Abstract
FKHRL1 (FOXO3a) and p53 are two potent stress-response regulators. Here we show that these two transcription factors exhibit “crosstalk” in vivo. In response to DNA damage, p53 activation led to FKHRL1 phosphorylation and subcellular localization change, which resulted in inhibition of FKHRL1 transcription activity. AKT was dispensable for p53-dependent suppression of FKHRL1. By contrast, serum- and glucocorticoid-inducible kinase 1 (SGK1) was significantly induced in a p53-dependent manner after DNA damage, and this induction was through extracellular signal-regulated kinase 1/2-mediated posttranslational regulation. Furthermore, inhibition of SGK1 expression by a small interfering RNA knockdown experiment significantly decreased FKHRL1 phosphorylation in response to DNA damage. Taken together, our observations reveal previously unrecognized crosstalk between p53 and FKHRL1. Moreover, our findings suggest a new pathway for understanding aging and the age dependency of human diseases governed by these two transcription factors.
Human longevity depends on genome stability. Several mouse models have revealed that an age-related decrease of DNA repair or an increase in DNA damage plays a role in mammalian aging (1–3). The idea that cellular responses to stress may be important in aging is supported by studies of p53, the functions of which are critical for both the apoptotic and senescence responses to DNA damage, telomeric shortening, and oxidative stress (4–6).
The mammalian FOXO family of forkhead transcription factors (including FKHR, FKHRL1, and AFX) have been proposed as antiaging factors based on evidence from their orthologues, DAF-16 in Caenorhabditis elegans and dFOXO in Drosophila melanogaster, which regulate longevity in response to reduced insulin/insulin-like growth factor I (IGF-I) signaling or by overexpressing constitutively active FOXO (7, 8). Growth factor signaling to FOXO family members through phosphatidylinositol 3-kinase (PI3K) and its downstream kinase, Akt, has been found to be evolutionarily conserved for FOXO phosphorylation, subcellular translocation, and inhibition of its transcriptional activity (9). However, the role of FOXO in response to DNA damage, as well as the signaling pathway upstream of FOXO, is, as yet, unclear. Overexpression of FKHRL1 can protect cells from oxidative stress-induced cell death, as described in refs. 10–12. Induction of a number of antioxidant enzymes and stress-related gene products has been proposed as a potential mechanism (9, 13–15). The fact that the precise biological consequences of FOXO activation are cell-type-specific and stress-type-dependent suggests that there might be crosstalk between FOXO and other stress regulators. How FOXO communicates and coordinates with other signaling pathways in response to genotoxic stress remains unknown.
The opposing functions of p53 and FKHRL1 with regard to the aging process suggest that a regulatory mechanism might exist to integrate these two signaling pathways. We sought to elucidate the mechanism underlying this crosstalk and its regulation in the hopes of better understanding how these two transcription factors communicate and coordinate with each other in controlling cell fate in response to genotoxic stress.
Experimental Procedures
Cell Culture. E1A/H-ras-V12-transformed p53+/+ and p53–/–mouse embryonic fibroblasts (MEFs) were kind gifts from Scott Lowe (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY). p53QS primary MEFs were a gift of Geoffrey Wahl (The Salk Institute for Biological Studies, San Diego). MEFs were cultured in DMEM (GIBCO) supplemented with 10% FCS (GIBCO). Primary MEFs were transformed with pLPC ras/E1A (a gift from Athena Lin, Roswell Park Cancer Institute, Buffalo, NY) and then subjected to puromycin selection. Pharmacological inhibitors PD98059 (50 μM), LY294002 (10 μM), and SB203580 (20 μM) were purchased from Calbiochem.
Constructs and Antibodies. The pSIRIPP retroviral small interfering RNA (siRNA) vector (a gift from Tyler Jacks, Massachusetts Institute of Technology, Cambridge) is described in ref. 16. Oligonucleotide sequences for murine serum- and glucocorticoid-inducible kinase 1 (SGK1) siRNA are available on request. The pBabe Akt-KD, human WT p53, p53R175H, and p53R273H expression plasmids were kindly provided by Arnold Levine (Institute for Advanced Study, Princeton). pECE-HA-FKHRL1, HA-FKHRL1-TM expression plasmids, and the anti-SGK antibody were generously provided by Michael Greenberg (Harvard Medical School, Boston). Anti-p53 antibody (FL393, Santa Cruz Biotechnology), anti-p21 and anti-p27 antibodies (BD Biosciences), antibodies against total FKHRL1 or phospho-Thr-32 FKHRL1 (Upstate Biotechnology, Lake Placid, NY), anti-total-Akt or phospho-Akt antibodies (Cell Signaling Technology, Beverly, MA), and anti-total-extracellular signal-regulated kinase (ERK) 1/2 or phospho-ERK1/2 antibodies (Cell Signaling Technology) were purchased commercially.
