15%) for pRW3619, 1,286 (0.62%) for pRW3620, and 369 (0.15%) for pRW3621] from a total of 707,742 CFUs. Thus, the full-length poly(R·Y) tract, but not its truncated counterpart, increased the frequency of plasmid instability that led to a loss of the fluorescent phenotype by ≈4-fold.
To identify the mechanisms involved, we analyzed in detail the 1,434 white CFUs obtained by transformation with pRW3619 and pRW3620 in all strains, except ΔMutL, in which the PKD1 tract exhibited no effect. There were 295/197,251 white CFUs for pRW3619 and 1,139/127,467 white CFUs for pRW3620. Further screening revealed that loss of fluorescence was caused by plasmid loss, by multimerization, or by the arrest of cell growth (Fig. 6 and Table 5, which are published as supporting information on the PNAS web site).
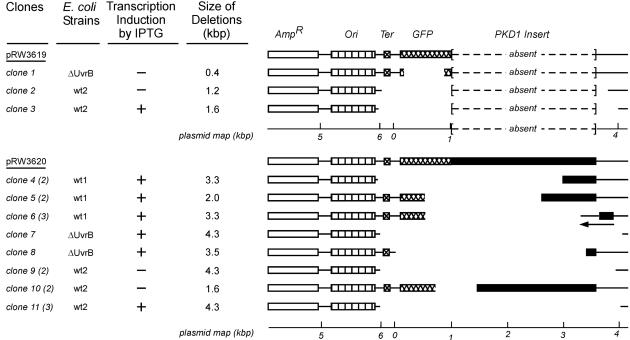
Restriction mapping and DNA sequence analyses revealed that 19/1,434 white CFUs (1.3%) contained a nonfunctional GFP gene (termed “mutant white CFUs”), and these clones were characterized in detail. Three were from pRW3619 and 16 from pRW3620, representing ratios of 1.5 × 10–5 and 12.6 × 10–5 mutants per all CFUs, respectively [P < 0.001, P(α)0.05 = 0.94]. All mutations were large (0.4–4.3 kbp) deletions (Fig. 2), and one (clone 6) contained an additional inversion. More than one clone with identical mutations was found from individual transformations with pRW3620. Because mutations may not have arisen independently, we considered 8/16 clones with pRW3620 as unique mutation events, representing a proportion of 6.3 × 10–5 [P = 0.049, P(α)0.05 = 0.50]. This value, which is 4 times greater than for the vector pRW3619, is a conservative estimate because no corrections were made for “nonunique” fluorescent CFUs.
Fig. 2.
Restriction mapping studies of the mutant white CFUs. The first column lists the individual clones; the numbers of clones with identical restriction patterns are given in parentheses. The second column provides the E. coli host strains before the plasmids were isolated and retransformed in the ΔUvrB strain. The third column indicates whether transcription was induced by IPTG. The fourth column indicates the size of the deletions. A schematic representation of the mutations is provided on the right. The space between segments indicates the size and location of each deletion. The symbols designating the regions of DNA are as in Fig. 1.
All three pRW3620 mutants isolated from wt1 cells (36,957 colonies were screened for pRW3620 from the wt1 strain), as well as one from wt2 (clone 10) and one from ΔUvrB (clone 8), contained a breakpoint within the poly(R·Y) tract. Moreover, 75% of pRW3620 mutants were from cells grown in the presence of IPTG (active transcription) and all mutants from the isogenic wt1 and ΔUvrB were obtained from transcription-induced cells.
These data indicate that the 2.5-kbp poly(R·Y) tract from intron 21 of the PKD1 gene induced several types of plasmid alterations, including long deletions, which caused loss of fluorescence.
