Abstract
AlkB repairs 1-alkyladenine and 3-methylcytosine lesions in DNA by directly reversing the base damage. Although repair studies with randomly alkylated substrates have been performed, the miscoding nature of these and related individually alkylated bases and the suppression of mutagenesis by AlkB within cells have not yet been explored. Here, we address the miscoding potential of 1-methyldeoxyadenosine (m1A), 3-methyldeoxycytidine (m3C), 3-ethyldeoxycytidine (e3C), 1-methyldeoxyguanosine (m1G), and 3-methyldeoxythymidine (m3T) by synthesizing single-stranded vectors containing each alkylated base, followed by vector passage through Escherichia coli. In SOS–, AlkB-deficient cells, m1A was only 1% mutagenic; however, m3C and e3C were 30% mutagenic, rising to 70% in SOS+ cells. In contrast, the mutagenicity of m1G and m3T in AlkB– cells dropped slightly when SOS polymerases were expressed (m1G from 80% to 66% and m3T from 60% to 53%). Mutagenicity was abrogated for m1A, m3C, and e3C in wild-type (AlkB+) cells, whereas m3T mutagenicity was only partially reduced. Remarkably, m1G mutagenicity was also eliminated in AlkB+ cells, establishing it as a natural AlkB substrate. All lesions were blocks to replication in AlkB-deficient cells. The m1A, m3C, and e3C blockades were completely removed in wild-type cells; the m1G blockade was partially removed and that for m3T was unaffected by the presence of AlkB. All lesions demonstrated enhanced bypass when SOS polymerases were induced. This work provides direct evidence that AlkB suppresses both genotoxicity and mutagenesis by physiologically realistic low doses of 1-alkylpurine and 3-alkylpyrimidine DNA damage in vivo.
Keywords: AlkB, DNA repair, DNA alkylation
Unrepaired endogenous and exogenous alkylation damage to DNA may cause mutations ultimately leading to cancer and other genetic diseases. Damage such as O6-alkylguanine, O4-alkylthymine, and methylphosphotriesters can be removed nonenzymatically by the Ada and Ogt proteins of Escherichia coli (1). In response to DNA alkylation damage, the ada regulon in E. coli is up-regulated whereby, in addition to the Ada DNA repair protein, the levels of AlkB, AlkA, and AidB are increased (2). Unlike Ada, the AlkA and AlkB DNA repair proteins are truly catalytic enzymes. AlkA is a double-stranded DNA glycosylase shown to exhibit a broad substrate specificity that includes 3- and 7-alkylpurines, O2-alkylpyrimidines, ethenopurines, as well as other lesions (3, 4). The resulting AP site must then be processed by AP endonucleases, followed by DNA synthesis and ligation to restore the predamaged state.
Unlike the Ada, Ogt and the human MGMT methyltransferases, AlkB and its human homologs hABH2 and hABH3 not only reverse alkylation base damage directly, but they do so catalytically and with a substrate specificity aimed at the base-pairing interface of the G:C and A:T base pairs (5–7). 1-Alkyladenine and 3-methylcytosine lesions possibly form when adenine and cytosine are in a single-stranded conformation (during replication or transcription) and are substrates for AlkB, hABH2, and hABH3 (7, 8). The mechanism by which AlkB affords protection to DNA from SN2 alkylators has recently been elucidated (9, 10). Computational protein folding sequence alignment studies have assigned AlkB as a dioxygenase (11). Biochemical evidence shows that AlkB uses the cofactors α-ketoglutarate, non-heme iron (II), and O2 to hydroxylate the rogue methyl group of 1-methyladenine or 3-methylcytosine, ultimately resulting in the release of dealkylated DNA, formaldehyde, succinate, and carbon dioxide (5, 6). This direct reversal of base damage mechanism is efficient, because an AP site intermediate, which would need further processing, is not formed. Remarkably, AlkB and hABH3 can repair not only DNA, but also RNA (8).
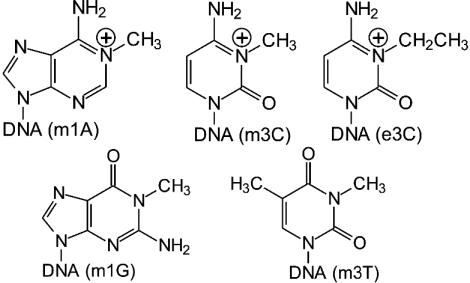
The studies above have implicated AlkB and homologs in repair of a host of DNA lesions, and one purpose in the work described here was to provide uniquely modified oligonucleotide and genome resources that would be useful for both biochemical and genetic studies. Herein, we describe a study whereby the contribution of a single 1-methyldeoxyadenosine (m1A), 3-methyldeoxycytidine (m3C), 3-ethyldeoxycytidine (e3C), 1-methyldeoxyguanosine (m1G), or 3-methyldeoxythymidine (m3T) lesion (Fig. 1) to genotoxicity and mutagenesis by normal or bypass polymerases, as well as the ease of adduct repair by AlkB within cells, was investigated. Because DNA that had been randomly alkylated to produce 1-ethyldeoxyadenosine lesions has been recently found to be repaired by AlkB (7), we additionally explored the capacity of AlkB to work on the pyrimidine 3-ethyldeoxycytidine. We also wanted to ascertain whether the uncharged lesions m1G and m3T, which are structurally analogous with respect to the alkylated position of m1A and m3C, would also be substrates for AlkB. The m1A, m3C, and e3C lesions in the present study are preferentially formed by alkylating agents in single-stranded DNA and have been detected in vitro (5, 6, 12–18) and in vivo (12, 17, 19–24). Although less prevalent, m1G lesions have also been found in vitro (16) and in vivo (19), and m3T in vitro (12, 13, 16, 18, 25) and in vivo (12, 25).
Fig. 1.
