Abstract
Target cell tropism of enveloped viruses is regulated by interactions between viral and cellular factors during transmission, dissemination, and replication within the host. Binding of viral envelope glycoproteins to specific cell-surface receptors determines susceptibility to viral entry. However, a number of cell-surface molecules bind viral envelope glycoproteins without mediating entry. Instead, they serve as capture receptors that disseminate viral particles to target organs or susceptible cells. We and others recently demonstrated that the C type lectins L-SIGN and DC-SIGN capture hepatitis C virus (HCV) by specific binding to envelope glycoprotein E2. In this study, we use an entry assay to demonstrate that HCV pseudoviruses captured by L-SIGN+ or DC-SIGN+ cells efficiently transinfect adjacent human liver cells. Virus capture and transinfection require internalization of the SIGN–HCV pseudovirus complex. In vivo, L-SIGN is largely expressed on endothelial cells in liver sinusoids, whereas DC-SIGN is expressed on dendritic cells. Capture of circulating HCV particles by these SIGN+ cells may facilitate virus infection of proximal hepatocytes and lymphocyte subpopulations and may be essential for the establishment of persistent infection.
Hepatitis C virus (HCV) is the etiologic agent of non-A non-B hepatitis in humans (1, 2). Only ≈15% of infected individuals clear the virus, and ≈170 million people worldwide are persistently infected with HCV (3, 4). These individuals may remain asymptomatic or may develop chronic hepatitis or cirrhosis, the latter often leading to hepatocellular carcinoma (5). Hepatocytes are the primary target cells for HCV infection (6–8). Virus-like particles have been visualized in liver biopsies of HCV+ individuals (9–11), and in vitro infection, albeit inefficient, of primary hepatocytes and hepatoma cells has been documented (12–14). The existence of extrahepatic reservoirs of HCV is suggested by the detection of viral RNA in serum and peripheral blood mononuclear cells of HCV+ individuals (15–17). Both B and T lymphocytes appear to be infected in vivo, which is supported by in vitro infection of B and T cell lines (7, 8, 18). One study, however, shows that replicating forms of HCV RNA are restricted to hepatocytes, whereas only nonreplicating forms are present in B lymphocytes, and none are in T lymphocytes (6).
HCV envelope glycoproteins E1 and E2 mediate entry into target cells. We and others recently demonstrated that unmodified E1E2 heterodimers reach the cell surface and are incorporated into retroviral pseudoparticles, which can infect primary hepatocytes and some hepatoma cell lines (19–22). Use of the soluble E2 ectodomain as a surrogate model for studying HCV interactions with cell-surface molecules has identified several potential HCV entry receptors, including CD81, scavenger receptor class B type 1, low-density lipoprotein receptor, and glycosaminoglycans (22–24). Several groups, including ours, have shown that CD81 is necessary but not sufficient for HCV pseudovirus entry into target cells (19, 25, 26). Furthermore, we recently demonstrated that CD81 functions as a postattachment entry coreceptor (26). Cellular factors that act in concert with CD81 to mediate HCV binding and entry remain to be identified.
Engagement of specific receptors is required for viral fusion and entry, but adsorption of viral particles to the cell surface can occur through envelope glycoprotein interactions with other molecules (27–33). The C type lectins, DC-SIGN (dendritic cell-specific intercellular adhesion molecule-3-grabbing nonintegrin; CD209) and L-SIGN (DC-SIGNR; liver and lymph node-specific; CD209L), function as capture receptors for several viruses, including HIV type 1 (HIV-1) (34), Ebola virus (35), cytomegalovirus (36), and dengue virus (37). Both L-SIGN and DC-SIGN have an extracellular C-terminal region that contains a calcium-dependent carbohydrate recognition domain (CRD) and a membrane-proximal heptad-repeat region important for oligomerization (38–41). Capture of viral particles is mediated by the CRD and promotes infection of target cells both in cis and in trans (34, 35, 42, 43). DC-SIGN also recognizes intercellular adhesion molecules 2 and 3, which function as cell-adhesion receptors that regulate transendothelial migration of dendritic cells (DC) from blood to tissues as well as DC–T cell interactions.
We and others have recently demonstrated that recombinant soluble E2, patient-derived HCV virions, and retroviruses pseudotyped with HCV envelope glycoproteins specifically bind to L-SIGN and DC-SIGN (44–46). HCV capture by SIGN molecules depends on the presence of the CRD, indicating that recognition of high mannose oligosaccharides in the viral envelope glycoproteins is critical for binding. The specificity of this interaction is underscored by observations that (i) other C type lectins, such as langerin, CD23, and CLEC-1/2, do not bind HCV E2 (45, 46); (ii) glycosylated envelope proteins of several viruses show little or no avidity for SIGN molecules (36, 47); and (iii) anti-L-SIGN and anti-DC-SIGN mAbs as well as mannan inhibit soluble E2 and HCV capture.
In this study, we used an HCV entry assay to demonstrate that capture of viral particles by L-SIGN+ and DC-SIGN+ cells promotes transinfection of human liver cells. Transinfection is specifically blocked by mannan and antibodies recognizing CRD of SIGN receptors. Similarly, we show that primary human DC mediate transinfection of target cells by a DC-SIGN-dependent mechanism that requires internalization of the receptor–pseudovirus complex. Our results suggest that HCV capture by SIGN molecules plays an important role in viral dissemination to host target organs. In particular, L-SIGN+ liver sinusoidal epithelial cells (LSEC) may facilitate infection of hepatocytes, whereas DC-SIGN+ DC may transmit HCV to hepatocytes as well as subpopulations of B and/or T lymphocytes.