Non-B DNA Structures at Breakpoints. We considered whether the PKD1-induced deletions might be caused by non-B DNA structures in the poly(R·Y) tract. First, analysis of the breakpoint sequences (Table 1) revealed the following. (i) Short (1–8 bp) homologies occurred at the breakpoints (interestingly, clones 6 and 11 had inverted homologies). (ii) Breakpoints did not occur at random positions (e.g., in mutants 2, 3, 4, 7, 9, and 11, deletions occurred within ≈150 bp of the replication origin), suggesting that structural features in the DNA were indeed involved in deletion mutagenesis. Second, we used complexity analysis (see Supporting Methods), a technique based on text-compression algorithms (28) (Fig. 7, which is published as supporting information on the PNAS web site) to search for any direct, inverted, or mirror (symmetric) repeats (capable of forming slipped structures and tetraplexes, cruciforms, and triplexes, respectively) (10, 12, 24–27) in the sequences flanking the breakpoints. This analysis revealed that such repeats were indeed present in all cases, both within the PKD1 tract and in the vector, and that the predicted non-B DNA structures involved a larger number of nucleotides (hence were likely more stable) in the PKD1 tract than in the vector.
Table 1. Sequences and locations of breakpoints.
| Mutant clone | First breakpoint | Second breakpoint | Junction | Break positions* |
|---|---|---|---|---|
| 1 | GAAGGTGAtgca | acatTGAAGATG | GAAGGTGAAGATG | 395/801 |
| 2 | GGAAGCAtaaa | tactAGTCGGC | GGAAGCAGTCGGC | 6087/3828 |
| 3 | GGCCTTTttacg | ttgtacTGAGAGT | GGCCTTTGAGAGT | 5931/4024 |
| 4 | CGCCAGCaacg | cctcCCCTTCT | CGCCAGCCCTTCT | 5919/3057 |
| 5 | ATACCCTTgtta | ccttCCTTTCCCC | ATACCCTTTCCCC | 645/2699 |
| 6 | GATACCCttgtt | ccagCCCCGACAC | GATACCCCGACAC | 643/3985 |
| GGGGAGagagga | tcccgGGAGACGG | GGGGAGGAGACGG | 3567/3902 | |
| 7 | TGGATAAccgt | tctaAATACATT | TGGATAATACATT | 6005/4215 |
| 8 | GCCTCTCCCCgc | ttCTCTCCCCTCC | GCCTCTCCCCTCC | 29/3517 |
| 9 | GCCTTTttacgg | tgtacTGAGAGTG | GCCTTTGAGAGTG | 5931/4024 |
| 10 | ATGGCCctgt | tcccCCTCCTTCT | ATGGCCTCCTTCT | 862/2423 |
| 11 | GGAAGCataa | tattTGTTTAT | GGAAGCTGTTTAT | 6086/4201 |
Uppercase letters indicate retained nucleotides, lowercase nucleotides were deleted or were beyond the breakpoint of inversion; underlined nucleotides were homologous between the first and second breakpoints and were retained. Sequences that were joined, but in an inverted orientation, are in boldface.
Breakpoint positions were the last 3′ and first 5′ retained nucleotides. Nucleotide numbering was that of pRW3620. In the second and third columns, the sequence of the top strand reads from low to high numbers in the plasmid map; for the inversion, the bottom strand is shown and map positions decrease.
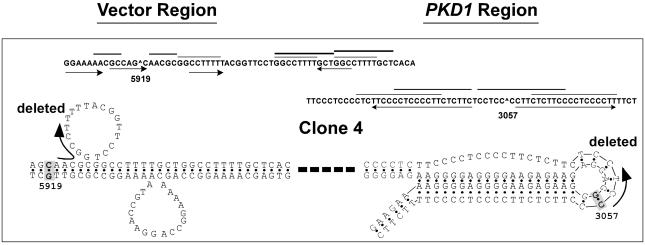
Third, we accommodated the repeats in their preferred alternative DNA conformations, and related their location to the breakpoints. In all cases, these two elements coincided. Fig. 3 depicts the results for clone 4, whereas Fig. 8, which is published as supporting information on the PNAS web site, displays the results for the remaining clones. We identified five additional deletions of pRW3620 by recultivation assays in which deleted clones became enriched in the cell cultures by virtue of their growth advantage (12). In these clones, we also confirmed the localization of deletion breakpoints at sites of non-B DNA structures (Supporting Results in Supporting Text and Figs. 9 and 10, which are published as supporting information on the PNAS web site).