Retroviral Infection. Phoenix cells were transfected with indicated retroviral constructs by calcium chloride transfection. At 24 h posttransfection, conditioned medium recovered from cultures of transfected Phoenix cells was filtered through a 0.45-μm filter and added to the MEF cultures. Spin infection was performed at 1,400 × g for 90 min. Cells were then cultured for ≈20 h and then subjected to puromycin selection.
Chromatin Immunoprecipitation (ChIP) Assays. ChIP assays were performed by using an Acetyl-Histone H3 Immunoprecipitation Assay Kit (17-245, Upstate Biotechnology). Briefly, 107 cells were fixed with 1% formaldehyde and then washed, harvested in SDS lysis buffer, and sonicated. Lysates containing soluble chromatin were prepared and incubated overnight with 2.5 μg of anti-FKHR antibody (H-144, Santa Cruz Biotechnology). DNA–protein immunocomplexes were collected with protein A-agarose beads, washed, eluted from the beads, and then incubated with sodium chloride (final concentration of 0.2 M) at 65°C for 4 h to reverse crosslink DNA–protein complex. Two microliters of proteinase K (10 mg/ml) was added to the samples and incubated for 1 h at 45°C. DNA samples were then purified with phenol/chloroform and precipitated with ethanol. Murine p27 ChIP primer sequences are available on request. Cyclin G2 primers were as described in ref. 17.
Immunofluorescent Staining. p53–/–MEFs were plated onto glass coverslips at a density of 3 × 105 per well in six-well dishes. The cells were transfected by using FuGENE reagent (Roche Diagnostics) with 1.5 μg of HA-FKHRL1 plus 7.5 μg of various p53 constructs. At 24 h posttransfection, cells were exposed to UV radiation (30 J/m2). Cells were harvested at 8 h postexposure, fixed for 10 min in 4% paraformaldehyde at room temperature, and permeabilized with 0.1% Triton X-100 for 5 min. Coverslips were washed with PBS, and nonspecific antibody binding sites were blocked by incubation with PBS containing 3% BSA. The coverslips were then incubated with anti-HA antibody diluted in PBS-BSA for 1 h and washed five times with PBS. Cells were then incubated for 1 h with a secondary antibody (Alexa Fluor 568 goat anti-mouse IgG, Molecular Probes) diluted in PBS-BSA containing 4′,6-diamidino-2-phenylindole. After extensive washes in PBS, coverslips were mounted and examined under a Zeiss LSM 510 META NLO two-photon fluorescence microscope. For quantification, 80–100 cells per coverslip were counted.
Results
DNA Damage Induces p53-Dependent Inhibition of FKHRL1. To investigate the potential for crosstalk between p53 and FKHRL1 in response to genotoxic stress, we treated transformed WT and p53–/–MEFs with various DNA-damaging agents that are known to activate p53 and then examined the status of FKHRL1. After UV radiation or etoposide treatment, the phosphorylation of FKHRL1 at Thr-32 increased significantly in WT MEFs but not in p53–/–MEFs (Fig. 1A). We also tested γ irradiation and found p53-dependent phosphorylation of FKHRL1 in response to γ irradiation (data not shown). To exclude the possibility that the increase in FKHRL1 phosphorylation in DNA-damaged WT cells was simply a consequence of more apoptosis in these cells, we treated WT and p53–/–MEFs with TNF-α. TNF-α has been reported to induce p53-independent apoptosis (18). Flow cytometric analysis confirmed that treatment with UV radiation or etoposide, but not TNF-α, triggered p53-dependent apoptosis in WT MEFs (Fig. 1B). By contrast, the majority of p53–/–cells arrested at the G2/M stage in response to all stress stimuli except TNF-α. Despite the fact that p53 is dispensable for TNF-α-induced apoptosis, FKHRL1 phosphorylation in TNF-α-treated cells increased in the presence of p53 but not in p53–/–cells (Fig. 1A). These data suggested that, in response to DNA damage, p53 regulates FKHRL1 by increasing its phosphorylation.
