Abstract
CD26 is a T cell costimulatory molecule with dipeptidyl peptidase IV activity in its extracellular region. We previously reported that recombinant soluble CD26 enhanced T cell proliferation induced by the recall antigen tetanus toxoid (TT). However, the mechanism involved in this enhancement is not yet elucidated. We now demonstrate that CD26 binds Caveolin-1 on antigen-presenting cells, and that residues 201–211 of CD26 along with the serine catalytic site at residue 630 contribute to binding to caveolin-1 scaffolding domain. In addition, after CD26–caveolin-1 interaction on TT-loaded monocytes, caveolin-1 is phosphorylated, which links to activate NF-κB, followed by up-regulation of CD86. Finally, reduced caveolin-1 expression on monocytes inhibits CD26-mediated CD86 up-regulation and abrogates CD26 effect on TT-induced T cell proliferation. Taken together, these results strongly suggest that CD26–caveolin-1 interaction plays a role in the up-regulation of CD86 on TT-loaded monocytes and subsequent engagement with CD28 on T cells, leading to antigen-specific T cell activation.
CD26 is a widely distributed, 110-kDa cell-surface glycoprotein with known dipeptidyl peptidase IV (DPPIV) (EC 3.4.14.5) activity in its extracellular domain (1, 2), capable of cleaving amino-terminal dipeptides with either l-proline or l-alanine at the penultimate position. The CD4+CD26high T cells respond maximally to recall antigens, such as tetanus toxoid (TT) (3). Crosslinking of CD26 and CD3 with solid-phase immobilized mAbs induces T cell costimulation and IL-2 production by either human CD4+ T cells or CD26 Jurkat transfectants (4, 5, 6). Importantly, DPPIV enzyme activity is required for CD26-mediated T cell costimulation (7). More recently, we have shown that internalization of CD26 after crosslinking is mediated in part by the mannose-6-phosphate/insulin-like growth factor II receptor (M6P/IGF-IIR), and that CD26–M6P/IGFIIR interaction plays a role in CD26-induced T cell costimulation (8).
Caveolin-1 was first identified as a major tyrosine phosphorylated protein in v-Src-transformed chicken embryo fibroblasts (9). Multiple lines of evidence suggest that caveolin-1 acts as a scaffolding protein capable of directly interacting with and modulating the activity of caveolin-bound signaling molecules. In support of this hypothesis, caveolin-1 binding can functionally modulate the activity of G protein-coupled proteins, membrane proteins, nonreceptor tyrosine and serine/threonine kinases, protein kinase C isoforms, epidermal growth factor receptor, and endothelial nitric oxide synthetase (10, 11). In immune cells, caveolin-1 on monocytes/macrophages regulates metabolism of scavenged lipids (12). However, it is unknown whether caveolin-1 also plays a role in signal transduction in antigen-presenting cells (APC).
Maximal T cell activation requires both an antigen-specific stimulus provided by an MHC peptide complex and a costimulatory signal (13). Engagement of CD28 on T cell surface by B7-1 (CD80) or B7-2 (CD86) expressed on APC provides a potent costimulatory signal, and CD28–B7 interactions lead to T cell proliferation, differentiation, and cytokine secretion (14–17). Despite the structural similarities and costimulatory functions of CD80 and CD86, CD86 may have a different role from that of CD80. Both molecules are expressed on activated B cells, activated monocytes, activated and resting dendritic cells, and activated T cells and are absent from resting B and T cells (16). CD86 molecules are up-regulated on resting monocytes and B and T cells after induction by CD40–CD154 interaction, IL-1, and IFN-γ (13, 18, 19).
In our previous report, we have shown that exogenous recombinant soluble CD26 (rsCD26) enhanced proliferative responses of peripheral blood lymphocytes to stimulation with the soluble antigen TT (7). More recently, we demonstrated that the target cells of rsCD26 were the CD14-positive monocytes in the peripheral blood and that rsCD26 could up-regulate CD86 expression but not CD80 or HLA-DR antigen levels on monocytes (20). Significantly, M6P/IGF-IIR was identified as a platform molecule for CD26 interaction with APC (20). However, although both DPPIV-positive and DPPIV-negative rsCD26 were taken up by monocytes by means of M6P/IGF-IIR, only DPPIV-positive rsCD26 affected CD86 up-regulation on monocytes, thus suggesting that additional key factors may interact with CD26 in this process. Moreover, the molecular mechanism for the maximal response of CD4+CD26high T cells to memory antigens has not yet been clarified.
In this study, we attempted to identify CD26-interacting molecules directly involved in the up-regulation of CD86. We identify caveolin-1 in APC as a functional receptor for CD26 and demonstrate that CD26 stimulation turns on the positive signaling effects of caveolin-1 by inducing tyrosine phosphorylation of caveolin-1.
Materials and Methods
Cells. HEK293 human embryonal kidney, COS-7 monkey fibroblast, and THP-1 human monocyte cell lines were used in this study. Human peripheral monocytes were purified from peripheral blood mononuclear cells collected from TT-sensitized healthy adult volunteers according to the methods described in ref. 20. Human peripheral monocytes and T cells were cultured in serum-free media, Macrophage-SFM medium, and AIM-V medium, respectively (GIBCO).
Constructions of Plasmids. GST-caveolin-1 and caveolin-1-GFP were made by inserting caveolin-1 cDNA into pGEX6p1 (Amersham Pharmacia) and pEB6-CAG-enhanced GFP (EGFP) (a kind gift from Y. Miwa, University of Tsukuba, Tsukuba, Japan) (21) vectors, respectively. A series of caveolin-1 deletion mutants was made by inserting cDNA fragments of mutated caveolin-1 generated by the PCR into pGEX6p1 and pEB6-CAG-EGFP. CD26-EGFP was made by inserting CD26 cDNA into pEB6-CAG-EGFP. Mutant CD26-EGFP constructions [deletions (del) 201–211] were generated by the site-directed mutagenesis method. GST-CD26 and its deletion mutants were made by inserting wild-type (wt) or mutant CD26 cDNA into a mammalian GST-expressing vector, pEBG vector (22).
