Abstract
Understanding the evolutionary relationship between genome content and ecological niche is one of the fundamental challenges of biology. The distinct physiologies of yeast species provide a window into how genomes evolve in concert with niche. Although the enzymes of the well studied yeast galactose utilization pathway are present in all domains of life, we have found that multiple genes of the GAL pathway are absent from four yeast species that cannot use galactose. Whereas three species lack any trace of the pathway except a single gene, Saccharomyces kudriavzevii, a close relative of Saccharomyces cerevisiae, retains remnants of all seven dedicated GAL genes as syntenic pseudogenes, providing a rare glimpse of an entire pathway in the process of degeneration. An estimate of the timing of gene inactivation suggests that pathway degeneration began early in the lineage and proceeded rapidly. S. kudriavzevii exhibits several other divergent physiological properties that are associated with a shift in ecological niche. These results suggest that rapid and irreversible gene inactivation and pathway degeneration are associated with adaptation to new ecological niches in natural populations. Inactivated genes may generally serve as markers of specific functions made dispensable by recent adaptive shifts.
Evolution takes place in an ecological context that shapes the contents of the genomes of species (1, 2). When a population adapts to a new ecological niche, it encounters a different selection regime that imposes altered physiological demands (1–3). Under new selection regimes, adaptations may evolve while established functions may become less important. Many components of genetic pathways are expected to be resistant to mutation because genes often have pleiotropic functions and complex interactions with one another and with other pathways. Pleiotropy may constrain how the evolution of genetic pathways proceeds and which genetic changes can be exploited. However, little is known about the effects that changes in selection pressure have on genetic pathways and the content of the genome.
Yeast species present an exceptional opportunity to study the fate of genetic pathways during evolution because numerous species have complete genome sequences that are available. The well characterized and diverse physiologies (4, 5) of these species allow strong inferences to be made about the importance of nutrients in their respective ecological niches. Some substrates, such as galactose, are commonly used, whereas other substrates, such as the plant-made fructose polymer inulin, are used by fewer species (4, 5).
As a model of the genetic effects of changing selection regimes, we examined the evolution of the Leloir galactose utilization pathway in yeast, one of the best-studied eukaryotic genetic pathways (6, 7). This pathway converts galactose into glucose-6-phosphate, which enters glycolysis (6, 7). The GAL pathway of Saccharomyces cerevisiae is a complex metabolic and genetic pathway that is composed of both regulatory components (GAL3, coinducer; GAL4, transcriptional activator; and GAL80, corepressor) and structural components (GAL1, galactokinase; GAL2, galactose permease; GAL7, galactose-1-phosphate uridyl transferase; and GAL10, UDP-glucose 4-epimerase) (6, 7). In the absence of galactose, Gal80p prevents transcriptional activation of the pathway by means of a physical interaction with Gal4p (6, 7). In the presence of galactose, Gal3p relieves this repression, and Gal4p activates the transcription of target genes, including some that are not involved directly in galactose catabolism (6–9).
Despite galactose utilization being widespread among the yeasts and the broad distribution of the GAL enzymes through all domains of life (6), several yeast species lack the ability to use galactose (4, 5, 10). Here, we have investigated the genetic basis of the inability to use galactose in several yeast species. We have found that at least three independent lineages of yeast have inactivated or lost most or all of the genes of the GAL pathway while leaving interacting genes intact. The parallel losses of this entire pathway are extreme examples of an emerging general principle that gene-inactivation events reveal specific functions affected by recent changes in the ecological pressures acting on a species.
Materials and Methods
Phylogenetic Reconstruction. The genomes of Candida glabrata, Kluyveromyces lactis, Kluyveromyces waltii, and Eremothecium gossypii (syn. Ashbya gossypii; refs. 11–13) were screened for annotated orthologs of 106 genes from a previous genome-scale phylogenetic analysis (14). Candida albicans (15) was used as the outgroup. Individual genes were aligned by codon by using clustal w (16) as implemented in bioedit (version 5.0.9) (17). All gene alignments were manually edited to exclude indels and areas of ambiguous alignment from further analysis. Because individual genes have been shown to have a high probability of supporting conflicting topologies (14), only the concatenated data set was analyzed. Phylogenetic analyses were performed on nucleotides under the optimality criteria of maximum likelihood (ML) and maximum parsimony (MP) as implemented in paup* (version 4.0b10) (18). In both ML and MP analyses, tree space was searched by using the branch-and-bound algorithm, which guarantees the finding of optimal tree(s). Tree reliability under ML and MP was assessed by using nonparametric bootstrap resampling of 100 replicates. With MP, all characters were equally weighted, whereas with ML, the model of sequence evolution was optimized by using likelihood-ratio tests (19) as implemented in modeltest (version 3.06) (20).