Fig. 1.
p53 negatively regulates FKHRL1 in response to genotoxic stress. (A) Increased phosphorylation of FKHRL1 after DNA damage in the presence of p53. Transformed WT and p53–/–MEFs were treated with UV radiation (30 J/m2), etoposide (0.5 μM), or TNF-α (10 ng/ml). Cell extracts were analyzed at the indicated time points by immunoblotting with antibodies directed against p53, p21, and phosphorylated FKHRL1. Actin was used as the loading control. For all figures, results shown are representative of at least three independent experiments. (B) p53-dependent and -independent apoptosis and G2/M arrest caused by various stimuli. Transformed WT and p53–/–MEFs were treated for 24 h with the same DNA-damaging agents as in A, followed by propidium iodide (PI) staining and fluorescence-activated cell sorter analysis. Data are shown for the sub-G1 (apoptosis) (Left) and G2–M(Right) phases. Data are the means and variances for two independent experiments performed in duplicate. (C) Nuclear exclusion of FKHRL1 induced by p53 in response to DNA damage. p53–/–MEFs were cotransfected with an expression plasmid encoding HA-tagged WT FKHRL1 plus a plasmid encoding WT p53, p53R175H, or p53R273H. At 24 h posttransfection, cells were treated with UV radiation (30 J/m2) for 8 h. FKHRL1 was detected by immunofluorescent staining with anti-HA antibody. Nuclear DNA was stained with 4′,6-diamidino-2-phenylindole. The images were merged to detect nuclear localization/exclusion of FKHRL1. Results shown are one trial representative of three experiments. Quantification of the means and variances from three experiments is shown. (D) Decreased p27 expression in WT cells after DNA damage treatment. WT and p53–/–MEFs were exposed to UV radiation (30 J/m2) for 10 h or etoposide (0.5 μM) for 5 or 10 h. Cell lysates were analyzed by Western blotting with antibodies against p27 and actin. (E) Reduced association of FKHRL1 with the endogenous p27 and cyclin G2 promoters in the presence of p53 upon UV radiation. ChIP assays were performed on WT or p53–/–MEFs left untreated or treated with UV radiation (30 J/m2) for 10 h. PCR was conducted by using primers flanking murine p27 or cyclin G2 promoter regions containing FKHRL1-binding motifs. DNA templates from a protein–DNA complex immunoprecipitated with either FKHRL1-specific antibody (Upper) or no antibody (Lower, –Ab) were used. Input DNA was amplified for normalization (Lower).
Phosphorylated FKHRL1 translocates into the cytoplasm, whereas unphosphorylated FKHRL1 remains in the nucleus. To determine the subcellular localization of FKHRL1 after DNA damage, we cotransfected p53–/–MEFs with HA-tagged FKHRL1 plus either WT p53 or one of two p53 mutants, p53R175H or p53R273H, both of which are hot-spot mutations found in the human p53 DNA-binding domain. At 24 h posttransfection, the transfected cells were exposed to UV radiation (30 J/m2) for 8 h. Immunofluorescent staining showed that the overexpression of WT p53, but not the two mutant p53s, significantly increased the cytoplasmic translocation of FKHRL1 after UV radiation exposure (Fig. 1C).
The state of phosphorylation of FKHRL1 and its subcellular localization are critical for its transcriptional activity. Because only WT p53 promoted the exit of FKHRL1 from the nucleus of UV-irradiated cells, we expected that p53-dependent phosphorylation and cytoplasmic localization of FKHRL1 might lead to a reduction in FKHRL1-dependent transcription. p27 has been identified as a direct downstream target of FKHRL1. We examined levels of endogenous p27 expression before and after DNA damage to see whether they correlated with FKHRL1 phosphorylation. Indeed, p27 protein decreased with time after UV radiation or etoposide treatment in WT MEFs but not in p53–/–cells (Fig. 1D). To determine whether p53 regulated the DNA-binding activity of FKHRL1, we performed ChIP assays by using primers flanking the FKHRL1-binding site in the p27 promoter region. FKHRL1 association with the p27 promoter in vivo decreased dramatically after UV damage in WT MEFs but not in p53–/–cells (Fig. 1E), indicating that p53 can inhibit native FKHRL1-targeted promoters. Examination by ChIP assay of the association of FKHRL1 with another FKHRL1 downstream target gene, cyclin G2, showed that there was a general inhibition of FKHRL1 DNA-binding capacity by p53 after DNA damage. Taken together, our data suggested that activation of p53 after DNA damage could increase the phosphorylation and nuclear exclusion of FKHRL1, leading to inhibition of its transcriptional activity.