Materials and Methods
Plasmids, Antibodies, and Inhibitors. Construct pcDNA3.1-ΔC-E1-E2 was used to express HCV envelope glycoproteins. Sequences encoding the full-length E1 and E2 (amino acids 132–746), starting with the last 60 amino acids of the capsid (ΔC), were PCR amplified from p90/HCV FL-long pU comprising the genome of infectious HCV isolate H77 (48) and subcloned into pcDNA3.1 (Invitrogen). Putative splice acceptor sites were modified by conservative mutagenesis, as described (21). mAbs 507D, 604L, and 612X recognizing the CRD of DC-SIGN, L-SIGN, or both lectins, respectively, were purchased fromR&D Systems. Anti-HCV E2 mAb 091b-5 was purchased from Austral Biological. Anti-CD81 mAb JS-81 was obtained from Pharmingen. Chloroquine and mannan were purchased from Sigma.
Cell Lines. Unless otherwise specified, cells were purchased from the American Type Culture Collection and grown in DMEM supplemented with 10% fetal bovine serum/1% penicillin/streptomycin/2 mM glutamine. HeLa cells stably expressing L-SIGN or DC-SIGN were generated as described (44) and maintained in DMEM supplemented with 600 μg/ml G418 (Life Technologies, Grand Island, NY). Primary human immature DC were differentiated from peripheral blood mononuclear cells with 1,000 units/ml granulocyte-macrophage colony-stimulating factor (R & D Systems) and 1,000 units/ml IL-4 (R & D Systems) in culture medium for 5 days, as described (49).
Viral Production and Transinfection. Standard calcium phosphate precipitation was used to transfect 293T cells (1.5 × 106) with NLluc+env-reporter vector (50) and pcDNA3.1-ΔC-E1-E2 in a 1:3 ratio. NLluc+env-encodes an HIV-1NL4.3 envelope-deficient genome expressing luc instead of nef. Cell culture supernatants, containing HIV-1 particles pseudotyped with HCV E1E2 envelope glycoproteins, were collected 48 h posttransfection and cleared of cellular debris by low-speed centrifugation.
Parental HeLa cells or L-SIGN+ or DC-SIGN+ transfectants (2 × 104) were incubated with 200 μl of viral supernatant for 2 h at 37°C. Alternatively, primary DC (105) were incubated under similar conditions, at 37°C or at 4°C, with 1 ml of viral supernatant. For inhibition experiments, cells were incubated with mAbs (10 μg/ml), sera from HCV+ or HCV– individuals (1:100), chloroquine (50 μM), or mannan (20 μg/ml) for 30 min at 37°C before addition of viral supernatants. Alternatively, anti-CD81 mAb JS-81 was added to cells after virus capture by DC. After washing three times with serum-free medium to remove unbound virus, cells were cocultured with Huh-7 target cells (4 × 104) for an additional 48 h at 37°C. Luciferase activity [relative light units (R.L.U.)] was measured in cell lysates by using the Luciferase Assay System (Promega) according to the manufacturer's instructions.
Virus-Binding Assay. Binding of HCV pseudoviruses to HeLa cells expressing L-SIGN or DC-SIGN or to DC was performed as described by Gardner et al. (44). Briefly, adherent target cells (104) were washed in buffer (20 mM Tris·HCL, pH 8.0/150 mM NaCl/1 mM CaCl2/2 mM MgCl2/0.5% BSA) and incubated with an equal volume of viral supernatant for 1 h at 37°C with gentle agitation every 15 min. Alternatively, cells were incubated with anti-L/DC-SIGN mAb 612X (10 μg/ml) or mannan (20 μg/ml) for 30 min at room temperature before addition of viral supernatants. After washing five times with adherence buffer to remove unbound virus, cells were lysed according to the manufacturer's instructions for the preparation of viral RNA (Qiagen, Valencia, CA). Cell lysates were analyzed for HIV RNA content with the HIV Ultrasensitive Amplicor Assay (LabCorp, Burlington, NC) or p24 content by using the Coulter HIV-1 p24 antigen assay (Beckman Coulter).
E2-Binding Assay. Binding of soluble E2 was performed as described (44). Briefly, primary DC (5 × 105) were preincubated with mannan (20 μg/ml) or mAbs (10 μg/ml) for 10 min at room temperature. E2-coated fluorescent beads were prepared with the anti-E2 091b-5 capture mAb and added to cells (20 beads per cell) for 30 min at 37°C. Binding was quantified by flow cytometry.
Results
Binding of HCV Pseudoviruses to DC-SIGN+ and L-SIGN+ Cells. HeLa cells were modified to stably express similar levels of cell-surface L-SIGN or DC-SIGN and were shown to specifically capture soluble E2 envelope glycoprotein and HCV virions from sera of infected individuals (44). In this study, we first confirmed the ability of L-SIGN+ and DC-SIGN+ HeLa cells to capture HIV-1 particles pseudotyped with HCV envelope glycoproteins E1E2 (referred to as HCV pseudoviruses from here on in the text). Cells were incubated with pseudovirus-containing supernatants, washed extensively, and the amount of cell-associated particles was determined by quantification of HIV-1 Gag protein (p24) in cell lysates. Significantly higher p24 values were found in lysates of L-SIGN+ and DC-SIGN+ cells, compared with parental HeLa (Table 1 and data not shown). Preincubation of cells with an anti-L/DC-SIGN mAb (10 μg/ml) recognizing CRDs of both lectins decreased L-SIGN+ and DC-SIGN+ HeLa cell-associated p24 by 59% and 30%, respectively, compared with control mouse IgG (10 μg/ml) (Table 1). Similarly, mannan (20 μg/ml), which specifically interacts with lectin CRDs, decreased cell-associated p24 by 84% and 73% (Table 1). Comparable results were obtained when HCV pseudovirus binding to HeLa cells and SIGN+ derivatives was quantified with the Amplicor HIV-1 Monitor test (Roche), which detects HIV-1 genomic RNA (data not shown). Taken together, these results indicate that L-SIGN and DC-SIGN molecules capture HCV pseudoviruses through specific interactions between envelope glycoprotein-associated carbohydrates and lectin CRDs.