Genome Searches. There were no annotated GAL genes for Saccharomyces kudriavzevii, C. glabrata, K. waltii, and E. gossypii, except for E. gossypii GAL4. To confirm gene absence, we performed blastp and tblastn searches (wu-blast 2.0, http://blast.wustl.edu) against the ORF annotations and genome contigs (11–13, 21), respectively. Except for the S. kudriavzevii GAL pseudogenes, we found no additional GAL genes or pseudogenes. S. kudriavzevii, C. glabrata, K. waltii, and E. gossypii homologs of MIG1, PGM1, PGM2, GCY1, MTH1, and PCL10 were also located by using this approach and confirmed by performing blastp searches against the annotated S. cerevisiae genome [Saccharomyces Genome Database (SGD), www.yeastgenome.org]. Because of a gap in the genome sequence, the existence of Saccharomyces kluyveri GAL1 was inferred from a previous PCR assay (22), whereas complete or partial sequences for other S. kluyveri GAL genes were present on contigs (21).
Relative-Rate Test. The accelerated rate of evolution of GAL4 in the lineage leading to E. gossypii was assessed by a relative-rate test (23) in which E. gossypii GAL4 was compared with S. kluyveri GAL4 (21), with K. lactis GAL4 (11) as the outgroup (X21 = 15.69, P = 0.000075). The relative-rate test (23), as implemented in the mega software (24) was applied to protein alignments produced with dialign 2 (25). Only nongapped sites aligned in all three species were used.
Sequencing. The sequences of existing S. kudriavzevii contigs (21) were used to design primers that amplified all five GAL loci such that at least one complete ORF on either side of the GAL pseudogene or pseudogene complex was included in the PCR product. Product-specific primers were then used to sequence the PCR products. At least three independent PCRs were pooled, and all bases were confirmed by a minimum of two unambiguous reads. The same method was used to complete and verify the sequences of S. kudriavzevii PCL10, PGM2, and PGM1 (21) and to acquire the sequence of a large gap surrounding Saccharomyces mikatae GAL2 (26), except that sequencing primers were designed progressively until the large gap was closed. We closed 11 gaps in the S. kudriavzevii genome sequence (21) and a 4,990-bp gap in the S. mikatae genome sequence (26), and we corrected minor assembly and sequencing artifacts, including a frameshift that truncated S. kudriavzevii PGM1 incorrectly (21). These changes have been submitted to SGD to curate and integrate into the current genome sequences (21, 26).
Gal4p Binding-Site Statistics. By using the neutral rate of evolution Dneu for the lineage leading to S. kudriavzevii as determined below, the probability that none of the 18 consensus nucleotides in the Gal4p upstream activating sequences (UASs) in GCY1, MTH1, and PCL10 would have changed was calculated as follows: p = (1 – Dneu)18 = 0.0044.
Modified Relative-Rate Test. Because of the difficulty of determining the beginning and end of the pseudogenes of S. kudriavzevii, the intergenic region between the two neighboring genes was used. For S. cerevisiae, S. mikatae, and Saccharomyces bayanus, the corresponding orthologous ORFs were used (refs. 21 and 26; SGD). Alignments were performed by using dialign 2 with the codon-alignment option (25), which generally agreed with blastx analysis. Only nongapped sites aligned in all four species were analyzed further.
The best-fit ML model was identified by using likelihood-ratio tests (19), as implemented in modeltest (version 3.0.6) (20). The optimal parameter values suggested by modeltest were used in the calculation of the branch lengths of the species tree (14) in a ML framework, as implemented in paup* (version 4.0b10) (18). Branch lengths were expressed in terms of the expected number of substitutions per nucleotide site (ENSNS).
We define DScer, DSmik, and DSkud as the ENSNSs from the node of the common ancestor of S. cerevisiae, S. mikatae, and S. kudriavzevii to the tips of the S. cerevisiae, S. mikatae, and S. kudriavzevii branches, respectively. DSkud is a composite of two ENSNSs, DSkud_sc and DSkud_neu; DSkud_sc represents the ENSNS during the period in which the gene was functionally active and presumably under selective constraint, whereas DSkud_neu reflects the ENSNS from the point in time in which function was lost, and mutation accumulation proceeded at the neutral rate. DSkud_sc was estimated under the assumption that DSkud_sc ≅ DScer ≅ DSmik. Fig. 3, which is published as supporting information on the PNAS web site, graphically depicts these values and their determination. This estimate of DSkud_neu was conservative because it underestimated the time since the S. kudriavzevii gene had been a pseudogene by subtracting DScer or DSmik and assuming that the gene was under selective constraint until very recently in evolutionary time. Under this assumption, the amount of the neutral ENSNS in the S. kudriavzevii sequence since loss of function occurred was calculated as DSkud_neu = DSkud – (DScer + DSmik)/2. The use of DScer or DSmik instead of their average produced very similar estimates. Confidence intervals (95%) were obtained by nonparametric bootstrap resampling of 100 replicates. For functionally constrained genes, negative values can be produced by this test because the neutral rate is slower in S. kudriavzevii [0.26 (0.02)] than in S. cerevisiae [0.48 (0.02)] and S. mikatae [0.46 (0.02), as calculated below].
