Abstract
Embryonal carcinoma (EC) cells have served as a model to study the relationship between cancer and cellular differentiation given their potential to produce tumors and, to varying degrees, participate in embryonic development. Here, nuclear transplantation was used to assess the extent to which the tumorigenic and developmental potential of EC cells is governed by epigenetic as opposed to genetic alterations. Nuclei from three independent mouse EC cell lines (F9, P19, and METT-1) with differing developmental and tumorigenic potentials all were able to direct early embryo development, producing morphologically normal blastocysts that gave rise to nuclear transfer (NT)-derived embryonic stem (ES) cell lines at a high efficiency. However, when tested for tumor or chimera formation, the resulting NT ES cells displayed an identical potential as their respective donor EC cells, in stark contrast to previously reported NT ES cells derived from transfer of untransformed cells. Consistent with this finding, comparative genomic hybridization identified previously undescribed genetic lesions in the EC cell lines. Therefore, nonreprogrammable genetic modifications within EC nuclei define the developmental and tumorigenic potential of resulting NT ES cells. Our findings support the notion that cancer results from the deregulation of stem cells and further suggest that the genetics of ECs will reveal genes involved in stem cell self-renewal and pluripotency.
Epigenetics plays an important role in development and disease. During development, cells become increasingly restricted in their developmental potential with most cells becoming locked into a single fate. The locking in of cell fate is maintained by modifications to DNA that are heritable, but do not involve a change in primary DNA sequence. These changes have, therefore, been coined epigenetic modifications (1). Well known examples are the methylation of CpG islands and the methylation/acetylation of histones (2). Analogously, disease can result from errors in epigenetic modifications. Epigenetic errors have become increasingly appreciated for their role in cancer (3). For example, it is now known that many tumor suppressor genes are silenced during tumorigenesis by erroneous methylation of their promoters (4). However, unlike genetic mutations, it remains unclear to what extent these and other cancer-associated epigenetic changes are causally involved in the tumorigenic process.
Transfer of a nucleus from a differentiated cell to an enucleated oocyte can reset the developmental state of the differentiated cell nucleus to that of a zygote with the potential to produce a new organism with an identical genetic constitution, i.e., a clone (5). This remarkable feat requires the reversal of the epigenetic modifications that were established during cellular differentiation. Similarly, transfer of a nucleus from a cancer cell may be able to reverse both aberrant and developmentally associated epigenetic modifications that had accumulated during tumorigenesis. Previous results in amphibians and mice have suggested that a cancer cell nucleus can indeed be reprogrammed by nuclear transfer (NT) and go on to direct early development (6–9). However, the exceedingly low efficiency at which they produced clones raises questions as to the origin of the donor cell used in these experiments (9, 10). Furthermore, resulting embryos were abnormal and arrested early in development. A two-step approach to cloning can overcome the difficulty of interpreting data resulting from the inefficient process of direct cloning (11, 12). Two-step cloning involves the derivation of embryonic stem (ES) cells from NT-derived blastocysts, followed by tests of ES cell potency including production of teratomas and chimeric animals. This approach has enabled the cloning of exceptionally inefficient donor cells such as lymphocytes and neurons (12, 13).
Teratocarcinomas provide an attractive tumor model system in which to test the reversibility of tumorigenic potential by NT (14). Teratocarcinomas can be induced with a high frequency and short latency in mice by transplanting early embryos under the testicular capsule of an adult host (15). Embryonal carcinoma (EC) cell lines derived from teratocarcinomas produce tumors when injected s.c. or i.p. into animals. Surprisingly, they can also be induced to differentiate into various cell types in vitro and can contribute to the somatic lineages of a developing embryo when injected into a blastocyst (16, 17). Therefore, the tumorigenic potential is not absolute and can be reversed in certain cellular contexts. The reversibility of the tumorigenic potential of EC cells suggests that epigenetic changes may play an important role in defining the EC phenotype. We sought to answer two questions: (i) can the nucleus from an EC cell be reprogrammed to produce a blastocyst that is able to generate an ES cell line and (ii) do the resulting ES cells manifest an altered developmental and/or tumorigenic potential relative to the parental EC cell line? The answers to these questions should functionally define the role of epigenetics in EC.
Materials and Methods
Cell Lines. F9 (passage no. unknown, kindly provided by François Jacob, Institute Pasteur, Paris), P19 (passages 20–25, kindly provided by Michael McBurney, Ottawa Regional Cancer Center, Ottawa, Canada), and METT-1 (passages 10–15) were grown in ES cell medium (DMEM plus 15% FBS, nonessential amino acids, 2-mercaptoethanol, and 1,000 units/ml lymphocyte inhibitory factor). All ES cells were grown in ES medium on irradiated mouse embryonic fibroblasts. GFP-labeled EC cell lines were produced by targeting the Rosa26 locus with enhanced GFP. The targeting construct was produced by subcloning enhanced GFP (Clontech) into a Rosa26-puroR targeting vector (18).