Luciferase chimera of the 5′ flanking region of the human CD86 gene was generated by inserting PCR fragments of the promoter regions into MluI-XhoI sites of pGL3-basic vector (Promega). PCR fragments of the 5′ flanking region of the human CD86 gene was made with a ResGen bacterial artificial chromosome RPC11 289N10 clone (Invitrogen) as a template.
Production of GST Fusion Protein and Pulldown Assay. To produce GST-caveolin-1 and its deletion mutants, the plasmid constructs were transformed into BL21 (DE3); pT-Trx Escherichia coli (a kind gift from S. Ishii, RIKEN Tsukuba Institute, Ibaraki, Japan) (23). GST fusion proteins and pulldown assay were generated as previously described (24). GST-CD26 and its deletion mutants were produced by the mammalian cell line COS-7, and purified as described elsewhere (25).
Purification and Separation of CD26 Interacting Proteins. Total cell lysate of the THP-1 monocyte cell line was applied to the CD26-adenosine deaminase (ADA) bead columns (26). CD26-ADA bead affinity-purified proteins were separated by SDS/PAGE and stained by silver. Peptide mass mapping was performed by recording peptide mass fingerprints of typical in-gel digests of the corresponding gel bands by using matrix-assisted laser desorption ionization–time-of-flight MS (AXIMA-CFR plus, Shimadzu) and subsequent use of the mascot search engine (Matrix Science, Tokyo).
Confocal Laser Microscopy. For fluorescent microscopy experiments with HEK293 cells, cells were preincubated in Lab-Tek four-well chamber slide glass (Nalge Nunc). GFP-fused constructs were transfected and incubated with Texas red-conjugated recombinant proteins. After being washed, cells were fixed in 4% paraformaldehyde, followed by mounting with an Antifade Prolong kit (Molecular Probes). T cell-APC conjugation assay was performed by the method described in ref. 27.
Small Interfering RNA (siRNA) Against Caveolin-1. We selected two target sequences from nucleotides 81–101 (sense 1) and 138–153 (sense 2) downstream of the start codon of caveolin-1 mRNA: sense1 siRNA, 5′ AACAACAAGGCCAUGG CAGACdTdT; and sense 2 siRNA, 5′ AAGGAGA UCGACCUGGUCAAC dTdT). Moreover, mismatched siRNA at four nucleotides was prepared to examine nonspecific effects of siRNA duplexes (mismatched siRNA, 5′ UACAAGAAGGGCATG GCAGACdTdT). These selected sequences were also submitted to a blast search against the human genome sequence to ensure that only one gene of the human genome was targeted. siRNAs were purchased from Qiagen (Valencia, CA). Sixty picomole of siRNA duplexes was transfected into 0.5 × 106 cells by using HVJ-E vector (Genome-One, kindly provided by Ishihara Sangyo Kaisha, Osaka).
Luciferase Assay, T Cell Proliferation Assay, and Statistics. HEK293 cells or COS-7 cells were used to assay for CD86 promoter activity after CD26–caveolin-1 interaction (28). Freshly isolated monocytes (0.5 × 106 cells per well) were transfected with siRNAs and preincubated with 0.5 μg/ml TT followed by 24-h incubation with 0.5 μg/ml rsCD26. After washing with PBS, preincubated monocytes (1.0 × 104 per well) were then subjected to the assay with purified T cells (1 × 105 per well) from the same donor. Proliferation of cells was monitored in all instances by measuring BrdUrd incorporation by ELISA BrdU Kit (Roche) on day 7 of culture. Student's t test was used to determine whether the difference between control and sample was significant (P < 0.05 being significant).
Results
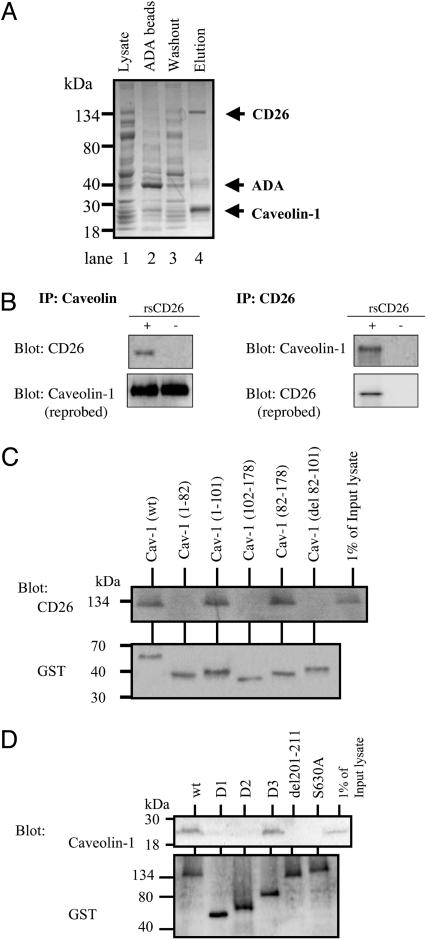
Identification of CD26-Binding Protein. We initially analyzed cell-surface expression of the CD26-binding proteins on various types of cells, such as Jurkat, Molt-4, and THP-1. Of these cells, sCD26-Oregon green bound to CD14-positive monocytes among peripheral blood mononuclear cells and the THP-1 cell line (Fig. 6, which is published as supporting information on the PNAS web site. This binding was inhibited by the 10× concentration of cold sCD26 (data not shown). Therefore, binding of CD26 to the cells was specific. To identify CD26-interacting proteins in monocytes, we next generated CD26-bound affinity columns with ADA-Sepharose, because ADA is a CD26-binding protein (29). Cellular extracts from THP-1 were applied to this CD26-ADA Sepharose column, and bound proteins were eluted. These proteins were then subjected to SDS/PAGE analysis. Nonspecific multiple bands were found in lanes of the lysate, elution of ADA (mock) column, and washout solution after eluting through CD26-ADA columns (Fig. 1A, lanes 1–3). To eliminate these random background proteins bound to ADA-Sepharose beads, the lysates were precleared by ADA-Sepharose beads and then applied to CD26-ADA columns. Three major bands were revealed in the elution of CD26-ADA columns (Fig. 1A, lane 4). The protein bands that were specifically bound to CD26 on ADA columns were subjected to peptide mass fingerprinting by matrix-assisted laser desorption ionization–time-of-flight MS. After searching the mascot database, we obtained masses and apparent molecular masses of the different polypeptides that revealed that the fraction eluted from CD26-ADA Sepharose columns contained three major bands, and these bands were determined to be CD26, ADA, and caveolin-1 (Fig. 1A). Caveolin-1 at ∼20–25 kDa was strongly stained with silver in the elution fraction (Fig. 1A, lane 4) and was not detected specifically in the fraction through the mock ADA bead column (Fig. 1A, lane 2). These findings suggested that caveolin-1 was associated with CD26.