To calculate the relative timing since gene inactivation of each S. kudriavzevii ortholog, we estimated the genome-wide neutral ENSNS for S. kudriavzevii Dneu by analysis of the 4-fold degenerate sites from a 106-gene data set for these four species (14) using the same analyses as described above. This method may underestimate the neutral rate by not taking into account codon bias but using subsets of genes with different degrees of codon bias produced similar results (data not shown). We calculated the DSkud_neu/Dneu ratio for each gene. Ratio values close to one suggest that gene inactivation occurred early in the evolution of the S. kudriavzevii lineage, whereas values close to zero suggest that gene inactivation occurred recently. Control genes and the concatenated seven-pseudogene GAL data set were treated in the same way.
Results and Discussion
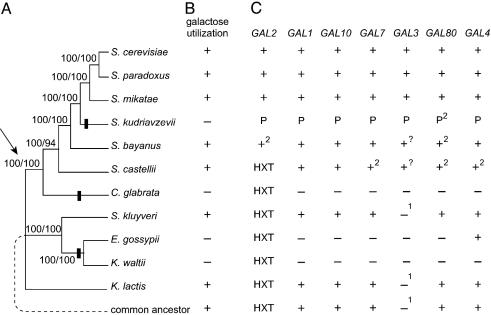
Parallel Losses of Galactose Utilization. To determine whether the inability to use galactose has evolved independently in different yeast species, we used available genomic data to examine the phylogenetic relationships of seven species that can use galactose (S. cerevisiae, Saccharomyces paradoxus, S. mikatae, S. bayanus, Saccharomyces castellii, S. kluyveri, and K. lactis) and four that cannot (S. kudriavzevii, C. glabrata, K. waltii, and E. gossypii) (refs. 4, 5, 10–13, 21, and 26 and SGD). Phylogenetic reconstruction using two different optimality criteria provides unequivocal support for the following two major clades: a clade that includes the closely related sensu stricto yeast species (S. cerevisiae, S. paradoxus, S. mikatae, S. kudriavzevii, and S. bayanus), S. castellii, and C. glabrata; and a clade that includes S. kluyveri, K. waltii, and E. gossypii (Fig. 1A). This phylogeny suggests that galactose utilization is ancestral and that there have been at least three parallel losses to account for the inability of S. kudriavzevii, C. glabrata, K. waltii, and E. gossypii to use galactose (Fig. 1 A and B).
Fig. 1.
Multiple evolutionary losses of the GAL pathway in yeast. (A) Phylogenetic relationships of the analyzed yeast species. Numbers at branches represent bootstrap support (ML/MP). Vertical bars represent GAL pathway losses inferred by parsimony. Arrow indicates the branch along which the whole genome duplication is inferred to have occurred (12, 13, 43). The exact placement of K. lactis is unresolved because the two methods of analysis produce conflicting results. Whereas ML suggests that K. lactis is an outgroup to all species shown (87% bootstrap support), MP suggests that it is the outgroup to the S. kluyveri/K. waltii/E. gossypii clade (100% bootstrap support). Also, analyses using the amino acid sequence data instead of nucleotides produce conflicting topologies regarding the placement of E. gossypii (data not shown). (B) Species that use galactose or species in which strains exhibit variation in galactose utilization are indicated as positive (+), whereas only species in which no isolates are capable of using galactose are indicated as negative (–) (4, 5, 10). (C) The presence (+), absence (–), or pseudogene status (P) is shown for GAL pathway components. 1, Species inferred to have a bifunctional GAL1 gene that also performs the function of GAL3 (7, 44).?, Species has a GAL3 ortholog syntenic with GAL3 but with ambiguous functional identity. Unlike S. cerevisiae, S. bayanus GAL3 contains a consensus GLSSSAA galactokinase motif (45). S. castellii GAL3 does not contain a consensus galactokinase motif (45) but has an alanine-to-serine substitution in the motif rather than the two-residue deletion present S. cerevisiae. 2, Species has two paralogs of the gene or pseudogene. S. castellii GAL4B lies on the Watson strand between SGS1 and SPG5. S. castellii and S. bayanus GAL80B lie on the Crick strand between HIT1 and YJR056C. S. kudriavzevii GAL80B is a pseudogene syntenic with the above GAL80B genes, but it has degenerated further than the other GAL pseudogenes. S. castellii has two GAL enzyme-encoding gene complexes: one that is syntenic with GAL7, GAL10, and GAL1 that contains GAL7 and GAL1; and one that is syntenic with GAL3 that contains GAL7B, GAL10B, and GAL3. S. bayanus GAL2B is a tandem duplicate. HXT, unambiguous GAL2 orthologs cannot be identified, but members of the large hexose transporter family, of which Gal2p is a member, are indicated.