NT and ES Cell Derivation. NT was performed as described (19). Oocytes were collected from superovulated (C57BL/6 × DBA/2 F1) females. Enucleation and NT were done with a piezo-driven micromanipulator system (Primetech, Ibaraki, Japan) on a Nikon microscope with inverted optics. One to three hours after NT, reconstructed oocytes were activated for 5 h with 10 mM Sr2+ in Ca2+-free medium in the presence of 5 μg/ml cytochalasin B. Resulting embryos were cultured to the blastocyst stage in KSOM medium (Specialty Media, Lavellette, NJ). Resulting blastocysts were treated with acid Tyrode's solution to remove the zona pellucida and then cultured in ES medium supplemented with 5 × 10–5 M PD98059 MEK1 inhibitor (Cell Signaling Technology, Beverly, MA) on mouse embryonic fibroblasts. Inner cell mass outgrowths were mechanically dissociated in the presence of trypsin and then replated on mouse embryonic fibroblasts in ES medium.
Pluripotency Assays. Teratomas were produced by injecting 3 × 106 cells s.c. in the flanks and anterior back of severe combined immunodeficient mice. Tumor tissue samples were fixed in 10% buffered formalin for 24 h and embedded in paraffin. Sections (5 μm) were stained with hematoxylin and eosin. At least two teratomas were analyzed per cell line. Chimeras were produced by injecting diploid blastocysts isolated from (C57BL/6 × DBA/2 F2) crosses. Three to eight cells were injected per blastocyst before transfer into day-2.5 pseudopregnant Swiss or (C57BL/6 × DBA/2 F1) females. Chimeras were isolated by Caesarean section at days 9.5, 14.5, and 19.5. Day-9.5 and -14.5 embryos were analyzed under fluorescence microscopy. Representative GFP-positive embryos were fixed, sectioned, and stained as described above. Immunostaining was performed by using an avidin-biotin immunoperoxidase technique. Primary antibody (anti-GFP; 1:1,000, Molecular Probes) was incubated overnight at 4°C. Samples were subsequently incubated with biotinylated secondary antibodies (Vector Laboratories) for 30 min, and then with avidin-biotin peroxidase complexes (Vector Laboratories) for 30 min. Diaminobenzadine was used as the chromogen and hematoxylin was used as the counterstain. For negative controls, primary antibodies were omitted. Ill-appearing day-19.5 pups were killed, and representative tumors were fixed and analyzed as above. Healthy-appearing day-19.5 pups were fostered with lactating BALB/c females. Cell lines were derived from representative tumors by mechanical dissociation with scalpel and culture in ES media.
Array-Comparative Genomic Hybridization (CGH) Profiling on Long Oligomer Microarrays and Spectral Karyotyping (SKY). Genomic DNA was fragmented with DpnII digest and random-prime-labeled after purification according to modified protocols (20). Two micrograms of digested DNA was used per labeling reaction, and each sample was dye-swap-labeled for two hybridizations against reference. Normal ES, V6.5, genomic DNA was used as reference. Labeled DNAs were hybridized onto mouse long oligonucleotide microarrays (Agilent Technologies, Chicago) for 18–20 h at 65°C. For detailed labeling and hybridization protocol, see http://genomic.dfci.harvard.edu. After hybridization, arrays were washed and scanned on an Agilent scanner, and scanned images were analyzed for spot-finding and flagging as well as reporting of Cy3 and Cy5 foreground and background signals for each spot by using standard Agilent software designed for the scanner. Custom analytical tools were used to calculate fluorescence ratio calculation (in log2) and oligomer-to-chromosome location mapping (40). The resultant raw array-CGH profiles were analyzed further by using a segmentation algorithm developed by Olshen and Venkatraman (21) that uses permutation to determine the significance of change points in the raw data to identify statistically significant transitions in copy number. In this study, significant copy number changes are determined based on segmented profiles only. SKY was performed as described (22) according to manufacturer's recommendations by using mouse chromosome paint probes (Applied Spectral Imaging, Vista, CA) on a Nikon Eclipse 800 microscope equipped with an Applied Spectral Imaging interferometer and workstation.