Fig. 3.
Representative DNA structures for clone 4. The nucleotides in boldface and shading correspond to homologous sequences. The arrows indicate the sequences that were deleted, and a thick dashed line denotes intervening DNA. Repetitive motifs are identified as in the legend to Fig. 7. The breakpoint in the PKD1 tract (G·C 3057) occurred within a 7-bp stretch that was flanked by two perfect 19-bp mirror repeats (TTCCCCTCCCCTTCTCTTC). The mirror symmetry and the R·Y composition are consistent with the formation of an intramolecular triplex with a DNA loop of 7 bp. The location of the breakpoint within the loop coincides with nucleotides known to have unusual torsion angles (26) and to be susceptible to chemical modification and nuclease cleavage in vitro. In the vector, the breakpoint (G·C 5919) was located 6 bp upstream of three direct repeats, GGCCTTTT followed after 9 bp by two tandem GGCCTTTTGCT repeats. In the looped structure resulting from slippage with the first repeat, the breakpoint is located in a region of perturbed helical DNA.
In summary, these analyses support our contention that non-B DNA conformations formed, which in turn served as substrates for the generation of gross deletions.
Involvement of Repair Proteins and Transcription in Instabilities. The supercoil-dependent induction of plasmid instability by the PKD1 tract (21) suggests that non-B DNA structures were involved in all white CFUs found (the 2,027 described above). Table 2 shows the distribution of all white CFUs. In the absence of transcription (–IPTG), the full-length PKD1 tract increased the fraction of white CFUs in all strains [P(α)0.01 = 1.00 between pRW3620 and pRW3619, and between pRW3620 and pRW3621], except ΔMutL [P(α)0.01 of 0.01 and 0.11, respectively], indicating that (i) the PKD1 tract exerted its mutagenic influence in association with DNA replication, (ii) the 2.5-kbp poly(R·Y) tract was more effective than the ≈1-kbp truncated tract, and (iii) a proficient mismatch-repair system was required.
Table 2. Influence of host cell genotype and transcription on number of white CFUs per 10,000 total CFUs.
| pRW3619
|
pRW3620
|
pRW3621
|
Statistical significance
|
|||||
|---|---|---|---|---|---|---|---|---|
| E. coli strain | -IPTG | +IPTG | -IPTG | +IPTG | -IPTG | +IPTG | pRW3619 vs. pRW3620 | pRW3620 vs. pRW3621 |
| wt1 | 7 | 3 | 30* | 92* | 10 | 4 | S | S |
| ΔUvrB | 14* | 2* | 104 | 103 | 30* | 5* | S | S |
| wt2 | 32 | 53 | 103* | 155* | 19 | 40 | S | S |
| ΔMutL | 11 | 18 | 18 | 19 | 20 | 12 | NS | NS |
| ΔRecA | 4 | 5 | 61 | 60 | 2 | 1 | S | S |
The statistical significance is [P < 0.01 with P(α)0.01 > 0.90]. The data that are statistically significantly different (between -IPTG and +IPTG) for a particular plasmid and E. coli strain are marked with asterisks. The results for the combined -IPTG and +IPTG data for each strain are shown in the last two columns. S, significant; NS, not significant.
In the presence of transcription (+IPTG), the full-length PKD1 tract increased the fraction of white CFUs in the wt strains [P(α)0.01 = 1.00 for wt1 and 0.95 for wt2 between –IPTG and +IPTG] but not in ΔMutL, ΔUvrB, and ΔRecA cells, indicating that transcription increased the frequency of the PKD1 tract-induced plasmid alterations in wt backgrounds.
