Abstract
An attack by a parasitic wasp activates a vigorous cellular immune response in Drosophila larvae. This response is manifested by an increased number of circulating cells, the hemocytes, and by the appearance of a specialized class of hemocyte, the lamellocytes, which participate in the encapsulation and killing of the parasite. To study the molecular mechanisms of this response, we have overexpressed different genes in the hemocytes, by using the GAL4-upstream activating sequence system and a hemocyte-specific Hemese-GAL4 driver. Multiple transgenes were tested, representing several important signaling pathways. We found that the proliferation response and the activation of lamellocyte formation are independent phenomena. A drastic increase in the number of circulating hemocytes is caused by receptor tyrosine kinases, such as Egfr, Pvr, and Alk, as well as by the downstream signaling components Ras85D and pointed, supporting the notion that the Ras–mitogen-activated protein kinase pathway regulates hemocyte numbers. In the case of Pvr and Alk, this phenotype also is accompanied by lamellocyte formation. By contrast, constitutively active hopscotch and hemipterous give massive activation of lamellocyte formation with little or no increase in total hemocyte numbers. This finding indicates that both the Jak/Stat and the Jun kinase pathways affect lamellocyte formation. Still other signals, mediated by aopACT, Toll10b, and Rac1 expression, cause a simultaneous increase in lamellocyte and total cell numbers, and the same effect is seen when WNT signaling is suppressed. We conclude that the activation of a cellular response is complex and affected by multiple signaling pathways.
The immune system in Drosophila has attracted much interest as a model for innate immunity. Receptors and signaling pathways involved in regulating the humoral defense are now relatively well known (for reviews, see refs. 1–3). However, the immune repertoire of the fly also includes an important cellular component (reviewed in refs. 4–7), which is thus far poorly understood. Circulating cells, referred to as hemocytes, participate in phagocytosis of bacteria and encapsulation of parasites, and they also may contribute to the humoral response.
Of particular interest is the cellular reaction against parasitoid wasps such as Leptopilina boulardi that deposit their eggs inside Drosophila larvae (4, 8). The total number of hemocytes increases in parasitized larvae, and a specialized class of hemocytes, the lamellocytes, appears en masse (5, 9). In a successful response, the lamellocytes form a melanized capsule around the parasite, which is eventually killed. Lamellocytes are relatively large (up to 60 μm or more), flat cells, with a characteristic appearance under the microscope. They adhere to larger foreign bodies, and the resulting multilayered sheet undergoes melanization. The melanization reaction is thought to depend on a second class of hemocytes, the crystal cells. Lamellocytes are essentially absent in healthy uninfected larvae (10), but they may appear in low numbers at the time of metamorphosis (4). Instead, a third class of hemocyte dominates under normal conditions, the phagocytically active plasmatocytes.
A major source of hemocytes in Drosophila is the so-called lymph glands, a multilobed structure near the anterior end of the aorta. They produce a population of hemocytes that is released at the time of metamorphosis. A second population of hemocytes derives directly from embryonic hemocytes, and the two populations originate from distinct anlagen in the cellular blastoderm (11). The lamellocytes' hematopoietic origin is unclear. They may come from uncommitted precursor cells, preplasmatocytes, located in the lymph glands or elsewhere (10). It also has been proposed that plasmatocytes can differentiate directly into lamellocytes (4).
Few genes that control hematopoiesis in Drosophila have been identified. The gene serpent is required for normal hematopoiesis (12), and Pvf2 recently has been shown to affect hemocyte proliferation (13). The gene glial cells missing determines plasmatocyte fate, whereas lozenge and Notch control the formation of crystal cells (14–17). These and other genetic interactions in Drosophila hematopoiesis are reviewed by Evans et al. (7). Even less is known about the cellular response to parasites. Carton and Nappi (8) have shown that the resistance to parasitic infestation is genetically controlled, but the genes involved have not yet been identified. Many mutants show increased lamellocyte numbers and form melanotic masses, “pseudotumors,” in what is probably an overreaction of the cellular immune response (18, 19). Notably, mutations that activate the Toll and Jak/Stat pathways show this phenotype, suggesting an important role for these pathways in the cellular response (20–23). Furthermore, Notch function is required for a normal cellular response (16). The Hemese gene encodes a hemocyte surface protein, which may play a modulatory role (24).
