Abstract
The T1R receptors, a family of taste-specific class C G proteincoupled receptors, mediate mammalian sweet and umami tastes. The structure–function relationships of T1R receptors remain largely unknown. In this study, we demonstrate the different functional roles of T1R extracellular and transmembrane domains in ligand recognition and G protein coupling. Similar to other family C G protein-coupled receptors, the N-terminal Venus flytrap domain of T1R2 is required for recognizing sweeteners, such as aspartame and neotame. The G protein coupling requires the transmembrane domain of T1R2. Surprisingly, the C-terminal transmembrane domain of T1R3 is required for recognizing sweetener cyclamate and sweet taste inhibitor lactisole. Because T1R3 is the common subunit in the sweet taste receptor and the umami taste receptor, we tested the interaction of lactisole and cyclamate with the umami taste receptor. Lactisole inhibits the activity of the human T1R1/T1R3 receptor, and, as predicted, blocked the umami taste of l-glutamate in human taste tests. Cyclamate does not activate the T1R1/T1R3 receptor by itself, but potentiates the receptor's response to l-glutamate. Taken together, these findings demonstrate the different functional roles of T1R3 and T1R2 and the presence of multiple ligand binding sites on the sweet taste receptor.
A family of class C G protein-coupled receptors (GPCRs), T1Rs, is selectively expressed in the taste buds (1–6). Functional expression of T1Rs in human embryonic kidney (HEK)-293 cells revealed that different combinations of T1Rs respond to sweet and umami taste stimuli (6, 7). T1R2 and T1R3, when coexpressed in HEK-293 cells, recognize diverse natural and synthetic sweeteners. Similarly, T1R1 and T1R3, when coexpressed in HEK-293 cells, respond to the umami taste stimulus l-glutamate. This response is enhanced by 5′ ribonucleotides, a hallmark of umami taste. Recent experiments with knockout mice confirmed that T1Rs indeed mediate mouse sweet and umami tastes (8, 9).
The class C GPCRs possess a large N-terminal extracellular domain, often referred to as the Venus flytrap domain (10), and are known to function as either homodimers, in the cases of metabotropic glutamate receptors (mGluRs) and calciumsensing receptor, or heterodimers, in the case of γ-aminobutyric acid type B receptor (GABABR) (10). The functional expression data suggest a heterodimer mechanism for T1Rs: both T1R1 and T1R2 need to be coexpressed with T1R3 to be functional, which is supported by the overlapping expression patterns of T1Rs in rodent tongue. Nonetheless, there has been no direct evidence that T1Rs function as heteromeric complexes. It is possible that T1R3 is not a functional component of sweet and umami taste receptors, but merely a chaperone protein, which facilitates the proper folding or intracellular translocation of T1R1 and T1R2. The distinct ligand specificities of T1R1/T1R3 and T1R2/T1R3 receptors suggest that T1R1 and T1R2 play more important roles in ligand binding in sweet and umami taste receptors than T1R3. Support for this hypothesis was provided recently by results from mouse genetics where human T1R2 transgenic mice, generated on the T1R2 knockout background, displayed sweetener taste preferences similar to those of humans (9). However, the functional role of T1R3 and the overall structure/function relationship of T1R taste receptors remain largely unknown.
Another intriguing observation about the T1R2/T1R3 receptor is the structural diversity of its ligands. This receptor is able to recognize every sweetener tested, including carbohydrate, amino acids and derivatives, proteins, and synthetic sweeteners (7). On the other hand, the receptor exhibits stereo-selectivity for certain molecules. For example, it responds to d-tryptophan but not l-tryptophan (7), which is in correlation with the sensory data. It is still a puzzle as to how this single receptor can recognize such a large collection of diverse chemical structures.
There are differences in human and rodent sweet taste in terms of the ligand specificity, G protein-coupling efficiency, and sensitivity to inhibitors. In this study, we use the species differences in T1R ligand specificity to demonstrate that the sweet taste receptor indeed functions as a heteromeric complex, and that there are likely more than one ligand binding sites on the receptor. Furthermore, we uncover a functional link between the sweet and umami taste receptors mediated by T1R3.
Materials and Methods
T1R1/T1R3 Stable Cell Line. Human T1R1/T1R3-expressing stable cell lines were generated by transfecting linearized pEAK10-derived T1R1 and pCDNA3.1/ZEO-derived (Invitrogen) T1R3 vectors into HEK/Gα15 cells. Cells were selected in 0.5 μg·ml–1 puromycin (Calbiochem) and 100 μg·ml–1 zeocin (Invitrogen) at 37°C in glutamine-free DMEM supplemented with GlutaMAX, 10% dialyzed FBS, and 3 μg·ml–1 blasticidin. Resistant colonies were expanded, and their responses to umami taste stimuli were evaluated by fluorescence microscopy.