The roles of the mismatch-repair (MutL, MutS, and MutH) and RecA proteins were investigated further, by analyzing PKD1- and transcription-induced deletions in pRW3620 as a function of successive 1-day recultivations in mutant cells (part B of Table 4). In the control wt2 (Fig. 11, which is published as supporting information on the PNAS web site), deletions accumulated at a constant rate of 4%/day in the absence of transcription. In the presence of transcription, the rate varied but peaked at 30% on day 2. These transcription-dependent rate changes were also accompanied by extensive plasmid loss (up to 98%) and by alterations in the restriction patterns of deleted products with time of recultivation. This finding suggests that either transcription increased the rates of plasmid alterations (including deletions) or deletions were simply a side-product of full-length plasmid loss. In ΔMutH cells, deletion rates also varied but attained lower values (15% on day 2) than wt2, whereas the restriction patterns of deletions changed, as in wt2. These data support the conclusion (from Table 2) that the postreplicative mismatch-repair pathway was involved in the PKD1-induced instabilities. In the ΔMutL and ΔMutS strains, the deletion rates were constant and lower than wt2 (4%/day and 2%/day, respectively) in the presence of transcription, whereas the plasmid restriction patterns were unchanged throughout the time of recultivation (as in wt2 without transcription). Thus, the MutL and MutS proteins were specifically involved in the transcription-induced selection pressure imposed by the PKD1 tract. Hence, these activities occurred outside the classical postreplicative mismatch repair pathway (31). In the absence of RecA, the rates of deletions were constant (5%/day) and restriction patterns were unchanged. In conclusion, these biochemical data confirm that both mismatch repair and RecA proteins were involved in the PKD1- and transcription-induced plasmid alterations, and they suggest that mutations may have arisen from recombination-repair activities. Plasmid loss may also have arisen from failure to repair non-B DNA structures and/or from repair events that disrupted either the replication origin or the ampicillin-resistance gene.
(RY·RY)n and Oligo(R·Y)n Tracts Are Located Near Human Rearrangement Breakpoints. Presuming that conclusions from E. coli can be extrapolated to mammalian systems, we determined whether non-B DNA-forming sequences also colocalize with breakpoints in human gene rearrangements. GRaBD represents a large repository of human germ-line and somatic breakpoints derived from chromosomal rearrangements, mostly causative of either inherited disease or cancer (32). We originally found that repetitive sequences were over-represented at these breakpoint junctions (32). To investigate whether breakpoint location and nearest repeating DNA motif were correlated, we determined the distances of the “nearest tract to breakpoint” [i.e., motifs located within 125 bp upstream (5′) or downstream (3′) of the breakpoint site] for the (RY·RY)n and oligo(R·Y)n tracts (for which complexity analysis parameters were well defined), capable of forming Z-DNA and slipped/triplex structures, respectively, in 126 gross deletions and 96 translocations. For the (RY·RY)n tracts and unambiguous breakpoints (i.e., no homologies between breakpoints; 37 deletions and 53 translocations), the distribution was significantly different at 1% and 5% levels for motifs both 5′ and 3′ to deletion breakpoints, when compared with distances from a set of reference DNA sequences (Table 3). A comparison of case and control mean distances revealed that the “nearest (RY·RY)n tract to breakpoint” was on average nearer the breakpoint junction than in matched control DNA sequences.
Table 3. Significance levels in the distance of the nearest (RY·RY)n or (R·Y)n tract to a human rearrangement breakpoint (n = 5–25).
| 5′ to breakpoint* | 3′ to breakpoint* | Significance |
|---|---|---|
| Deletions | ||
| (RY·RY)13-25 | (RY·RY)9-25 | 0.01 |
| (RY·RY)5,7,11 | (RY·RY)7 | 0.05 |
| Translocations | ||
| (R·Y)13-21 | - | 0.01 |
| (R·Y)11,23,25 | - | 0.05 |
Range applies only to motifs of odd lengths. Average distance to breakpoint was shorter in breakpoint junctions than in control sequences.