Inactivation or Loss of Multiple GAL Pathway Genes. The regulation and function of the GAL pathway of S. cerevisiae can be disrupted by mutations in several genes (6, 7). In addition to PGM2 (GAL5), which is shared with other pathways, the GAL pathway contains seven genes dedicated to galactose catabolism or regulating the transcriptional response to galactose levels (Fig. 2 A and B) (6, 7), but it is not known whether any of these genes are involved in naturally occurring interspecific differences in the ability to use galactose. To analyze and compare possible genetic bases of the inability to use galactose, we searched several genomes (refs. 11–13, 21, and 26; SGD) for ORFs representing GAL pathway components. We found a tight correlation between the ability to use galactose and the presence of GAL pathway members (Fig. 1 B and C).
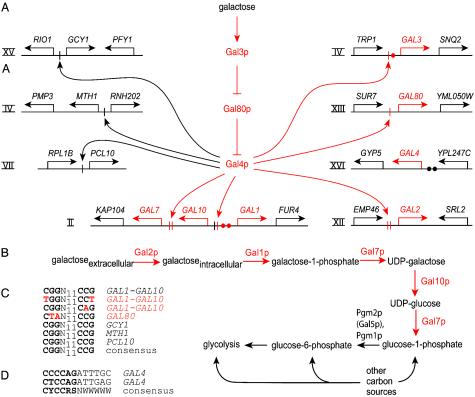
Fig. 2.
The S. cerevisiae GAL pathway and its degeneration in S. kudriavzevii. Pseudogenes and degenerate or missing components of S. kudriavzevii are shown in red. (A) Regulation of the GAL pathway and other Gal4p target genes in S. cerevisiae (refs. 6–9 and 27–29; SGD) and corresponding S. kudriavzevii loci. Genes are represented by arrows for genes on the Watson strand (pointing right) and genes on the Crick strand (pointing left). Roman numerals indicate chromosome number. A single Gal4p UAS (6, 9) from S. cerevisiae is indicated by a single bar, and multiple UASs for a given gene are indicated by a double bar. A single Gal4p UAS allows moderate activation of transcription in the presence of galactose and low-level activation in the absence of galactose, whereas multiple Gal4p UASs allow strong activation in the presence of galactose and almost no activation without galactose (6, 9, 29). Gal4p UASs activate downstream promoters, but both divergent promoters are not always affected; GAL1, GAL10, GCY1, and MTH1 are activated by Gal4p, but RIO1 and RNH202 are not (6, 8, 9). Consensus Mig1p binding sites (27) responsible for catabolite repression (28) are indicated by filled circles. Arrows within the regulatory pathway indicate activation, and lines ending against a perpendicular line indicate repression. (B) Galactose import and catabolism in S. cerevisiae (6, 7). Pgm2p is also known as Gal5p because of its mutant phenotype and role in galactose catabolism (6, 7). Alignable sequences of S. kudriavzevii that are orthologous to S. cerevisiae Gal4p UASs (C) and Mig1p binding sites (D) are compared with the consensus sites (6, 9, 27). N11, the internal 11 nucleotides of Gal4p UASs. The residues that are most important for function are shown in bold (6, 9, 27). Y, C or T; R, A or G; S, C or G; W, A or T.
In all species that can use galactose, we found enzymatic and regulatory components of the GAL pathway. In contrast, we found neither functional GAL enzymes nor the corepressor GAL80 in the genomes of the four species that cannot use galactose. The precise fates of GAL pathway genes differ between lineages. All dedicated GAL genes are absent and untraceable in C. glabrata and K. waltii. Whereas the other dedicated GAL genes are also absent and untraceable in E. gossypii, a syntenic GAL4 ortholog has been retained (Fig. 1C). E. gossypii GAL4 has undergone an accelerated rate of evolution in this lineage (P < 10–4), suggesting it has been exposed to a novel selection regime and may have acquired new biological roles. In contrast, S. kudriavzevii, a sensu stricto species closely related to S. cerevisiae, retains pseudogenes at syntenic loci for all seven dedicated members of the GAL pathway (Figs. 1C and 2 A and B). This finding provides a rare opportunity to study an entire pathway in the process of degeneration.
GAL Pseudogenes and Pathway Degeneration. We completed and verified the sequences of each S. kudriavzevii GAL pseudogene and their adjacent ORFs and examined them for the footprints left by the process of pathway degeneration. All GAL pseudogenes contain multiple stop codons, frameshifts, and deletions that are predicted to render them nonfunctional (Fig. 2 A). We examined the degree of degeneration of the pseudogenes, reasoning that pseudogenes that were more advanced in the process of degeneration might have been inactivated first. Analysis of the upstream regulatory sequences of the GAL pseudogenes might further suggest which genes had degenerated most and allow us to infer whether additional constraints were acting on the regulatory sequences during pathway degeneration. We also examined the upstream regulatory sequences of non-GAL Gal4p target genes to determine how the loss of functional Gal4p might have affected their Gal4p UASs.