Regulation of FKHRL1 by p53 Is Akt-Independent. To investigate the molecular mechanism underlying the p53-mediated inhibition of FKHRL1, we evaluated the status of two kinases, Akt and SGK1, that had been previously reported as responsible for FKHRL1 phosphorylation in response to growth factor stimuli (19). However, there was no change in the phosphorylation of Akt in WT or p53–/–MEFs after either UV or γ radiation treatment (Fig. 2A). To further rule out a role for Akt in FKHRL1 regulation by p53, we used retroviral infection to introduce into WT MEFs a kinase-dead form of Akt (Akt-DN) that can also inhibit endogenous Akt activity (20). A stable Akt-DN cell line and the corresponding retroviral control line were generated by using puromycin selection and were subjected to UV radiation. Compared with the introduction of the empty retroviral vector, expression of Akt-DN did not reduce the phosphorylation of FKHRL1 after UV damage (Fig. 2B). These results indicated that Akt was not involved in the p53-dependent phosphorylation of FKHRL1 that occurs in response to genotoxic stress.
Fig. 2.
Regulation of FKHRL1 by p53 in response to genotoxic stress is Akt-independent. (A) Unaltered Akt phosphorylation after DNA damage. WT and p53–/–MEF cells were treated with UV (30 J/m2) or γ (10 Gy) radiation for 10 or 24 h. Total Akt and phospho-Akt were assessed by Western blotting. (B) Akt-DN failed to reduce FKHRL1 phosphorylation after UV irradiation. p53+/+ stable MEF cell lines expressing either the control pBabe vector or the pBabe Akt-KD vector were treated with UV radiation (30 J/m2) for 8 h. Western blotting of cell lysates was carried out by using antibodies against phospho-FKHRL1, p53, and actin.
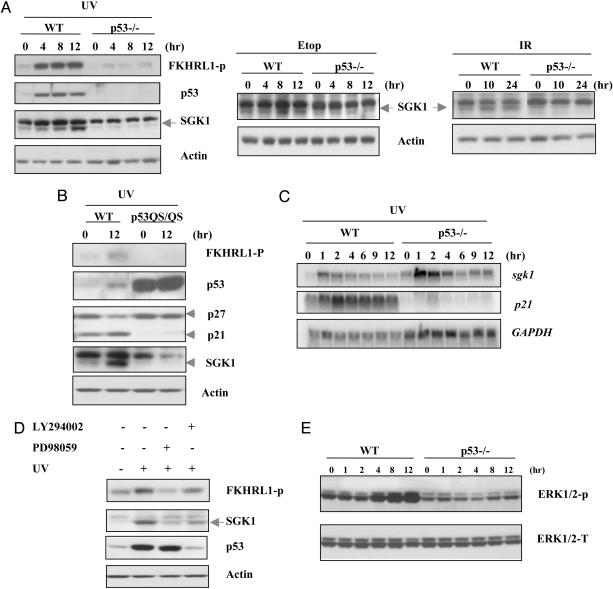
p53 Up-Regulates SGK1 Expression in Response to DNA Damage Through the ERK1/2 Signaling Pathway. In contrast to unchanged total or phosphorylated AKT levels, there was a significant induction of SGK1 in WT (but not in p53–/–) MEFs after UV radiation, etoposide, or γ radiation treatment (Fig. 3A). To explore the p53 dependence of SGK1 induction, we took advantage of a mutant p53 called Trp-53L25Q,W26S (referred to as p53QS) that contains two point mutations in the transactivation domain and displays constitutive nuclear localization but is transcriptionally inactive (21, 22). Transformed mutant MEFs bearing a knock-in mutation of p53QS were treated with UV radiation for 12 h, and the induction of various molecules was examined. Although phosphorylated FKHRL1 and total SGK1 were induced in UV-treated WT MEFs, neither molecule could be detected in p53QS knock-in MEFs (Fig. 3B). This result indicated that p53 transactivation domain was required for induction of SGK1 in response to UV irradiation.