The distribution of oligo(R·Y)n tracts was found to cluster specifically at translocation breakpoints and predominantly on the 5′ side. For the nearest 5′ oligo(R·Y)n tract to translocation breakpoint, a comparison of case and control mean distances revealed that these motifs were, on average, closer to the breakpoint in translocations than in matched control DNA sequences. An examination of the set of ambiguous breakpoint sequences revealed that 40 overlapped with an (RY·RY)n or oligo(R·Y)n tract (30%), whereas 55–72% contained one or the other tract within 20 bp (Table 6, which is published as supporting information on the PNAS web site). Thus, we conclude that a significant association exists between the location of (RY·RY)n or oligo(R·Y)n tracts and the sites of DNA breakage, supporting a causative role for these sequences in the destabilization and cleavage of duplex DNA.
To verify whether breakpoints occurred at short homologous nucleotide sequences located within, or adjacent to, potential non-B DNA structures, as in the bacterial cases, we analyzed the DNA sequences of 11 gross deletions associated with autosomal dominant polycystic kidney disease, early-onset Parkinsonism, Menkes syndrome, α+-thalassemia, adrenoleukodystrophy, and hydrocephalus, taken from five randomly selected studies (Table 7 and Fig. 12, which are published as supporting information on the PNAS web site). Twelve LINE and SINE elements were noted in eight cases, nine of which spanned the breakpoint junctions; microhomologies and short repetitive motifs were present in all cases. Significantly, by folding the repetitive motifs into their preferred non-B conformations, all breakpoint junctions could be explained by their location at, or near, sites of expected DNA distortion (Fig. 4 shows three examples). Hence, we found a consistent correspondence between the location of a DNA breakage and the site of an alternative DNA conformation.
Fig. 4.
Representative DNA structures for human deletion breakpoints. (a) Sequences A and B flanking the first breakpoint share extensive inverted homology with sequences C and D flanking the second breakpoint in the PKD1 deletion (Fig. 12 A) such that a stable hairpin may form. An ≈70-bp (R·Y) tract is present just downstream of the first breakpoint, whereas several TGCC and oligo(C) motifs flank the second breakpoint, suggesting that slipped, triplex, and/or tetraplex structures could destabilize the duplex DNA and favor the long-range hairpin interaction. (b) The breakpoints affecting the PARK7 gene (Table 7 and Fig. 12B) occurred between an AluSx and an AluJo element, respectively, that share direct homology over ≈50 bp and may form two cruciforms with the breakage site in the loop. An additional AluSx element is present downstream of AluJo. Among the features that may have synergized with the AluJo sequence are two flanking A+T-rich regions, a TGGTG tandem repeat, and an oligo(C) tract, whose low melting temperature and slipping potential may have contributed to duplex destabilization. (c) Several oligo(G) tracts flank the first breakpoint in a 4-kbp deletion affecting the ALD gene (Fig. 12I), which is drawn in a tetraplex structure. An ≈20-bp-long cruciform containing the second breakpoint is also possible. Interestingly, two mirror (GGGGAG) repeats 5′ to the first break and 3′ to the second break may fold into a short triplex structure (arrow 1), thereby stabilizing DNA looping between the breakpoint junctions. Alternatively (arrow 2), two inverted repeats may hybridize to achieve stabilization. Features that contribute to the destabilization of duplex DNA at the second breakpoints are slipped structures [TGGGGTGG and GGCA tandem repeats and GAGG(GGC) direct repeats] and triplexes [two oligo(R·Y) tracts] within ≈200 bp of the breakpoint.
