Abstract
Box C/D RNAs are small, noncoding RNAs that function in RNA modification in eukaryotes and archaea. Here, we report that box C/D RNAs exist in the rare biological form of RNA circles in the hyperthermophilic archaeon Pyrococcus furiosus. Northern analysis of box C/D RNAs reveals two prominent RNA species of different electrophoretic mobilities in total P. furiosus RNA preparations. Together, the results of Northern, ribozyme, RT-PCR, and lariat debranching analyses indicate that the two species are circular and linear RNAs of similar length and abundance. It seems that most, if not all, species of box C/D RNAs exist as circles in P. furiosus. In addition, the circular RNAs are found in complexes with proteins required for box C/D RNA function. Our finding places box C/D RNAs among the extremely few circular RNAs known to exist in nature. Moreover, the unexpected discovery of circular box C/D RNAs points to the existence of a previously unrecognized biogenesis pathway for box C/D RNAs in archaea.
Box C/D RNAs function as the guide components of RNA–protein complexes [box C/D ribonucleoproteins (RNPs)] that 2′-O-methylate rRNA and small nuclear RNA in eukaryotes and tRNA in archaea (1–7). The RNAs are characterized by the presence of one or two sets of the short, conserved sequence elements termed box C and box D. Base-pairing of the guide region of a box C/D RNA to the target RNA establishes a site of 2′-O-methylation of the target RNA. The modification is catalyzed by fibrillarin, one of three protein components of the box C/D RNP in archaea (2, 4, 8). The other two box C/D RNP proteins are L7Ae (a multifunctional RNA binding protein that is also a component of the ribosome and of box H/ACA pseudouridylation guide RNPs) and Nop56/58 (2, 4, 8, 9). More than 50 box C/D RNAs are encoded in the genomes of humans and Pyrococcus furiosus (1, 2, 4).
In eukaryotes, box C/D modification guide RNAs are made by two primary pathways (2, 5, 10, 11). Some are found in the introns of host genes, and the linear box C/D RNAs are processed from debranched lariats. In other cases, linear box C/D RNAs are processed from mono- or polycistronic transcripts synthesized from independent promoters. Box C/D RNAs were found only recently in archaea (12, 13). The pathway by which archaeal box C/D RNAs are made is unknown. The archaeal box C/D RNAs are located primarily in intergenic regions of the genome, sometimes overlapping the 3′ or 5′ end of flanking ORFs (1, 4, 12, 13). Independent promoters have not been identified.
In the course of analyzing a cDNA library of small RNAs from P. furiosus, we observed an interesting incongruence between the linear sequence of some box C/D RNA clones (sR2, -11, -14, -27, -29, and -44) and the P. furiosus genome sequence (N.G.S., S.M., L.S.J., S.R.E., R.M.T., and M.P.T., unpublished data). For example, if the genomic sequence of a box C/D RNA is represented as 1–2–3–4 (corresponding to the four conserved sequence boxes found in box C/D RNAs, C–D′–C′–D), incongruent clones took the form 3–4–1–2, 2–3–4–1, etc. One potential source of the incongruent clones that we considered was broken circles, wherein RNAs were transcribed linearly from the gene (as 1–2–3–4), subsequently circularized, and then randomly broken during experimental isolation (for example, between points 2 and 3). The experiments described here were designed to address the possible existence of circular box C/D RNAs in P. furiosus.
Methods
Preparation of RNA and Cell Extracts. RNA was isolated from P. furiosus by acid-phenol extraction and treated additionally with DNase I (DNA-Free, Ambion, Austin, TX) before RT-PCR was performed. P. furiosus cell extract was prepared by sonication in 50 mM Tris·HCl (pH 7.4)/100 mM NaCl/1 mM MgCl2/1× protease inhibitor mixture (Roche)/10 units/ml RQ 1 RNase-free DNase (Promega)/3 mM DTT/0.2 units/μl RNasin (Promega), and then centrifugation for 10 min at 20,000 × g. The extract was dialysed against Ipp 300 (10 mM Tris·HCl, pH 8.0/300 mM NaCl/0.05% Igepal), supplemented with 0.1 mM PMSF, 1 mM EDTA, and 2 mM DTT.