To identify genes that regulate the cellular immune response, we have systematically overexpressed different genes in hemocytes, by using the GAL4-upstream activating sequence (UAS) system (25), and assayed their effects on hemocyte activation. For the limited screen described here, we tested a selected set of genes that represent important cellular signaling pathways, focusing on those potentially involved in immunity or cell fate. The results indicate that several signaling systems are able to activate a lamellocyte response.
Materials and Methods
Fly Strains and Crosses. Flies were kept on a standard mashed-potato diet. The binary UAS-GAL4 system (25) was used to create specific gain-of-function phenotypes in larval hemocytes. Most UAS fly stocks included in this study were obtained from the Bloomington Stock Center (Bloomington, IN), and the references are given in Flybase (http://fbserver.gen.cam.ac.uk:7081). The P{UAS-Tl.10b:11} stock was constructed by J.-M. Reichhart (Centre National de la Recherche Scientifique, Strasbourg, France) and carries a Toll10b insert on the X chromosome (J.-M. Reichhart, personal communication). Several independent transgenic UAS lines, containing wild-type Smox (dSmad2), were constructed by T. Haerry (University of Minnesota, Minneapolis) and M. B. O'Connor (University of Minnesota, Minneapolis; personal communication).
New transgenic stocks were generated by P element-mediated transformation (26). Full sequence information about these constructs can be obtained upon request. P{UAS-Alk.act} is a ligand-independent, constitutively active Alk transgenic fly line. Briefly, the entire cytoplasmic domain of Alk was cloned in frame with a 169-aa-long stably disulfide-bonded and hemagglutinin-tagged extracellular and transmembrane domain derived from the receptor-like protein tyrosine phosphatase-α (27).
To generate a Hemese-GAL4 driver line, we used the PCR to synthesize an ∼1-kb-long fragment from the promoter of the Hemese gene (24), by using the following primers with additional EcoRI digestion sites: CGGGATCCATTTGTTGGTAA-TGTCCTCAAGC and GTTTACATAAGTCACTAAGAAT-TCCG. The resulting PCR fragment includes the 5′ untranslated leader of Hemese and all-intergenic sequence between Hemese and the neighboring gene CG8942. This fragment was subcloned by TA cloning (Invitrogen), cleaved out with BamHI and EcoRI, and cloned upstream of the GAL4 gene in the modified P vector, p{2/17-Hal4 RSBSK} (28). Of several transgenic lines obtained, a third chromosome insertion, P{Hemese-GAL4}85, was selected as the strongest expressing line. We will refer to it here as Hemese-GAL4. In most experiments, we used a Hemese-GAL4 recombinant stock that also carries the GFP reporter P{UAS-GFP.nls}8 on the third chromosome, for convenient detection of hemocyte phenotypes.
To create a transgenic UAS-Hemese strain, Hemese cDNA (24) was inserted into the pUAST vector (25). Transgenic flies, carrying the Hemese gene under the control of UAS, were subsequently generated.
Hemocyte Collection, Counting, and Statistics. Hemese-GAL4 UAS-GFP.nls virgin females were crossed to the indicated transgenic UAS line. The females were allowed to lay eggs at 21°C for 2 days before the vial was transferred to the indicated rearing temperature. Larvae were staged according to procedures described in ref. 29. A red household food dye was added to the food to allow for visualization of the gut contents. The emptying of the gut marks the difference between early- and late-wandering third-instar larvae. Staged larvae were first washed in water and then bled into 20 μl of PBS (137 mM NaCl/2.7 mM KCl/6.7 mM Na2HPO4/1.5 mM KH2PO4), ripping the cuticle by using two fine forceps. The cells then were redistributed gently, and diluted if necessary, before they were transferred to a Neubauer improved hemocytometer for counting. Hemocytes were counted and classified as either plasmatocytes or lamellocytes, based on their morphology. Circulating crystal cells were not counted separately. Instead, we counted sessile crystal cells in the last two posterior dorsal segments of third-instar larvae (16). They were visualized by heating the larvae for 10 min at 60°C in a water bath (30).