Discussion
Rearrangement breakpoint junctions occurred at sequences known to form non-B DNA structures in the PKD1 poly(R·Y) tract. All 16 bacterial deletions, 11 human gene deletions, and a significant portion of GRaBD translocation/deletion breakpoint junctions were also close to, or overlapped with, inverted, direct, and mirror repeats, and (RY·RY)n and oligo(R·Y)n tracts, respectively, similarly known to form non-B DNA structures; thus, a causative relationship may exist between alternative DNA conformations formed at these sequences and the occurrence of breakpoints. Recent work on human constitutional translocations involving a common locus on chromosome 22q11.2 (15–18) has implied the formation of large cruciform structures in the generation of breakpoint junctions. Our results have greatly expanded these observations, validated previous hypotheses (Table 8, which is published as supporting information on the PNAS web site) and implicated the formation of most, if not all, identified non-B DNA structures (10, 12, 21, 23–27, 33) in rearrangement mutagenesis. A role for transcription in instabilities was also revealed, which is consistent with the non-B DNA structure-stabilizing effect of the negative supercoil tension associated with RNA elongation by the polymerase complex (23, 34–36).
Non-B DNA structures served to determine the end points for gross deletions. This behavior may originate from weakened chemical stability and/or increased solvent or enzyme accessibility, as evidenced by the susceptibility of nucleotides within or adjacent to regions of alternative DNA conformations to chemical modification and enzymatic cleavage (10, 24–26). Several elements provide useful insights into the nature of the underlying mutational mechanism(s) leading to rearrangements. First, the effectors of our bacterial deletions are also found in class-switch recombination (CSR), the process that potentiates the production of IgG, IgA, or IgE antibodies from IgM-expressing B lymphocytes (37). These effectors include (i) non-B DNA-forming sequences at breakpoints, (ii) short homologies, (iii) induction by transcription, and (iv) dependence on mismatch repair (38–40). Second, the increase in plasmid dimerization, also observed for the (GAA·TTC)n repeat associated with Friedreich ataxia (33), is consistent with the stimulation of both intra- and intermolecular recombination. Third, deleted DNA fragments (products of the deletion process) may be recovered as circular molecules (6, 41). Fourth, the parental sequence is generally reconstituted from the products' ends with little, if any, nucleotide loss. Fifth, fragments from other chromosomal regions are occasionally inserted at the breakpoint junctions. These elements serve to identify the following substrate and product species from the sequences flanking the first (AΛB) and second (CΛD) breakpoints for a simple interstitial deletion or translocation: AΛB + CΛD → AΛD + BΛC (Fig. 7). This reaction scheme is consistent with the putative recombination repair mechanism outlined for segmental duplications (42), rather than with slipped mispairing.
Double-strand breaks, which are generated by several mechanisms, are involved in recombination repair and genetic instability (43, 44). Our findings provide a strong rationale for the initiating events of genomic rearrangements, because the non-B DNA configurations that are formed at these sequences are expected to increase the rate of single-strand lesions, and hence contribute to their conversion to double-strand breaks (22, 45). Thus, this work has important implications both for the biology of targeted homologous recombination and for the optimization of mutation search strategies.
The non-B DNA structure-dependent mutations required mismatch repair functions. The involvement of MutS and MutL, whose activities have been conserved throughout evolution, strongly suggests the existence of conserved pathways for the recognition and processing of non-B DNA conformations. Therefore, it is possible that chromosomal architectural features, such as noncanonical DNA conformations, have played a crucial role in mediating genome plasticity, driving not only evolution and speciation but also heritable disease and cancer (46, 47).
Supplementary Material
Acknowledgments
We thank Bozenna Jaworska for technical assistance. This research was supported by grants from the National Institutes of Health (NS37554 and ES11347), the Robert A. Welch Foundation, the Muscular Dystrophy Foundation, and the Seek-a-Miracle Foundation (to R.D.W.). D.N.C. acknowledges the financial support of Celera Genomics and International Research Funding.
Abbreviations: IPTG, isopropyl β-d-thiogalactoside; wt, wild-type; CFUs, colony-forming units.