Table 1. HCV pseudovirus binding to SIGN+ HeLa cells.
| L-SIGN
|
DC-SIGN
|
|||
|---|---|---|---|---|
| p24, pg/ml | Inhibition, % | p24, pg/ml | Inhibition, % | |
| IgG control | 121 | 0 ± 5 | 160 | 0 ± 4 |
| mAb 612X | 43 | 59 ± 8 | 109 | 30 ± 11 |
| Mannan | 23 | 84 ± 13 | 49 | 73 ± 6 |
L-SIGN+, DC-SIGN+, or parental HeLa cells were incubated with a nonspecific murine IgG (10 μg/ml), anti-L/DC-SIGN mAb 612X (10 μg/ml), or mannan (20 μg/ml), followed by incubation with HCV pseudovirus-containing supernatants. Cell-associated p24 (pg/ml) was measured by using a Coulter HIV-1 p24 antigen assay and corrected for background binding to parental HeLa cells. Percentages of binding inhibition were calculated and adjusted for background by using the formula 100-[(p24 with inhibitors - p24 HeLa with IgG)/(p24 without inhibitor - p24 HeLa with IgG) × 100]%. Negative values are represented as 0 for ease of interpretation. One representative experiment of three independent experiments is shown. The percentages of binding inhibition are means of three independent experiments ± SD.
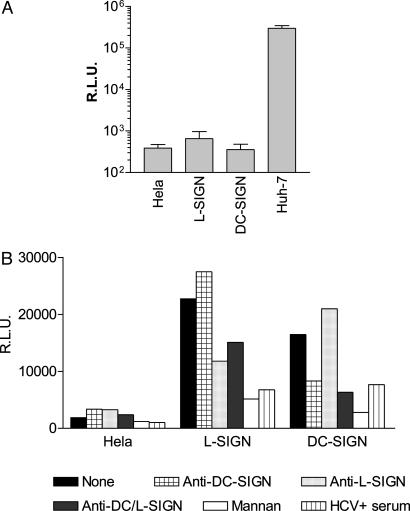
SIGN-Mediated Transinfection of Huh-7 Hepatoma Cells with HCV Pseudoviruses. HCV pseudoviruses comprise an HIV-1 core and genome, which is envelope glycoprotein-deficient and expresses the luc reporter gene. HCV E1E2-mediated entry of these viral particles into primary human hepatocytes and certain hepatoma cells is inhibited by sera from HCV+ individuals as well as an anti-CD81 mAb and certain anti-E2 mAbs (19, 20, 26). This system therefore authentically reproduces the early steps of HCV replication, and we used it to investigate the ability of HeLa cells expressing L-SIGN or DC-SIGN to transfer captured HCV pseudoviruses to entry-permissive human liver (hepatoma) cells. HeLa cells as well as SIGN+ transfectants are resistant to E1E2-mediated entry, because infection with HCV pseudoviruses resulted only in background levels of luciferase activity (Fig. 1A). Huh-7 hepatoma cells, however, are highly permissive to HCV pseudovirus entry, with luciferase activities typically in the 105–106 relative light unit (R.L.U.) range (Fig. 1A). Preincubation of L-SIGN+ or DC-SIGN+ HeLa with HCV pseudovirus supernatants, followed by removal of unbound particles and coculture with Huh-7 hepatoma cells, resulted in significant transinfection of these cells. Luciferase activities were typically in the 104–105 R.L.U. range, which corresponds to ≈10% of entry levels obtained by direct infection (Fig. 1B). Transinfection mediated by L-SIGN was consistently more efficient than transinfection mediated by DC-SIGN. Coculture of HeLa cells with Huh-7 cells did not result in transinfection (Fig. 1B), further demonstrating that viral particles must be captured by SIGN molecules to remain infectious and to be transmitted to target cells.
Fig. 1.
L-SIGN- and DC-SIGN-mediated transinfection of HCV pseudoviruses into Huh-7 cells. L-SIGN+, DC-SIGN+, parental HeLa cells, or Huh-7 cells were infected with HCV pseudoviruses. Luciferase activities [relative light units (R.L.U.)] were measured in cell lysates 48 h postinfection. Values are means of three independent experiments ± SD. (A) L-SIGN+, DC-SIGN+, or parental HeLa cells were incubated with HCV pseudovirus-containing supernatants, washed, and cocultured with Huh-7 cells. Luciferase activity in cell lysates was measured 48 h postinfection. (B) Alternatively, DC-SIGN+, L-SIGN+, or parental HeLa cells were preincubated with anti-DC-SIGN mAb 507D (crosshatched bars), anti-L-SIGN mAb 604L (dotted bars), anti-DC/L-SIGN mAb 612X (gray bars), mannan (white bars), HCV+ sera (striped bars), or HCV – sera (data not shown), before addition of pseudoviral supernatants. One representative experiment of three independent experiments is shown.