The degree of degeneration varied among the upstream regulatory sequences of the GAL pseudogenes (Fig. 2 A, C, and D). The entire GAL7 upstream sequence was deleted, along with the 5′ end of the GAL7 ORF and the 3′ end of the GAL10 ORF. No alignable traces of the GAL2 and GAL3 regulatory sequences were found. The GAL1–GAL10 regulatory region retained only a 60- to 70-bp remnant in S. kudriavzevii that aligned with three adjacent Gal4p UASs found in other sensu stricto yeasts (22, 26), but only one of the S. kudriavzevii UASs would be predicted to be bound by Gal4p (Fig. 2 A and C) (6, 9). The GAL80 and GAL4 upstream regulatory sequences aligned well with those of other species and contained only small deletions and mutations, including a degenerated Gal4p UAS in GAL80 (Fig. 2 A, C, and D). The remnants of these regulatory sequences suggest that purifying selection may have been acting on these promoters longer than on the other genes of the pathway. Furthermore, the GAL4 regulatory sequence retains the Mig1p binding sites (27), which are responsible for catabolite repression of the GAL pathway (Fig. 2 A and D) (28), suggesting there may have been selection for transcriptional repression of the GAL4 pseudogene during pathway degeneration.
Gal4p also positively regulates genes that are not a direct part of the GAL pathway, and a single Gal4p UAS allows for weak activation even in the absence of galactose (6, 8, 9, 29). This interaction raises the question of how these regulatory sequences might be affected by the loss of Gal4p function. A functional Gal4p UAS occurs in GCY1 (8), and both chromatin immunoprecipitation and galactose-dependent increases in transcription suggest that MTH1 and PCL10 are Gal4p targets (9). In the genome of S. cerevisiae, we located putative Gal4p UASs upstream of MTH1 and PCL10. The S. kudriavzevii upstream sequences of GCY1, MTH1, and PCL10 retain orthologous consensus Gal4p UASs (Fig. 2 A and C) (9), despite the fact that Gal4p is no longer present to regulate these genes. Assuming a neutral rate of evolution for these sites during the entire S. kudriavzevii lineage, at least 1 of the 18 consensus nucleotides in these three sites would be expected to have changed (P < 0.005). The retention of these sites suggests that the GAL4 gene was functional for some length of time in this lineage or that other factors were constraining the evolution of these sites during the evolution of the S. kudriavzevii lineage.
Early and Rapid Pathway Degeneration. A classic model for the evolution of anabolic pathways predicts enzymatic steps are added in reverse order of the pathway (30). Although no explicit models of pathway degeneration have been proposed, one possible corollary for the degeneration of catabolic pathways would be that enzymatic steps might be lost in the forward order of the pathway. Another possibility is that more pleiotropic genes might be retained longer during pathway degeneration to allow time for other genes to compensate for their pleiotropic roles. Because Gal4p regulates genes that are not dedicated members of the GAL pathway (8, 9) and Gal80p and Gal3p play important roles in regulating Gal4p (6, 7), we hypothesized that regulatory components of the GAL pathway might be more pleiotropic and retained longer than structural components during pathway degeneration. The retention of Gal4p UASs in non-GAL Gal4p targets, the retention of only GAL4 in E. gossypii, the relatively conserved GAL4 and GAL80 regulatory sequences, and the absence of major deletions in the pseudogenes of GAL4 and GAL80 are consistent with this scenario.
To determine whether the sequences of the pseudogenes revealed any evidence for ordered gene inactivation, we used a modified relative-rate test to estimate the relative time at which each gene became a pseudogene. All seven dedicated GAL genes showed an excess of substitutions far beyond what is expected for functionally constrained genes (Table 1). The excess substitutions present in individual pseudogenes were not significantly different from one another, and they provide no statistical support for ordered gene inactivation (Table 1). Although we cannot exclude that there was a historical order of gene inactivation, we conclude that pathway degeneration was so rapid that it is beyond resolution by this method. In contrast, several genes that interact with the GAL pathway show no elevated rate of evolution in S. kudriavzevii and are also present in C. glabrata, K. waltii, and E. gossypii, suggesting that the degeneration of the GAL pathway was gene-specific and had no overt effect on more pleiotropic genes (Table 1 and Fig. 2 A and B) (6–9, 28). Comparison of the rate of evolution of the pseudogenes with the S. kudriavzevii neutral rate yields strong support for pathway degeneration beginning early in the evolution of the lineage leading to S. kudriavzevii (Table 1).