Fig. 3.
p53-dependent induction of SGK1 in response to DNA damage through the ERK1/2 pathway. (A) WT and p53–/–MEFs were treated with UV radiation (30 J/m2), etoposide (Etop, 0.5 μM), or γ radiation (10 Gy). Western blotting of cell lysates was carried out to detect phospho-FKHRL1 (which only showed in UV treatment), p53, and SGK1 at the indicated time points. Actin was used as the loading control. (B) p53 transactivation domain was required for SGK1 induction. Transformed WT and p53QS/QS MEFs were exposed to UV radiation (30 J/m2). Cell lysates were prepared 12 h after treatment, and Western blotting was performed by using antibodies against phospho-FKHRL1, p53, p27, p21, and SGK1. (C) p53-dependent SGK1 induction in response to UV radiation is through posttranslational regulation. WT and p53–/–MEFs were exposed to UV radiation (30 J/m2) for indicated times, and Northern blotting of total RNA was carried out to detect murine sgk1 and p21 transcripts. Full-length murine sgk1 was used as a hybridization probe. GAPDH was used as the loading control. (D) ERK1/2 dependence of posttranslational SGK1 induction by p53. WT MEFs were pretreated with the PI3K inhibitor LY294002 (10 μM) or the MEK1 inhibitor PD98059 (50 μM) for 1 h before treatment with UV radiation (30 J/m2) for another 8 h. Cell lysates were analyzed by Western blotting by using antibodies specifictothe indicated proteins. (E) Time course of ERK1/2 activation after UV damage (30 J/m2) in WT and p53–/–MEFs. Cell lysates were prepared at the indicated time points after UV exposure, and phosphorylated ERK (ERK1/2-P) was detected by using phospho-specific anti-ERK antibody. Total ERK (ERK1/2K-T) was assessed as the loading control.
We next determined whether p53-dependent induction of SGK1 was through direct transcriptional regulation or posttranslational regulation. WT and p53–/–MEFs were subjected to UV radiation for a total of 12 h, and RNA samples collected at various points during the exposure were examined by Northern blotting. Consistent with previous reports that UV treatment can induce SGK1 transcription (23), we observed an immediate induction of SGK1 mRNA within 1 h of UV treatment in both WT and p53–/–MEFs (Fig. 3C). The level of SGK1 mRNA then gradually declined. Given that the p53 transactivation domain was required for SGK1 up-regulation after UV irradiation but that the kinetics of SGK1 mRNA induction were no different in WT and p53–/–MEFs, these results suggested that p53-dependent induction of SGK1 after UV treatment was through posttranslational regulation.
To elucidate the signaling pathway mediating p53-dependent regulation of SGK1, we tested several pharmacological inhibitors, including the PI3K inhibitor LY294002, the MEK1 inhibitor PD98059, and the p38 inhibitor SB203580, for their effects on SGK1 induction in WT MEFs. PD98059 blocked SGK1 expression almost completely, whereas LY294002 and SB203580 exhibited partial impairments (Fig. 3D). Because inhibition of PI3K or p38 also impairs p53 accumulation, the decreased SGK1 induction in the presence of LY294002 or SB203580 may be secondary to p53 inhibition. However, the blocking of ERK1/2 signaling not only inhibited SGK1 induction but also significantly decreased phosphorylation of FKHRL1 (Fig. 3D). To confirm that ERK1/2 was involved in p53-dependent SGK1 induction, we examined the kinetics of ERK1/2 phosphorylation after UV radiation damage. We found a significant difference in ERK1/2 phosphorylation in WT and p53–/–cells. In WT cells, ERK1/2 dramatically activated 4 h after UV radiation treatment, and the kinetics of ERK1/2 activation correlated well with those of SGK1 induction in WT MEFs. In contrast, there was only a slight activation of ERK1/2 in p53–/–MEFs (Fig. 3E). Taken together, these results show that activation of p53 upon DNA damage leads to induction of the SGK1 kinase via the ERK1/2 pathway. Given that SGK1 has been reported as an upstream kinase that could phosphorylate FKHRL1 in vitro, we hypothesized that p53-dependent inhibition of FKHRL1 after DNA damage might be SGK1-dependent.