Constructs. T1R2 chimeras were constructed by introducing an XhoI site with a silent mutation at human T1R2 amino acid 560 and rat T1R2 amino acid 564. T1R3 chimeras were constructed by introducing XhoI sites with point mutations (human T1R3 A564E and rat T1R3 A569E). All chimeras were cloned into pEAK10 expression vector. T1R mutants were generated by using standard PCR-based mutagenesis protocol.
T1R Assays. The T1R assays in transiently transfected cells were performed as described (7). HEK-293T and an HEK-293 derivative that stably expresses Gα15 (Invitrogen) were grown and maintained at 37°C in DMEM supplemented with 10% FBS and MEM nonessential amino acids (Invitrogen); media for Gα15 cells also contained 3 μg·ml–1 blasticidin (Invitrogen). For calcium-imaging experiments, cells were first seeded onto 48-well tissue-culture plates (≈30,000 cells per well) and transfected by using Mirus TransIt-293 (Invitrogen). Transfection efficiencies, which were estimated by cotransfection with a red fluorescent protein expression vector, were typically ≈60%. To minimize variations, data reported in the same panel were obtained on the same day by using the same batch of cell, which was transfected under the same conditions. To minimize glutamate-induced and glucose-induced desensitization, supplemented DMEM was replaced with low-glucose DMEM supplemented with GlutaMAX and 10% dialyzed FBS (Invitrogen) ≈24 h after transfection. After an additional 24 h, cells were loaded with the calcium dye fluo-4-AM (Molecular Probes), 3 μM in Dulbecco's PBS buffer (DPBS, Invitrogen), for 1.5 h at room temperature. After replacement with 100 μl of DPBS, stimulation was performed at room temperature by addition of 100 μl of DPBS supplemented with taste stimuli. Calcium mobilization was monitored on an Axiovert S100 microscope equipped with an inverted ×10/0.5 long working distance plano fluor objective (Zeiss) and a cooled charge-coupled device camera (Princeton Instruments, Trenton, NJ). Fluorescence images were acquired at 480-nm excitation and 535-nm emission and analyzed with imaging workbench 4.0 software (Axon Instruments, Union City, CA). T1R receptor activity was quantitated by counting the number of responding cells 30 s after stimulus addition.
The stable T1R2/T1R3- and T1R1/T1R3-expressing cell lines were manipulated as described (7). For calcium-imaging experiments, cells were seeded onto 48-well plates (≈50,000 cells per well) and incubated for 24 h. Cells were then loaded with the calcium dye fluo-4-AM (Molecular Probes), 5 μM in PBS, for 1 h at room temperature. After replacement with 100 μl of PBS, stimulation was performed at room temperature by the addition of 100 μl of PBS supplemented with stimuli.
Fluorescence Imaging Plate Reader (FLIPR) Protocols. For automated fluorometric imaging on FLIPR-I instrumentation (Molecular Devices), T1R1/T1R3 stable cells were first seeded onto 96-well plates (≈15,000 cells per well). After 24 h, cells were loaded with the calcium dye fluo-4-AM (Molecular Probes), 5 μM in PBS, for 1 h at room temperature. After replacement with 50 μl of PBS, stimulation was performed at room temperature by the addition of 50 μl of PBS supplemented with taste stimuli. Peak fluorescence (480-nm excitation and 535-nm emission) responses ≈20–30 s after compound addition were corrected for and normalized to background fluorescence.
Taste Tests. Detection threshold effects of lactisole on umami, sweet, and salt taste were determined by tasting dilution series of these taste stimuli in the presence and absence of lactisole as described (11). Detection threshold values were averaged over four trials for three subjects.