Fig. 1.
Purification and identification of CD26-binding proteins. (A) The indicated fractions were subjected to SDS/PAGE followed by silver staining. Proteins eluted from CD26-ADA Sepharose columns were identified by MS analysis, indicated on the right. (B) THP-1 cells cocultured with rsCD26 were immunoprecipitated (IP) with either anti-caveolin-1 (Left) or anti-CD26 Abs (Right). (C) The caveolin-1 SCD is necessary for binding to CD26. (D) Amino acids 201–211 and DPPIV of CD26 were required for binding of CD26 to caveolin-1.
CD26 was detected by its specific Ab in lysates coimmunoprecipitated with caveolin-1-specific Ab after the addition of rsCD26-wt to THP-1 cells (Fig. 1B Upper Left). Endogenous caveolin-1 was detected in THP-1 cells (Fig. 1B Lower Left). Caveolin-1 was detected specifically with Western blots of lysates coimmunoprecipitated with CD26-specific Ab (Fig. 1B Upper Right). Endogenous CD26 was not detected in THP-1 cells (Fig. 1B Lower Right). The above results thus indicated that caveolin-1 was the binding protein to CD26 in THP-1 cells.
GST pull-down assays using a series of GST-fused deletion mutants of caveolin-1 (Fig. 7, which is published as suppporting information on the PNAS web site) and CD26-transfected Jurkat T cells showed that CD26 was coprecipitated with GST-caveolin-1 wt, caveolin-1 (residues 1–101), and caveolin-1 (residues 82–178) but not with GST-caveolin-1 (residues 1–82), caveolin-1 (residues 102–178), and caveolin-1 (del 82–101) (Fig. 1C), indicating that residues 82–101 of caveolin-1, known as the scaffolding domain (SCD) of caveolin-1 (11), were the binding domain to CD26. To determine the region(s) in CD26 responsible for binding to caveolin-1, we performed a GST-pulldown assay by using a series of GST-fused deletion mutants of CD26 produced by COS-7 cells (Fig. 8, which is published as supporting information on the PNAS web site). As shown in Fig. 1D, caveolin-1 was coprecipitated with wt-GST-CD26 and CD26 D3 (residues 31–429) but did not coprecipitate with GST-CD26 D1 (residues 507–766), CD26 D2 (residues 267–584), and CD26 del 201–211. These results suggested that amino acids 201–211 of CD26 were required for the binding of CD26 to caveolin-1. This region in CD26 contains a caveolin-binding domain (CBD) (ΦXΦXXXXΦXXΦ; Φ and X depict aromatic residue and any amino acid, respectively) (11), specifically WVYEEEVFSAY in CD26 (4). It should be noted that caveolin-1 was not detected in complexes with GST-CD26 S630A, with DPPIV enzymatic activity being deleted (Fig. 1D).
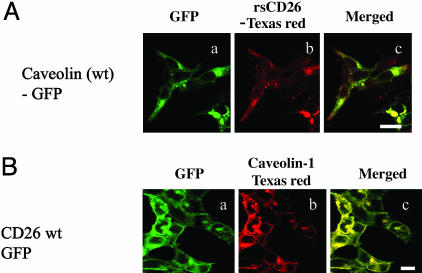
Caveolin-1 Interacts with DPPIV Active Site in CD26. To confirm the above findings in living cells, GFP-fused full-length caveolin-1 and caveolin-1 lacking the SCD (del 82–101) were transfected into HEK293 cells. Texas red-conjugated rsCD26 (rsCD26-TR) was added to the transfectants. Full-length caveolin-1-GFP, which was detected in surface membrane and perinuclear area (Fig. 2Aa), was clearly merged with rsCD26-TR (Fig. 2Ab and c). On the other hand, with caveolin residues 61–101 being necessary for oligomerization (11), caveolin (del 82–101)-GFP lacking the SCD was localized diffusely, with no evidence of interaction with rsCD26-TR (Fig. 9A, which is published as supporting information on the PNAS web site). Although rsCD26-TR was slightly detected in HEK293 cells transfected with GFP alone, rsCD26-TR was not associated with GFP (Fig. 9A). In addition, GFP-fused full-length CD26 mutant lacking the CBD (del 201–211) and a mutant lacking DPPIV enzymatic activity (S630A) were transfected into HEK293 cells, with Texas red-conjugated caveolin-1 (cav-TR) being added to the transfectants. CD26 wt-GFP, CD26 del201–211-GFP and CD26 S630A-GFP were localized in cell-surface membrane (Figs. 2Ba and 9B). Cav-TR was colocalized in HEK293 cells transfected with CD26-wt-GFP (Fig. 2B b and c). However, cav-TR was not detected in CD26 (del 201–211) or CD26 S630A-GFP-transfected cells (Fig. 9B). Cav-TR was not observed in HEK293 cells transfected with GFP alone (Fig. 9B). To evaluate the possibility that M6P/IGF-IIR modulates the binding of CD26 to caveolin-1, CD26 wt-GFP-transfected HEK293 cells were pretreated with M6P followed by staining with cav-TR, with no changes in the binding of CD26 to caveolin-1 being observed (data not shown). Taken together, CD26 was demonstrated to bind to caveolin-1 through the CBD as well as the serine residue 630 in CD26 and the SCD in caveolin-1.