Knockdown of SGK1 by SiRNA Impairs p53-Dependent FKHRL1 Phosphorylation. To elucidate the role of SGK1 as a p53 effector molecule that regulates FKHRL1, we introduced SGK1 siRNA into WT MEFs. To this end, we designed two pairs of oligonucleotides, namely con siRNA and SGK1 siRNA. We established two stable MEF lines expressing con siRNA or SGK1 siRNA and treated these cells with IGF-I. Northern blot analysis and RT-PCR showed that SGK1 mRNA was significantly reduced in SGK1 siRNA cells but not in con siRNA cells with or without IGF-I stimulation (Fig. 4A and data not shown). To test the specificity of SGK1 siRNA oligonucleotides, we measured expression levels of cytokine-independent survival kinase (CISK), another SGK family member that had been identified previously as an FKHRL1 upstream kinase in vitro (24), in con siRNA and SGK1 siRNA stable MEFs by RT-PCR. Neither siRNA oligonucleotides affected the CISK expression level, indicating that the SGK1 siRNA specifically targeted SGK1 (Fig. 4B). More intriguingly, phosphorylation of FKHRL1 in SGK1 siRNA cells was largely blocked when cells were exposed to UV radiation or etoposide treatment (Fig. 4C). These data imply that SGK1 is the critical kinase responsible for FKHRL1 phosphorylation after DNA damage.
Fig. 4.
Essential role of SGK1 in p53-dependent inhibition of FKHRL1 in response to genotoxic stress. (A) Reduced SGK1 mRNA expression by SGK1 siRNA. Northern blot was performed by using total RNA isolated from con siRNA and SGK1 siRNA cells before and after exposure to IGF-I. (B) Specificity of SGK1 siRNA. RT-PCR of CISK mRNA from con siRNA or SGK1 siRNA stable MEF cells was carried out by using two independent primers (A and B) specific for murine CISK. hprt was used as the loading control. (C) Knockdown SGK1 by SGK1 siRNA impaired p53-dependent FKHRL1 phosphorylation upon DNA damage treatment. WT, con siRNA, and SGK1 siRNA stable MEF cells were exposed to UV radiation (30 J/m2) or etoposide (0.5 μM) for 8 h. Phospho-FKHRL1, p53, and actin expression were analyzed by Western blotting. Results shown are one trial representative of three independent experiments. (D) Signaling pathway of p53-dependent regulation of FKHRL1 in response to DNA damage.
In summary, we reported evidence that activation of p53 upon DNA damage led to increased expression of SGK1 kinase via the ERK1/2 pathway. This p53-dependent up-regulation of SGK1 induced FKHRL1 phosphorylation after DNA damage treatment and subsequent nuclear exclusion, which led to suppression of FKHRL1 transcriptional activity (Fig. 4D).
Discussion
The forkhead transcription factor FKHRL1 serves as a substrate for both SGK1 and Akt, two kinases with many similarities. A question thus arises: Why should there be two families of PI3K-regulated kinases that phosphorylate FKHRL1? A clue may lie in in vitro studies showing that SGK1 and Akt preferentially phosphorylate FKHRL1 at overlapping but not identical sites (19), suggesting that SGK1 and Akt have complementary, rather than redundant, roles in promoting cell survival. Our demonstration in this study that genotoxic stress that induces SGK1 has no effect on Akt phosphorylation indicates that these two kinases are regulated by different pathways in a cell-type- and stress-specific manner. Interestingly, a recent report from C. elegans found that SGK-1, not AKT-1 or AKT-2, is the crucial kinase for the regulation of life span and stress response through regulating the phosphorylation, intracellular localization, and activity of DAF-16 (25). In our study, we show that the stress-response function of SGK1 through FOXO factors is conserved between worms and mammals.
In our system, we tested Thr-32, a common site of FKHRL1 that could be phosphorylated by both SGK1 and Akt. Our data showed that Akt was dispensable for p53-dependent inhibition of FKHRL1 in response to genotoxic stress. In contrast, WT MEFs (but not p53 null cells) showed SGK1 induction upon treatment with DNA-damaging agents. Northern blots showed that this p53-dependent up-regulation of SGK1 was not directed through transcriptional regulation by p53 where the p53 transactivation domain was required. Pharmacological inhibitor experiments revealed that signaling via the ERK1/2 pathway led to SGK1 induction. p53-dependent activation of ERK1/2 in response to DNA damage has been previously reported in regulation of the NF-κB pathway (26). Thus, we suggest that p53 activation leads to ERK1/2 signaling that induces SGK1 and that up-regulated SGK1, in turn, phosphorylates FKHRL1, thereby inhibiting FKHRL1 transcriptional activity. To test this hypothesis, we examined FKHRL1 phosphorylation in cells in which the SGK1 expression had been inhibited. Acute loss of SGK1 induced by siRNA knockdown experiments largely reduced FKHRL1 phosphorylation in response to both UV radiation and etoposide treatment. Thus, SGK1 appears to be the key kinase responsible for p53-dependent FKHRL1 phosphorylation in vivo for the cell types and stress examined in this study.