Ribozyme Analysis. Oligo HH-sR29 (AGCTAATACCCTGATGAGTCCGTGAGGACGAAACATCATCAC) and primers SP6-HH-sR29 (ATTTAGGTGACACTATAGAGCTAATACCCTGATGAGTC) and 3′ HH-sR29 (GTGATGATGTTTCGTCCTC) were used in PCR to generate a template for in vitro transcription of anti-sR29 ribozyme. The sequences that encode the hybridization arms of the ribozyme (complementary to sR29) are underlined. Cleavage was in 40 mM Tris·HCl (pH 7.4)/2 mM spermidine/10 mM MgCl2 at 37°C for 2 h. Cleavage products were visualized by using direct autoradiography (in vitro transcribed sR29) or Northern blotting.
RT-PCR Analysis. The Titan One Tube RT-PCR System (Roche) and the following primers were used: sR2 outward primers, TAGGGATGAGGAGCCGATGC and TCAGAGTGAGGGAAAAACTC; sR14 inward primers, CCAATGATGATGGATCAACC and CCCCTCAGGTGATGCGGACC; sR14 outward primers, ATGATGAGGTCCGCATCACC and TTCAGCCGGTTGATCCATC; and sR29 outward primers, CCTGAAAGGTGATGATGTCG and GGGCAAGGCACATCATCCG. sR2 and sR29 inward primers are described in ref. 14.
Debranching Assay. Slow-migrating sR29 was gel-purified from 10 μg of total RNA. Radiolabeled group II intron lariat of yeast mitochondrial RNA was generated by using pJD20 (kindly provided by Kevin Jarrell, Modular Genetics, Woburn, MA) as described in ref. 15. Debranching was performed with HeLa S100 extract (kindly provided by Adrian Krainer, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY) as described in ref. 16.
Immunoprecipitation. Rabbit anti-P. furiosus Nop56/58 and preimmune sera were coupled to protein A-Sepharose CL-4B (Sigma). P. furiosus extract was supplemented with 1 vol of Ipp 300/1× protease inhibitor cocktail (Roche)/0.2 units/μl RNasin (Promega)/5 mM DTT. Washes were performed with Ipp 300.
Results and Discussion
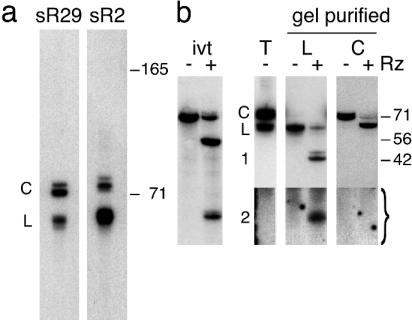
Circular RNAs are known to migrate more slowly than linear RNAs of the same length on denaturing PAGE (17, 18), and, interestingly, multiple bands are visible in a previous Northern analysis of box C/D RNAs from the closely related species Pyrococcus abyssi (13). Fig. 1a shows a Northern analysis of two of the P. furiosus box C/D RNAs for which we had found incongruent clones, sR29 and sR2. The detectable species of these RNAs fall into two major classes (Fig. 1a, C and L). Within each class, at least two bands are discernible that our data suggest reflect minor length heterogeneity within the forms. The two forms are present at similar levels in total P. furiosus RNA preparations (Fig. 1a and see also Fig. 4b, T lanes). We estimated the size of the faster migrating form (primary band) to be ≈60 nt for sR29 and ≈62 nt for sR2, close to the computationally predicted lengths of the linear RNAs (12). The mobility of the slow-migrating form (corresponding to linear RNA of ≈72 nt for sR29 and ≈74 nt for sR2) is consistent with several possibilities, including a longer linear form (e.g., a linear precursor), a lariat, or a circular RNA.
Fig. 1.