Results and Discussion
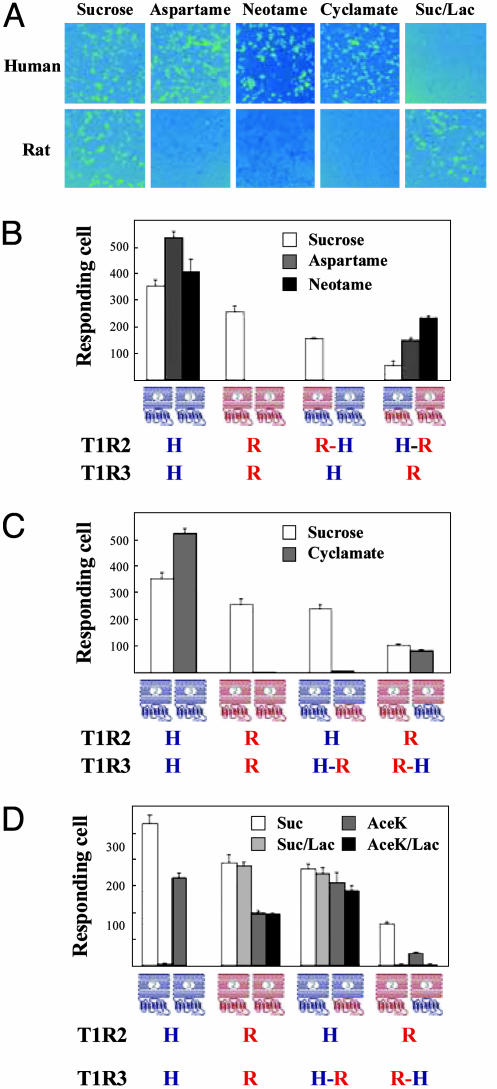
Mapping of Ligand Interaction Sites on the Sweet Taste Receptor. The agonist specificities of human and rat T1R2/T1R3 were previously characterized by functional expression of the receptors in HEK-293 cells. Both human and rat sweet taste receptors can efficiently couple to a chimeric Gα15 with the C-terminal tail sequence from Gαi1 (Gα15/i1) (7). Consistent with the sensory/behavioral data, human but not rat T1R2/T1R3 selectively responds to a group of sweeteners, including aspartame, neotame, and cyclamate (7). These differences in agonist specificity can be used to map their binding sites on the receptor. We generated chimeric T1Rs between human and rat genes, with the junction located immediately before the proposed transmembrane domain. Each T1R chimera therefore consists of two halves, the N-terminal extracellular domain and the C-terminal transmembrane and intracellular domain, from different species. For example, a chimeric T1R2, termed T1R2H-R, has a sequence from the N terminus of human T1R2 fused to rat T1R2 C-terminal sequence. We transfected HEK-293 cells with Gα15/i1 and different combinations of the chimeric receptors and tested their responses to aspartame, neotame, and cyclamate (Fig. 1).
Fig. 1.
Sweeteners map to different domains/subunits of the human sweet taste receptor. (A) Responses of human and rat sweet taste receptors to sucrose (200 mM), aspartame (10 mM), neotame (0.1 mM), cyclamate (10 mM), and sucrose (200 mM) in the presence of lactisole (1 mM) (Suc/Lac). HEK-293T cells were transiently transfected with human or rat T1R2, T1R3, and a Gα15 chimera, Gα15/i1 (7), and assayed for intracellular calcium increases in response to sweeteners. (B) Aspartame and neotame were mapped to the N-terminal extracellular domain of human T1R2. Combinations of T1R chimeras were transiently transfected into HEK-293T cells with Gα15/i1 and assayed for responses to sweeteners at the concentrations listed in A. The T1R2H-R/rat T1R3 combination generated a significantly weaker response to the control sweetener sucrose than did the WT receptors, possibly because of less than perfect folding of the artificial receptor subunit. Nonetheless, the same receptor responds well to aspartame and neotame. Because of the potential differences in folding, surface targeting, and coupling efficiency, we avoided comparing the relative activities of different combinations. Instead, we looked for the presence or absence of response to different sweeteners within each combination. (C) Cyclamate was mapped to the C-terminal transmembrane domain of human T1R3. (D) Lactisole was mapped to the transmembrane domain of human T1R3. Different combinations of T1R chimeras were transiently transfected into HEK-293T cells with Gα15/i1 and assayed for responses to sucrose (200 mM) and AceK (10 mM) in the absence or presence of lactisole (1 mM). The activities in B–D represent the mean ± SE of the number of responding cells for four imaged fields of ≈1,000 confluent cells. H, Human; R, rat.
Through coexpression of T1R2R-H with human T1R3, we replaced part of the human sweet taste receptor (the N-terminal domain of T1R2) with rat protein sequence, in which case, the responses to aspartame and neotame are abolished, suggesting the N-terminal domain of human T1R2 is required for recognizing aspartame and neotame. Similarly, we can also replace the rat T1R2 N-terminal domain with human protein sequence by coexpressing T1R2H-R with rat T1R3, in which case the chimeric receptor gains the ability to respond to aspartame and neotame. This finding suggests that the same domain of human T1R2 is also sufficient (in the context of sweet taste receptors) to enable activation by those two sweeteners (Fig. 1B). These functional expression data confirm the mouse genetics results that T1R2 mediates the taste of some sweeteners (9) and further indicate that the important interaction determinants for aspartame and neotame are located in the N-terminal extracellular domain.