Recent studies find there are striking similarities between p53 and FKHRL1. Both of these transcription factors can be modified by acetyl transferases such as p300/CBP and deacetylases such as Sirt1, the mammalian homologue of yeast sirt2 (27–29). p53 and FKHRL1 also have downstream target genes in common, including GADD45, Wip1, and the Fas ligand (15). Lastly, up-regulation of either p53 or FKHRL1 leads to cell-cycle arrest or apoptosis, depending on cellular context and type of stress. These findings suggest that p53 may parallel in its functions with FOXO3a, and our data clearly demonstrate that these two transcription factors crosstalk with each other. p53 inhibits FKHRL1 transcription activity in vivo after stress treatment through the SGK1 protein kinase. The biological consequence of this regulation is currently under investigation.
Acknowledgments
We thank A. Levine, M. Greenberg, G. Wahl, S. Lowe, T. Jacks, B. Burgering (University Medical Center Utrecht, Utrecht, The Netherlands), and S. Benchimol (Ontario Cancer Institute, Toronto) for reagents; S. Benchimol and V. Stambolic for critical discussion; and M. Saunders for scientific editing. This work was supported by the Terry Fox Cancer Foundation of the National Cancer Institute of Canada. H.Y. is the recipient of a postdoctoral fellowship from the Cancer Research Institute, New York.
Abbreviations: Akt-DN, kinase-dead form of Akt; ChIP, chromatin immunoprecipitation; CISK, cytokine-independent survival kinase; ERK, extracellular signal-regulated kinase, IGF-I, insulin-like growth factor I; MEF, mouse embryonic fibroblast; PI3K, phosphatidylinositol 3-kinase; SGK1, serum- and glucocorticoid-inducible kinase 1; siRNA, small interfering RNA; TNF-α, tumor necrosis factor α.
References
- 1.de Boer, J., Andressoo, J. O., de Wit, J., Huijmans, J., Beems, R. B., van Steeg, H., Weeda, G., van der Horst, G. T., van Leeuwen, W., Themmen, A. P., et al. (2002) Science 296, 1276–1279. [DOI] [PubMed] [Google Scholar]
- 2.Vogel, H., Lim, D. S., Karsenty, G., Finegold, M. & Hasty, P. (1999) Proc. Natl. Acad. Sci. USA 96, 10770–10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trifunovic, A., Wredenberg, A., Falkenberg, M., Spelbrink, J. N., Rovio, A. T., Bruder, C. E., Bohlooly, Y. M., Gidlof, S., Oldfors, A., Wibom, R., et al. (2004) Nature 429, 417–423. [DOI] [PubMed] [Google Scholar]
- 4.Chin, L., Artandi, S. E., Shen, Q., Tam, A., Lee, S. L., Gottlieb, G. J., Greider, C. W. & DePinho, R. A. (1999) Cell 97, 527–538. [DOI] [PubMed] [Google Scholar]
- 5.Rudolph, K. L., Chang, S., Lee, H. W., Blasco, M., Gottlieb, G. J., Greider, C. & DePinho, R. A. (1999) Cell 96, 701–712. [DOI] [PubMed] [Google Scholar]
- 6.Sharpless, N. E. & DePinho, R. A. (2002) Cell 110, 9–12. [DOI] [PubMed] [Google Scholar]
- 7.Lin, K., Hsin, H., Libina, N. & Kenyon, C. (2001) Nat. Genet. 28, 139–145. [DOI] [PubMed] [Google Scholar]
- 8.Hwangbo, D. S., Gersham, B., Tu, M. P., Palmer, M. & Tatar, M. (2004) Nature 429, 562–566. [DOI] [PubMed] [Google Scholar]
- 9.Brunet, A., Bonni, A., Zigmond, M. J., Lin, M. Z., Juo, P., Hu, L. S., Anderson, M. J., Arden, K. C., Blenis, J. & Greenberg, M. E. (1999) Cell 96, 857–868. [DOI] [PubMed] [Google Scholar]