Hemocytes were counted from at least 15 larvae of each genotype. For the statistical analysis, the data for total hemocyte number and percentage of lamellocytes were log-transformed. An initial ANOVA indicated that the UAS constructs and their overexpression significantly affect total hemocyte number and the percentage of lamellocytes, independently of other variables. The crystal cell number depended only on the UAS construct. To study specific interactions between certain genotypes and their corresponding crosses, multiple t tests were performed. In detail, each cross was compared with its corresponding UAS construct, with the driver, and with the mean of all UAS constructs at the corresponding temperature. The risk of the type I error was set at a = 0.01. Because multiple comparisons may yield a significant result by chance, the threshold for significance was adjusted by using a sequential Bonferroni correction. Differences in hemocyte number were considered to be significant only if both the comparison of the cross to the driver and the comparison of the cross to the corresponding UAS construct were significant. All computations were performed with spss (Version 11.5, SPSS, Chicago).
Hemocyte Imaging. For studies on living cells, hemolymph from late-wandering larvae was bled into 8 μl of PBS/PTU solution (PBS with a small amount of phenylthiocarbamide, a phenoloxidase inhibitor that blocks melanization reactions; see ref. 31) on a 12-well slide (SM-011, Hendley-Essex, Essex, U.K.). Cells were allowed to settle at room temperature for 30 min in a humid chamber before examination by light and UV microscopy (Zeiss). Digital pictures were taken with a Hamamatsu C4742-95 video unit, controlled by the openlab program (Improvision, Coventry, U.K.). photoshop (Version 6.0, Adobe Systems, San Jose, CA) and appleworks (Version 6, Macintosh) were used for digital editing.
For immunohistochemistry, larvae were bled, the carcass was removed, and the hemocytes were allowed to settle as described above. Then the cells were fixed for 7 min by the addition of 20 μl of 3.7% paraformaldehyde, washed in PBS/PTU solution, and blocked in PBS containing 5% normal goat serum (DAKO). The samples then were incubated overnight in a humid chamber at 4°C, with 20 μl of monoclonal antibody of either panhemocyte (H1) or lamellocyte (L1) specificity (24). The primary antibody was detected by a Cy3-conjugated goat antibody to mouse IgG (Jackson ImmunoResearch) under UV light.
To visualize hemocytes inside living larvae, wandering third-instar larvae were washed in water and immobilized by chilling at 8°C for 3 h. The larvae then were transferred to a drop of chilled glycerol on an object slide.
Results
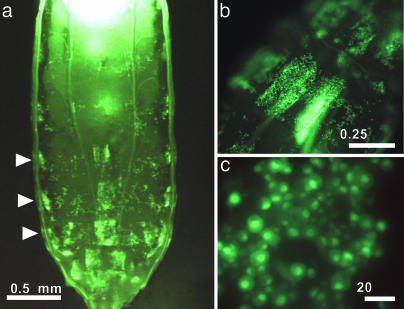
Creation of a Hemocyte-Specific Hemese-GAL4 Driver. We have described previously a transmembrane protein, Hemese, which is specifically expressed in Drosophila hemocytes and seems to play a modulatory role in their activation/recruitment (24). The Hemese gene normally is expressed in all classes of larval hemocytes as well as in the major hematopoietic organs, the lymph glands (24). To create a construct for hemocyte-specific expression, we fused the Hemese promoter to the yeast GAL4 gene and introduced this fusion construct into transgenic flies. The resulting Hemese-GAL4 transgenic line drives strong GFP expression in circulating hemocytes, which can be observed through the cuticle of the living larva. Strong GFP expression also is seen in a population of sessile hemocytes, which are found segmentally arranged under the epidermis and in large clusters in the posterior end of the larva (Fig. 1). Morphologically, this sessile population is indistinguishable from circulating plasmatocytes. This pattern, which reflects the expression of the endogenous Hemese protein, was observed with several independent inserts of the P{Hemese-GAL4} transgene (data not shown).
Fig. 1.
GFP reporter gene expression in vivo. Strong GFP expression in sessile and circulating hemocytes can be observed through the cuticle of third-instar Hemese-GAL4/UAS-GFP.nls larvae. (a) Posterior part of a larva, showing segmentally arranged sessile hemocytes (arrowheads). (b) Close-up of a posterior segmental cluster. (c) Single hemocytes in a posterior segmental cluster.