Transinfection was also measured in the presence of specific inhibitors. Mannan (20 μg/ml) blocked HCV pseudovirus transmission to Huh-7 cells by L-SIGN+ and DC-SIGN+ HeLa by 75% and 85%, respectively (Fig. 1B). Pseudoviral entry was also significantly reduced by the anti-L/DC-SIGN mAb 612X (10 μg/ml), with a greater effect on DC-SIGN-mediated transinfection (Fig. 1B). As expected, anti-L-SIGN mAb 604L (10 μg/ml) and anti-DC-SIGN mAb 507D (10 μg/ml) inhibited only transinfection mediated by L-SIGN and DC-SIGN, respectively (Fig. 1B). Generally, transinfection was inhibited by ≈30–60% with the different mAbs, all of which recognize the CRDs of the lectin receptors. We also ascertained that transinfection depended on HCV envelope glycoproteins by showing that it is inhibited by ≈80% with sera from HCV+, but not HCV–, donors (1:100) (Fig. 1B and data not shown). Taken together, these findings show that HCV particles captured by L-SIGN+ or DC-SIGN+ cells via interactions with E1E2 remain infectious and are efficiently transmitted to target cells.
Human DC Bind to HCV via E2 and Mediate Transinfection of Liver Cells. We next tested whether DC-SIGN+ primary human DC could specifically capture soluble E2 envelope glycoprotein and HCV pseudoparticles. Peripheral blood mononuclear cells were isolated from an HCV– donor, and monocytes were differentiated into immature DC by treatment with granulocyte–macrophage colony-stimulating factor and IL-4. Soluble E2 was coated onto fluorescent beads conjugated to anti-E2 mAb 091b-5, and binding to DC was analyzed by flow cytometry, as described (44). Over 50% of cells bound E2-coated beads, and binding was inhibited with anti-L/DC-SIGN mAb 612X (10 μg/ml) and mannan (20 μg/ml) by 71% and 66%, respectively (Table 2). In addition, binding of HCV pseudoparticles to DC was detected by RT-PCR with the Ultrasensitive Amplicor Assay. Similar to inhibition of soluble E2 binding, HCV pseudovirus attachment to DC was inhibited with anti-L/DC-SIGN mAb 612X and mannan by 74% and 49%, respectively (Table 2). These results demonstrate that both soluble E2 and HCV pseudoviruses are captured onto primary DC by specifically interacting with DC-SIGN. Variations in potency of inhibition in Tables 1 and 2 could be due to differences in cellular backgrounds, levels of DC-SIGN expression, and use of different assays to detect HCV pseudoparticle binding to cells.
Table 2. Soluble E2 and HCV pseudovirus binding to primary dendritic cells.
| Fluorescent cells, % | Inhibition, % | Units/ml | Inhibition, % | |
|---|---|---|---|---|
| IgG control | 55 | — | 65,000 | — |
| 612X mAb | 13 | 71 ± 15 | 17,000 | 74 ± 8 |
| Mannan | 22 | 66 ± 12 | 32,000 | 49 ± 12 |
Primary DC isolated from an HCV- donor were incubated with a control murine IgG (10 μg/ml), anti-L/DC-SIGN mAb 612X (10 μg/ml), or mannan (20 μg/ml), followed by incubation with fluorescent beads coupled with soluble E2. Binding was measured by flow cytometry and expressed as a percentage of fluorescently labeled cells. Alternatively, DC were incubated with HCV pseudovirus-containing supernatants, and the number of cell-associated HIV-1 RNA copies was quantified by real-time PCR. Percentages of inhibition were calculated as described for Table 1. One representative experiment of three independent experiments is shown. The percentages of binding inhibition are means of three independent experiments ± SD.
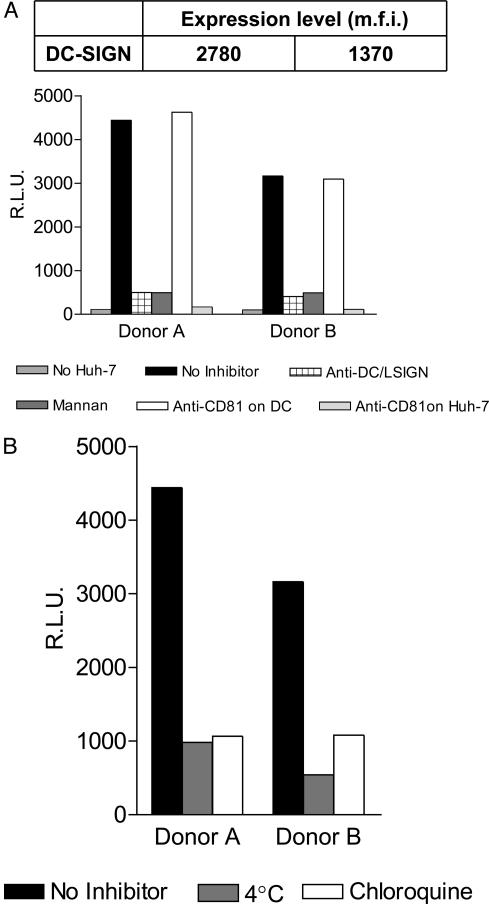
Transinfection of target cells by DC-captured HCV pseudoviruses was also evaluated by using DC derived from two HCV– donors. DC from donor A expressed twice as much DC-SIGN compared with cells derived from donor B, as determined by flow cytometry analyses after labeling with mAb 612X (Fig. 2A Upper). DC from both donors were incubated with HCV pseudoviruses followed by washing and coculture with Huh-7 cells. We note that only background luciferase activities were observed in the absence of target cells, confirming that DC are not susceptible to E1E2-mediated pseudovirus entry (Fig. 2A). DC derived from donor A were ≈30% more efficient at mediating transinfection of Huh-7 cells than DC derived from donor B (Fig. 2A), consistent with higher levels of DC-SIGN expression. Moreover, transinfection was inhibited by both mannan (20 μg/ml) and the anti-DC/L-SIGN mAb 612X (10 μg/ml), confirming that it occurs through a DC-SIGN-dependent mechanism (Fig. 2A). When DC were incubated with the anti-CD81 mAb JS-81 (10 μg/ml) before HCV pseudovirus capture, transinfection of Huh-7 cells was unaffected. However, addition of the anti-CD81 mAb JS-81 (10 μg/ml) to mixtures of DC and target cells after HCV pseudoparticle capture by the former completely inhibited transinfection (Fig. 2B). Therefore, CD81 does not play a role in SIGN-mediated virus capture but is still necessary for transinfection of target cells.