Results
Derivation of ES Cells After Transfer of EC Cell Nuclei into Oocytes. In this study, three different EC cell lines (F9, P19, and METT-1) were chosen based on their varying degrees of tumorigenic and developmental potential. Whereas F9 and P19 cells are highly tumorigenic (23, 24), METT-1 cells rarely produce tumors in chimeric mice (25). The tumorigenic potential of these three lines is inversely related to their developmental potential as F9 cells have no, and P19 cells a limited, developmental potential (nullipotent and restricted potency, respectively), whereas METT-1 cells can differentiate into multiple cell types and generate viable chimeras (pluripotent EC cells) (23–26). Nuclei from the three EC cell lines were transferred into enucleated oocytes, and the reconstituted eggs were activated and monitored for their capacity to develop in vitro to the blastocyst stage. Nuclei from all three EC cell lines were able to direct preimplantation development, resulting in normal appearing blastocysts (Table 1). Resulting blastocysts were explanted onto irradiated mouse embryonic fibroblasts to derive ES cell lines. Previous work has shown a strong correlation between the differentiation state of the donor nucleus and efficiency of postblastocyst development in vivo (5). Similarly, the efficiency of ES cell derivation from blastocysts in vitro is low after NT of differentiated cells (Table 1) (12, 13, 27). Here, we find that EC and ES cell donors produce ES cell lines from NT-derived blastocysts at a high efficiency (Table 1). Blastocysts derived from the three EC donor cell lines generated ES cells at an efficiency of 44–57% and from the ES donor cell line at 50%, much higher efficiencies than observed in the cloning of somatic donor nuclei such as cumulus cells (14%) (27), tail tip fibroblasts (5.2%) (27), lymphocytes (5%) (12), and neurons (7.8%) (13). These results indicate that the epigenetic state of the EC cell genome can be efficiently reprogrammed to support preimplantation development and generate ES cells. Furthermore, the high efficiency of ES cell derivation appears to reflect a similar developmental and epigenetic state of EC and ES cells as compared with differentiated somatic cells.
Table 1. Cloning efficiency of EC cell lines.
| Cell line (ref.) | No. of pronuclei | No. of blastocysts (% of PN) | No. of ES cell lines (% of blastocysts) |
|---|---|---|---|
| F9 | 141 | 9 (6.4) | 4 (44) |
| P19 | 256 | 21 (8.2) | 12 (57) |
| METT-1 | 38 | 2 (5.3) | 1 (50) |
| V6.5 ES* | 65 | 4 (6) | 2 (50) |
| Lymphocytes (12) | 758 | 41 (5.4) | 2 (4.9) |
| Neurons (13) | 493 | 64 (13) | 5 (7.8) |
| Cumulus cells (27) | 120 | 17 (14) | |
| Tail tip fibroblasts (27) | 344 | 18 (5.2) |
V6.5 represents a WT ES cell line (39).
Developmental and Tumorigenic Potential of EC Cells. We used the formation of teratomas and chimeric mice as criteria to compare the tumorigenic and developmental potential of the donor EC cells with that of the respective ES cells derived by NT. Teratoma formation is a measure of spontaneous differentiation, whereas generation of chimeras after injection of the cells into blastocysts requires organized differentiation under the controlled environment of mouse development. First, we reassessed the developmental and tumorigenic potential of the donor EC cell lines. Each EC cell line as well as multiple subclones were injected s.c. into severe combined immunodeficient mice to produce tumors (Table 2 and Fig. 1). The rate of tumor formation differed between the three lines. F9 and P19 EC cells produced visible tumors rapidly (1–2 weeks and 2–3 weeks, respectively), whereas METT-1 EC cells produced them at a rate similar to that of WT ES cells (3–4 weeks) (Table 2). Histology of resulting tumors showed patterns of differentiation consistent with the published developmental potentials of these differing lines (23, 28, 29). Tumors derived from F9 EC cells showed no evidence of differentiation (nullipotent teratocarcinomas, Fig. 1 A) whereas the tumors derived from P19 EC cells exhibited differentiation into immature neural tissue, cartilage, and unspecified glandular tissue (characteristic of teratocarcinomas with restricted differentiation, Fig. 1C). METT-1 EC cells produced tumors that differentiated into a broad range of cell types, including mature neuronal tissue, cartilage, bone, ciliated glandular tissue, and melanocytes (Table 2 and Fig. 1E). However, tumors derived from METT-1 EC cells contained a higher fraction of undifferentiated cells than those derived from WT ES cells, showing that although they have greater developmental potential than the other EC cell lines, they are not equivalent to ES cell lines (Table 2).