Two forms of P. furiosus box C/D RNAs distinguished by Northern and ribozyme analysis. (a) Fast- and slow-migrating forms of sR2 and sR29 (L and C) are apparent in the Northern analysis of total P. furiosus RNA. Positions of standard 165- and 71-nt RNAs are indicated. (b) Anti-sR29 ribozyme (Rz) cleaves in vitro-transcribed 32P-labeled sR29 RNA into fragments of 48 and 21 nt (ivt, ± Rz) and gel-purified fast-migrating sR29 into fragments of ≈42 (or ≈45) and ≈20 nt (L, ± Rz, bands 1 and 2). Slow-migrating sR29 is cleaved to a product of ≈63 nt. The bracketed region is from a separate Northern analysis performed with higher specific activity probe and hybridization conditions optimized for detection of very small RNAs. The positions of a standard 71-nt RNA and 56- and 42-nt DNAs are indicated.
Fig. 4.
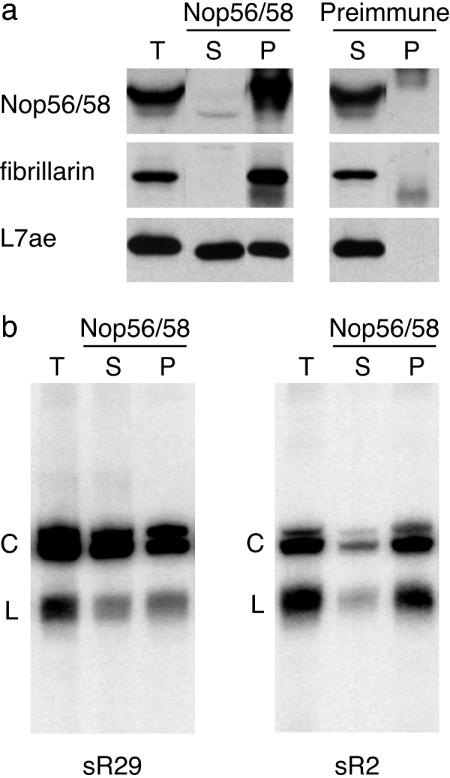
Coimmunoprecipitation of circular sR2 and sR29 with box C/D protein complexes. (a) Antibodies against Nop56/58 coimmunoprecipitate Nop56/58, fibrillarin, and L7Ae proteins. T, total P. furiosus cellular extract; S and P, supernatant and pellet of anti-Nop56/58 and preimmune antibody immunoprecipitations, respectively. Proteins were detected by Western blotting. (b) Northern detection of sR2 and sR29 in complexes immunoprecipitated with Nop56/58. Preimmune antibodies do not coimmunoprecipitate box C/D RNAs (data not shown).
To assess the nature of the two forms of sR29 found in P. furiosus, we used a hammerhead ribozyme designed to cleave the linear RNA into two fragments of unequal length: ≈40 and 20 nt (depending on the actual length and ends of sR29 in P. furiosus, which have not been determined). The ribozyme cleaved an in vitro-transcribed, 32P-labeled sR29 RNA of 69 nt into fragments of 48 and 21 nt as expected (Fig. 1b, ivt).
The two native forms of sR29 (C and L) each were gel-purified, and ribozyme cleavage products were detected by Northern blotting. Cleavage of the fast-migrating form produced a fragment of ≈42 nt (with a second minor band at ≈45 nt reflecting observed heterogeneity of the linear form) and a fragment of ≈20 nt (Fig. 1b, L), indicating that the fast-migrating form is linear sR29 RNA with a length of ≈62–65 nt. Conversely, the ribozyme converted the slow-migrating sR29 to RNA with a mobility similar to that of the fast-migrating form (Fig. 1b, C), suggesting linearization of a circular RNA of ≈63 nt. The results are consistent with the hypothesis that the slow-migrating RNA is a lariat or circular form of sR29.