Fig. 2.
CD26 on T cells interacts with caveolin-1 on monocytes. (A) rsCD26-TR was incubated with HEK293 cells transfected with GFP-fused wt caveolin-1. (Scale bar, 10 μm.) (B) Cav-TR was incubated with HEK293 cells transfected with GFP-fused wt CD26 (Scale bar, 10 μm.)
CD26 on T Cells Interacts with Caveolin-1 on Monocytes. Because TT was taken up by APC through caveolae (30, 31), we next examined caveolin-1 trafficking in monocytes after treatment with TT. By using flow cytometry, caveolin-1 was detected on the cell surface of monocytes 12–24 h after treatment with TT (Fig. 10, which is published as supporting information on the PNAS web site), whereas caveolin-1 on untreated monocytes was not detected after 0–48 h of culture. To further examine the TT effect on caveolae, we used monocytes treated with filipin, which inhibits caveolae trafficking (32). As shown in Fig. 10, caveolin-1 was not detected on monocytes treated with filipin, even after TT was loaded. These data demonstrated that certain populations of TT-loaded monocytes were found to express caveolin-1 on the cell surface.
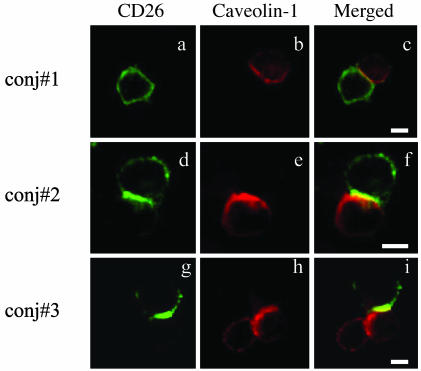
We next examined potential colocalization of CD26 and caveolin-1 at the T cell–monocyte contact site. CD26 and caveolin-1 were recruited to the contact area of activated T cells and TT-loaded monocytes (Fig. 3). Quantification of cell conjugation between T cells and monocytes is shown in Fig. 11, which is published as supporting information on the PNAS web site. As shown in the caveolin-1 expression study (Fig. 10), conjugation formation of activated T cells and TT-loaded monocytes was increased in monocytes with TT loaded for 12–24 h (Fig. 11). Cell conjugation was not detected in TT-untreated monocytes or with filipin-treated monocytes (Fig. 11). These data suggested that memory CD26-positive T cells interacted with antigen-loaded monocytes through the interaction of CD26 on T cells and caveolin-1 expressed on monocytes.
Fig. 3.
Caveolin-1 in monocytes was exposed to the cell surface after TT treatment and interacted with CD26 on activated T cells. To form cell–cell conjugation, activated T cells and TT-loaded monocytes were mixed and centrifuged. Conjugates were fixed without permeabilization and stained with anti-CD26 (FITC) and anti-cav-TR Abs. Three representative conjugates are shown (conj#1, conj#2, and conj#3). (Scale bars, 10 μm.)
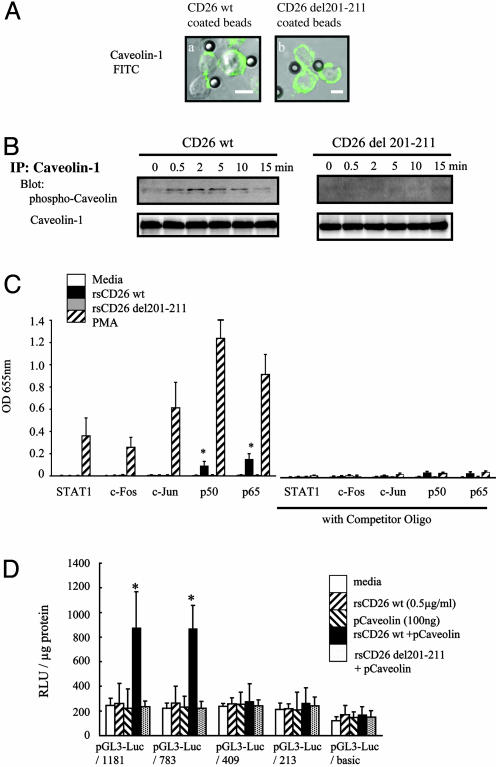
Phosphorylation of Caveolin-1 Leads to Up-Regulation of CD86 in Monocytes by Means of NF-κB Activation. We next focused on caveolin-1-mediated signal transduction events to determine whether such events up-regulate CD86 expression. For this purpose, we used CD26-coated polystyrene latex beads to mimic the physiological interaction of CD26 expressed on T cells and TT-loaded monocytes. Fig. 4A Left shows that beads coated with wt CD26 engaged caveolin-1 on TT-loaded monocytes, whereas beads coated with mutant CD26 lacking the CBD did not alter caveolin-1 expression on TT-loaded monocytes (Fig. 4A Right). Because phosphorylation of caveolin-1 was implicated (11) at various time periods after stimulation of TT-loaded monocytes by these beads, we determined the phosphorylation status of caveolin-1. As shown in Fig. 4B, 0.5–10 min after stimulation with CD26 wt-coated beads, caveolin-1 was phosphorylated (Fig. 4B Upper Left). On the other hand, caveolin-1 was not phosphorylated after stimulation with mutant CD26 (del 201–211) beads (Fig. 4B Upper Right). It should be noted that equal expression of caveolin-1 was determined by reprobing with anti-caveolin-1 Ab (Fig. 4B Lower). These results suggested that CD26 induced phosphorylation of caveolin-1 by its binding.
Fig. 4.