Unlike the endogenous Hemese gene (24), the Hemese-GAL4 driver shows very little expression in the lymph glands, affecting at most a few scattered cells. Furthermore, only ∼80% of the circulating hemocytes express GFP (data not shown). Thus, it is possible that the promoter fragment used for the Hemese-GAL4 construct does not include all control elements required for full hemocyte expression of Hemese. In addition to the Hemese-driven hemocyte expression of the transgene, strong ectopic expression also is seen in salivary glands and in sections of the midgut. Similar observations also have been reported for other GAL4 constructs and may be due to residual promoter activity from the hsp70 gene in the vector (32, 33).
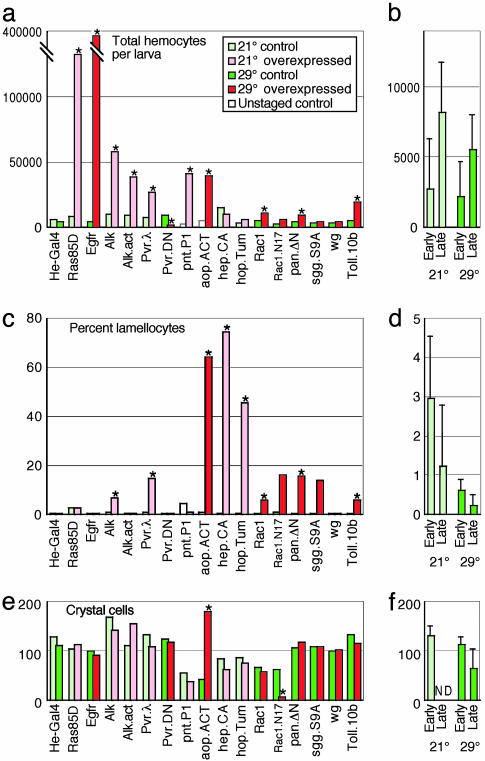
Expression of Selected UAS Transgenes Affects Lamellocyte Formation and Total Hemocyte Numbers. To study the effects of several important signaling pathways on hemocyte activation, we crossed a recombinant GFP-expressing Hemese-GAL4 fly line to 57 selected transgenic UAS stocks, as listed in Table 1, and Table 2, which is published as supporting information on the PNAS web site. For optimal expression by the GAL4-UAS system, the standard rearing temperature was 29°C. However, overexpression of some constructs was lethal at 29°C, and these therefore were assayed at 21°C (Fig. 2). We estimated the hemocyte numbers in wandering third-instar larvae. Because the number of circulating hemocytes increases rapidly during development, we further staged the wandering larvae according to the presence or absence of food in the gut. By this criterion, late-wandering larvae have on average 2–3 times more hemocytes than early wandering larvae and show less relative variation in cell numbers (Fig. 2b). The background of lamellocytes is also lower in late larvae (Fig. 2d). For these reasons, we show the hemocyte counts in late larvae, although qualitatively similar results were obtained for the earlier stage (data not shown).
Table 1. Constructs that give hemocyte phenotypes.
| Gene | Construct | Type | Prol. | Lam. | Crys. | Other |
|---|---|---|---|---|---|---|
| Alk | P{UAS-Alk} | WT | + | + | 0 | |
| P{UAS-Alk.ACT} | CA | + | 0 | 0 | ||
| aop | P{UAS-aop.ACT} | CA | + | + | + | Melanotic masses |
| Egfr | P{UAS-Egfr.B}32-26-1 | WT | + | 0 | 0 | |
| hep | P{UAS-hep.CA} | CA | 0 | + | 0 | Melanotic masses |
| hop | P{UAS-hop.Tum} | CA | 0 | + | 0 | Melanotic masses |
| pan | P{UAS-pan.dTCFΔN}4 | DN | + | + | 0 | Melanotic masses |
| pnt | P{UAS-pnt.P1} | WT | + | 0 | 0 | Melanotic masses |
| Pvr | P{UAS-Pvr.λ} | CA | + | + | 0 | Melanotic masses |
| P{UAS-Pvr.DN} | DN | - | 0 | 0 | ||
| Rac1 | P{UAS-Rac1.L} | WT | + | + | 0 | Melanotic masses |
| P{UAS-Rac1.N17}1 | Mut | 0 | * | - | Multinucleate cells | |
| P{UAS-Rac1.V12} | Mut | Lethal | ||||
| Ras85D | P{UAS-Ras85D.V12}TL1 | WT | + | 0 | 0 | Melanotic masses |
| sgg | P{UAS-sgg.S9A}MB14 | WT | 0 | (+) | 0 | Melanotic masses |
| Toll | P{UAS-Tl.10b:11} | CA | + | + | 0 | Melanotic masses |
Table 2 lists 41 additional tested constructs, representing 27 different genes, which do not give hemocyte phenotypes. Prol., proliferation; Lam., lamellocytes; Crys., crystal cells; WT, wild-type; CA, constitutively active; DN, dominant-negative; Mut, other mutant; +, increase; 0, no changes; -, decrease; (+), increase with low penetrance; *, multinucleate cells that do not express lamellocyte marker.