Structures of alkylated bases studied in this work.
Materials and Methods
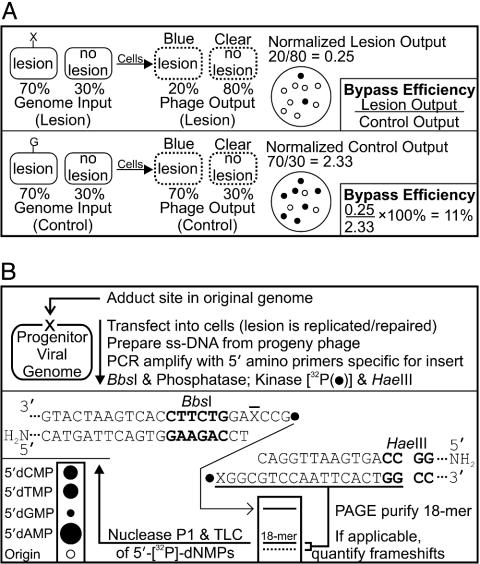
Lesion Bypass Analysis [Competitive Replication of Adduct Bypass (CRAB) Assay]. Solid-phase oligonucleotide synthesis and characterization by mass spectrometry and HPLC, as well as the construction and normalization of genomes and determination of ligation efficiency, are given as supporting information, which is published on the PNAS web site. The CRAB assay, which measures the ability of the DNA polymerase to traverse a lesion in vivo, is outlined in Fig. 2A. The CRAB assay relies on the accurate normalization (supporting information) of constructed genomes, which are mixed with a competitor genome before transfection and growth in liquid culture. A drop in lesion-derived output signifies a blockade to the DNA polymerase.
Fig. 2.
Methodology for determining lesion bypass and mutagenesis in vivo. (A) CRAB assay for quantification of lesion bypass in vivo. The power of a DNA lesion to block replication by normal and bypass polymerases, as well as lesion blockade relief by AlkB, was addressed by using the CRAB assay introduced here. In this assay, an arbitrary mixture of genome containing a lesion (70%, X) and an M13 wild-type competitor (30%, no lesion) are mixed together, electroporated, and allowed to grow in solution before analyzing the progeny phage output (dotted oval). If the lesion is a block to replication, more progeny from the “no lesion” competitor will occur. Progeny phage from the lesion (blue) and competitor (clear) are easily scored on indicator plates by means of the LacZα complementation peptide. Here, the lesion output (20%) is normalized to the competitor output (80%), and bypass efficiency is calculated by dividing this ratio by that obtained from the identical experiment employing a normal G as the lesion. The competitor also acts as an internal standard, removing unequal transformation efficiency as a source of error. (B) REAP assay for quantification of lesion mutagenesis at X. The mutations elicited by m1A, m3C, e3C, m1G, and m3T were scored by the REAP assay. The base composition at the lesion site (mutation frequency) is obtained by labeling the 5′ phosphate of the site that originally housed the lesion (X), and separating the radioactive 5′ monophosphates by means of TLC. This is accomplished by PCR amplification of the viral progeny, digestion with BbsI (recognition site in bold, cleaves outside its recognition sequence), dephosphorylation with alkaline phosphatase, 32P-labeling and HaeIII trimming. The liberated 18-mer is PAGE purified, followed by nuclease P1 digestion to 5′ nucleotide monophosphates. These 5′ [32P]dNMPs are separated on a TLC plate. The amount of ±1 and ±2 frameshift mutagenesis can also be scored at the PAGE step.
Preparation and electroporation of cells. E. coli HK81 (AB1157, nalA) and HK82 (as HK81 but alkB22) (26, 27) were from L. Samson (Massachusetts Institute of Technology, Cambridge, MA) and were made electrocompetent as follows. Each cell type was prepared in triplicate. Briefly, three flasks containing 150 ml of LB were each inoculated with 1.5 ml of independently grown overnight cultures, and incubated at 37°C with agitation to an OD600 of 0.40. Cells were kept at 0°C to 4°C, pelleted, and resuspended in 25 ml of cold 10 mM MgSO4, and each pellet was transferred to a 150 × 15 mm Petri dish. The AlkB–/SOS+ condition was achieved by irradiation of each dish (lid off) containing HK 82 cells with 254 nm of light at 45 J/m2. Contents from the three dishes (with or without irradiation) were each immediately transferred to 125 ml of 2× YT medium, grown at 37°C with agitation for 40 min, placed on ice and pelleted. At this point, the three pellets were combined and washed twice with 175 ml of 4°C water, resuspended in 6 ml of 10% glycerol, and stored at 4°C overnight for use the next day. Wild-type M13mp7(L2) was used as a competitor and was quantified by UV absorbance (1 A260 = 33 μg/ml, 2.36 μg/pmol). A mixture of 8 μl of M13mp7(L2) (40 fmol) and 5 μl of normalized genome construct was electroporated with 100 μl of electrocompetent cells at 2.5 kV and 129 ohms in a 0.2-cm cuvette (2.6 kV and 4.9 ms time constant delivered), providing ≈70% signal from the unmodified control genome construct containing G at the lesion site, and between 2 × 104 and 2 × 106 initial replicative events (28). The cells were immediately transferred to 10 ml of LB, incubated at 23°C for at least 30 min, and then grown at 37°C for 4 h with agitation, thus amplifying the initial events by >2,000-fold.