CD26 induced phosphorylation of caveolin-1 in TT-loaded monocytes, followed by NF-κB activation. (A) Caveolin-1 on TT-loaded monocytes was bound to polystyrene latex beads coated with wt CD26 (Left) or deletion mutant CD26 lacking the CBD (del 201–211) (Right). (Scale bars, 10 μm.) (B) Caveolin-1 on TT-loaded monocytes was phosphorylated by stimulation with wt CD26-coated beads. (C) NF-κB level was increased in nuclear extracts from TT-loaded monocytes stimulated with CD26-coated beads. Data represent mean of OD at 655 nm ± SE from triplicate experiments. Asterisks show points of significant increase. STAT1, signal transducer and activator of transcription 1; PMA, phorbol 12-myristate 13-acetate. (D) After HEK293 cells were cotransfected with CD86-promoter luciferase constructs and wt caveolin-1-expressing vectors, wt rsCD26 or mutant rsCD26 del 201–211 was added to the culture media. Luciferase activity is shown as being relative to 1 μg of applied protein. Data represent mean ± SE from triplicate experiments. Asterisks indicate points of significant increase.
We next identified the transcriptional factors activated by CD26 in the presence of TT-loaded monocytes. By using an ELISA-based DNA-binding detection method, we detected significant levels of p50 and p65 NF-κB components in nuclear extracts of TT-loaded monocytes stimulated with wt CD26 (Fig. 4C, left side of graph). The increase in p50 and p65 NF-κB levels was inhibited by the specific competitor oligos (Fig. 4C, right side of graph). Levels of AP-1 (c-Fos and c-Jun) and signal transducer and activator of transcription 1 (STAT1) were not detected in nuclear extracts of TT-loaded monocytes stimulated with wt CD26 (Fig. 4C). These results suggested that NF-κB was activated by means of caveolin-1 phosphorylation after stimulation with CD26.
We next examined whether NF-κB binding sites were required in the human CD86 promoter regions for activation, because previous reports revealed that IFN-γ activation site elements and NF-κB binding sites were present and required for activation of CD86 transcription (33). For this purpose, we constructed a series of luciferase chimera mutants of 5′ flanking promoter region of human CD86 (Fig. 12, which is published as supporting information on the PNAS web site). By using these luciferase mutants, we assayed for CD86 promoter activity following CD26–caveolin-1 interaction. In the presence or absence of IFN-γ elements (pGL3-Luc/1181 and pGL3-Luc/783), luciferase activity was not affected after stimulation of TT-loaded monocytes with CD26 (Fig. 4D). On the other hand, two NF-κB binding sites in the promoter regions were required for activation of CD86 transcription after CD26 treatment in caveolin-1-expressed HEK293 cells (Fig. 4D). In contrast, significant activity in a single NF-κB luciferase construct (pGL3-Luc/409) was not detected (Fig. 4D). These results showed that NF-κB activation downstream of caveolin-1 resulted in the up-regulation of CD86 in TT-loaded monocytes stimulated with CD26. We next examined whether the caveolin-1 phosphorylation site plays a role in CD86 promoter activity after CD26 treatment. As shown in Fig. 13, which is published as supporting information on the PNAS web site, an enhancement in luciferase activity was observed with increasing doses of caveolin-1 wt transfection in HEK293 cells cotransfected with pGL3-Luc/1181, followed by rsCD26 treatment. This dose-dependent luciferase activity was not observed in caveolin-1 Y14 deletion mutant transfection. These results suggested that tyrosine phosphorylation of caveolin-1 played an essential role in CD86 up-regulation triggered by CD26.
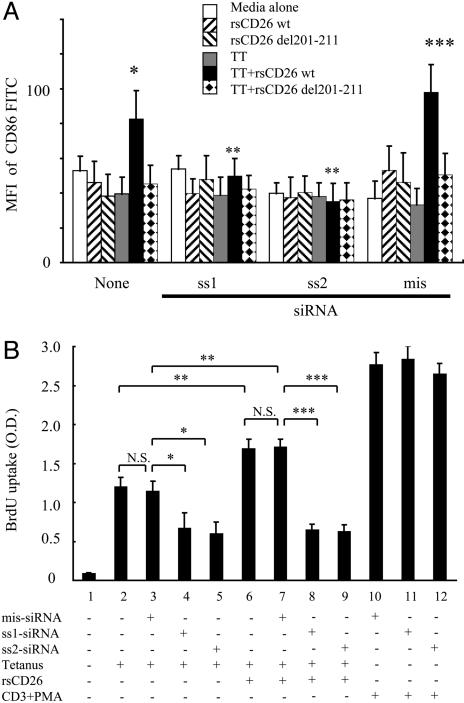
siRNA Against Caveolin-1 in Monocytes Attenuates CD26-Mediated Up-Regulation of CD86 and Inhibits T Cell Proliferation Driven by TT. To define CD26–caveolin-1 interaction and its functional consequences more directly, we performed siRNA experiments by using freshly isolated monocytes from TT-immunized healthy adults. We prepared two sets of specific siRNA against caveolin-1 as described in Materials and Methods, and both of these siRNA effectively knocked down caveolin-1 expression in monocytes (Fig. 14, which is published as supporting information on the PNAS web site). Because caveolin-1 in monocytes was not significantly knocked down by mismatched siRNA or HVJ-E vector alone (Fig. 14), this inhibitory effect by siRNA was specific. We next examined whether CD26 exerted its effect on monocytes in which caveolin-1 expression was knocked down by siRNA. CD86 was up-regulated among a significant population of TT-loaded monocytes stimulated with CD26 wt-coated beads (* in Fig. 5A). On the other hand, sense siRNA (sense 1 and sense 2) inhibited this effect on CD86 up-regulation in TT-loaded monocytes (** in Fig. 5A). Mismatched siRNA did not exhibit this inhibitory effect (*** in Fig. 5A). These results indicated that caveolin-1 played an important role in signal transduction following CD26 binding to TT-loaded monocytes, leading to the up-regulation of CD86 in monocytes. Finally, we examined whether T cell proliferation pulsed with TT was suppressed in the presence of caveolin-1 knock-down monocytes. As shown in Fig. 5B, TT-induced T cell proliferation was observed without caveolin-1 knockdown (Fig. 5B, bar graphs 2 and 3) and this proliferation was significantly inhibited by caveolin-1 knockdown in monocytes (Fig. 5B, bar graphs 4 and 5). On the other hand, as described in ref. 20, TT-induced T cell proliferation was significantly enhanced in the presence of rsCD26 (Fig. 5B, bar graphs 6 and 7 versus 2 and 3, respectively). This effect was more profoundly suppressed by caveolin-1 knockdown (Fig. 5B, bar graphs 6 and 7 versus 8 and 9, respectively) than the inhibition observed with TT alone (bar graphs 2 and 3 versus 4 and 5, respectively). The knockdown of caveolin-1 in monocytes did not have an effect of anti-CD3 plus phorbol 12-myristate 13-acetate-stimulated T cell proliferation (bar graphs 10–12). These results demonstrated that caveolin-1 had an important role in the enhancement of CD26 in TT-induced T cell proliferation.