Table 2. Teratomas from EC and NT ES cell lines.
| Cell type | No. of lines tested* | Latency, weeks | Degree of differentiation† | Types of tissues seen |
|---|---|---|---|---|
| F9 EC | 3 | 1-2 | 0 | Homogenous undifferentiated |
| F9 NT ES | 4 | 1-2 | 0 | Homogenous undifferentiated |
| P19 EC | 3 | 2-3 | + | Immature neuroepithelium, glandular epithelium, connective, adipose, and cartilage |
| P19 NT ES | 10 | 2-3 | + | Immature neuroepithelium, glandular epithelium, connective, adipose, and cartilage |
| METT-1 EC | 2 | 3-4 | ++ | Mature neuroepithelium and broad range of epithelial and mesodermal tissues |
| METT-1 NT ES | 1 | 3-4 | ++ | Mature neuroepithelium and broad range of epithelial and mesodermal tissues |
| V6.5 ES‡ | 1 | 3-4 | +++ | Broad range of epithelial and mesodermal tissues |
EC lines include parental line as well as GFP-targeted subclones. NT ES lines represent lines resulting from independent NTs.
Degree of differentiation was determined by percent area of undifferentiated cells in tumors. 0, 100%; +, > 50%; ++, 20∼50%; +++, < 20%.
V6.5 represents a WT ES cell line (39).
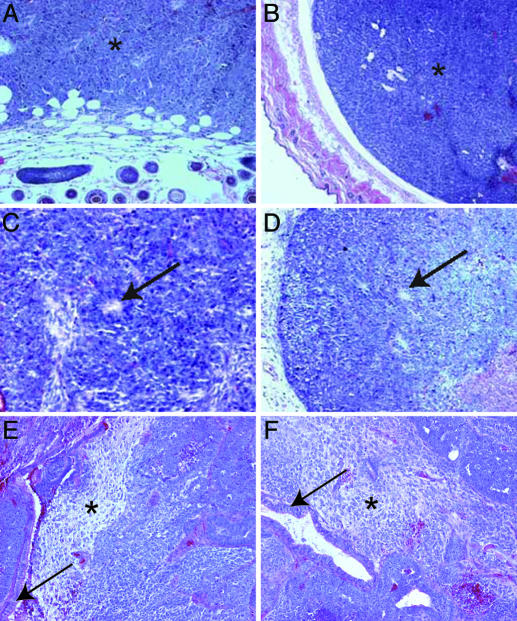
Fig. 1.
Teratoma analysis of EC and corresponding NT ES cell lines. Teratomas from EC (A, C, and E) and NT ES (B, D, and F) cell lines. (A and B) F9ECandNT ES teratomas remain completely undifferentiated (*). Host dermal and epidermal tissue is seen in bottom and lower left of A and D, respectively. (C and D) P19 EC and NT ES cells differentiate into immature neuroepithelium distinguished by rosette formation (arrow). (E and F) METT-1 EC and NT ES cells differentiate into a variety of tissues, including mature neural and glandular tissue (asterisk and arrow, respectively).
To test the potential of the three EC cell lines to produce chimeric mice, the cells were transfected with a ubiquitously expressed GFP construct and microinjected into C57BL/6 × DBA/2 F1 blastocysts to generate embryonic chimeras. Fig. 2 shows examples from embryonic day 14.5 chimeric embryos (summarized in Table 3). F9 cells failed to produce chimeric embryos. Instead, when surrogate mothers were analyzed at embryonic day 14.5, mostly aborted fetuses were observed, signifying a failure of the host embryo to alter the developmental potential of F9 EC cells. P19 EC cells were able to contribute to midgestation embryos, most clearly documented by the presence of GFP-positive cells in the skeleton and CNS (Fig. 2 A). However, most chimeric midgestation embryos showed morphological defects of the head and/or spinal cord, suggesting defective neural development, similar to previously published results (24) (Table 3 and Fig. 2 A). In contrast, METT-1 EC cells contributed broadly to all lineages, and the midgestation embryos appeared morphologically normal (Table 3 and Fig. 2B). Thus, the potential to differentiate as measured by teratoma and chimera formation was consistent with the original descriptions for each cell line, confirming that their distinct developmental and tumorigenic potentials have been preserved.
Fig. 2.
Chimera analysis of EC and corresponding NT ES cell lines. (A–D) Midgestation (embryonic day 14.5) chimeras. (A and C) P19 EC and NT ES cells contribute to developing bone (best seen in ribcage) and brain. Head and spinal morphogenesis is abnormal. (B and D) METT-1 EC and NT ES cells show a broad contribution to embryos, including bone, soft tissue, and skin, as seen here. (E–H) P19 EC and NT ES chimeras are born with head and neck embryocarcinomas. Tumors are obvious on external evaluation of P19EC (E) and P19 NT ES (F) pups (arrows). Hematoxylin/eosin staining (G) and GFP immunostaining (H) of a representative P19 NT ES chimera confirm tumors are ECs of donor cell origin.