CRAB data analysis and controls. More than 1,500 plaques were counted from each transformation to provide the number of blue and clear plaques (28). The blue plaques arise from the lesion-bearing oligonucleotide being in-frame within the coding region of the LacZα complementation peptide (28); the clear plaques from M13 wild-type DNA are out-of-frame. The percentage of clear plaques using 100% alkylated m1A, m3C, and e3C genomes for the three cell types was equal to the G control construct (≈1%) and was thus not corrected; however, in SOS–/AlkB–, SOS–/AlkB+, and SOS+/AlkB– cells, the respective percentage of clear plaques from pure genomes was 6.1%, 1.3%, and 1.8% (m1G) and 5.8%, 5.6%, and 1.6% (m3T). Correction to the bypass data for these lesions was made by expressing these percentages as a ratio of clear to blue plaques, which was multiplied by the respective number of blue plaques counted in the CRAB assay to obtain the expected number of lesion-derived clear plaques. Although negligible, this number was nevertheless added to the “blue” plaques and was subtracted from the “clear” plaques to obtain the adjusted adduct “output mixture” in Fig. 2 A. In a concurrent experiment (data not shown), a positive correlation existed between the percentage of clear plaques for a tetrahydrofuran abasic site analog control (processed in SOS–/AlkB– cells) and the 32P intensity of a –1 frameshift band from denaturing polyacrylamide gel electrophoresis (PAGE) obtained by means of the assay described below. However, the lack of such correlation (no appreciable –1 band, data not shown) for the m1G and m3T lesions suggests that targeted deletions/insertions were not the primary cause of the small percentage of clear plaques for m1G and m3T genomes.
Lesion Mutagenesis Analysis [Restriction Endonuclease and Postlabeling Analysis of Mutation Frequency (REAP) Assay]. The REAP assay is outlined in Fig. 2B, whereby the site that originally housed the lesion is 32P-labeled, and the liberated 5′ [32P]dNMPs are fractionated by means of TLC. The same batch of cells and transformation technique used for the CRAB assay were used for the REAP assay (5 μl of genome construct transformed), which determines the mutation frequency of the lesion (28). Template preparation. Mutagenesis results came from events within the cell and were not due to Pfu amplification of lesion-bearing genome template resident in the phage supernatant. A phage to original genome construct ratio of >99.9% was achieved by infecting ≈ 7 × 106 SCS110 cells (Stratagene) with 100 μl of supernatant containing >2 × 107 progeny phage in 10 ml of LB with agitation at 37°C for 6 h. Single-stranded viral template DNA was prepared by using 700 μl of the SCS110 phage supernatant (QIAPrep spin M13 kit, Qiagen, Valencia, CA), and was eluted in 100 μl of 10 mM Tris·HCl, pH 8.5.
PCR amplification. PCR was performed in a PTC-200 DNA Engine thermocycler (MJ Research, Waltham, MA) with Turbo Pfu thermostable polymerase (Stratagene). The 25-μl PCR mix included 10 mM KCl, 10 mM (NH4)2SO4, 20 mM Tris·HCl (pH 8.8), 2 mM MgSO4, 0.1% Triton X-100, 100 μg/ml BSA, 1.0 μM each primer, 0.2 mM each dNTP, and 0.05 unit/μl Pfu. The amplification cycle consisted of denaturation at 94°C for 0.5 min, annealing at 64°C for 1 min, and extension at 72°C for 1 min. After 30 cycles, samples were incubated at 72°C for 5 min and stored at 4°C until further use. The primers used to generate the 61-mer PCR product were 5′-YCAGCTATGACCATGATTCAGTGGAAGAC-3′ and 5′-YTGTAAAACGACGGCCAGTGAATTGGACG-3′ [Y is an aminoethoxyethyl ether group from a “5′-amino-modifier 5” phosphoramidite (Glen Research, Sterling, VA), which cannot be enzymatically 32P-labeled]. Addition of 85 μl of water to the PCR mix was followed by extraction with 100 μl of phenol:chloroform:isoamyl alcohol (25:24:1). After inactivation of the Pfu exonuclease domain, traces of organics and dNTPs inhibitory to the phosphatase and kinase steps were removed by passage of the aqueous layer through a homemade spin column (column, PerkinElmer Wallac; Sephadex G-50 Fine, Amersham Pharmacia) with packed resin dimensions of 7 × 30 mm.
Restriction endonuclease digestion. Ten microliters of eluant was incubated in 20 μl of final volume containing 1× buffer 2 [50 mM NaCl/10 mM Tris·HCl/10 mM MgCl2/1 mM DTT, pH 7.9], 5 units of BbsI (New England Biolabs), and 1 unit of shrimp alkaline phosphatase (Roche) at 37°C for 4 h, 80°C for 5 min, then cooled to 20°C at 0.2°C/s. Four microliters of cut, dephosphorylated DNA solution was incubated in a 6-μl final volume containing 1× buffer 2, 10 mM DTT, 3.33 μM cold ATP, 0.278 μM[γ-32P]ATP [6,000 Ci/mmol, PerkinElmer (1 Ci = 37 GBq)], and 5 units of T4 polynucleotide kinase (Amersham Pharmacia). This mixture was incubated at 37°C for 15 min, 65°C for 20 min, then cooled to 23°C at 0.1°C/s, after which 4 μl of 1× buffer 2 containing 10 units of HaeIII (New England Biolabs) was added and incubated at 37°C for 2 h.
Gel purification. The 18-mers containing 32P at the 5′ end for base composition analysis were gel purified on 20% denaturing polyacrylamide gels as described (28), but run for 3.3 h. The xylene cyanol dye marker ran 10.5 cm from the well, and the faster running 18-mer was accurately excised with the aid of mailing tape containing radioactive spots and ink applied with a ball-point pen [dipped in a 1/15 dilution of [32P]ATP (6,000 Ci/mmol) for each spot flanking the 18-mer band]. After exposure of the gel to a PhosphorImager (Amersham Biosciences) plate for 1 min, a transparency print-out at ×1 magnification was used as an excision template. The 18-mer was extracted from the gel, desalted (G-50 Fine Sephadex resin substituted for G-25), and lyophilized as described (28).