Fig. 5.
siRNA against caveolin-1 inhibited effect of CD26 on CD86 up-regulation in TT-loaded monocytes. (A) CD86 expression of caveolin-1 knock-down monocytes was not increased by CD26 stimulation. Data represent mean ± SE of five independent experiments. **, points of no significant change by sense siRNA. * and ***, points of significant increase. MFI, mean fluorescence intensity; SS1, sense 1; SS2, sense 2; mis, mismatched. (B) Purified monocytes were transfected with or without siRNA, followed by treatment with TT. After pulse with rsCD26, the preincubated monocytes were incubated with purified T cells from the same donor. Proliferation of T cells was monitored by measuring BrdUrd (BrdU) incorporation by ELISA. The degree of proliferation is indicated at OD450 with reference at OD690. The experiments represent mean values ± SE calculated from five independently performed experiments each from different donors. N.S., points of no significant change by mismatched siRNA. *, **, and ***, points of significant increase.
Discussion
In this study, we showed that CD26 on activated memory T cells interacted with caveolin-1 on TT-loaded monocytes. In this interaction, caveolin-1 residues 82–101, the SCD, were associated with CD26 residues 201–211, the CBD.
APC–T cell interaction plays a key role in triggering T cell proliferative response against a recall antigen, leading eventually to the expression of the T cell biological program (13–17). Our previous studies showed that the enhancing effect of rsCD26 on TT-induced T cell proliferation occurred in the early stages of the immune response (20). Moreover, we demonstrated that the target cells of rsCD26 were the CD14-positive monocytes in the peripheral blood and that rsCD26 could up-regulate CD86 expression, but not that of CD80, on monocytes (20). M6P/IGF-IIR is thought to be one of the platform molecules for CD26 interaction with APC. However, both DPPIV-positive and DPPIV-negative rsCD26 were taken up by monocytes by M6P/IGF-IIR, but only uptake of DPPIV-positive rsCD26 resulted in CD86 up-regulation. Furthermore, rsCD26 was taken up by 293 cells without caveolin-1 (Fig. 9A) and by resting monocytes (20). Although it has been reported that the 300-kDa protein M6P/IGF-IIR was linked with CD26 on the T cell surface to enhance CD26-mediated costimulation (8), no protein with a molecular mass of ∼300 kDa was detected by our purification method. An explanation may be that M6P/IGF-IIR might compete with ADA to bind CD26 because we used CD26-ADA Sepharose bead columns to purify potential CD26-associated molecules. Alternatively, our detection approach may miss this protein because of the relatively high molecular mass of M6P/IGF-IIR (34).
Caveolin-1 is the primary coat protein of caveolae, and it is involved as a regulator of signal transduction through the binding of the SCD to key signaling molecules in various cells (11, 32, 35). Although CD26 was present in caveolae of fibroblast-like synoviocytes (36), its direct binding or signaling event was not demonstrated in immune cells. We showed here that CD26 was directly bound to caveolin-1 by using a series of CD26 and caveolin-1 deletion mutants and that caveolin-1 was phosphorylated after binding to CD26. Caveolin-1 was reported to be an integral membrane protein with a cytoplasmic N-terminal domain and a cytoplasmic C-terminal domain (11). Our data showed that the N-terminal domain of caveolin-1 was expressed on the cell surface of monocytes 12–24 h after TT was loaded (Fig. 10). Because TT was trafficked in cells through caveolae (30, 31), caveolin-1 may be transported with the peptide-MHC complex developed in APC and may be expressed on the cell surface by antigen-processing machinery for T cell contact (37, 38). The data shown in Fig. 3 indicated that CD26 on activated memory T cells directly faced caveolin-1 on TT-loaded monocytes in the contact area, which was revealed as the immunological synapse for T cell–APC interaction. It is conceivable that the interaction of CD26 with caveolin-1 on antigen-loaded monocytes resulted in CD86 up-regulation, therefore enhancing the subsequent interaction of CD86 and CD28 on T cells to induce antigen-specific T cell proliferation and activation.
Crystal structure analysis of CD26/DPPIV showed the horizontal helix of residues 201–207 to be situated in front of the DPPIV enzyme active site at the serine residue 630. This small horizontal cavity allowed substrate amino acids to reach the active-site serine residue 630 and is involved in the DPPIV activity of CD26 (39). This horizontal cavity therefore has an essential role in caveolin-1 binding as well as DPPIV enzyme activity. In particular, CD26 mutants del 201–211 and S630A, in which this cavity was destroyed, lost the ability to associate with caveolin-1 (Figs. 1C and 2B) and did not exert an effect on CD86 up-regulation on monocytes (Fig. 5A) (20). In addition, binding of CD26 to caveolin-1 was inhibited by the competitive inhibitor of DPPIV, valine-pyrrolidide (data not shown). The valine-pyrrolidide inhibitor binds to a smaller pocket within the DPPIV enzymatic active site, with two glutamic acids in the horizontal helix of CD26, Glu-205 and Glu-206, forming salt bridges to the free amino group of valine-pyrrolidide (39). These findings explain our previous work showing that CD26 lacking DPPIV enzymatic activity could neither enhance TT-mediated T cell proliferation nor up-regulate CD86.