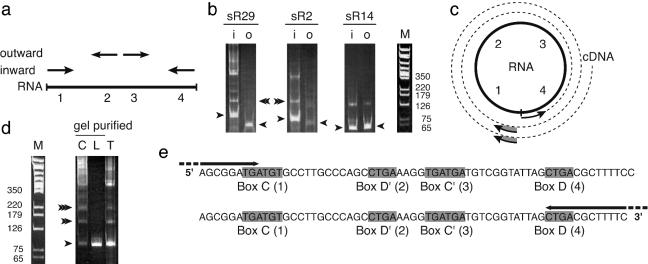
As another test of the circular nature of the box C/D RNAs in P. furiosus, we performed RT-PCR analysis. We synthesized two sets of primers for each sR29, sR2, and sR14: an inward-directed set located at the termini of the linear RNA sequence and an outward-directed set located internally (Fig. 2a). The inward primers are expected to amplify a fragment from either standard linear or circular RNA. The outward primers are not expected to generate a product from linear RNA but would be expected to generate a product from a circular RNA and possibly also from a lariat (19, 20).
Fig. 2.
RT-PCR analysis of box C/D RNAs. (a) Arrows, inward- and outward-directed primers for RT-PCR analysis. Conserved RNA box elements C, D′, C′, and D are represented as 1, 2, 3, and 4, respectively. (b) RT-PCR with inward (i) or outward (o) primers and total P. furiosus RNA. The primary (single arrowhead) and secondary (double arrowhead) products indicated were isolated and sequenced from the gel that is shown. M, DNA size standards. (c) Potential for multiple rounds of RT around a circular RNA template and subsequent PCR amplification of single and double products is illustrated, in this case with inward primers. Arrows, primers; broken line, RT product; gray shading, potential sites of PCR priming from the extended RT product. (d) RT-PCR with gel-purified, slow- and fast-migrating sR29 and total P. furiosus RNA (C, L, and T) by using inward-directed primers. Single, double, and triple arrowheads represent products found to consist of one, two, and three tandem copies of sR29, respectively. (e) Sequence of RT-PCR product marked with double arrowhead in d. Arrows, primers. Positions of conserved box elements in each of the two copies of sR29 present in the sequence are indicated.
First, we performed RT-PCR analysis with total RNA isolated from P. furiosus. Inward primers for sR29, sR2, and sR14 produced PCR products of the expected size (Fig. 2b, i, single arrowheads). In addition, outward primers for each of the RNAs produced PCR products (Fig. 2b, o, single arrowheads), suggesting the presence of circles or perhaps lariats in the total RNA preparations. Furthermore, consistent ladders of additional larger products were observed with both inward and outward primers (Fig. 2b), suggesting multiple rounds of reverse transcription (RT) around a circular RNA, followed by PCR amplification from the resulting concatameric cDNA (Fig. 2c).
To verify the composition of the RT-PCR products (Fig. 2b), we gel-purified, cloned, and sequenced primary and secondary products from all six reactions. The primary product of the inward primer reaction (Fig. 2b, i, single arrowheads) consisted of a simple linear box C/D RNA sequence with the configuration 1–2–3–4, as expected. The primary product generated with the outward primers (Fig. 2b, o, single arrowheads) included the junction of the termini of the linear RNA sequence (i.e., the circle ligation point). Specifically, the 3′ end of the linear RNA sequence was directly followed by the sequence of the 5′ end of the RNA in the configuration 3–4–1–2, as expected. The termini joined in the sequenced clones corresponded to the ends of linear RNAs of 62 or 63 nt for sR29, 64 nt for sR2, and 62 nt for sR14. Finally, the second product from ladders obtained with both inward and outward primers (including the products indicated with double arrowheads in Fig. 2b, which were isolated from the gels shown) consisted of two complete copies of the box C/D RNA in the configuration expected for RT-PCR from a circular RNA template (Fig. 2c). The results indicate that box C/D RNA circles or lariats are present in total RNA isolated from P. furiosus.