Nuclease P1 digestion and TLC analysis. After the DNA was resuspended in 5 μl of 30 mM NaOAc (pH 5.3), 10 mM ZnCl2, and 1 μg of nuclease P1 (Roche), and incubated at 50°C for 1 h, 0.5-μl samples were spotted 1 cm apart onto a 20 × 20 cm polyethyleneimine cellulose TLC plate (PEI-TLC, J. T. Baker) that had been prewashed in water for 2 min the previous night and air dried. Liquid from a saturated solution of (NH4)2HPO4 was transferred to a flask and pH adjusted to 5.8 with H3PO4, and 200 ml was used to develop the plate. After drying, the separated radiolabeled nucleotides were quantified by PhosphorImager analysis, thus providing the fractional base composition of each nucleotide at the lesion site, from which the mutation frequency and type were determined.
m1A Lesion Integrity Analysis. The modified nucleobase composition at the lesion site was quantified essentially as described for the REAP mutation frequency assay (Fig. 2B Lower), but with constructed m1A-genomes that had not been processed within E. coli. If the lesion did not remain 100% intact, multiple 5′ [32P]d(lesion)MP spots would be seen by means of TLC. A duplex region recognizable to BbsI was made by annealing 150 fmol of 36-mer 5′-CCAGTGAATTGGACGCCTAGGTCTTCCACTGAATCA-3′ with 100 fmol m1A-genome. Control oligonucleotides containing 5′ m1A or 5′ m6A lesions were 32P-labeled and used as 5′ dNMP markers. The Dimroth rearrangement was induced in a control reaction by heating the desalted, BbsI/HaeIII-liberated 32P-18-mer from three pooled m1A-genome constructs in 200 μl of concentrated aqueous NH4OH at 80°C for 3 h, followed by lyophilization before nuclease P1 digestion. Control reactions were performed where a 50/50 mixture of m1A and m6A 16-mer oligonucleotides was ligated into a larger oligonucleotide construct, taken through the entire assay, and both 5′ dNMPs were equally represented by means of TLC. Alkaline phosphatase was inactivated by incubation at 80°C for 5 min vs. phenol extraction at ambient temperature for the pure m1A genomes, with no difference in the ratio of 5′ dm1AMP to 5′ dm6AMP (data not shown).
Results
Bypass Efficiency of m1A, m3C, e3C, m1G, and m3T Lesions. To determine lesion bypass in vivo, it was necessary to quantify the ligation efficiency of the modified oligonucleotide into the genome and the concentration of viable (covalently closed circular) genomes. Rather than performing a Southern blot of electrophoresed genomes (29) or following the fate of a radio-labeled oligonucleotide to be incorporated into the genome during construction (30), we took a new route to achieve high sensitivity by simply annealing a 32P-labeled 30-mer to the nonradioactive single-stranded constructed genome in solution, running the isoforms out on an agarose gel, air-drying the gel to prevent differential 32P-leaching, and quantifying by Phosphor-Imager analysis. Because genomes were constructed by using two scaffolds in which no base was paired opposite the lesion (28), the ligation efficiency was equal (27.3 ± 1.7%) for all genome constructs, regardless of lesion.
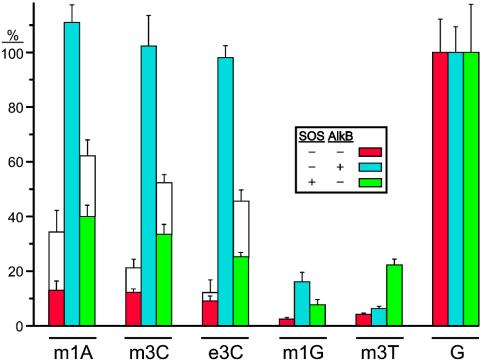
We addressed the lethality of m1A, m3C, e3C, m1G, and m3T alkylated DNA bases (Fig. 1) using the CRAB assay introduced in this work, which does not require an equal genome transfection efficiency to accurately score survival. The crux of this assay is based on transforming cells with a fixed ratio of lesion-bearing genome and unmodified competitor genome (Fig. 2 A), where an increase in progeny from the competitor signifies hindered lesion replication. The lacZα complementation peptide sequence was out-of-frame for the competitor (clear plaques) and, barring frameshift mutations, in-frame for the constructed genomes (blue plaques). The results from the CRAB assay (Fig. 3) reveal that all alkylated lesions were blocks to DNA replication, which were partially overcome by SOS bypass polymerases. These blockades were totally removed in cells expressing AlkB for m1A, m3C, and e3C, partially removed for m1G, but hardly removed for m3T.
Fig. 3.
Bypass of m1A, m3C, e3C, m1G, and m3T in E. coli. Modified and control genomes were constructed in triplicate and were normalized to one another before being mixed with competitor genome. Each mixture was transformed into SOS–/AlkB– (red), SOS–/AlkB+ (blue), and SOS+/AlkB– (green) E. coli, and bypass was scored as described (Fig. 2 A), with error bars representing one standard deviation of propagated error. Transformation mixtures were allowed to grow in solution before plating (colored bars) or, for m1A, m3C, and e3C, were additionally plated immediately after transformation (open bars), which can underestimate the genotoxicity of DNA lesions (see Discussion).