Fig. 2.
Hemocyte counts after overexpression of selected UAS transgenes. Hemese-GAL4 UAS-GFP driver flies were crossed with UAS constructs to express wild-type, constitutively active, or dominant-negative mutant forms of the indicated genes. Hemocytes were counted from at least 15 individual larval offspring of these crosses and from the corresponding parental UAS strain as a control. (a and b) The average estimated total number of circulating hemocytes per larva. (c and d) The corresponding percentage of lamellocytes. The control larvae of the UAS-pnt.P1 and UAS-aop.ACT strains were not staged and therefore have a variable fraction of lamellocytes. (e and f) The crystal cell counts from the sessile population of two segments. (a, c, and e) The average number of hemocytes in the different overexpressing (red) and control (green) late-wandering larvae. (b, d, and f) The combined average and SD of all of the nonoverexpressing control genotypes. See Table 1 for complete designation of the overexpressed constructs. An asterisk indicates a significant difference (t test, P < 0.01) compared with the parental UAS and Hemese-GAL4 strains. The penetrance of the lamellocyte phenotype is variable, sometimes leading to large deviations from a log-normal distribution. This observation explains why the t test does not detect significance for the Rac1.N17 and shaggy overexpression phenotypes.
Several of the tested constructs cause overproliferation of hemocytes and/or activation of lamellocyte formation, but these two effects do not seem to be correlated (Fig. 2 a and c and Table 1). For instance, a dramatic increase in the hemocyte number is seen with UAS-Ras85D and UAS-Egfr, and a smaller increase is seen with UAS-pointed.P1, without significant effect on the lamellocyte fraction. Conversely, UAS-hep and UAS-hop.Tum give very strong activation of lamellocyte formation but do not cause an increase in total hemocyte number. Other constructs, such as UAS-aop.act, UAS-Alk, UAS-Pvr.λ, and UAS-Toll.10b, stimulate both hemocyte proliferation and lamellocyte formation. A lamellocyte response with only a minor effect on cell number is seen with UAS-pan.TCFΔN, UAS-shaggy, and UAS-Rac1.L. Finally, a significant (7-fold) decrease in total hemocyte number is seen with a dominant-negative form of Pvr, UAS-Pvr.DN. It should be pointed out that the quantitative differences in phenotype strength should be interpreted with caution, because expression levels may vary between constructs and because different crosses had to be tested at different temperatures.
We also assayed the effect of the constructs on sessile crystal cells. In this case, we obtained more consistent results with early rather than with late-wandering larvae (Fig. 2f), presumably because later in development the sessile cells are released into circulation. Significant effects were seen only in UAS-Rac1.N17 larvae, which had much-reduced numbers of crystal cells, and UAS-aop.ACT, which had increased numbers (Fig. 2e).
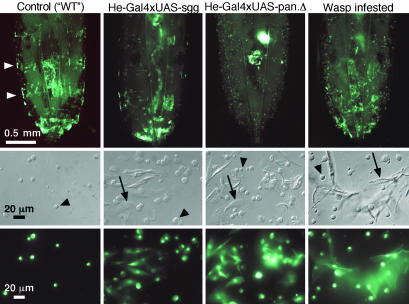
Further Characterization of the Overexpression Phenotypes. The expression of GFP in the hemocytes allowed us to follow these cells in vivo. We found that the formation of lamellocytes correlated with a disappearance of the banded pattern of sessile cells, as shown for UAS-pan.TCFΔN and UAS-sgg.S9A in Fig. 3. The sessile population is similarly reduced when lamellocyte formation is triggered by wasp infestation. It is possible that these sessile hemocytes act as a source of lamellocyte precursors.
Fig. 3.