In contrast, replacing either half of human T1R2 with rat protein sequence does not affect its response to cyclamate. Instead, the C-terminal domain of human T1R3 is required and sufficient, when coexpressed with T1R2, to recognize cyclamate (Fig. 1C). The transmembrane domain of family C GPCRs has been known to contain binding sites for allosteric modulators (12). In this case, cyclamate binds directly to the transmembrane domain and activates the receptor in the absence of another ligand. Recently, Zhao et al. (9) described the taste behavior of a transgenic mouse expressing human T1R2 in the background of a mouse T1R2 knockout. Although cyclamate was not tested, based on our data, human T1R3 but not human T1R2 would be expected to convey cyclamate preference to mouse taste.
Lactisole, an aralkyl carboyxlic acid, is a human-specific sweet taste inhibitor, which has no effect on the rodent sweet taste. Consistent with the behavioral observations, lactisole inhibits the human, but not rat, T1R2/T1R3 response to sucrose in our assay system (Fig. 1 A). We performed the same kind of mapping experiments to determine the lactisol interaction site by using the T1R chimeras. Like cyclamate, lactisole requires the human T1R3 C-terminal domain to inhibit the receptor's response to sucrose and acesulfame K (AceK) (Fig. 1D). This result further demonstrates the importance of the T1R3 C-terminal domain in the sweet taste receptor function. We tested the chimeras in all 16 possible combinations (Fig. 6, which is published as supporting information on the PNAS web site), and all functional combinations generated results consistent with our model.
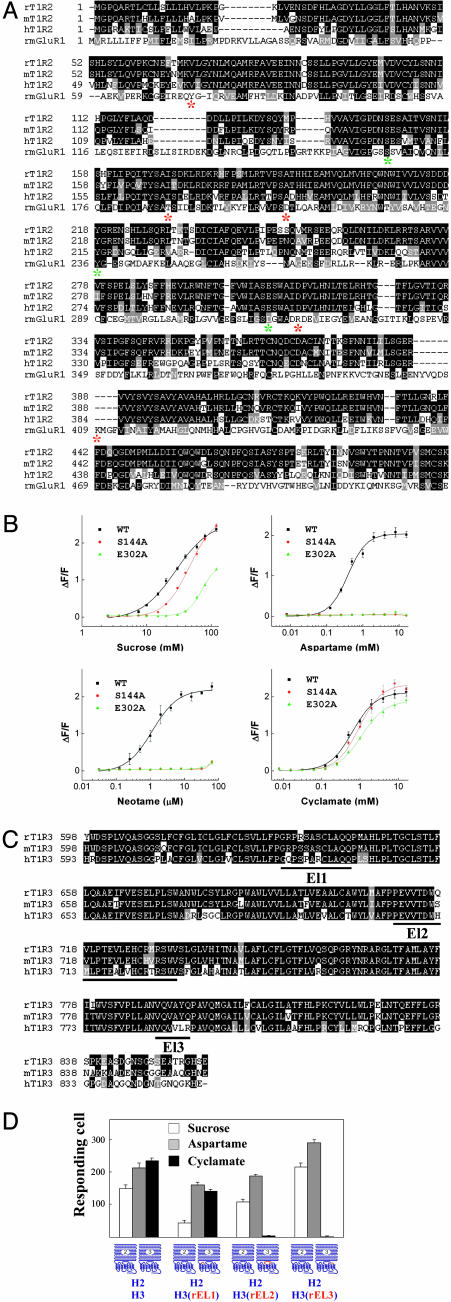
We conducted mutagenesis studies on both T1R2 and T1R3 to narrow down the essential amino acids in the recognition of aspartame, neotame, and cyclamate. If T1R2 and T1R3 are responsible for recognizing different sweeteners, we would expect mutations in the T1R2 N-terminal domain to affect responses to aspartame and neotame, but not cyclamate. In addition, we would predict mutations in T1R3 C-terminal domain to have the opposite effect. To select the crucial amino acid residues in the T1R2 N-terminal domain, we aligned the sequence of T1R2 with mGluR1 (Fig. 2A). Among the eight residues that are crucial in ligand binding in mGluR1 (13), three are conserved in human T1R2 (S144, Y218, and E302). We mutated each of the three residues and tested the resulting receptors for their response to different sweeteners. Substitution of Y218 to A abolished the responses to all sweeteners tested including cyclamate (data not shown). Y218 might be important for the overall conformation of the sweet taste receptor, but it is also possible that that Y218A failed to express or target to the cell surface, considering that equivalent substitutions in mGluR1 (14) and mGluR8 (15) led to partially functional receptors. However, the two other human T1R2 variants, containing S144A and E302A, selectively affected the response to aspartame and neotame but not cyclamate. Stable cell lines expressing S144A and E302A human T1R2 variants (coexpressed with WT human T1R3 and Gα15) did not respond to aspartame or neotame at the physiologically relevant concentrations, but did respond to cyclamate (Fig. 2B).