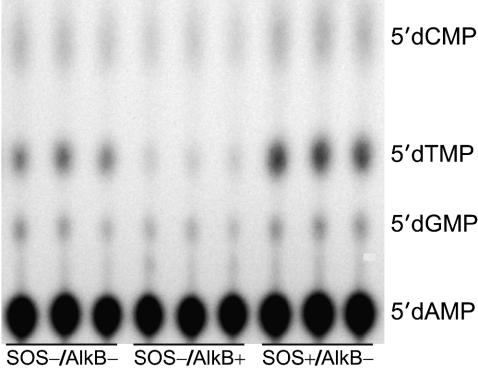
Mutagenicity of m1A, m3C, e3C, m1G, and m3T Lesions. Lesion mutagenicity was ascertained by using the REAP assay (Fig. 2B). The base composition at the lesion site is found by PCR amplification of progeny, 32P-labeling the site that had originally contained the lesion, and ultimately separating the 5′ [32P]dNMPs by means of TLC. Use of PCR primers specific for the inserted oligonucleotide-vector juncture allowed for the selective amplification of progeny that had contained the modified oligonucleotide, thereby eliminating competing amplification of genetic engineering artifacts that lack the BbsI site, which may result in weak 32P-PAGE bands. Because the primers do not anneal with a five-base tract that housed the centrally located lesion, the most common mutagenesis events (point mutations, and insertions or deletions of one or two bases) can be scored. We saw no appreciable frameshift mutagenesis for the five lesions in the three cellular scenarios tested. A representative thin-layer chromatogram for m1A mutagenesis is shown (Fig. 4), with the base composition at the lesion site providing the mutation frequency. This TLC demonstrates that, although m1A is not very mutagenic, the REAP assay is sensitive enough to discern subtle yet reproducible differences in m1A mutagenesis (A to T mutagenesis was suppressed by AlkB but enhanced by SOS polymerases). The base composition for A vs. T was respectively 99.0 ± 0.15% vs. 0.61 ± 0.18% in SOS–/AlkB– cells, 99.7 ± 0.05% vs. 0.06 ± 0.00% in SOS–/AlkB+ cells, and 98.6 ± 0.21% vs. 1.0 ± 0.16% in SOS+/AlkB– cells.
Fig. 4.
Mutagenesis of m1A in E. coli. The base composition at the lesion site for m1A processed in the designated cell strains was obtained by the REAP assay. This polyethyleneimine TLC plate reveals that m1A is weakly mutagenic. The assay reproducibly detected subtle differences in mutagenicity, such as the suppression of A to T mutations by AlkB (center lanes), and the enhancement of such mutations by SOS polymerases (right lanes).
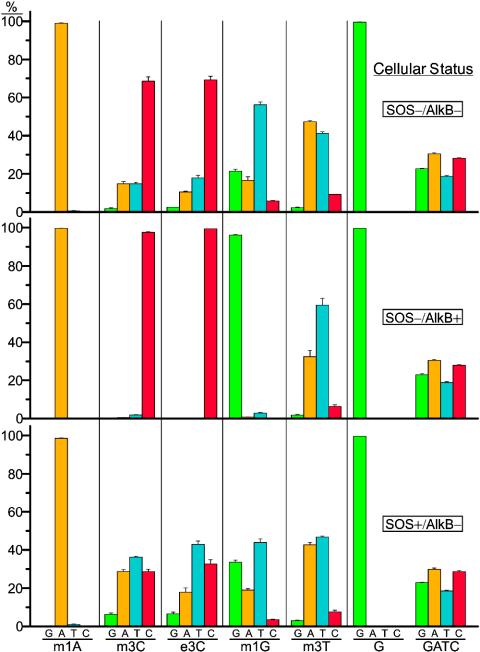
Results from the REAP assay on all lesions are compiled in Fig. 5. In the absence of AlkB and SOS bypass polymerases, m3C and e3C were 30% mutagenic, with the predominant mutations being C to T and C to A. Basal expression of AlkB abrogated the mutagenicity of m3C and e3C, whereas expression of SOS bypass polymerases in the absence of AlkB increased m3C and e3C mutagenicity to 70%. The m1G adduct was very mutagenic (80%) in SOS–/AlkB– cells providing G to T (57%), G to A (17%), and G to C (6%) mutations, whereas m1G mutagenicity fell from 80% to 4% in SOS–/AlkB+ cells. In SOS+/AlkB– cells, the SOS bypass polymerases were slightly antimutagenic, yielding a mutation frequency of 66%. Incorporation of A opposite m1G was disfavored with respect to the normal polymerases, giving G to T (44%), G to A (19%), and G to C (4%) mutations. The m3T adduct was ≈60% mutagenic in SOS–/AlkB– cells, providing mostly T to A (47%) and T to C (9%) mutations, whereas the mutation frequency dropped to 40% in cells expressing AlkB (with the mutational specificities proportionately scaled-down). The SOS bypass polymerases were also somewhat anti-mutagenic, yielding a mutation frequency of 53% with a decrease in T to A mutations (to 42%) with respect to normal DNA polymerases. A genome was also constructed with oligonucleotides containing a nearly equal mixture of G, A, T, and C at the lesion site, which was transformed into E. coli and processed with the REAP assay. Because all bases were well represented (Fig. 5), there was neither bias in ligation, nor in any subsequent enzymatic steps of the REAP assay, assuring that each type of mutation was accurately represented. In fact, a reanalysis of the concentration of each oligonucleotide as determined by 32P-labeling revealed an exact match between the input “GATC” oligonucleotide mixture and the REAP output from the “GATC” genome, which was also used to generate 5′ [32P]dNMP TLC markers.
Fig. 5.
Mutagenesis of m1A, m3C, e3C, m1G, and m3T in E. coli. Genomes were constructed in triplicate, and each was transformed into the designated cell strain. The output from the REAP assay depicted in Fig. 2B from all lesions in the designated cell strains is reported with one standard deviation of error. The percentage of G (green), A (amber), T (turquoise), and C (crimson) at the lesion site reveals that the mutagenicity of the 3-alkylated cytosines and 1-methylguanine is expunged by AlkB, but exacerbated by SOS bypass polymerases for the 3-alkylated cytosines.