Table 1. Excess ENSNS and relative timing of inactivation of dedicated GAL genes and interacting genes.
| Gene | Length, bp | Dskud_neu (95% C.I.) | Dskud_neu/Dneu (95% C.I.) |
|---|---|---|---|
| GAL2 | 644 | 0.34 (0.13) | 1.31 (0.48) |
| GAL1 | 728 | 0.30 (0.12) | 1.17 (0.47) |
| GAL10 | 547 | 0.17 (0.11) | 0.66 (0.43) |
| GAL7 | 390 | 0.19 (0.13) | 0.72 (0.49) |
| GAL3 | 625 | 0.13 (0.09) | 0.52 (0.36) |
| GAL80 | 1,088 | 0.24 (0.06) | 0.95 (0.25) |
| GAL4 | 1,583 | 0.19 (0.07) | 0.74 (0.26) |
| All GAL genes | 5,605 | 0.21 (0.03) | 0.82 (0.13) |
| GCY1 | 921 | -0.07 (0.06) | -0.26 (0.25) |
| MTH1 | 1,287 | -0.05 (0.05) | -0.21 (0.21) |
| PCL10 | 1,284 | -0.05 (0.06) | -0.20 (0.23) |
| PGM2 (GAL5) | 1,710 | -0.04 (0.05) | -0.16 (0.20) |
| PGM1 | 1,699 | -0.09 (0.05) | -0.35 (0.20) |
| MIG1 | 1,476 | -0.09 (0.05) | -0.34 (0.19) |
Dskud_neu, ML estimate of the excess ENSNS that have occurred since gene inactivation. Dskud_neu/Dneu, relative timing of the inactivation of each gene with lineage divergence, where 1 suggests early inactivation and 0 suggests recent inactivation. C.I., confidence interval established by nonparametric bootstrap resampling.
The Divergent Ecology of S. kudriavzevii. The early timing, extent, and rapidity of GAL pathway degeneration in the S. kudriavzevii lineage suggests that a change in niche may have relieved the purifying selection acting on the GAL pathway genes. In addition to being the only characterized sensu stricto yeast species that cannot use galactose, S. kudriavzevii exhibits several unusual properties (10). It is the only sensu stricto species that readily utilizes the plant-made fructose polymer inulin or that secretes starch-like compounds, and it is one of two sensu stricto species that readily utilizes galactitol (4, 5, 10). In liquid culture, S. kudriavzevii exhibits unusually heavy aggregation and sedimentation. Of the four known S. kudriavzevii strains, three were isolated on decaying leaves and one was isolated from the soil (10) (Japan National Institute of Technology and Evaluation Biological Resource Center, www.nbrc.nite.go.jp), in contrast with the sugar-rich substrates where most other sensu stricto strains have been isolated (4, 5). The rapid degeneration of the GAL pathway of this species may represent only one set of the physiologically relevant genetic changes that occurred within this lineage. Therefore, S. kudriavzevii may provide a model of niche specialization in which the full complement of yeast genetic tools can be applied.
Lineage-Specific Pseudogenes Mark Adaptive Shifts. This study demonstrates that the parallel loss of multiple members of a genetic pathway correlates with the loss of its physiological function. The loss of genes and pathways through reductive evolution has been inferred for many organisms that have adapted to pathogenic or endosymbiotic lifestyles (11, 31–37). Although the losses of the GAL pathway in E. gossypii and C. glabrata (11) may be related to a pathogenic lifestyle, S. kudriavzevii and K. waltii are not known pathogens. In extreme cases, reductive evolution has been observed as a genome-wide phenomenon (31–33, 35, 36). In contrast, the losses of the GAL pathway in the yeast species studied here were gene-specific because interacting genes with more pleiotropic functions (such as PGM2 (GAL5), GCY1, and MIG1) remained intact, with no elevated rate of evolution. With few exceptions (33, 34), bacterial pseudogenes are rare, presumably because of a strong deletion bias (32). The absence of GAL pseudogenes in three of the yeast species that we studied suggests that there may be a similar deletion bias in yeasts or that the losses of their GAL pathways are ancient.
It has been shown that adaptation to a new ecological niche may result in a “cost” in terms of lost ancestral capabilities (1–3). These capabilities may be lost either because they are no longer under selection (neutral) or because of a deleterious effect on fitness in a new niche (1–3). The parallel losses of the ability to use galactose provide clear examples of the cost of adaptation in genetic terms, namely, the irreversible degeneration of multiple genes of the GAL pathway such that these genes are no longer available for future deployment. Studies of evolution in laboratory strains have demonstrated that gene inactivation can occur rapidly (3, 38). The inactivation of seven pathway members early in the lineage leading to S. kudriavzevii suggests that gene inactivation can also occur rapidly in natural populations. Nonetheless, the specificity of gene inactivation argues that pleiotropy constrains which genes are affected. In no case did we find the inactivation of non-GAL Gal4p-regulated genes or the inactivation of genes involved in cross-pathway interactions with the GAL pathway. The degeneration of the E. gossypii GAL pathway appears to have been more constrained because GAL4 was retained, presumably because it plays important roles in regulating other biological processes.