Fig. 2.
Mutations in T1R2 or T1R3 selectively affect the activity of different sweeteners. (A) Sequence alignment of the N-terminal ligand binding domain of rat mGluR5 with human and rodent T1R2s. The eight critical amino acids involved in ligand binding in mGluR5 are labeled with *; three of the eight amino acids are conserved in T1R2 and labeled with green *. (B) Two point mutations in the human T1R2 N-terminal extracellular domain abolish response to aspartame and neotame without affecting cyclamate. Stable cell lines of human T1R2/human T1R3 (WT), human T1R2 S144A/human T1R3 (S144A), and E302A/human T1R3 (E302A) were generated as described (7). The dose–responses of these stable lines were determined by FLIPR for sucrose, aspartame, neotame, and cyclamate. The activities represent the mean ± SE of fold increases in fluorescence intensities for four recorded wells. (C) Sequence alignment of human and rodent T1R3 transmembrane domains. The three extracellular loops are underlined and labeled EL1, EL2, or EL3, according to their order in the protein sequences. (D) Mutations in the extracellular loop of human T1R3 abolish response to cyclamate without affecting aspartame. Each of the three extracellular loops of human T1R3 was replaced with rat protein sequence separately, and the resulting human T1R3 mutants were transiently transfected into HEK-293T cells together with Gα15/i1 and assayed for responses to sucrose (200 mM), aspartame (10 mM), and cyclamate (10 mM), and sucrose (200 mM) in the presence of lactisole (1 mM). The activities represent the mean ± SE of the number of responding cells for four imaged fields of ≈1,000 confluent cells.
To further map the cyclamate-binding site, we focused on the three extracellular loops in the T1R3 C-terminal domain. Alignment of human and rodent T1R3s reveals multiple amino acid differences in the three extracellular loops (Fig. 2C). Replacing extracellular loop 2 or loop 3 with rat sequences abolished the cyclamate response without affecting the sucrose or aspartame responses. In contrast, replacing extracellular loop 1 had no obvious effect on the response to cyclamate, suggesting important roles for extracellular loops 2 and 3 in recognizing cyclamate (Fig. 2D). Interestingly, none of those loop replacements affected the inhibition effect of lactisole (Fig. 2D), suggesting a different binding mechanism. In summary, amino acid substitutions in T1R2 or T1R3 result in selective interference of activities induced by different sweeteners, consistent with the chimeric receptor results.
Taken together, the above results demonstrate that human sweet taste receptor functions as a heteromeric complex of T1R2 and T1R3. Both subunits are required for recognizing different sweeteners, and our data indicate the existence of multiple binding pockets on the receptor for different classes of agonists. The presence of multiple ligand-binding sites provides a possible explanation for the structural diversity of sweeteners.