Table 3. Midgestation (embryonic day 14.5) chimeras.
| Cell type | No. of blastocysts injected | No. of GFP+ embryos | GFP contribution | Phenotype |
|---|---|---|---|---|
| F9-GFP EC | 80 | 0 | ||
| F9-GFP NT ES | 60 | 0 | ||
| P19-GFP EC | 80 | 4 | Neural and developing bone | Three embryos with abnormal CNS morphogenesis |
| P19-GFP NT ES | 120 | 6 | Neural and developing bone | Four embryos with abnormal CNS morphogenesis |
| METT-1-GFP EC | 200 | 2 | Broad contribution | Appear normal |
| METT-1-GFP NT ES | 68 | 2 | Broad contribution | Appear normal |
Developmental and Tumorigenic Potential of ES Cells Derived After NT of EC Cell Nuclei. Next, we evaluated the developmental and tumorigenic potential of the ES cells derived after NT of the three EC cell lines. Strikingly, each NT ES cell line generated phenotypes that were indistinguishable from their corresponding donor EC cell line. Specifically, F9 NT ES cells showed neither spontaneous differentiation into teratomas nor contribution to midgestation embryos (Tables 2 and 3 and Fig. 1B). P19 NT ES cells exhibited limited spontaneous differentiation into teratomas and produced predominantly abnormal midgestation chimeras with obvious head and spinal cord defects (Tables 2 and 3 and Figs. 1D and 2C). As seen with the P19 EC chimeras, some P19 NT ES chimeras developed to term, but all were afflicted with head and neck ECs and were inviable (Fig. 2 E–H). METT-1 NT ES cells showed a broad differentiation potential in teratomas and also a broad contribution to normal-appearing midgestation embryos (Tables 2 and 3 and Figs. 1F and 2D). Furthermore, normal-appearing chimeric adult mice were derived from blastocysts injected with METT-1 NT ES cells (data not shown). Thus, whereas reprogramming the EC cell nuclei by NT did expand early embryonic developmental potential as evidenced by phenotypically normal blastocysts that were able to produce NT ES cell lines, the NT ES postimplantation development potential was restricted and similar to that of the respective donor EC cells. These data suggest that the tumorigenic and restricted developmental potential of EC cells is determined either by genetic changes or epigenetic modifications that are not reversed by the reprogramming activity of the oocyte.
CGH Analysis of EC Cells. We sought to determine the spectrum of genetic changes that may influence the developmental and tumorigenic potential of the three EC cell lines. To that end, we performed CGH to generate a genomewide view of deletions and amplifications in each of the EC cell lines (Fig. 3). F9 EC cells possessed deletions of 4D3-4E1 and 6B3-6C3 and gains of 8, 12A2–12ter, and 15 (Fig. 3A). P19 EC cells harbored deletions of 4E1-4E2 and 14D1-14qter as well as gain of 6D2-qter (Fig. 3B), the latter two represented a nonreciprocal translocation involving chromosomes 6 and 14 as documented by SKY (Fig. 3B Lower). Finally, a subpopulation of METT-1 EC cells showed a deletion of 14C2-14ter (Fig. 3C). Importantly, the CGH profiles of the ES cell lines derived after NT of the EC lines showed similar CGH patterns, providing definitive evidence that these ES cells were indeed derived from the nuclei of the transferred parental EC cells (Fig. 3). F9 NT ES cells did accumulate an extra copy of chromosome 14 sometime during the cloning and ES derivation process, suggesting ongoing chromosomal instability in this cell line.
Fig. 3.
CGH analysis of EC and corresponding NT ES cell lines. CGH profiles of F9 (A), P19 (B), and METT-1 (C) EC and NT ES cell lines. The x axis represents chromosomes 1–19 (sex chromosomes are not included). The y axis represents relative copy numbers of genetic material along chromosomes. (B Lower) SKY image of chromosomes 6 and 14 in a P19 EC cell, showing two normal chromosomes 6, one normal chromosome 14, and a nonreciprocal translocation of tip of chromosome 6 to chromosome 14, resulting in gain of 6q and loss of 14q. (Left) SKY images. (Center) Reverse 4′,6-diamidino-2-phenylindole images. (Right) Computer-generated representations of SKY images.