Fig. 2.
DC-mediated transinfection of HCV pseudoviruses into Huh-7 cells. Immature human DC were differentiated from peripheral blood mononuclear cells of two HCV– donors (A and B) by using granulocyte–macrophage colony-stimulating factor and IL-4. (A) Expression of DC-SIGN was quantified by flow cytometry and represented as mean fluorescent intensity (m.f.i.) of an anti-DC-SIGN mAb. DC were incubated with HCV pseudovirus-containing supernatants, washed, and cocultured with Huh-7 cells. Luciferase activity was measured in cell lysates 48 h postinfection. Direct infection of DC by HCV pseudoviruses was measured in the absence of Huh-7 cells (dotted bars). DC were also preincubated with HCV pseudoviruses alone (black bars) or in the presence of anti-DC/L-SIGN 612X (crosshatched bars), mannan (gray bars), or anti-CD81 mAb JS-81 (white bars). Alternatively, JS-81 was added to DC and Huh-7 cells after pseudovirus capture (dotted bars). (B) Alternatively, DC were incubated with HCV pseudovirus-containing supernatants at 37°C (black bars), 4°C (gray bars), or in the presence of chloroquine (white bars), washed, and cocultured with Huh-7 cells. Luciferase activity was measured in cell lysates 48 h postinfection.
Immature DC efficiently internalize and process viruses for presentation to the immune system. DC-SIGN is believed to play a role in virus escape from this process. Consequently, we also investigated the role of virus internalization in DC-mediated transinfection of HCV to target cells. DC were incubated with HCV pseudoviruses at 4°C, which is permissive for attachment but not internalization of virus–receptor complexes. Alternatively, DC-SIGN-mediated capture of HCV pseudoviruses was carried out in the presence of chloroquine, which is a weak base that prevents acidification of endosomes and blocks receptor recycling from the endocytic pathway. DC were then washed and cocultured with Huh-7 cells at 37°C and entry was quantified 48 h later. Incubation at 4°C or in the presence of chloroquine during HCV pseudovirus captures strongly inhibited (>80%) transinfection mediated by DC from both donors (Fig. 2B). These results indicate that endocytosis of the DC-SIGN–virus complex plays a critical role in transinfection.
Discussion
The HCV E2 envelope glycoprotein has 11 N-linked glycosylation sites, and the majority of oligosaccharides on E2 are high-mannose structures. C type lectin CRDs specifically bind high-mannose N-linked oligosaccharides associated with surface components of viruses and bacteria, which are then internalized and routed to lysosomes and MHC class II positive endosomes. Certain viruses, however, are able to escape targeting to lysosomes that occurs for other pathogens (42). Instead, captured viral particles are recycled from nonlysosomal intracellular compartments and are transferred to target cells, where virus-cell fusion is mediated by envelope glycoprotein interactions with bona fide entry receptors. The best-characterized example is HIV-1, which is captured by DC-SIGN+ DC at sites of mucosal exposure. DC-SIGN-mediated internalization of HIV-1 particles into nonlysosomal compartments appears to protect the virus from degradation during transport to lymphoid organs, where it is transmitted to CD4+ CCR5+ lymphocytes (34, 42, 43). Other viruses, such as Ebola, dengue, and Sindbis, may use similar mechanisms to infect target cells either in cis or in trans (35, 37, 51).
HCV replicates in hepatocytes, and several studies have documented HCV replication in B and T lymphocytes (9, 10, 52–55), but the determinants of HCV tropism are unknown. We recently demonstrated that CD81 serves as an obligate entry coreceptor for HCV (26). However, CD81 is ubiquitously expressed on human cells and cannot account for the restricted tropism of HCV (56). Instead, other HCV receptors may be expressed on hepatocytes and some lymphocytes, thereby restricting viral entry into these cell types. We and others have shown (44–46) that HCV envelope glycoprotein E2 as well as HCV virions and pseudoviruses specifically bind to L-SIGN+ and DC-SIGN+ cells. In this study, we demonstrate that expression of SIGN receptors does not render cells permissive to infection by HCV pseudoviruses but does enable them to mediate infection of human liver cells in trans. The specificity of this process is underscored by inhibition of pseudovirus capture and transinfection with mannan and mAbs against CRDs of L-SIGN and DC-SIGN. Moreover, primary human DC also mediate DC-SIGN-dependent transinfection of liver cells, which requires internalization of the DC-SIGN–pseudovirus complex. An additional determinant of HCV tropism therefore may be L-SIGN- and DC-SIGN-mediated targeting of the virus to the liver and lymph nodes.