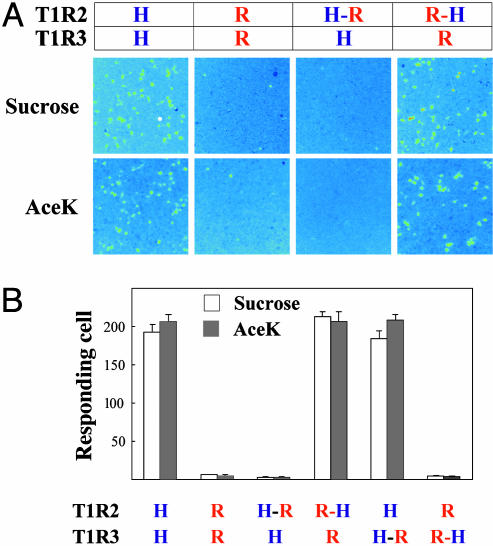
Mapping of Receptor–G Protein Interactions. The human and rat sweet taste receptors are also different in their G protein-coupling efficiency. Even though both human and rat receptors can couple efficiently to Gα15/i1, only the human receptor can couple efficiently to Gα15 (7) (Fig. 3A). This species difference allows us to map the receptor G protein interactions by using the same chimeric receptors as described above. T1R2 but not T1R3 appears to be critical for Gα15 coupling, because replacing the C terminus of human T1R2 with the corresponding rat sequence abolished coupling, and replacing the rat T1R2 C-terminal half with human sequence enabled the receptor to couple to Gα15 and respond to sucrose and AceK (Fig. 3). Swapping the T1R3 C-terminal sequences had no effect on Gα15 coupling (Fig. 3B). This observation demonstrates the important role of T1R2 in G protein coupling in our functional expression system. Gustducin (16) has been proposed to be an endogenous G protein for the sweet taste receptor, and we speculate that T1R2 should be the subunit responsible for coupling in taste cells. GABABR is the other example of heteromeric family C GPCR, where one subunit (GABABR1) is responsible for ligand binding, and the other (GABABR2) for G protein coupling (17–20). The sweet taste receptor is different from GABABR in that T1R2 is required for both ligand recognition and G protein coupling.
Fig. 3.
Human T1R2 is required for Gα15 coupling. (A) Responses of human (H), rat (R), and chimeric sweet taste receptors to sucrose (200 mM) and AceK (10 mM). Stable Gα15 cells were transiently transfected with human, rat, or chimeric T1Rs and assayed for intracellular calcium increases in response to sweeteners. (B) Gα15 coupling is mediated by human T1R2. The activities represent the mean ± SE of the number of responding cells for four imaged fields of ≈1,000 confluent cells.
Lactisole Antagonizes Human T1R1/T1R3 and Inhibits Human Umami Taste. We reasoned that T1R1/T1R3 may function as a heteromeric receptor just like T1R2/T1R3, and that lactisole should have a similar effect on T1R1/T1R3 activity, because T1R3 is a common subunit between the two receptors. Consistent with this logic, we found that lactisole antagonizes human T1R1/T1R3 (Fig. 4A). Lactisole acts as a noncompetitive inhibitor of T1R1/T1R3, because the IC50 values apparently do not depend on glutamate concentration (Fig. 4B), and lactisole reduces the maximal activities of the receptor without significantly changing the EC50 of agonists (Fig. 4C). These results demonstrate that lactisole binds to a different site from l-glutamate and are consistent with our hypothesis that the glutamate-binding pocket is located in T1R1 (7). Interestingly, lactisole appears to be a competitive inhibitor of the sweet taste receptor, as its IC50s depend on the concentrations of the sweeteners, and it increases the EC50s of the sweeteners without significantly affecting the maximal activities (Fig. 6). Further investigation is needed to explain the apparent difference in its inhibition patterns for the sweet and umami taste receptors.
Fig. 4.
The effect of lactisole and cyclamate on the human T1R1/T1R3 umami taste receptor. (A) Response of the human T1R1/T1R3 stable cell line tol-glutamate (5 mM) and l-glutamate/IMP (1/0.2 mM) in the absence and presence of lactisole (5 mM). (B) The lactisole dose-dependent inhibition curves were determined for l-glutamate (Glu) and l-glutamate with 0.2 mM IMP (Glu/IMP), each at two different concentrations. The IC50s are 0.19 ± 0.02 and 0.21 ± 0.01 mM for l-glutamate at 8 and 80 mM, respectively, and 0.35 ± 0.03 and 0.82 ± 0.06 mM for l-glutamate with IMP at 0.8 and 8 mM, respectively. (B) The dose–responses for l-glutamate, with or without 0.2 mM IMP, were determined in the presence of different concentrations of lactisole. In the presence of 0, 25, or 50 μM lactisole, the EC50s are 9.9 ± 1.5, 7.9 ± 0.5, and 7.0 ± 0.3 mM, respectively, for l-glutamate. In the presence of 0, 100, or 200 μM lactisole, the EC50s are 0.53 ± 0.04, 0.71 ± 0.10, and 0.84 ± 0.10 mM, respectively, for l-glutamate with IMP. Values represent the mean ± SE for four independent responses. (D) The detection thresholds for sweet, umami, and salty taste stimuli were determined in the presence or absence of lactisole. The inhibition effect of lactisole is shown as fold increases in detection thresholds. The detection threshold values were averaged over four trials for three subjects. (E) The responses of the human T1R1/T1R3 stable cell line to threshold level of l-glutamate (4 mM) and endogenous M2 receptor agonist carbachol were assayed on FLIPR in the absence and presence of various concentrations of cyclamate. (F) Dose–responses of the human T1R1/T1R3 stable cell line were determined on FLIPR for l-glutamate with or without 0.2 mM IMP in the absence and presence of cyclamate (8 mM). The activities in B, C, E, and F represent the mean ± SE of fold increases in fluorescence intensities for four recorded wells. The dose–responses in B, C, E, and F were reproduced at least six times independently.