Interestingly, these high-resolution CGH profiles on the various EC cell lines differ from previously described karyotypes that were analyzed by conventional banding cytogenetics (28–31). F9 EC cells were described as being aneuploid, with monosomy 14, trisomy 8, and a Robertsonian translocation (2:8), whereas P19 and METT-1 were described as being euploid. The apparent discrepancies between the published karyotypes and the CGH profiles may stem from changes that emerged in the course of tissue culture. Because the resolution used for array-CGH in this study is ≈50 kB, it is also possible that subchromosomal alterations involving small portions of a chromosome were missed previously by low-resolution banding analysis. That aside, the documentation of these irreversible chromosomal aberrations in the EC cells and their NT ES derivatives suggests that the developmental and tumorigenic potential of EC cells after NT are restricted by these irreversible genetic changes.
Discussion
In this study, cloning by NT was used to address the role of epigenetic modifications in determining the developmental and tumorigenic potential of EC cells. We find that, whereas the transfer of nuclei from EC cells results in morphologically normal blastocysts from which ES cell lines can be produced, the resulting ES cells have the same developmental and tumorigenic potential as the parental EC cell lines. Strikingly, the nuclei from the three EC cell lines used in this study conferred their distinctive developmental and tumorigenic potentials to the resulting NT ES cells. The failure to reprogram the EC phenotype suggested that genetic alterations were responsible for limiting the developmental potential of the NT ES lines. Consistent with this, CGH showed shared and unique genetic alterations between the different EC cell lines. These results show that whereas the genomes of EC cells can redirect normal preimplantation development, genetic changes transmitted from the EC nuclei determine the developmental and tumorigenic potential of the NT ES cells.
ES cell lines were derived at a high efficiency after NT of the EC donors, which is in stark contrast to lymphocyte, neuron, and fibroblast donors. Accumulating data show a strong correlation between the differentiation state of the donor nucleus and the efficiency of producing full-term pups and ES cell lines after NT (see Table 1). EC and ES cells express markers of pluripotency, including Oct4, Nanog, and SSEA (32–34). Therefore, the high cloning efficiency of these cells may be caused by the expression of a set of pluripotency genes that are sufficient to direct early development. On the other hand, nuclei from differentiated donor cells must reactivate pluripotency genes and, because this process is often incomplete, the cloning of somatic cell types is less efficient than from ES and EC cells (35). Consistent with the notion, we have found that the derivation of ES cells after NT of a differentiated tumor model occurs at a low efficiency if at all (41).
Although the EC cell lines have multiple genetic changes as seen by CGH, these changes did not interfere with preimplantation development or ES cell derivation. The abnormal phenotype of clones became obvious when the resulting NT ES cells were tested in developmental and tumorigenic assays. In these assays, the phenotypes of the NT ES cell lines appeared identical to the corresponding donor EC lines and, therefore, very different from each other (i.e., METT-1 vs. P19 vs. F9). This finding suggests that each EC cell line harbors its own unique set of unreprogrammable modifications that function within the epigenetic context of a pluripotent stem cell, limiting its developmental potency. Consistent with this finding, CGH showed unique genetic changes in F9, P19, and METT-1 EC cells. Interestingly, there were also overlapping changes among the cell lines, including the loss of 4E1 in F9 and P19 and loss of 14D1-qter in P19 and METT-1. It is therefore tempting to speculate that an accumulation of genetic changes underlies the increasingly limited differentiation potential and, hence, increasing tumorigenic potential of the EC cell lines. Identifying genes that cause EC cells to become locked into their fates should provide insights into the general mechanisms governing tumorigenesis and developmental potential. That is, the genetics of EC should reveal the genetics of self-renewal and pluripotency.
Interestingly, the 6D2-qter region that is amplified in P19 cells is syntenic to chromosome 12p in humans, which is the most common genetic modification seen in human embryonic carcinomas (36). In particular, two minimal regions of amplification of 12p have been described in human EC, 12p11.2–12.1 and 12p13. Furthermore, expression profiling has identified an overrepresentation of highly expressed genes in the 12p13 region in numerous human ECs (37). The combination of genomic amplification and transcriptional overexpression suggests that these genes are causally involved in the tumorigenic and/or developmental potential of EC cells. Consistent with this notion is the recent demonstration that the misexpression of nanog, which maps to the 12p13 region, inhibits ES cell differentiation producing an EC-like phenotype (32, 38).
Although EC appeared to be a good candidate for an epigenetic model of tumorigenesis, the failure to alter their developmental and tumorigenic phenotype by NT argues that genetic lesions rather than epigenetic restrictions are responsible. This does not exclude the potential to reprogram other tumor types. For example, abnormal E7 mouse embryos were produced after transfer of nuclei from a medulloblastoma cell line, but it was not possible to analyze later phenotypes because embryonic development arrested and ES cells were not derived (9). We have found that the genome of a malignant melanoma cell can be reprogrammed to pluripotency after nuclear transplantation although the tumorigenic potential appeared unchanged in chimeric animals derived from the NT ES cells (41).