Next, we separately tested the two native forms of sR29 RNA. Gel-purified fast- and slow-migrating sR29 were subjected to RT-PCR with inward-directed primers. A single product of the expected size was obtained with the fast-migrating form (Fig. 2d, L) and was confirmed by sequencing to consist of a simple linear box C/D RNA sequence, as expected from a linear RNA. [We also have confirmed the sequence composition of the linear forms of sR29, sR2, and sR14 by RT-primer extension analysis (S.M., R.M.T., and M.P.T., unpublished data).] Conversely, RT-PCR with the slow-migrating form of sR29 produced a ladder of products like that observed with total RNA (Fig. 2d, C and T), consistent with its hypothesized circular conformation. Sequence analysis confirmed that the first three products in the ladder consist of one, two, and three copies of sR29, respectively. The sequence of the second ladder product is shown in Fig. 2e. Analysis of RT-PCR products from the fast- and slow-migrating forms of sR2 produced similar results (data not shown).
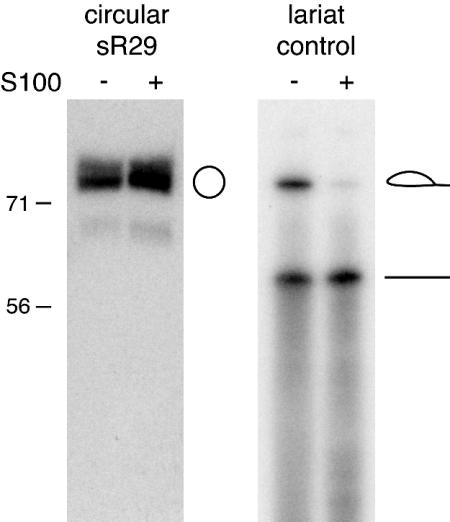
Taken together, our results strongly indicate that some box C/D RNAs exist in two prominent forms in P. furiosus: simple linear RNAs and RNA circles or lariats. Whereas circular RNAs are known to exist in archaea (21, 22), RNA lariats have not been described. To distinguish the possibilities, we tested the sensitivity of the RNA to debranching activity. The debranching activity present in HeLa S100 extract linearizes lariats (with tails as short as 1 nt) but not RNA circles (23). The slow-migrating form of sR29 and a positive control RNA (the lariat form of a yeast mitochondrial RNA group II intron) were incubated with S100 extract and analyzed by PAGE (Fig. 3). sR29 was resistant to debranching (Fig. 3), indicating that the slow-migrating form of sR29 is not likely a lariat.
Fig. 3.
Resistance of circular sR29 to lariat debranching activity. Slow-migrating sR29 and lariat control (group II intron lariat of yeast mitochondrial RNA) incubated with (+) and without (–) HeLa S100 extract. As observed in ref. 16, random nicking generates some linearized lariat RNA in the absence of S100 extract. Positions of a standard 71-nt RNA and 56-nt DNA relative to sR29 are indicated.
We originally had isolated incongruent clones (i.e., broken circles) for six P. furiosus box C/D RNAs and have analyzed three of these RNAs by RT-PCR. To objectively test the prevalence of the circular form among box C/D RNAs, we analyzed four RNAs for which we had not observed incongruent clones. Outward-directed primers for sR8, sR37, sR46, and sR55 were used in RT-PCR with total P. furiosus RNA. In each case, a ladder of products was observed, and sequencing confirmed the configuration of the primary RT-PCR product (i.e., 3–4–1–2). (In addition, we have observed electrophoretic mobility variants suggestive of circular forms in Northern analysis of every RNA that we have examined.) Our results indicate that most, if not all, of the box C/D RNAs exist as circles in P. furiosus.
The discovery of circular box C/D RNAs indicates that box C/D RNAs arise by a novel pathway in P. furiosus, one that produces significant amounts of circular RNA. An obvious possibility is the pathway that generates other RNA circles in archaea. Some of the few circular RNAs known to exist in nature are generated as byproducts and intermediates of tRNA and rRNA processing in archaea. The mechanism involves recognition of a bulge–helix–bulge motif formed by RNA sequences at the boundaries of a tRNA or rRNA intron (21, 22, 24). Cleavage occurs within the bulges, and the ends of the intron (as well as of the tRNA or rRNA) are subsequently ligated, resulting in a circularized, excised intron. Interestingly, one unusual box C/D RNA (sR40, not analyzed in this work) is located within the intron of pre-tRNATrp in euryarchaeotes, including P. furiosus, and thus likely exists in a circular form upon intron excision (12, 25). This unusual box C/D RNA is thought to act in cis (before excision) on its host RNA, pre-tRNATrp (25). However, the majority of box C/D RNAs are located in intergenic regions in archaea (12, 13). Moreover, we (the laboratories of R.M.T., M.P.T., and S.R.E.) and Christian Marck (Gif-sur-Yvette, France) have been unable to identify plausible bulge–helix–bulge motifs that could account for the generation of most circular box C/D RNAs, even by using the relaxed consensus motifs described in ref. 24, suggesting that box C/D RNAs may be generated by an uncharacterized pathway in P. furiosus.