L-SIGN is expressed mainly by endothelial cells in liver and lymph node sinusoids, whereas DC-SIGN is expressed mostly on myeloid-lineage DC (38, 39, 57, 58). The endothelium of liver sinusoids (specialized capillaries) serves as an active barrier between circulating blood and hepatocytes. LSEC lack a basal lamina and comprise unique pores (fenestrae), which act as filters for fluids, solutes, and particles that are exchanged between the sinusoidal lumen and hepatocytes (59). Another functional characteristic of LSEC is their effective uptake of a wide variety of substances from the blood by receptor-mediated endocytosis. Moreover, some of these substances are actively transferred across the endothelial barrier to surrounding tissues by transcytosis. We postulate that L-SIGN-mediated capture of circulating HCV particles by LSEC results in transcytosis of the virus across the endothelial barrier, thereby concentrating infectious particles and placing them in direct contact with entry-permissive hepatocytes. Similarly, DC-SIGN+ DC, which are present in mucosal compartments and circulating blood, may capture HCV at these sites of primary infection. DC are antigen-presenting cells and interact directly with both B and T lymphocytes (60–63), some of which may be susceptible to HCV infection. Also, DC have been shown to migrate from the blood to the liver by translocation across the endothelium of hepatic sinusoids and may thus transport HCV to hepatocytes (64).
The precise mechanism of SIGN-mediated transinfection is unknown. We show that, similar to HIV-1, HCV transinfection mediated by DC depends on internalization of viral particles. Treatment of DC with chloroquine results in inhibition of transinfection, indicating that pH changes within intracellular compartments are critical for the process. This finding is highlighted by the observation that HCV pseudovirions that have been captured but not internalized (by incubation at 4°C) are not transmitted to target cells. It is possible that acidification of the SIGN–virus complex leads to its dissociation, thereby facilitating transfer of infectious particles to receptor-expressing target cells. Even though HCV pseudoviruses enter cells by low pH-dependent receptor-mediated endocytosis (20), it is remarkable that SIGN-mediated internalization does not prematurely expose cryptic E1E2 fusion domains and inactivate the virus. This may indicate that HCV envelope glycoproteins are insensitive to low pH-induced modifications in the absence of receptor-induced conformational changes. In addition, DC can selectively retain antigens in their native form inside neutral to mildly acidic vesicles (65, 66).
Conclusion
HCV envelope glycoproteins participate in a complex cascade of interactions with specific cell-surface molecules to target and enter host cells. Binding of E1E2 to the CD81 coreceptor and other molecules on hepatocytes results in viral fusion and entry. In addition, HCV envelope glycoproteins bind to at least two C type lectins, L-SIGN and DC-SIGN, expressed on LSEC and DC, respectively, which interact with HCV target cells. SIGN-mediated transinfection of hepatocytes with HCV, which are not in direct contact with circulating blood, may be essential for establishment of persistent infection.
Acknowledgments
We thank Dr. Steven Porcelli (Albert Einstein College of Medicine, New York) for kindly providing primary human DC for this study. This work was supported by National Institutes of Health Grant AI060390 and the Speaker's Fund for Biomedical Research Young Investigators' Award (to T.D.), National Institutes of Health Grant AI051134 (to J.P.G.), and Progenics Pharmaceuticals, Incorporated. This work was also supported in part by National Institute of Allergy and Infectious Disease Centers for AIDS Research Grant AI051519 to Albert Einstein College of Medicine.
Abbreviations: HCV, hepatitis C virus; HIV-1, HIV type 1; CRD, carbohydrate recognition domain; DC, dendritic cells; LSEC, liver sinusoidal epithelial cells.
References
- 1.Alter, H. J. & Seef, L. B. (2000) Semin. Liver Dis. 20, 17–35. [DOI] [PubMed] [Google Scholar]
- 2.Lauer, G. M. & Walker, B. D. (2001) N. Engl. J. Med. 345, 41–52. [DOI] [PubMed] [Google Scholar]
- 3.Cooper, S., Erickson, A. L., Adams, E. J., Kansopon, J., Weiner, A. J., Chien, D. Y., Houghton, M., Parham, P. & Walker, C. M. (1999) Immunity 10, 439–449. [DOI] [PubMed] [Google Scholar]
- 4.Lechner, F., Wong, D. K., Dunbar, P. R., Chapman, R., Chung, R. T., Dohrenwend, P., Robbins, G., Phillips, R., Klenerman, P. & Walker, B. D. (2000) J. Exp. Med. 191, 1499–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fry, D. E. & Flint, L. M., Jr. (1997) Bull. Am. Coll. Surg. 82, 8–13. [PubMed] [Google Scholar]
- 6.Boisvert, J., He, X. S., Cheung, R., Keeffe, E. B., Wright, T. & Greenberg, H. B. (2001) J. Infect. Dis. 184, 827–835. [DOI] [PubMed] [Google Scholar]