Acknowledgments
We thank Christopher Leo for help with CGH, Alexei Protopopov for SKY analysis, Jessie Dausman for help with mouse work, Caroline Beard for help building the GFP-expressing construct, Michael McBurney for P19 cells, and Laurie Jackson-Grusby, Caroline Beard, and Heinz Linhart for critiquing the manuscript. Array-CGH profiling was performed at the Arthur and Rochelle Belfer Cancer Genomics Center at the Dana–Farber Cancer Institute. R.H.B. is supported by a Lance Armstrong Foundation Grant. K.H. is supported by a Ph.D. fellowship from the Boehringer Ingelheim Fonds. C.B. is supported by National Institutes of Health Grant T32 CA09382 and the LeBow Fund for Myeloma Cure. L.C. is supported by National Institutes of Health Grant RO1 CA93947, and R.J. is supported by National Institutes of Health Grant ROI-CA-87-869.
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on April 29, 2003.
Abbreviations: NT, nuclear transfer; ES, embryonic stem; EC, embryonal carcinoma; CGH, comparative genomic hybridization; SKY, spectral karyotyping.
See accompanying Biography on page 13982.
References
- 1.Jaenisch, R. & Bird, A. (2003) Nat. Genet. 33, Suppl., 245–254. [DOI] [PubMed] [Google Scholar]
- 2.Li, E. (2002) Nat. Rev. Genet. 3, 662–673. [DOI] [PubMed] [Google Scholar]
- 3.Jones, P. A. & Baylin, S. B. (2002) Nat. Rev. Genet. 3, 415–428. [DOI] [PubMed] [Google Scholar]
- 4.Herman, J. G. & Baylin, S. B. (2003) N. Engl. J. Med. 349, 2042–2054. [DOI] [PubMed] [Google Scholar]
- 5.Hochedlinger, K. & Jaenisch, R. (2002) Curr. Opin. Cell Biol. 14, 741–748. [DOI] [PubMed] [Google Scholar]
- 6.Diberardino, M. A. & King, T. J. (1965) Dev. Biol. 11, 217–242. [DOI] [PubMed] [Google Scholar]
- 7.King, T. J. & DiBerardino, M. A. (1965) Ann. N.Y. Acad. Sci. 126, 115–126. [DOI] [PubMed] [Google Scholar]
- 8.McKinnell, R. G., Deggins, B. A. & Labat, D. D. (1969) Science 165, 394–396. [DOI] [PubMed] [Google Scholar]
- 9.Li, L., Connelly, M. C., Wetmore, C., Curran, T. & Morgan, J. I. (2003) Cancer Res. 63, 2733–2736. [PubMed] [Google Scholar]
- 10.Carlson, D. L., Sauerbier, W., Rollins-Smith, L. A. & McKinnell, R. G. (1994) J. Comp. Pathol. 111, 197–204. [DOI] [PubMed] [Google Scholar]
- 11.Rideout, W. M., III, Hochedlinger, K., Kyba, M., Daley, G. Q. & Jaenisch, R. (2002) Cell 109, 17–27. [DOI] [PubMed] [Google Scholar]
- 12.Hochedlinger, K. & Jaenisch, R. (2002) Nature 415, 1035–1038. [DOI] [PubMed] [Google Scholar]
- 13.Eggan, K., Baldwin, K., Tackett, M., Osborne, J., Gogos, J., Chess, A., Axel, R. & Jaenisch, R. (2004) Nature 428, 44–49. [DOI] [PubMed] [Google Scholar]
- 14.Martin, G. R. (1980) Science 209, 768–776. [DOI] [PubMed] [Google Scholar]
- 15.Stevens, L. C. (1970) Dev. Biol. 21, 364–382. [DOI] [PubMed] [Google Scholar]
- 16.Andrews, P. W. (2002) Philos. Trans. R. Soc. London B 357, 405–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mintz, B. (1978) Harvey Lect. 71, 193–246. [PubMed] [Google Scholar]
- 18.Wutz, A., Rasmussen, T. P. & Jaenisch, R. (2002) Nat. Genet. 30, 167–174. [DOI] [PubMed] [Google Scholar]
- 19.Wakayama, T., Perry, A. C., Zuccotti, M., Johnson, K. R. & Yanagimachi, R. (1998) Nature 394, 369–374. [DOI] [PubMed] [Google Scholar]