Box C/D RNAs function as part of a complex with fibrillarin, Nop56/58, and L7Ae. As an indication of whether the circular box C/D RNAs could function in P. furiosus (or alternatively whether circularization of the box C/D RNAs might preclude function), we determined whether the circular RNAs were found in complexes with the box C/D proteins. Antibodies against P. furiosus Nop56/58 that coimmunoprecipitate fibrillarin and L7Ae were used to immunoprecipitate RNP complexes (Fig. 4a). We tested for the presence of box C/D RNAs in the complexes by Northern hybridization and found that both the linear and circular forms of sR29 and sR2 are associated with Nop56/58 complexes (Fig. 4b). RT-PCR analysis confirmed that circular sR29, sR2, and sR14 were coimmunoprecipitated with Nop56/58 (data not shown). Nop56/58 interacts with box C/D RNAs by means of the L7Ae protein and mediates the binding of fibrillarin (8). Thus, the circular box C/D RNAs are associated with box C/D protein complexes and could function to guide RNA modifications in P. furiosus.
Box C/D RNAs are noncoding RNAs, which, like rRNAs, tRNAs, and microRNAs, do not encode proteins but function directly in diverse cellular processes (26, 27). Many stable noncoding RNAs possess significant secondary structure. Maintaining RNA secondary structure is a particular challenge for hyperthermophiles like P. furiosus, which grows at 100°C. RNA secondary structure can be stabilized by enrichment in GC base pairs, and indeed the GC content of rRNA and tRNA increases with the optimal growth temperature of an organism (28). Klein et al. (29) successfully exploited the high GC content of structured noncoding RNAs to identify new noncoding RNAs in hyperthermophiles. Box H/ACA RNAs and all of the known tRNAs, rRNAs, RNase P RNA, and 7S RNA were identified, but none of the >50 known box C/D RNAs in P. furiosus were identified (unless they were directly adjacent to another structured noncoding RNA). One structural element found in box C/D RNAs and recognized for its importance in the stability and function of the box C/D RNAs in eukaryotes is the terminal stem (1, 2, 4, 5, 7, 30). The 5′ and 3′ termini of linear box C/D RNAs are normally tethered by base-pairing. This terminal stem contributes to the box C/D motif (a K turn), which is required for binding by L7Ae and assembly of a functional complex (8, 31, 32). However, the terminal stem is less, not more, prominent in the sequences of box C/D RNAs of the hyperthermophilic archaea including P. furiosus (12, 13). Perhaps a function of circularization is to secure the termini and essential secondary structure of box C/D RNAs in hyperthermophilic archaea.
The findings presented here place the box C/D RNAs among the rare circular RNAs known in nature. Known circular RNAs include the genomes of a very few subviral agents such as hepatitis delta virus and the viroids (33, 34). In addition, in cellular organisms, some RNA processing pathways give rise to circular RNA byproducts or intermediates. Perhaps the best known of these is the excised Group I intron of Tetrahymena preribosomal RNA, which was shown to be circularized after self-splicing in vitro by the Cech lab in 1981 (18). Both Group I introns (found in eukaryotes and bacteria) and bulge–helix–bulge-derived introns (found in archaea) can be circularized after excision from precursor tRNA and rRNA (18, 21, 22, 35, 36). In addition, in rare cases aberrant mRNA processing in eukaryotes can generate circular mRNA fragments (37–40). The previously described circular RNAs are not known to function directly in cellular processes, making the possibility that circular box C/D RNAs guide RNA modification in archaea even more provocative.