References
- 1.Kolomietz, E., Meyn, M. S., Pandita, A. & Squire, J. A. (2002) Genes Chromosomes Cancer 35, 97–112. [DOI] [PubMed] [Google Scholar]
- 2.Roth, D. B. & Wilson, J. H. (1986) Mol. Cell. Biol. 6, 4295–4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albertini, A. M., Hofer, M., Calos, M. P. & Miller, J. M. (1982) Cell 29, 319–328. [DOI] [PubMed] [Google Scholar]
- 4.Chuzhanova, N., Abeysinghe, S. S., Krawczak, M. & Cooper, D. N. (2003) Hum. Mutat. 22, 245–251. [DOI] [PubMed] [Google Scholar]
- 5.Chedin, F., Dervyn, E., Dervyn, R., Ehrlich, S. D. & Noirot, P. (1994) Mol. Microbiol. 12, 561–569. [DOI] [PubMed] [Google Scholar]
- 6.Pont, G., Degroote, F. & Picard, G. (1988) Nucleic Acids Res. 16, 8817–8833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones, R. S. & Potter, S. S. (1985) Proc. Natl. Acad. Sci. USA 82, 1989–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robinson, E. K., Cohen, P. D. & Blackburn, E. H. (1989) Cell 58, 887–900. [DOI] [PubMed] [Google Scholar]
- 9.Singh, K. & Modak, M. J. (2003) J. Biol. Chem. 278, 11289–11302. [DOI] [PubMed] [Google Scholar]
- 10.Sinden, R. R. (1994) DNA Structure and Function (Academic, San Diego).
- 11.Majumdar, A. & Patel, D. J. (2002) Acc. Chem. Res. 35, 1–11. [DOI] [PubMed] [Google Scholar]
- 12.Bowater, R. P. & Wells, R. D. (2000) Prog. Nucleic Acid Res. Mol. Biol. 66, 159–202. [DOI] [PubMed] [Google Scholar]
- 13.Ripley, L. S. & Glickman, B. W. (1983) Cold Spring Harbor Symp. Quant. Biol. 47, Part 2, 851–861. [DOI] [PubMed] [Google Scholar]
- 14.Nalbantoglu, J., Hartley, D., Phear, G., Tear, G. & Meuth, M. (1986) EMBO J. 5, 1199–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gotter, A. L., Shaikh, T. H., Budarf, M. L., Rhodes, C. H. & Emanuel, B. S. (2004) Hum. Mol. Genet. 13, 103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurahashi, H., Shaikh, T. H., Hu, P., Roe, B. A., Emanuel, B. S. & Budarf, M. L. (2000) Hum. Mol. Genet. 9, 1665–1670. [DOI] [PubMed] [Google Scholar]
- 17.Nimmakayalu, M. A., Gotter, A. L., Shaikh, T. H. & Emanuel, B. S. (2003) Hum. Mol. Genet. 12, 2817–2825. [DOI] [PubMed] [Google Scholar]
- 18.Edelmann, L., Spiteri, E., Koren, K., Pulijaal, V., Bialer, M. G., Shanske, A., Goldberg, R. & Morrow, B. E. (2001) Am. J. Hum. Genet. 68, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rossetti, S., Strmecki, L., Gamble, V., Burton, S., Sneddon, V., Peral, B., Roy, S., Bakkaloglu, A., Komel, R., Winearls, C. G. & Harris, P. C. (2001) Am. J. Hum. Genet. 68, 46–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ariyurek, Y., Leeuwen, I. L., Spruit, L., Ravine, D., Breuning, M. H. & Peters, D. J. (2004) Hum. Mutat. 23, 99. [DOI] [PubMed] [Google Scholar]
- 21.Bacolla, A., Jaworski, A., Connors, T. D. & Wells, R. D. (2001) J. Biol. Chem. 276, 18597–18604. [DOI] [PubMed] [Google Scholar]
- 22.Patel, H. P., Lu, L., Blaszak, R. T. & Bissler, J. J. (2004) Nucleic Acids Res. 32, 1460–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rahmouni, A. R. & Wells, R. D. (1992) J. Mol. Biol. 223, 131–144. [DOI] [PubMed] [Google Scholar]
- 24.Wells, R. D., Collier, D. A., Hanvey, J. C., Shimizu, M. & Wohlrab, F. (1988) FASEB J. 2, 2939–2949. [PubMed] [Google Scholar]
- 25.Frank-Kamenetskii, M. D. & Mirkin, S. M. (1995) Annu. Rev. Biochem. 64, 65–95. [DOI] [PubMed] [Google Scholar]
- 26.Soyfer, V. N. & Potaman, V. N. (1996) Triple-Helical Nucleic Acids (Springer, New York).