- 20.Pollack, J. R., Perou, C. M., Alizadeh, A. A., Eisen, M. B., Pergamenschikov, A., Williams, C. F., Jeffrey, S. S., Botstein, D. & Brown, P. O. (1999) Nat. Genet. 23, 41–46. [DOI] [PubMed] [Google Scholar]
- 21.Olshen, A. B. & Venkatraman, E. S. (2004) J. Biostatistics, in press. [DOI] [PubMed]
- 22.Artandi, S. E., Chang, S., Lee, S. L., Alson, S., Gottlieb, G. J., Chin, L. & DePinho, R. A. (2000) Nature 406, 641–645. [DOI] [PubMed] [Google Scholar]
- 23.Berstine, E. G., Hooper, M. L., Grandchamp, S. & Ephrussi, B. (1973) Proc. Natl. Acad. Sci. USA 70, 3899–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rossant, J. & McBurney, M. W. (1982) J. Embryol. Exp. Morphol. 70, 99–112. [PubMed] [Google Scholar]
- 25.Stewart, T. A. & Mintz, B. (1981) Proc. Natl. Acad. Sci. USA 78, 6314–6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pierce, G. B., Pantazis, C. G., Caldwell, J. E. & Wells, R. S. (1982) Cancer Res. 42, 1082–1087. [PubMed] [Google Scholar]
- 27.Wakayama, T., Tabar, V., Rodriguez, I., Perry, A. C., Studer, L. & Mombaerts, P. (2001) Science 292, 740–743. [DOI] [PubMed] [Google Scholar]
- 28.McBurney, M. W. & Rogers, B. J. (1982) Dev. Biol. 89, 503–508. [DOI] [PubMed] [Google Scholar]
- 29.Mintz, B. & Cronmiller, C. (1981) Somatic Cell Genet. 7, 489–505. [DOI] [PubMed] [Google Scholar]
- 30.Alonso, A., Breuer, B., Steuer, B. & Fischer, J. (1991) Int. J. Dev. Biol. 35, 389–397. [PubMed] [Google Scholar]
- 31.Hogan, B. L. M., Constantini, F. & Lacy, E. (1986) Manipulating the Mouse Embryo (Cold Spring Harbor Lab. Press, Plainview, NY).
- 32.Chambers, I., Colby, D., Robertson, M., Nichols, J., Lee, S., Tweedie, S. & Smith, A. (2003) Cell 113, 643–655. [DOI] [PubMed] [Google Scholar]
- 33.Okamoto, K., Okazawa, H., Okuda, A., Sakai, M., Muramatsu, M. & Hamada, H. (1990) Cell 60, 461–472. [DOI] [PubMed] [Google Scholar]
- 34.Solter, D. & Knowles, B. B. (1978) Proc. Natl. Acad. Sci. USA 75, 5565–5569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bortvin, A., Eggan, K., Skaletsky, H., Akutsu, H., Berry, D. L., Yanagimachi, R., Page, D. C. & Jaenisch, R. (2003) Development (Cambridge, U.K.) 130, 1673–1680. [DOI] [PubMed] [Google Scholar]
- 36.Skotheim, R. I. & Lothe, R. A. (2003) Apmis 111, 136–150; discussion 50–51. [DOI] [PubMed] [Google Scholar]
- 37.Sperger, J. M., Chen, X., Draper, J. S., Antosiewicz, J. E., Chon, C. H., Jones, S. B., Brooks, J. D., Andrews, P. W., Brown, P. O. & Thomson, J. A. (2003) Proc. Natl. Acad. Sci. USA 100, 13350–13355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitsui, K., Tokuzawa, Y., Itoh, H., Segawa, K., Murakami, M., Takahashi, K., Maruyama, M., Maeda, M. & Yamanaka, S. (2003) Cell 113, 631–642. [DOI] [PubMed] [Google Scholar]
- 39.Eggan, K., Akutsu, H., Loring, J., Jackson-Grusby, L., Klemm, M., Rideout, W. M., III, Yanagimachi, R. & Jaenisch, R. (2001) Proc. Natl. Acad. Sci. USA 98, 6209–6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brennan, C., Zhang, Y., Leo, C., Feng, B., Cauwels, C., Aguirre, A. J., Kim, M., Protopopov, A. & Chin, L. (2004) Cancer Res. 4, 4744–4748. [DOI] [PubMed] [Google Scholar]
- 41.Hochedlinger, K., Blleloch, R., Brennan, C., Yamada, Y., Kim, M., Chin, L. & Jaenisch, R. (2004) Genes Dev., in press. [DOI] [PMC free article] [PubMed]