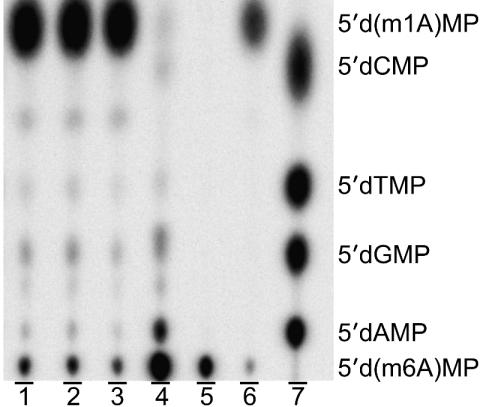
Integrity of m1A. 1-Methyldeoxyadenosine is known to be unstable, undergoing a base-catalyzed Dimroth rearrangement in which an N1 alkyl group migrates to the exocyclic N6 position of adenine (31). Matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry would not distinguish whether m1A within the oligonucleotide had converted to m6A during storage or under genome construction conditions, because both species would have the same mass for a given charged state. We therefore devised a means to verify lesion purity after genome construction. The method is similar to the REAP method (Fig. 2B Lower), which was performed on m1A genome constructs before their being introduced to cells; instead of separating 5′ dNMPs of G, A, T, and C, however, the 5′ d(lesion)MP was separated from its conversion products, if they were present. The TLC plate (Fig. 6) shows that the m1A genomes contained 96% m1A and 4% m6A at the lesion site just before electroporation (lanes 1–3). The Dimroth rearrangement was promoted in a control reaction whereby an 18-mer containing a 5′ m1A was liberated from the genome construct and incubated in concentrated aqueous ammonium hydroxide at 80°C for 3 h, resulting in 94% conversion to m6A and, interestingly, 6% conversion to A (lane 4). Lanes 5 and 6 are markers generated by labeling an oligonucleotide containing the appropriate 5′ lesion, followed by gel purification and nuclease P1 digestion. The 5′ d(m1A)MP migrates faster than 5′ d(m6A)MP on the weakly basic polyethyleneimine TLC plate, because it has one fewer negative charge at pH 5.8 [pKa of the N6 amino group of m1A is 9.3 (32)].
Fig. 6.
Integrity of m1A lesion: lesion integrity assay. This assay monitors lesion purity after genome construction, before passage through cells. This result is accomplished by annealing an oligonucleotide to span the lesion site, and applying the techniques used in Fig. 2B (starting from the BbsI digestion step). Unlike the REAP mutagenesis assay, which ultimately uses TLC to separate the four normal 5′ dNMPs at the lesion site after biological processing (providing the mutation frequency), the REAP lesion integrity assay separates the modified 5′ dNMP at the lesion site from its conversion products, if present, before biological processing. Lanes 1–3 are the analysis of triplicate m1A genome constructs, lane 4 is an induced Dimroth rearrangement, lanes 5 and 6 are markers made from oligonucleotides containing m6A or m1A lesions at the 5′ end, whereas lane 7 is a marker from a “GATC” genome.
Discussion
To provide an understanding of the impact of specifically alkylated DNA lesions on mutagenesis and repair by AlkB within cells, we placed m1A, m3C, e3C, m1G, and m3T at a unique site within a single-stranded vector, which was replicated in cells that were AlkB-deficient, wild-type, or AlkB-deficient expressing SOS bypass polymerases.
The m1A, m3C, and e3C lesions were strong blocks to replication (Fig. 3), which were removed in AlkB+ cells. This finding is in agreement with previous studies whereby AlkB had conferred increases in cell survival (33), single-stranded phage DNA reactivation (34), and primer extension (6) by using globally methylated substrates. Our discovery that the 3-ethyldeoxycytidine blockade is removed by AlkB further extends the substrate specificity of AlkB, which has recently been shown to repair 1-ethyldeoxyadenosine (7), and derivatives thereof, as well as the monophosphate 5′ d(m1A)MP (35). The removal of replication blockades in AlkB+ cells (Fig. 3) correlated well with the disappearance of mutation (Fig. 5), consistent with the lesions being repaired in single-stranded DNA before their being bypassed. We also demonstrate for these lesions that the widely used method for determining lesion genotoxicity, i.e., immediately plating cells transfected with lesion vs. lesion-free single-stranded genomes and comparing the difference in survival, tends to underestimate lethality (Fig. 3, open bars). A lesion can be an impediment to replication, but, if it is eventually traversed before cellular degradation of the genome inside a cell fixed in agar, a successful bypass survival event is recorded as a plaque. In contrast, growth of lesion and competitor genomes in solution allows for the accurate scoring of intermediate blocks to DNA polymerases, because a genome that contains a lesion that slows down replication has a competitive growth disadvantage.
Although studies involving replication past m1A and m1G by DNA polymerases are lacking, it was previously shown in vitro that m3C within a poly(dC) template inhibits replication by DNA polymerase I and does not cause mutation (14, 36, 37); however, in an in vitro study done under Mn2+ error-prone permissive conditions, some bypass was reported with the incorporation of dAMP and dTMP opposite m3C (14). In our study done in vivo, m3C was also genotoxic, and we saw this same dNTP incorporation preference in SOS–/AlkB– cells, with an appreciable mutation frequency of 30% even under normal growth conditions (Fig. 5). Mutations increased dramatically to 70% in SOS+/AlkB– cells, thus mirroring the in vitro study whereby dNTP incorporation selectivity was relaxed with Mn2+ (14). It is left to speculation why the replicative polymerase within E. coli is more mutagenic toward m3C than DNA polymerase I. Studies performed in vitro have also shown m3T to be a powerful block to the Klenow fragment of DNA polymerase I, which promotes a “slight” increase in dTTP incorporation by using a poly(dC-dm3T) template (38); however, exclusive incorporation of T opposite the analogous 3-ethyldeoxythymidine adduct is observed from the translesion synthesis product (39). Our results of m3T genotoxicity and mutagenicity performed in vivo are in general agreement with these in vitro findings, as the T to A mutation was substantial at 47% in SOS–/AlkB– cells (although we additionally detected T to C mutations at 9%). Furthermore, the T to A mutation dropped to 32% in cells expressing normal levels of AlkB, suggestive of AlkB repairing m3T (although poorly).