The cloning and functional analysis of a 1.3-kbp fragment in the 5′ flanking region upstream of the transcriptional site of the CD86 gene indicated that two NF-κB binding sites were required for the up-regulation of CD86 after CD26–caveolin-1 interaction (Fig. 4D). Moreover, in a transcription factor assay of TT-loaded monocytes stimulated with CD26, levels of NF-κB (p50 and p65) were detected to be significantly higher than those of signal transducer and activator of transcription 1 or AP-1 (c-Fos and c-Jun) (Fig. 4C). Although several factors, such as IFN-γ, tumor necrosis factor α, or CD40-CD154 ligation, were also reported to be involved in the up-regulation of CD86 (13, 18, 19, 33), our present data demonstrated that CD26 on T cells triggered up-regulation of CD86 on TT-loaded monocytes by association with caveolin-1.
Because loss of caveolae in monocytes was not reported in a caveolin-1 knockout mouse model (40) and the role and distribution of CD26 in humans is different from that in mice (1), we used the RNA interference method to directly analyze the function of native caveolin-1 in purified human monocytes. Caveolin-1 is expressed constitutively in monocytes and other human tissues, including endothelia, fibroblasts, and adipocytes, and treatment of purified human monocytes with siRNA resulted in knockdown of caveolin-1 and inhibition of CD86 up-regulation after stimulation with CD26-coated beads (Figs. 5A and 14). TT-induced T cell proliferation was inhibited by siRNA against caveolin-1 in monocytes (Fig. 5B), consistent with previous findings showing that TT was trafficked by means of caveolae (30, 31). However, more profound inhibition by siRNA was observed in TT-induced T cell proliferation after treatment of rsCD26 (Fig. 5B). Thus, our findings strongly suggested that caveolin-1 was directly involved in the immuno-enhancement effect of CD26 by means of CD86 up-regulation in monocytes.
On the basis of our results and previously reports, we propose a model to describe the signaling events in monocytes triggered by CD26–caveolin-1 interaction. With regards to T cell–APC local interaction and immune response, entry of recall antigens by means of caveolae into APC leads to presentation of antigen peptides on MHC class II molecules and exposure of caveolin-1. Then, APC induces the activation of memory T cells through T cell antigen receptor (TCR) and costimulatory molecules, such as CD86/CD80-CD28, leading to formation of mature immunological synapse. After the association between caveolin-1 on APC and CD26 on memory T cells, CD86 is up-regulated on the APC surface, and memory T cells are subsequently activated by the costimulatory effect of CD26 on TCR activation (6). By enhancing TCR activation by means of CD26–caveolin-1 interaction, prolongation of the immunological synapse may be maintained (41). Finally, CD86 up-regulation results in potent T cell–APC interaction, leading to the development of activated memory T cells locally and activation of the immune response as well as the consequence of various inflammatory diseases. Our results may provide an approach to the treatment of autoimmune diseases or other immune-mediated disorders by directly interfering with activated memory T cell and APC interaction.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant AR33713 and by grants-in-aid from the Ministry of Education, Science, Sports, and Culture and the Ministry of Health, Labor, and Welfare, Japan (to C.M.). K.O. is a recipient of a grant from the Japan Society for the Promotion of Science. N.H.D. is the recipient of grants from the M. D. Anderson Cancer Center Physician-Scientist Program and the Gillson Longenbaugh Foundation.
Abbreviations: ADA, adenosine deaminase; APC, antigen-presenting cells; cav-TR, Texas red-conjugated caveolin-1; CBD, caveolin-binding domain; DPPIV, dipeptidyl peptidase IV; EGFP, enhanced GFP; M6P/IGF-IIR, mannose 6-phosphate/insulin-like growth factor II receptor; rsCD26, recombinant soluble CD26; SCD, scaffolding domain; siRNA, small interfering RNA; rsCD26-TR, Texas red-conjugated rsCD26; TT, tetanus toxoid; wt, wild-type; del, deletions.
References
- 1.Morimoto, C. & Schlossman, S. F. (1998) Immunol. Rev. 161, 55–70. [DOI] [PubMed] [Google Scholar]
- 2.von Bonin, A., Huhn, J. & Fleischer, B. (1998) Immunol. Rev. 161, 43–53. [DOI] [PubMed] [Google Scholar]
- 3.Morimoto, C., Torimoto, Y., Levinson, G., Rudd, C. E., Schrieber, M., Dang, N. H., Letvin, N. L. & Schlossman, S. F. (1989) J. Immunol. 143, 3430–3439. [PubMed] [Google Scholar]
- 4.Tanaka, T., Camerini, D., Seed, B., Torimoto, Y., Dang, N. H., Kameoka, J., Dahlberg, H. N., Schlossman, S. F. & Morimoto, C. (1992) J. Immunol. 149, 481–486. [PubMed] [Google Scholar]
- 5.Fleischer, B. (1994) Immunol. Today 15, 180–184. [DOI] [PubMed] [Google Scholar]
- 6.Hegen, M., Kameoka, J., Dong, R. P., Schlossman, S. F. & Morimoto, C. (1997) Immunology 90, 257–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanaka, T., Duke-Cohan, J. S., Kameoka, J., Yaron, A., Lee, I., Schlossman, S. F. & Morimoto, C. (1994) Proc. Natl. Acad. Sci. USA 91, 3082–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ikushima, H., Munakata, Y., Ishii, T., Iwata, S., Terashima, M., Tanaka, H., Schlossman, S. F. & Morimoto, C. (2000) Proc. Natl. Acad. Sci. USA 97, 8439–8444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glenney, J. R., Jr. (1989) J. Biol. Chem. 264, 20163–20166. [PubMed] [Google Scholar]