- 10.Kops, G. J., Dansen, T. B., Polderman, P. E., Saarloos, I., Wirtz, K. W., Coffer, P. J., Huang, T. T., Bos, J. L., Medema, R. H. & Burgering, B. M. (2002) Nature 419, 316–321. [DOI] [PubMed] [Google Scholar]
- 11.Nemoto, S. & Finkel, T. (2002) Science 295, 2450–2452. [DOI] [PubMed] [Google Scholar]
- 12.Leenders, H., Whiffield, S., Benoist, C. & Mathis, D. (2000) Eur. J. Immunol. 30, 2980–2990. [DOI] [PubMed] [Google Scholar]
- 13.Kops, G. J., Medema, R. H., Glassford, J., Essers, M. A., Dijkers, P. F., Coffer, P. J., Lam, E. W. & Burgering, B. M. (2002) Mol. Cell. Biol. 22, 2025–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stahl, M., Dijkers, P. F., Kops, G. J., Lens, S. M., Coffer, P. J., Burgering, B. M. & Medema, R. H. (2002) J. Immunol. 168, 5024–5031. [DOI] [PubMed] [Google Scholar]
- 15.Tran, H., Brunet, A., Grenier, J. M., Datta, S. R., Fornace, A. J., Jr., DiStefano, P. S., Chiang, L. W. & Greenberg, M. E. (2002) Science 296, 530–534. [DOI] [PubMed] [Google Scholar]
- 16.Sage, J., Miller, A. L., Perez-Mancera, P. A., Wysocki, J. M. & Jacks, T. (2003) Nature 424, 223–228. [DOI] [PubMed] [Google Scholar]
- 17.Martinez-Gac, L., Marques, M., Garcia, Z., Campanero, M. R. & Carrera, A. C. (2004) Mol. Cell. Biol. 24, 2181–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lanni, J. S., Lowe, S. W., Licitra, E. J., Liu, J. O. & Jacks, T. (1997) Proc. Natl. Acad. Sci. USA 94, 9679–9683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brunet, A., Park, J., Tran, H., Hu, L. S., Hemmings, B. A. & Greenberg, M. E. (2001) Mol. Cell. Biol. 21, 952–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dudek, H., Datta, S. R., Franke, T. F., Birnbaum, M. J., Yao, R., Cooper, G. M., Segal, R. A., Kaplan, D. R. & Greenberg, M. E. (1997) Science 275, 661–665. [DOI] [PubMed] [Google Scholar]
- 21.Lin, J., Chen, J., Elenbaas, B. & Levine, A. J. (1994) Genes Dev. 8, 1235–1246. [DOI] [PubMed] [Google Scholar]
- 22.Jimenez, G. S., Nister, M., Stommel, J. M., Beeche, M., Barcarse, E. A., Zhang, X. Q., O'Gorman, S. & Wahl, G. M. (2000) Nat. Genet. 26, 37–43. [DOI] [PubMed] [Google Scholar]
- 23.Leong, M. L., Maiyar, A. C., Kim, B., O'Keeffe, B. A. & Firestone, G. L. (2003) J. Biol. Chem. 278, 5871–5882. [DOI] [PubMed] [Google Scholar]
- 24.Liu, D., Yang, X. & Songyang, Z. (2000) Curr. Biol. 10, 1233–1236. [DOI] [PubMed] [Google Scholar]
- 25.Hertweck, M., Gobel, C. & Baumeister, R. (2004) Dev. Cell 6, 577–588. [DOI] [PubMed] [Google Scholar]
- 26.Ryan, K. M., Ernst, M. K., Rice, N. R. & Vousden, K. H. (2000) Nature 404, 892–897. [DOI] [PubMed] [Google Scholar]
- 27.Nasrin, N., Ogg, S., Cahill, C. M., Biggs, W., Nui, S., Dore, J., Calvo, D., Shi, Y., Ruvkun, G. & Alexander-Bridges, M. C. (2000) Proc. Natl. Acad. Sci. USA 97, 10412–10417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brunet, A., Sweeney, L. B., Sturgill, J. F., Chua, K. F., Greer, P. L., Lin, Y., Tran, H., Ross, S. E., Mostoslavsky, R., Cohen, H. Y., et al. (2004) Science 303, 2011–2015. [DOI] [PubMed] [Google Scholar]
- 29.Motta, M. C., Divecha, N., Lemieux, M., Kamel, C., Chen, D., Gu, W., Bultsma, Y., McBurney, M. & Guarente, L. (2004) Cell 116, 551–563. [DOI] [PubMed] [Google Scholar]