- 7.Sung, V. M., Shimodaira, S., Doughty, A. L., Picchio, G. R., Can, H., Yen, T. S., Lindsay, K. L., Levine, A. M. & Lai, M. M. (2003) J. Virol. 77, 2134–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kato, N., Nakazawa, T., Mizutani, T. & Shimotohno, K. (1995) Biochem. Biophys. Res. Commun. 206, 863–869. [DOI] [PubMed] [Google Scholar]
- 9.De Vos, R., Verslype, C., Depla, E., Fevery, J., Van Damme, B., Desmet, V. & Roskams, T. (2002) J. Hepatol. 37, 370–379. [DOI] [PubMed] [Google Scholar]
- 10.Bosman, C., Valli, M. B., Bertolini, L., Serafino, A., Boldrini, R., Marcellini, M. & Carloni, G. (1998) Res. Virol. 149, 311–314. [DOI] [PubMed] [Google Scholar]
- 11.Iacovacci, S., Manzin, A., Barca, S., Sargiacomo, M., Serafino, A., Valli, M. B., Macioce, G., Hassan, H. J., Ponzetto, A., Clementi, M., Peschle, C. & Carloni, G. (1997) Hepatology 26, 1328–1337. [DOI] [PubMed] [Google Scholar]
- 12.Ikeda, M., Sugiyama, K., Mizutani, T., Tanaka, T., Tanaka, K., Sekihara, H., Shimotohno, K. & Kato, N. (1998) Virus Res. 56, 157–167. [DOI] [PubMed] [Google Scholar]
- 13.Fournier, C., Sureau, C., Coste, J., Ducos, J., Pageaux, G., Larrey, D., Domergue, J. & Maurel, P. (1998) J. Gen. Virol. 79, 2367–2374. [DOI] [PubMed] [Google Scholar]
- 14.Castet, V., Fournier, C., Soulier, A., Brillet, R., Coste, J., Larrey, D., Dhumeaux, D., Maurel, P. & Pawlotsky, J. M. (2002) J. Virol. 76, 8189–8199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roque Afonso, A. M., Jiang, J., Penin, F., Tareau, C., Samuel, D., Petit, M. A., Bismuth, H., Dussaix, E. & Feray, C. (1999) J. Virol. 73, 9213–9221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cabot, B., Martell, M., Esteban, J. I., Sauleda, S., Otero, T., Esteban, R., Guardia, J. & Gomez, J. (2000) J. Virol. 74, 805–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laskus, T., Radkowski, M., Wang, L. F., Nowicki, M. & Rakela, J. (2000) J. Virol. 74, 1014–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shimizu, Y. K., Iwamoto, A., Hijikata, M., Purcell, R. H. & Yoshikura, H. (1992) Proc. Natl. Acad. Sci. USA 89, 5477–5481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartosch, B., Dubuisson, J. & Cosset, F. L. (2003) J. Exp. Med. 197, 633–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsu, M., Zhang, J., Flint, M., Logvinoff, C., Cheng-Mayer, C., Rice, C. M. & McKeating, J. A. (2003) Proc. Natl. Acad. Sci. USA 100, 7271–7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dumonceaux, J., Cormier, E. G., Kajumo, F., Donovan, G. P., Roy-Chowdhury, J., Fox, I. J., Gardner, J. P. & Dragic, T. (2003) J. Virol. 77, 13418–13424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartosch, B., Vitelli, A., Granier, C., Goujon, C., Dubuisson, J., Pascale, S., Scarselli, E., Cortese, R., Nicosia, A. & Cosset, F. L. (2003) J. Biol. Chem. 278, 41624–41630. [DOI] [PubMed] [Google Scholar]
- 23.Flint, M., Quinn, E. R. & Levy, S. (2001) Clin. Liver Dis. 5, 873–893. [DOI] [PubMed] [Google Scholar]
- 24.Scarselli, E., Ansuini, H., Cerino, R., Roccasecca, R. M., Acali, S., Filocamo, G., Traboni, C., Nicosia, A., Cortese, R. & Vitelli, A. (2002) EMBO J. 21, 5017–5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang, J., Randall, G., Higginbottom, A., Monk, P., Rice, C. M. & McKeating, J. A. (2004) J. Virol. 78, 1448–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cormier, E. G., Tsamis, F., Kajumo, F., Durso, R. J., Gardner, J. P. & Dragic, T. (2004) Proc. Natl. Acad. Sci. USA 101, 7270–7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, J. & Thorp, S. C. (2002) Med. Res. Rev. 22, 1–25. [DOI] [PubMed] [Google Scholar]
- 28.Reddi, H. V. & Lipton, H. L. (2002) J. Virol. 76, 8400–8407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rostand, K. S. & Esko, J. D. (1997) Infect. Immun. 65, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shukla, D. & Spear, P. G. (2001) J. Clin. Invest. 108, 503–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Summerford, C. & Samulski, R. J. (1998) J. Virol. 72, 1438–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ugolini, S., Mondor, I. & Sattentau, Q. J. (1999) Trends Microbiol. 7, 144–149. [DOI] [PubMed] [Google Scholar]
- 33.WuDunn, D. & Spear, P. G. (1989) J. Virol. 63, 52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geijtenbeek, T. B., Kwon, D. S., Torensma, R., van Vliet, S. J., van Duijnhoven, G. C., Middel, J., Cornelissen, I. L., Nottet, H. S., KewalRamani, V. N., Littman, D. R., et al. (2000) Cell 100, 587–597. [DOI] [PubMed] [Google Scholar]
- 35.Alvarez, C. P., Lasala, F., Carrillo, J., Muniz, O., Corbi, A. L. & Delgado, R. (2002) J. Virol. 76, 6841–6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Halary, F., Amara, A., Lortat-Jacob, H., Messerle, M., Delaunay, T., Houles, C., Fieschi, F., Arenzana-Seisdedos, F., Moreau, J. F. & Dechanet-Merville, J. (2002) Immunity 17, 653–664. [DOI] [PubMed] [Google Scholar]