Circular RNAs could be more prevalent in biological systems than currently recognized. RNA circles normally would not be found by using standard RNA cloning approaches, where termini are required for cloning. In electrophoretic analyses such as Northern hybridization, circular RNAs can be misinterpreted as variant linear forms. The unexpected discovery of circular box C/D RNAs in P. furiosus has revealed a previously unrecognized physical form and biogenic pathway for this class of noncoding RNAs.
Acknowledgments
We thank Mike Adams, Gerti Schut, and Frank Jenney (University of Georgia) for generously sharing P. furiosus resources; Zhu-Hong Li for assistance with ribozyme experiments; Adrian Krainer for HeLa S100 extract; Kevin Jarrell for pJD20; Christian Marck and Henri Grosjean for scientific discussion; and Jim Dahlberg and Elsebet Lund (University of Wisconsin) and Claiborne V. C. Glover III (University of Georgia) for comments on the manuscript. This work was supported by a grant from the National Science Foundation (to M.P.T. and R.M.T.). S.R.E. and L.S.J. were supported by the Howard Hughes Medical Institute, National Institutes of Health, and Alvin Goldfarb.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: RNP, ribonucleoprotein; RT, reverse transcription.
References
- 1.Dennis, P. P., Omer, A. & Lowe, T. (2001) Mol. Microbiol. 40, 509–519. [DOI] [PubMed] [Google Scholar]
- 2.Terns, M. P. & Terns, R. M. (2002) Gene Expression 10, 17–39. [PMC free article] [PubMed] [Google Scholar]
- 3.Kiss, T. (2002) Cell 109, 145–148. [DOI] [PubMed] [Google Scholar]
- 4.Bachellerie, J. P., Cavaille, J. & Huttenhofer, A. (2002) Biochimie 84, 775–790. [DOI] [PubMed] [Google Scholar]
- 5.Filipowicz, W. & Pogacic, V. (2002) Curr. Opin. Cell Biol. 14, 319–327. [DOI] [PubMed] [Google Scholar]
- 6.Decatur, W. A. & Fournier, M. J. (2003) J. Biol. Chem. 278, 695–698. [DOI] [PubMed] [Google Scholar]
- 7.Omer, A. D., Ziesche, S., Decatur, W. A., Fournier, M. J. & Dennis, P. P. (2003) Mol. Microbiol. 48, 617–629. [DOI] [PubMed] [Google Scholar]
- 8.Omer, A. D., Ziesche, S., Ebhardt, H. & Dennis, P. P. (2002) Proc. Natl. Acad. Sci. USA 99, 5289–5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rozhdestvensky, T. S., Tang, T. H., Tchirkova, I. V., Brosius, J., Bachellerie, J. P. & Huttenhofer, A. (2003) Nucleic Acids Res. 31, 869–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tycowski, K. T. & Steitz, J. A. (2001) Eur. J. Cell Biol. 80, 119–125. [DOI] [PubMed] [Google Scholar]
- 11.Brown, J. W., Echeverria, M. & Qu, L. H. (2003) Trends Plant Sci. 8, 42–49. [DOI] [PubMed] [Google Scholar]
- 12.Omer, A. D., Lowe, T. M., Russell, A. G., Ebhardt, H., Eddy, S. R. & Dennis, P. P. (2000) Science 288, 517–522. [DOI] [PubMed] [Google Scholar]
- 13.Gaspin, C., Cavaille, J., Erauso, G. & Bachellerie, J. P. (2000) J. Mol. Biol. 297, 895–906. [DOI] [PubMed] [Google Scholar]
- 14.Speckmann, W. A., Li, Z. H., Lowe, T. M., Eddy, S. R., Terns, R. M. & Terns, M. P. (2002) Curr. Biol. 12, 199–203. [DOI] [PubMed] [Google Scholar]
- 15.Daniels, D. L., Michels, W. J., Jr., & Pyle, A. M. (1996) J. Mol. Biol. 256, 31–49. [DOI] [PubMed] [Google Scholar]