Recent studies (11, 31–42) are beginning to suggest that adaptation to new ecological niches may be associated with gene inactivation. For example, associations have been made between gene inactivation in the morning glory pigmentation pathway and the evolution of pollinator preference (40), excess olfactory receptor pseudogenes and the evolution of true trichromatic vision in primates (41), and the inactivation of a gene encoding a tissue-specific myosin heavy chain and an evolutionary decrease in jaw musculature in the human lineage (42). Although gene inactivation is more likely to be a consequence than a cause of adaptation (3), inactivated genes and pseudogenes are evidence that an adaptive shift in niche might have occurred. In general, the inactivation of genes that have been conserved in closely related taxa provides conspicuous clues as to which ancestral functions became dispensable as changing ecological demands imposed altered selective constraints. The application of this basic principle to the wealth of incoming genomic data may identify genetic changes associated with ecological shifts in recent evolutionary lineages.
Supplementary Material
Acknowledgments
We thank M. Johnston (Washington University, St. Louis) for providing the S. kudriavzevii IFO 1802T and S. mikatae IFO 1815T strains used in this study, L. Olds for assistance with graphics, B. L. Williams and J. H. Yoder for critical reading of the manuscript, and J. F. Crow and Carroll Laboratory members for helpful discussions. C.T.H. is a Howard Hughes Medical Institute Predoctoral Fellow, A.R. is a Human Frontier Science Program Long-Term Fellow, and S.B.C. is an Investigator of the Howard Hughes Medical Institute.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ENSNS, expected number of substitutions per nucleotide site; SGD, Saccharomyces Genome Database; UAS, upstream activating sequence; ML, maximum likelihood; MP, maximum parsimony.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AY740026–AY740033 (S. kudriavzevii) and AY740034 (S. mikatae)].
References
- 1.Bell, G. (1997) The Basics of Selection (Chapman & Hall, New York).
- 2.Kassen, R. (2002) J. Evol. Biol. 15, 173–190. [Google Scholar]
- 3.MacLean, R. C. & Bell, G. (2002) Am. Nat. 160, 569–581. [DOI] [PubMed] [Google Scholar]
- 4.Kurtzman, C. P. & Fell, J. W. (1998) The Yeasts, a Taxonomic Study (Elsevier, Amsterdam).
- 5.Barnett, J. A., Payne, R. W. & Yarrow, D. (2000) Yeasts: Characteristics and Identification (Cambridge Univ. Press, Cambridge, U.K.).
- 6.Johnston, M. (1987) Microbiol. Rev. 51, 458–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhat, P. J. & Murthy, T. V. S. (2001) Mol. Microbiol. 40, 1059–1066. [DOI] [PubMed] [Google Scholar]
- 8.Magdolen, V., Oechsner, U., Trommler, P. & Bandlow, W. (1990) Gene 90, 105–114. [DOI] [PubMed] [Google Scholar]
- 9.Ren, B., Robert, F., Wyrick, J. J., Aparicio, O., Jennings, E. G., Simon, I., Zeitlinger, J., Schreiber, J., Hannett, N., Kanin, E., et al. (2000) Science 290, 2306–2309. [DOI] [PubMed] [Google Scholar]
- 10.Naumov, G. I., James, S. A., Naumova, E. S., Louis, E. J. & Roberts, I. N. (2000) Int. J. Syst. Evol. Microbiol. 50, 1931–1942. [DOI] [PubMed] [Google Scholar]
- 11.Dujon, B., Sherman, D., Fischer, G., Durrens, P., Casaregola, S., Lafontaine, I., de Montigny, J., Marck, C., Neuveglise, C., Talla, E., et al. (2004) Nature 430, 35–44.15229592 [Google Scholar]
- 12.Kellis, M., Birren, B. W. & Lander, E. S. (2004) Nature 428, 617–624. [DOI] [PubMed] [Google Scholar]
- 13.Dietrich, F. S., Voegeli, S., Brachat, S., Lerch, A., Gates, K., Steiner, S., Mohr, C., Pohlmann, R., Luedi, P., Choi, S., et al. (2004) Science 304, 304–307. [DOI] [PubMed] [Google Scholar]
- 14.Rokas, A., Williams, B. L., King, N. & Carroll, S. B. (2003) Nature 425, 798–804. [DOI] [PubMed] [Google Scholar]
- 15.Jones, T., Federspiel, N. A., Chibana, H., Dungan, J., Kalman, S., Magee, B. B., Newport, G., Thorstenson, Y. R., Agabian, N., Magee, P. T., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 7329–7334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall, T. A. (1999) Nucleic Acids Symp. Ser. 41, 95–98. [Google Scholar]
- 18.Swofford, D. L. (2002) paup*, Phylogenetic Analysis Using Parsimony (*and Other Methods) (Sinauer, Sunderland, MA).