- 27.Sinden, R. R. & Wells, R. D. (1992) Curr. Opin. Biotechnol. 3, 612–622. [DOI] [PubMed] [Google Scholar]
- 28.Gusev, V. D., Nemytikova, L. A. & Chuzhanova, N. A. (1999) Bioinformatics 15, 994–999. [DOI] [PubMed] [Google Scholar]
- 29.Fukuda, H., Arai, M. & Kuwajima, K. (2000) Biochemistry 39, 12025–12032. [DOI] [PubMed] [Google Scholar]
- 30.Van Raay, T. J., Burn, T. C., Connors, T. D., Petri, L. R., Germino, G. G., Klinger, K. W. & Landes, G. M. (1996) Microb. Comp. Genomics 1, 317–327. [DOI] [PubMed] [Google Scholar]
- 31.Harfe, B. D. & Jinks-Robertson, S. (2000) Annu. Rev. Genet. 34, 359–399. [DOI] [PubMed] [Google Scholar]
- 32.Abeysinghe, S. S., Chuzhanova, N., Krawczak, M., Ball, E. V. & Cooper, D. N. (2003) Hum. Mutat. 22, 229–244. [DOI] [PubMed] [Google Scholar]
- 33.Vetcher, A. A. & Wells, R. D. (2004) J. Biol. Chem. 279, 6434–6443. [DOI] [PubMed] [Google Scholar]
- 34.Liu, L. F. & Wang, J. C. (1987) Proc. Natl. Acad. Sci. USA 84, 7024–7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen, D. & Lilley, D. M. (1999) J. Mol. Biol. 285, 443–448. [DOI] [PubMed] [Google Scholar]
- 36.Bowater, R. P., Chen, D. & Lilley, D. M. (1994) Biochemistry 33, 9266–9275. [DOI] [PubMed] [Google Scholar]
- 37.Kenter, A. L. (1999) J. Exp. Med. 190, 307–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ehrenstein, M. R., Rada, C., Jones, A. M., Milstein, C. & Neuberger, M. S. (2001) Proc. Natl. Acad. Sci. USA 98, 14553–14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schrader, C. E., Edelmann, W., Kucherlapati, R. & Stavnezer, J. (1999) J. Exp. Med. 190, 323–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ehrenstein, M. R. & Neuberger, M. S. (1999) EMBO J. 18, 3484–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tracy, R. B., Hsieh, C.-L. & Lieber, M. R. (2000) Science 288, 1058–1061. [DOI] [PubMed] [Google Scholar]
- 42.Emanuel, B. S. & Shaikh, T. H. (2001) Nat. Rev. Genet. 2, 791–800. [DOI] [PubMed] [Google Scholar]
- 43.Zucman-Rossi, J., Legoix, P., Victor, J. M., Lopez, B. & Thomas, G. (1998) Proc. Natl. Acad. Sci. USA 95, 11786–11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elliott, B. & Jasin, M. (2002) Cell. Mol. Life Sci. 59, 373–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vilenchik, M. M. & Knudson, A. G. (2003) Proc. Natl. Acad. Sci. USA 100, 12871–12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kost-Alimova, M., Kiss, H., Fedorova, L., Yang, Y., Dumanski, J. P., Klein, G. & Imreh, S. (2003) Proc. Natl. Acad. Sci. USA 100, 6622–6627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bacolla, A. & Wells, R. D. (2004) J. Biol. Chem. 279, in press. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.