- 16.Peebles, C. L., Perlman, P. S., Mecklenburg, K. L., Petrillo, M. L., Tabor, J. H., Jarrell, K. A. & Cheng, H. L. (1986) Cell 44, 213–223. [DOI] [PubMed] [Google Scholar]
- 17.Bruce, A. G. & Uhlenbeck, O. C. (1978) Nucleic Acids Res. 5, 3665–3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grabowski, P. J., Zaug, A. J. & Cech, T. R. (1981) Cell 23, 467–476. [DOI] [PubMed] [Google Scholar]
- 19.Lorsch, J. R., Bartel, D. P. & Szostak, J. W. (1995) Nucleic Acids Res. 23, 2811–2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vogel, J., Hess, W. R. & Borner, T. (1997) Nucleic Acids Res. 25, 2030–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kjems, J. & Garrett, R. A. (1988) Cell 54, 693–703. [DOI] [PubMed] [Google Scholar]
- 22.Salgia, S. R., Singh, S. K., Gurha, P. & Gupta, R. (2003) RNA 9, 319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruskin, B. & Green, M. R. (1985) Science 229, 135–140. [DOI] [PubMed] [Google Scholar]
- 24.Marck, C. & Grosjean, H. (2003) RNA 9, 1516–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clouet d'Orval, B., Bortolin, M. L., Gaspin, C. & Bachellerie, J. P. (2001) Nucleic Acids Res. 29, 4518–4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eddy, S. R. (2001) Nat. Rev. Genet. 2, 919–929. [DOI] [PubMed] [Google Scholar]
- 27.Storz, G. (2002) Science 296, 1260–1263. [DOI] [PubMed] [Google Scholar]
- 28.Galtier, N. & Lobry, J. R. (1997) J. Mol. Evol. 44, 632–636. [DOI] [PubMed] [Google Scholar]
- 29.Klein, R. J., Misulovin, Z. & Eddy, S. R. (2002) Proc. Natl. Acad. Sci. USA 99, 7542–7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiss, T. (2001) EMBO J. 20, 3617–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watkins, N. J., Dickmanns, A. & Luhrmann, R. (2002) Mol. Cell. Biol. 22, 8342–8352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marmier-Gourrier, N., Clery, A., Senty-Segault, V., Charpentier, B., Schlotter, F., Leclerc, F., Fournier, R. & Branlant, C. (2003) RNA 9, 821–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kos, A., Dijkema, R., Arnberg, A. C., van der Meide, P. H. & Schellekens, H. (1986) Nature 323, 558–560. [DOI] [PubMed] [Google Scholar]
- 34.Sanger, H. L., Klotz, G., Riesner, D., Gross, H. J. & Kleinschmidt, A. K. (1976) Proc. Natl. Acad. Sci. USA 73, 3852–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tabak, H. F., Van der Horst, G., Osinga, K. A. & Arnberg, A. C. (1984) Cell 39, 623–629. [DOI] [PubMed] [Google Scholar]
- 36.Daros, J. A. & Flores, R. (1996) RNA 2, 928–936. [PMC free article] [PubMed] [Google Scholar]
- 37.Hensgens, L. A., Arnberg, A. C., Roosendaal, E., van der Horst, G., van der Veen, R., van Ommen, G. J. & Grivell, L. A. (1983) J. Mol. Biol. 164, 35–58. [DOI] [PubMed] [Google Scholar]
- 38.Capel, B., Swain, A., Nicolis, S., Hacker, A., Walter, M., Koopman, P., Goodfellow, P. & Lovell-Badge, R. (1993) Cell 73, 1019–1030. [DOI] [PubMed] [Google Scholar]
- 39.Cocquerelle, C., Mascrez, B., Hetuin, D. & Bailleul, B. (1993) FASEB J. 7, 155–160. [DOI] [PubMed] [Google Scholar]
- 40.Zaphiropoulos, P. G. (1996) Proc. Natl. Acad. Sci. USA 93, 6536–6541. [DOI] [PMC free article] [PubMed] [Google Scholar]