Dynamics of the sessile hemocyte population. Examples of the disappearance of the sessile hemocyte population in wasp-infested larvae and in larvae that are subjected to Hemese-GAL4-driven overexpression of shaggy or dominant-negative pangolin. White arrowheads indicate two of the segmental hemocyte clusters in a control larva. Pictures show the posterior half of third-instar larvae (Top) or bled hemocytes (Middle and Bottom) from the indicated crosses. Examples of plasmatocytes and lamellocytes are marked with black arrowheads and arrows, respectively. The control larva and the larva subjected to wasp infestation are offspring of the cross Hemese-GAL4 UAS-GFP × w1118. UAS-sgg, UAS-shaggy.S9A; UAS-pan.Δ, UAS-pan.dTCFΔN.
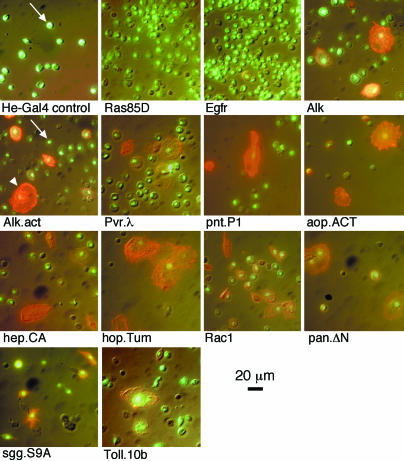
To verify the formation of lamellocytes, we stained fixed cell smears with the lamellocyte-specific antibody L1 (24). Fig. 4 confirms that the large, flattened cells are indeed lamellocytes. Larvae that overexpress Rac1.N17 produce large multinucleate cells that do not stain with the L1 marker (data not shown). The lamellocyte-producing crosses also give a considerable number of plasmatocyte-shaped cells and intermediate forms that stain positive for the lamellocyte marker, suggesting the presence of lamellocyte precursors in the pool of circulating cells in these larvae. Such cells are rare in the controls and also in the UAS-Ras85D.V12 and UAS-EGFR.B crosses. The cells observed from these and other crosses are hemocytes (data not shown), as confirmed with the monoclonal pan-hemocyte antibody H1 (24).
Fig. 4.
Hemocyte phenotypes caused by overexpression of the indicated genes. Pictures of lamellocyte antibody staining (red) and nuclear GFP fluorescence (green) were merged to a Nomarski picture of the same field. {UAS-GFP.nls}8 is a marker of Hemese-GAL4 driver activity. Examples of plasmatocytes and lamellocytes are marked with white arrows and arrowheads, respectively.
Many of the constructs that give a hemocyte phenotype also promote the formation of melanotic masses in the larvae (Table 1). The penetrance of this phenotype is variable and does not reach 100% for any of the crosses. Constructs that give an increased number of lamellocytes typically also cause formation of melanotic masses, except in the UAS-Alk and UAS-Alk.act animals. However, melanization was seen in other experiments with UAS-Alk.act. Furthermore, a few small melanotic nodules were observed among the larvae that overexpress Ras85D or pointed. Although the percentage of lamellocytes is not significantly increased with the latter constructs, the absolute numbers are 16-fold with Ras85D and 3-fold with pointed.
We considered the possibility that some of the hemocyte responses could be indirect, triggered by toxic effects of ectopic Hemese-GAL4 expression in the salivary glands. Indeed, we observed that the salivary glands were reduced in size with some of the constructs. To address this possibility, we repeated the experiments with the salivary gland-specific driver SaGa 49 E (A. Hofmann, personal communication). Overexpression of UAS-Alk.act and UAS-pan.dTCFΔN with this driver had no effect on hemocytes. Furthermore, we also tested a second, recently described hemocyte-specific driver, Hemolectin-GAL4 (34). This driver is considerably weaker when compared with Hemese-GAL4, but it is not expressed in the salivary glands. The resulting hemocyte phenotypes are also correspondingly weaker but are essentially the same. This observation was confirmed for one tyrosine kinase (UAS-Alk.act), for one member of the wingless pathway (UAS-pan.TCFΔN), for UAS-hep.CA, and for the Rac1 and pointed constructs (data not shown). Thus, we conclude that the hemocyte responses we observe are indeed caused by expression of the various transgenes in the hemocytes themselves.