The fact that AlkB could not appreciably remove the m3T blockade but could still manifest a drop in T to A transversions can be explained if the lesion is a very strong block to replication, which can be partially overcome when the lesion within single-stranded DNA is partially repaired by AlkB. In line with this notion, the distribution of noncomplementary bases was proportionately scaled down in AlkB+ cells. This argument also holds for m1G, which seems to be a better substrate for AlkB than m3T, because m1G bypass increased 8-fold from 2% in AlkB– cells to 16% in AlkB+ cells (Fig. 3), with a remarkable reduction in mutations from 80% in AlkB– cells to 4% in AlkB+ cells (Fig. 5).
Methyl methanesulfonate mutagenesis of F′ episomes within alkB E. coli demonstrates a small, but reproducible, increase in G:C to A:T, G:C to T:A, and A:T to T:A base substitutions (34) with respect to wild-type E. coli. Consistent with our site-specific mutagenesis results, the G:C to A:T mutation could result from m1G:T or A:m3C pairings, whereas the G:C to T:A mutation may arise from m1G:A or T:m3C pairings. Because m1A was not very mutagenic (≈1% in AlkB–/SOS– cells, Figs. 4 and 5), the A:T to T:A mutation is ascribed to m3T.
The primary role of AlkB for m1A lesions seems to be the prevention of genotoxicity, because m1A was not very mutagenic; however, the appreciable mutagenesis of the 3-alkylcytosine and m1G lesions ascribes AlkB as defender against both genotoxicity and mutagenicity from alkylative genomic insults. Lesions in this study consisted of positively charged bases [the ribonucleosides of m1A and m3C have respective pKas of 9.3 and 9.6 (32)], and neutral bases (m1G and m3T). Because m1G (and to a lesser extent, m3T) was repaired by AlkB, a formal positive charge on the base is not a prerequisite for AlkB to function. However, lesions that contained a positive charge were repaired better than the uncharged bases. At this point, we do not know whether this result is due to better recognition of positively charged bases by means of electrostatics with AlkB, or whether the positively charged base simply makes the DNA a better leaving group after hydroxylation of the methyl group.
Although the charged 3-alkylcytosine lesions exhibited enhanced mutagenicity under SOS-induced conditions (Fig. 5, increasing from 30% to 70%), induction of SOS bypass polymerases seemed to be somewhat anti-mutagenic toward the uncharged m1G and m3T lesions; mutagenicity for m1G fell from 80% in SOS– cells to 66% in SOS+ cells, whereas for m3T fell from 60% in SOS– cells to 53% in SOS+ cells. These uncharged lesions were, incidentally, also stronger blocks to the replicative DNA polymerases than were the positively charged m1A, m3C, and e3C lesions.
The potently genotoxic and mutagenic m1G lesion yields both G to T and G to A mutations, which are those most widely observed in organisms subjected to alkylation damage. It remains to be seen whether other repair systems can clear m1G, as well as the 3-alkylcytosine, m1A, and m3T lesions.
Supplementary Material
Acknowledgments
We thank William L. Neeley for statistical advice concerning the bypass assay. This work was supported by National Institutes of Health Grants CA80024 and ES11399.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: m1A, 1-methyldeoxyadenosine; m3C, 3-methyldeoxycytidine; e3C, 3-ethyldeoxycytidine; m1G, 1-methyldeoxyguanosine; m3T, 3-methyldeoxythymidine; CRAB, competitive replication of adduct bypass; REAP, restriction endonuclease and postlabeling analysis of mutation frequency.
Note. While this manuscript was in preparation, a paper (40) appeared demonstrating that m3T is subject to modest repair by AlkB in vitro, which is in agreement with what we found in vivo. Also, in addition to m1A, we performed the lesion integrity assay on m1G, m3T, m3C, and e3C genomes. Although m1G and m3T gave predominantly one 5′ [32P]d(lesion)MP spot, the m3C genome gave 9%, and the e3C genome gave 3% of a slower moving 5′ [32P]d(converted lesion)MP spot. This minor amount of conversion was most likely due to deamination during deprotection to give 3-methyldeoxyuridine and 3-ethyldeoxyuridine (41). Accordingly, we have avoided reaching a conclusion on whether AlkB works better on e3C than on m3C. If formed, the small amount of 3-alkyluridine (the deamination product of 3-alkylcytosine) has no bearing on any of our major findings.
References
- 1.Sassanfar, M., Dosanjh, M. K., Essigmann, J. M. & Samson, L. (1991) J. Biol. Chem. 266, 2767–2771. [PubMed] [Google Scholar]
- 2.Volkert, M. R. (1988) Environ. Mol. Mutagen. 11, 241–255. [DOI] [PubMed] [Google Scholar]
- 3.Sedgwick, B. & Lindahl, T. (2002) Oncogene 21, 8886–8894. [DOI] [PubMed] [Google Scholar]
- 4.Thomas, L., Yang, C. H. & Goldthwait, D. A. (1982) Biochemistry 21, 1162–1169. [DOI] [PubMed] [Google Scholar]
- 5.Trewick, S. C., Henshaw, T. F., Hausinger, R. P., Lindahl, T. & Sedgwick, B. (2002) Nature 419, 174–178. [DOI] [PubMed] [Google Scholar]
- 6.Falnes, P. O., Johansen, R. F. & Seeberg, E. (2002) Nature 419, 178–182. [DOI] [PubMed] [Google Scholar]
- 7.Duncan, T., Trewick, S. C., Koivisto, P., Bates, P. A., Lindahl, T. & Sedgwick, B. (2002) Proc. Natl. Acad. Sci. USA 99, 16660–16665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aas, P. A., Otterlei, M., Falnes, P. O., Vagbo, C. B., Skorpen, F., Akbari, M., Sundheim, O., Bjoras, M., Slupphaug, G., Seeberg, E., et al. (2003) Nature 421, 859–863. [DOI] [PubMed] [Google Scholar]
- 9.Kataoka, H., Yamamoto, Y. & Sekiguchi, M. (1983) J. Bacteriol. 153, 1301–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Begley, T. J. & Samson, L. D. (2003) Trends Biochem. Sci. 28, 2–5. [DOI] [PubMed] [Google Scholar]
- 11.Aravind, L. & Koonin, E. V. (2001) Genome Biol. 2, research0007.1–0007.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singer, B. & Grunberger, D. (1983) in Molecular Biology of Mutagens and Carcinogens (Plenum, New York), pp. 45–96.