- 37.Tassaneetrithep, B., Burgess, T. H., Granelli-Piperno, A., Trumpfheller, C., Finke, J., Sun, W., Eller, M. A., Pattanapanyasat, K., Sarasombath, S., Birx, D. L., et al. (2003) J. Exp. Med. 197, 823–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pohlmann, S., Soilleux, E. J., Baribaud, F., Leslie, G. J., Morris, L. S., Trowsdale, J., Lee, B., Coleman, N. & Doms, R. W. (2001) Proc. Natl. Acad. Sci. USA 98, 2670–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bashirova, A. A., Geijtenbeek, T. B., van Duijnhoven, G. C., van Vliet, S. J., Eilering, J. B., Martin, M. P., Wu, L., Martin, T. D., Viebig, N., Knolle, P. A., et al. (2001) J. Exp. Med. 193, 671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Geijtenbeek, T. B., Torensma, R., van Vliet, S. J., van Duijnhoven, G. C., Adema, G. J., van Kooyk, Y. & Figdor, C. G. (2000) Cell 100, 575–585. [DOI] [PubMed] [Google Scholar]
- 41.Mitchell, D. A., Fadden, A. J. & Drickamer, K. (2001) J. Biol. Chem. 276, 28939–28945. [DOI] [PubMed] [Google Scholar]
- 42.Engering, A., Geijtenbeek, T. B., van Vliet, S. J., Wijers, M., van Liempt, E., Demaurex, N., Lanzavecchia, A., Fransen, J., Figdor, C. G., Piguet, V., et al.. (2002) J. Immunol. 168, 2118–2126. [DOI] [PubMed] [Google Scholar]
- 43.Kwon, D. S., Gregorio, G., Bitton, N., Hendrickson, W. A. & Littman, D. R. (2002) Immunity 16, 135–144. [DOI] [PubMed] [Google Scholar]
- 44.Gardner, J. P., Durso, R. J., Arrigale, R. R., Donovan, G. P., Maddon, P. J., Dragic, T. & Olson, W. C. (2003) Proc. Natl. Acad. Sci. USA 100, 4498–4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lozach, P. Y., Lortat-Jacob, H., de Lacroix de Lavalette, A., Staropoli, I., Foung, S., Amara, A., Houles, C., Fieschi, F., Schwartz, O., Virelizier, J. L., et al. (2003) J. Biol. Chem. 278, 20358–20366. [DOI] [PubMed] [Google Scholar]
- 46.Pohlmann, S., Zhang, J., Baribaud, F., Chen, Z., Leslie, G. J., Lin, G., Granelli-Piperno, A., Doms, R. W., Rice, C. M. & McKeating, J. A. (2003) J. Virol. 77, 4070–4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hong, P. W., Flummerfelt, K. B., De Parseval, A., Gurney, K., Elder, J. H. & Lee, B. (2002) J. Virol. 76, 12855–12865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kolykhalov, A. A., Agapov, E. V., Blight, K. J., Mihalik, K., Feinstone, S. M. & Rice, C. M. (1997) Science 277, 570–574. [DOI] [PubMed] [Google Scholar]
- 49.Caux, C., Dezutter-Dambuyant, C., Schmitt, D. & Banchereau, J. (1992) Nature 360, 258–261. [DOI] [PubMed] [Google Scholar]
- 50.Connor, R. I., Chen, B. K., Choe, S. & Landau, N. R. (1995) Virology 206, 935–944. [DOI] [PubMed] [Google Scholar]
- 51.Klimstra, W. B., Nangle, E. M., Smith, M. S., Yurochko, A. D. & Ryman, K. D. (2003) J. Virol. 77, 12022–12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Muller, H. M., Kallinowski, B., Solbach, C., Theilmann, L., Goeser, T. & Pfaff, E. (1994) Arch. Virol. Suppl. 9, 307–316. [DOI] [PubMed] [Google Scholar]
- 53.Morsica, G., Tambussi, G., Sitia, G., Novati, R., Lazzarin, A., Lopalco, L. & Mukenge, S. (1999) Blood 94, 1138–1139. [PubMed] [Google Scholar]
- 54.Muller, H. M., Pfaff, E., Goeser, T., Kallinowski, B., Solbach, C. & Theilmann, L. (1993) J. Gen. Virol. 74, 669–676. [DOI] [PubMed] [Google Scholar]
- 55.Hu, Y., Shahidi, A., Park, S., Guilfoyle, D. & Hirshfield, I. (2003) Am. J. Clin. Pathol. 119, 95–100. [DOI] [PubMed] [Google Scholar]
- 56.Levy, S., Todd, S. C. & Maecker, H. T. (1998) Annu. Rev. Immunol. 16, 89–109. [DOI] [PubMed] [Google Scholar]
- 57.Jameson, B., Baribaud, F., Pohlmann, S., Ghavimi, D., Mortari, F., Doms, R. W. & Iwasaki, A. (2002) J. Virol. 76, 1866–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Soilleux, E. J., Morris, L. S., Lee, B., Pohlmann, S., Trowsdale, J., Doms, R. W. & Coleman, N. (2001) J. Pathol. 195, 586–592. [DOI] [PubMed] [Google Scholar]
- 59.Braet, F. & Wisse, E. (2002) Comp. Hepatol. 1, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wykes, M., Pombo, A., Jenkins, C. & MacPherson, G. G. (1998) J. Immunol. 161, 1313–1319. [PubMed] [Google Scholar]
- 61.Kushnir, N., Liu, L. & MacPherson, G. G. (1998) J. Immunol. 160, 1774–1781. [PubMed] [Google Scholar]
- 62.Dubois, B., Vanbervliet, B., Fayette, J., Massacrier, C., Briere, F., Banchereau, J. & Caux, C. (1997) Adv. Exp. Med. Biol. 417, 329–334. [DOI] [PubMed] [Google Scholar]
- 63.Dubois, B., Massacrier, C. & Caux, C. (2001) J. Leukocyte Biol. 70, 633–641. [PubMed] [Google Scholar]
- 64.Kudo, S., Matsuno, K., Ezaki, T. & Ogawa, M. (1997) J. Exp. Med. 185, 777–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lutz, M. B., Rovere, P., Kleijmeer, M. J., Rescigno, M., Assmann, C. U., Oorschot, V. M., Geuze, H. J., Trucy, J., Demandolx, D., Davoust, J., et al. (1997) J. Immunol. 159, 3707–3716. [PubMed] [Google Scholar]
- 66.Ignatius, R., Mahnke, K., Rivera, M., Hong, K., Isdell, F., Steinman, R. M., Pope, M. & Stamatatos, L. (2000) Blood 96, 3505–3513. [PubMed] [Google Scholar]