- 19.Huelsenbeck, J. P. & Rannala, B. (1997) Science 276, 227–232. [DOI] [PubMed] [Google Scholar]
- 20.Posada, D. & Crandall, K. A. (1998) Bioinformatics 14, 817–818. [DOI] [PubMed] [Google Scholar]
- 21.Cliften, P., Sudarsanam, P., Desikan, A., Fulton, L., Fulton, B., Majors, J., Waterston, R., Cohen, B. A. & Johnston, M. (2003) Science 301, 71–76. [DOI] [PubMed] [Google Scholar]
- 22.Cliften, P. F., Hillier, L. W., Fulton, L., Graves, T., Miner, T., Gish, W. R., Waterston, R. H. & Johnston, M. (2001) Genome Res. 11, 1175–1186. [DOI] [PubMed] [Google Scholar]
- 23.Tajima, F. (1993) Genetics 135, 599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar, S., Tamura, K., Jakobsen, I. B. & Nei, M. (2001) Bioinformatics 17, 1244–1245. [DOI] [PubMed] [Google Scholar]
- 25.Morgenstern, B. (1999) Bioinformatics 15, 211–218. [DOI] [PubMed] [Google Scholar]
- 26.Kellis, M., Patterson, N., Endrizzi, M., Birren, B. & Lander, E. S. (2003) Nature 423, 241–254. [DOI] [PubMed] [Google Scholar]
- 27.Lundin, M., Nehlin, J. O. & Ronne, H. (1994) Mol. Cell. Biol. 14, 1979–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnston, M. (1999) Trends Genet. 15, 29–33. [DOI] [PubMed] [Google Scholar]
- 29.Melcher, K. & Xu, H. E. (2001) EMBO J. 20, 841–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horowitz, N. H. (1945) Proc. Natl. Acad. Sci. USA 31, 153–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andersson, S. G. & Kurland, C. G. (1998) Trends Microbiol. 6, 263–268. [DOI] [PubMed] [Google Scholar]
- 32.Lawrence, J. G., Hendrix, R. W. & Casjens, S. (2001) Trends Microbiol. 9, 535–540. [DOI] [PubMed] [Google Scholar]
- 33.Cole, S. T., Eiglmeier, K., Parkhill, J., James, K. D., Thomson, N. R., Wheeler, P. R., Honore, N., Garnier, T., Churcher, C., Harris, D., et al. (2001) Nature 409, 1007–1011. [DOI] [PubMed] [Google Scholar]
- 34.Xie, G., Bonner, C. A. & Jensen, R. A. (2002) Genome Biol. 3, research0051.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harrison, P. M. & Gerstein, M. (2002) J. Mol. Biol. 318, 1155–1174. [DOI] [PubMed] [Google Scholar]
- 36.Moran, N. A. (2003) Curr. Opin. Microbiol. 6, 512–518. [DOI] [PubMed] [Google Scholar]
- 37.Bungard, R. A. (2004) BioEssays 26, 235–247. [DOI] [PubMed] [Google Scholar]
- 38.Cooper, V. S., Schneider, D., Blot, M. & Lenski, R. E. (2001) J. Bacteriol. 183, 2834–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cocca, E., Ratnayake-Lecamwasam, M., Parker, S. K., Camardella, L., Ciaramella, M., di Prisco, G. & Detrich, H. W., III (1995) Proc. Natl. Acad. Sci. USA 92, 1817–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zufall, R. A. & Rausher, M. D. (2004) Nature 428, 847–850. [DOI] [PubMed] [Google Scholar]
- 41.Gilad, Y., Wiebe, V., Przeworski, M., Lancet, D. & Paabo, S. (2004) PLoS Biol. 2, 120–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stedman, H. H., Kozyak, B. W., Nelson, A., Thesier, D. M., Su, L. T., Low, D. W., Bridges, C. R., Shrager, J. B., Minugh-Purvis, N. & Mitchell, M. A. (2004) Nature 428, 415–418. [DOI] [PubMed] [Google Scholar]
- 43.Wolfe, K. H. & Shields, D. C. (1997) Nature 387, 708–713. [DOI] [PubMed] [Google Scholar]
- 44.Meyer, J., Walker-Jonah, A. & Hollenberg, C. P. (1991) Mol. Cell. Biol. 11, 5454–5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Platt, A., Ross, H. C., Hankin, S. & Reece, R. J. (2000) Proc. Natl. Acad. Sci. USA 97, 3154–3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.