The inhibition effect of lactisole is mediated by the T1R receptors because it had no effect on the endogenous muscarinic acetylcholine receptor in HEK cells or on a mouse bitter receptor, mouse T2R5, transiently expressed in HEK cells (data not shown). As was the case for the T1R2/T1R3 receptor, lactisole inhibition of the T1R1/T1R3 response to umami taste stimuli was reversible after washout and restimulation (data not shown).
To correlate the receptor activity with behavior, we tested the effect of lactisole on human umami taste. As we predicted, millimolar concentrations of lactisole dramatically increased detection thresholds for sweet and umami but not salt taste stimuli (Fig. 4D). Lactisole was previously not known to be an umami taste inhibitor. The correlation between receptor activity and taste results demonstrates a crucial role of T1Rs in human umami taste.
Cyclamate Enhances Human T1R1/T1R3 Receptor Activities. Based on the same heteromeric model of T1Rs (Fig. 5), we predicted that cyclamate would also modulate the activity of the human T1R1/T1R3 umami taste receptor by acting on T1R3. Although cyclamate alone had no effect on T1R1/T1R3, it enhanced the activity of the receptor in the presence of l-glutamate (Fig. 4E). This effect is specific for the human T1R1/T1R3, as cyclamate had no effect on the activities of the endogenous muscarinic acetylcholine receptor in the presence of carbachol (Fig. 4E). It is noteworthy that cyclamate has comparable EC50s for the sweet taste receptor (Fig. 2B) and umami taste receptor. Cyclamate reproducibly left-shifts the dose–response curves for l-glutamate by ≈2-fold in the presence or absence of IMP (Fig. 4F). IMP has a more dramatic effect of enhancing the receptor, and the effect of cyclamate is observed in the presence of IMP (Fig. 4F), suggesting a different mechanism from IMP in enhancing the receptor. We speculate that IMP binds to T1R1, because it has no effect on the sweet taste receptor (7). Other sweeteners, including sucrose, aspartame, saccharin, and d-tryptophan, had no effect on the human T1R1/T1R3 activities (data not shown). Because of the intense sweet taste of cyclamate, we could not determine its effect on umami taste.
Fig. 5.
A working model for the sweet and umami taste receptor structure–function relationships. Filled arrows indicate direct activation, open arrows indicate enhancement, and bar heads indicate inhibition. Solid lines indicate proposed mechanisms based on experimental evidence; broken lines indicate mechanisms based on our speculations.
In summary, we demonstrate that both T1R2 and T1R3 are required in a functional sweet taste receptor, that aspartame and neotame require the N-terminal extracellular domain of T1R2, that G protein coupling requires the C-terminal half of T1R2, and that cyclamate and lactisole require the transmembrane domain of T1R3. These findings demonstrate the different functional roles of T1R subunits in a heteromeric complex and the presence of multiple sweetener interaction sites on the sweet taste receptor. Because T1R3 is the common subunit in the sweet and the umami taste receptors, we predicted and confirmed the effect of cyclamate and lactisole on the umami taste receptor. Furthermore, we were able to correlate the lactisole effect on the receptor activities with human taste. Based on these observations, we propose a working model (Fig. 5) for the structure–function relationships of the T1R family of taste receptors. We speculate that natural carbohydrate sweeteners bind to the N-terminal domain of T1R2, similar to aspartame and neotame, and there may be other ligand-binding sites on the sweet taste receptor, for example, the transmembrane domain of T1R2. The umami taste receptor most likely functions similarly as a heteromeric complex, and we speculate that l-glutamate and IMP each bind to the T1R1 subunit, because neither has any effect on the sweet taste receptor (7) (data not shown), and that the transmembrane domain of T1R1 is responsible for coupling to G proteins.
Supplementary Material
Acknowledgments
We thank A. Watkins, A. Chang, and M. Qi for their scientific contributions and L. Stryer, B. Moyer, G. Servant, and A. Pronin for critical reading of this manuscript.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: GPCR, G protein-coupled receptor; HEK, human embryonic kidney; mGluR, metabotropic glutamate receptor; GABABR, γ-aminobutyric acid type B receptor; FLIPR, fluorescence imaging plate reader; AceK, acesulfame K.