- 13.Beranek, D. T., Weis, C. C. & Swenson, D. H. (1980) Carcinogenesis 1, 595–606. [DOI] [PubMed] [Google Scholar]
- 14.Boiteux, S. & Laval, J. (1982) Biochimie 64, 637–641. [DOI] [PubMed] [Google Scholar]
- 15.Gomes, J. D. & Chang, C. J. (1983) Anal. Biochem. 129, 387–391. [DOI] [PubMed] [Google Scholar]
- 16.Chang, C. J., Gomes, J. D. & Byrn, S. R. (1983) J. Org. Chem. 48, 5151–5160. [Google Scholar]
- 17.Kawasaki, H., Ninomiya, S. & Yuki, H. (1985) Chem. Pharm. Bull. 33, 1170–1174. [DOI] [PubMed] [Google Scholar]
- 18.Ashworth, D. J., Baird, W. M., Chang, C. J., Ciupek, J. D., Busch, K. L. & Cooks, R. G. (1985) Biomed. Mass. Spectrom. 12, 309–318. [DOI] [PubMed] [Google Scholar]
- 19.Culp, L. A., Dore, E. & Brown, G. M. (1970) Arch. Biochem. Biophys. 136, 73–79. [DOI] [PubMed] [Google Scholar]
- 20.Margison, G. P., Margison, J. M. & Montesano, R. (1976) Biochem. J. 157, 627–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frei, J. V., Swenson, D. H., Warren, W. & Lawley, P. D. (1978) Biochem. J. 174, 1031–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faustman, E. M. & Goodman, J. I. (1980) J. Pharmacol. Methods 4, 305–312. [DOI] [PubMed] [Google Scholar]
- 23.Beranek, D. T., Heflich, R. H., Kodell, R. L., Morris, S. M. & Casciano, D. A. (1983) Mutat. Res. 110, 171–180. [DOI] [PubMed] [Google Scholar]
- 24.Faustman-Watts, E. M. & Goodman, J. I. (1984) Biochem. Pharmacol. 33, 585–590. [DOI] [PubMed] [Google Scholar]
- 25.Den Engelse, L., Menkveld, G. J., De Brij, R. J. & Tates, A. D. (1986) Carcinogenesis 7, 393–403. [DOI] [PubMed] [Google Scholar]
- 26.Kondo, H., Nakabeppu, Y., Kataoka, H., Kuhara, S., Kawabata, S. & Sekiguchi, M. (1986) J. Biol. Chem. 261, 15772–15777. [PubMed] [Google Scholar]
- 27.Wei, Y. F., Chen, B. J. & Samson, L. (1995) J. Bacteriol. 177, 5009–5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delaney, J. C. & Essigmann, J. M. (1999) Chem. Biol. 6, 743–753. [DOI] [PubMed] [Google Scholar]
- 29.Tan, X., Suzuki, N., Grollman, A. P. & Shibutani, S. (2002) Biochemistry 41, 14255–14262. [DOI] [PubMed] [Google Scholar]
- 30.Bailey, E. A., Iyer, R. S., Harris, T. M. & Essigmann, J. M. (1996) Nucleic Acids Res. 24, 2821–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Engel, J. D. (1975) Biochem. Biophys. Res. Commun. 64, 581–586. [DOI] [PubMed] [Google Scholar]
- 32.Kettani, A., Gueron, M. & Leroy, J.-L. (1997) J. Am. Chem. Soc. 119, 1108–1115. [Google Scholar]
- 33.Chen, B. J., Carroll, P. & Samson, L. (1994) J. Bacteriol. 176, 6255–6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dinglay, S., Trewick, S. C., Lindahl, T. & Sedgwick, B. (2000) Genes Dev. 14, 2097–2105. [PMC free article] [PubMed] [Google Scholar]
- 35.Koivisto, P., Duncan, T., Lindahl, T. & Sedgwick, B. (2003) J. Biol. Chem. 278, 44348–44354. [DOI] [PubMed] [Google Scholar]
- 36.Abbott, P. J. & Saffhill, R. (1979) Biochim. Biophys. Acta 562, 51–61. [DOI] [PubMed] [Google Scholar]
- 37.Saffhill, R. (1984) Carcinogenesis 5, 691–693. [DOI] [PubMed] [Google Scholar]
- 38.Huff, A. C. & Topal, M. D. (1987) J. Biol. Chem. 262, 12843–12850. [PubMed] [Google Scholar]
- 39.Grevatt, P. C., Donahue, J. M. & Bhanot, O. S. (1991) J. Biol. Chem. 266, 1269–1275. [PubMed] [Google Scholar]
- 40.Koivisto, P., Robins, P., Lindahl, T. & Sedgwick, B. (July 20, 2004) J. Biol. Chem., 10.1074/jbc.M407960200. [DOI] [PubMed]
- 41.Leutzinger, E. E., Miller, P. S. & Kan, L. S. (1982) Biochim. Biophys. Acta 697, 243–251. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.