- 10.Razani, B., Woodman, S. E. & Lisanti, M. P. (2002) Pharmacol. Rev. 54, 431–467. [DOI] [PubMed] [Google Scholar]
- 11.Smart, E. J., Graf, G. A., McNiven, M. A., Sessa, W. C., Engelman, J. A., Scherer, P. E., Okamoto, T. & Lisanti, M. P. (1999) Mol. Cell. Biol. 19, 7289–7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lei, M. G. & Morrison, D. C. (2000) Infect. Immun. 68, 5084–5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lenschow, D. J., Sperling, A. I., Cooke, M. P., Freeman, G., Rhee, L., Decker, D. C., Gray, G., Nadler, L. M., Goodnow, C. C. & Bluestone, J. A. (1994) J. Immunol. 153, 1990–1997. [PubMed] [Google Scholar]
- 14.Yokochi, T., Holly, R. D. & Clark, E. A. (1982) J. Immunol. 128, 823–827. [PubMed] [Google Scholar]
- 15.Azuma, M., Ito, D., Yagita, H., Okumura, K., Phillips, J. H., Lanier, L. L. & Somoza, C. (1993) Nature 366, 76–79. [DOI] [PubMed] [Google Scholar]
- 16.Freeman, G. J., Gribben, J. G., Boussiotis, V. A., Ng, J. W., Restivo, V. A., Jr., Lombard, L. A., Gray, G. S. & Nadler, L. M. (1993) Science 262, 909–911. [DOI] [PubMed] [Google Scholar]
- 17.Chambers, C. A. (2001) Trends Immunol. 22, 217–223. [DOI] [PubMed] [Google Scholar]
- 18.Larsen, C. P., Ritchie, S. C., Hendrix, R., Linsley, P. S., Hathcock, K. S., Hodes, R. J., Lowry, R. P. & Pearson, T. C. (1994) J. Immunol. 152, 5208–5219. [PubMed] [Google Scholar]
- 19.Berberich, I., Shu, G. L. & Clark, E. A. (1994) J. Immunol. 153, 4357–4366. [PubMed] [Google Scholar]
- 20.Ohnuma, K., Munakata, Y., Ishii, T., Iwata, S., Kobayashi, S., Hosono, O., Kawasaki, H., Dang, N. H. & Morimoto, C. (2001) J. Immunol. 167, 6745–6755. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka, J., Miwa, Y., Miyoshi, K., Ueno, A. & Inoue, H. (1999) Biochem. Biophys. Res. Commun. 264, 938–943. [DOI] [PubMed] [Google Scholar]
- 22.Sanchez, I., Hughes, R. T., Mayer, B. J., Yee, K., Woodgett, J. R., Avruch, J., Kyriakis, J. M. & Zon, L. I. (1994) Nature 372, 794–798. [DOI] [PubMed] [Google Scholar]
- 23.Yasukawa, T., Kanei-Ishii, C., Maekawa, T., Fujimoto, J., Yamamoto, T. & Ishii, S. (1995) J. Biol. Chem. 270, 25328–25331. [DOI] [PubMed] [Google Scholar]
- 24.Ishii, T., Ohnuma, K., Murakami, A., Takasawa, N., Kobayashi, S., Dang, N. H., Schlossman, S. F. & Morimoto, C. (2001) Proc. Natl. Acad. Sci. USA 98, 12138–12143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen, P., Hutter, D., Liu, P. & Liu, Y. (2002) Protein Expression Purif. 24, 481–488. [DOI] [PubMed] [Google Scholar]
- 26.Tanaka, T., Kameoka, J., Yaron, A., Schlossman, S. F. & Morimoto, C. (1993) Proc. Natl. Acad. Sci. USA 90, 4586–4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, K. H., Holdorf, A. D., Dustin, M. L., Chan, A. C., Allen, P. M. & Shaw, A. S. (2002) Science 295, 1539–1542. [DOI] [PubMed] [Google Scholar]
- 28.Makino, Y., Nakamura, H., Ikeda, E., Ohnuma, K., Yamauchi, K., Yabe, Y., Poellinger, L., Okada, Y., Morimoto, C. & Tanaka, H. (2003) J. Immunol. 171, 6534–6540. [DOI] [PubMed] [Google Scholar]
- 29.Kameoka, J., Tanaka, T., Nojima, Y., Schlossman, S. F. & Morimoto, C. (1993) Science 261, 466–469. [DOI] [PubMed] [Google Scholar]
- 30.Montesano, R., Roth, J., Robert, A. & Orci, L. (1982) Nature 296, 651–653. [DOI] [PubMed] [Google Scholar]
- 31.Pelkmans, L. & Helenius, A. (2002) Traffic 3, 311–320. [DOI] [PubMed] [Google Scholar]
- 32.Peiro, S., Comella, J. X., Enrich, C., Martin-Zanca, D. & Rocamora, N. (2000) J. Biol. Chem. 275, 37846–37852. [DOI] [PubMed] [Google Scholar]
- 33.Li, J., Liu, Z., Jiang, S., Cortesini, R., Lederman, S. & Suciu-Foca, N. (1999) J. Immunol. 163, 6386–6392. [PubMed] [Google Scholar]
- 34.Scopes, R.K. (1993) in Protein Purification (Springer, Berlin), 3rd Ed., pp. 146–186.
- 35.Carver, L. A. & Schnitzer, J. E. (2003) Nat. Rev. Cancer 3, 571–581. [DOI] [PubMed] [Google Scholar]
- 36.Riemann, D., Hansen, G. H., Niels-Christiansen, L., Thorsen, E., Immerdal, L., Santos, A. N., Kehlen, A., Langner, J. & Danielsen, E. M. (2001) Biochem. J. 354, 47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grakoui, A., Bromley, S. K., Sumen, C., Davis, M. M., Shaw, A. S., Allen, P. M. & Dustin, M. L. (1999) Science 285, 221–227. [DOI] [PubMed] [Google Scholar]
- 38.Turley, S. J., Inaba, K., Garrett, W. S., Ebersold, M., Unternaehrer, J., Steinman, R. M. & Mellman, I. (2000) Science 288, 522–527. [DOI] [PubMed] [Google Scholar]
- 39.Rasmussen, H. B., Branner, S., Wiberg, F. C. & Wagtmann, N. (2003) Nat. Struct. Biol. 10, 19–25. [DOI] [PubMed] [Google Scholar]
- 40.Drab, M., Verkade, P., Elger, M., Kasper, M., Lohn, M., Lauterbach, B., Menne, J., Lindschau, C., Mende, F., Luft, F. C., et al. (2001) Science 293, 2449–2452. [DOI] [PubMed] [Google Scholar]
- 41.Huppa, J. B., Gleimer, M., Sumen, C. & Davis, M. M. (2003) Nat. Immunol. 4, 749–755. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.