See Commentary on page 13972.
References
- 1.Hoon, M. A., Adler, E., Lindemeier, J., Battey, J. F., Ryba, N. J. & Zuker, C. S. (1999) Cell 96, 541–551. [DOI] [PubMed] [Google Scholar]
- 2.Bachmanov, A. A., Li, X., Reed, D. R., Ohmen, J. D., Li, S., Chen, Z., Tordoff, M. G., de Jong, P. J., Wu, C., West, D. B., et al. (2001) Chem. Senses 26, 925–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Montmayeur, J. P., Liberles, S. D., Matsunami, H. & Buck, L. B. (2001) Nat. Neurosci. 4, 492–498. [DOI] [PubMed] [Google Scholar]
- 4.Max, M., Shanker, Y. G., Huang, L., Rong, M., Liu, Z., Campagne, F., Weinstein, H., Damak, S. & Margolskee, R. F. (2001) Nat. Genet. 28, 58–63. [DOI] [PubMed] [Google Scholar]
- 5.Kitagawa, M., Kusakabe, Y., Miura, H., Ninomiya, Y. & Hino, A. (2001) Biochem. Biophys. Res. Commun. 283, 236–242. [DOI] [PubMed] [Google Scholar]
- 6.Nelson, G., Hoon, M. A., Chandrashekar, J., Zhang, Y., Ryba, N. J. & Zuker, C. S. (2001) Cell 106, 381–390. [DOI] [PubMed] [Google Scholar]
- 7.Li, X., Staszewski, L., Xu, H., Durick, K., Zoller, M. & Adler, E. (2002) Proc. Natl. Acad. Sci. USA 99, 4692–4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Damak, S., Rong, M., Yasumatsu, K., Kokrashvili, Z., Varadarajan, V., Zou, S., Jiang, P., Ninomiya, Y. & Margolskee, R. F. (2003) Science 301, 850–853. [DOI] [PubMed] [Google Scholar]
- 9.Zhao, G. Q., Zhang, Y., Hoon, M. A., Chandrashekar, J., Erlenbach, I., Ryba, N. J. & Zuker, C. S. (2003) Cell 115, 255–266. [DOI] [PubMed] [Google Scholar]
- 10.Pin, J. P., Galvez, T. & Prezeau, L. (2003) Pharmacol. Ther. 98, 325–354. [DOI] [PubMed] [Google Scholar]
- 11.Schiffman, S. S., Frey, A. E., Luboski, J. A., Foster, M. A. & Erickson, R. P. (1991) Physiol. Behav. 49, 843–854. [DOI] [PubMed] [Google Scholar]
- 12.Gasparini, F., Kuhn, R. & Pin, J. P. (2002) Curr. Opin. Pharmacol. 2, 43–49. [DOI] [PubMed] [Google Scholar]
- 13.Kunishima, N., Shimada, Y., Tsuji, Y., Sato, T., Yamamoto, M., Kumasaka, T., Nakanishi, S., Jingami, H. & Morikawa, K. (2000) Nature 407, 971–977. [DOI] [PubMed] [Google Scholar]
- 14.Sato, T., Shimada, Y., Nagasawa, N., Nakanishi, S. & Jingami, H. (2003) J. Biol. Chem. 278, 4314–4321. [DOI] [PubMed] [Google Scholar]
- 15.Bessis, A. S., Rondard, P., Gaven, F., Brabet, I., Triballeau, N., Prezeau, L., Acher, F. & Pin, J. P. (2002) Proc. Natl. Acad. Sci. USA 99, 11097–11102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong, G. T., Gannon, K. S. & Margolskee, R. F. (1996) Nature 381, 796–800. [DOI] [PubMed] [Google Scholar]
- 17.Kniazeff, J., Galvez, T., Labesse, G. & Pin, J. P. (2002) J. Neurosci. 22, 7352–7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galvez, T., Duthey, B., Kniazeff, J., Blahos, J., Rovelli, G., Bettler, B., Prezeau, L. & Pin, J. P. (2001) EMBO J. 20, 2152–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Margeta-Mitrovic, M., Jan, Y. N. & Jan, L. Y. (2001) Proc. Natl. Acad. Sci. USA 98, 14643–14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Margeta-Mitrovic, M., Jan, Y. N. & Jan, L. Y. (2001) Proc. Natl. Acad. Sci. USA 98, 14649–14654. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.