Discussion
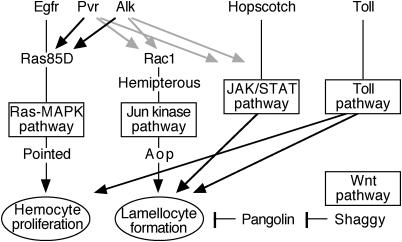
A surprising result of our screen is that several very different signaling pathways cause similar phenotypes upon ectopic activation in Drosophila hemocytes, as summarized in Fig. 5. The involvement of Toll and Jak/Stat signaling in lamellocyte formation was expected from previous studies on different mutant phenotypes (7, 23). We now find that a similar effect is caused by two receptor tyrosine kinases, Pvr and Alk, as well as by two negative regulators of wingless signaling and several genes associated with mitogen-activated protein kinase (MAPK) signaling. The most striking lamellocyte phenotype was seen with an activated form of Hemipterous, the Drosophila Jun kinase kinase. Conversely, a strong proliferative response was seen with Ras and with the receptor tyrosine kinases, in particular Egfr, but other signaling pathways also influenced the hemocyte numbers to a lesser extent. We will now discuss these effects in detail.
Fig. 5.
Signaling pathways implicated in hemocyte activation. For simplicity, all pathways are assumed to act in parallel, although our data are also consistent with models in which one or more pathways are connected in series. Possible cross-talk between the ETS factors Pointed and Aop are not shown. Alternative interpretations of our data are discussed in the text.
The Control of Hemocyte Number. As shown by Asha et al. (35), overexpression of Ras85D promotes cell division, causing a tumorous-like phenotype. Interestingly, similar effects also are seen with three receptor protein tyrosine kinases, Pvr, Alk, and Egfr. These kinases are involved in many biological processes that require cell proliferation and/or differentiation, and they have been linked to Ras-MAPK signaling (for review, see Rebay, ref. 36).
The Pvr gene encodes the recently described Drosophila homolog of the human platelet-derived growth factor and vascular endothelial growth factor receptors (37). A proliferative response to this receptor is not unexpected, because Munier et al. (13) showed that Pvr protein is expressed on the hemocyte surface and that overexpression of one of its putative ligands, Pvf2, induces a dramatic increase of the number of circulating hemocytes. Pvf2 also has been shown to be necessary for control of hemocyte migration in embryos (38, 39).
Alk encodes the Drosophila homolog of the human anaplastic lymphoma kinase (ALK) (40). This protein tyrosine kinase originally was identified in the human chromosomal translocation NPM-ALK 2;5, which is associated with non-Hodgkin's lymphomas. Misexpression of ALK in T cells, caused by the translocation, is known to cause malignant transformation of these lymphomas (41–43). Our results indicate that ectopic expression of Drosophila Alk in hemocytes has a similar effect, resulting in a dramatic hemocyte phenotype and tumor formation. It is not clear whether Alk functions in hemocytes under normal conditions, but antibody staining suggested that the protein is expressed in hemocytes (data not shown). Recent experiments indicated that a functional Alk gene is crucial for gut formation (44), but because embryonic lethality of Alk mutant animals precludes study of the larval hemocyte population, a hemocyte function cannot be excluded.
Whether the Egfr gene has any function in hemocytes in wild-type flies is even more uncertain. It has been implicated in a number of developmental processes in Drosophila (reviewed in ref. 45) but so far not in hematopoiesis or hemocyte function. It is possible that ectopic expression of Egfr may mimic, in part, the effect of other receptor tyrosine kinases that are normally present in the hemocytes, such as Pvr or perhaps Alk. Egfr overexpression gives an exclusively proliferative response, without stimulating lamellocyte formation. Ras85D and the pointed P1 transcript give a similar response, suggesting that they act in the same pathway. This finding is consistent with observations in other systems, where Egfr is known to stimulate the Drosophila ERK homolog Rolled by means of a Ras-MAPK pathway (46, 47). The ETS transcription factor encoded by pointed is a target of this pathway (48). Our results indicate that this pathway may be a main regulator of the number of circulating hemocytes, under the control of one or more of the receptor tyrosine kinases. A direct role for Pvr in this regulation is further supported by our finding that the hemocyte number is severely reduced on overexpression of a dominant-negative form of Pvr.
The Lamellocyte Response. In addition to their effect on total hemocyte counts, Pvr and Alk also stimulate lamellocyte formation. This observation suggests that a second signaling pathway is activated by these kinases. It is possible that Pvr and Alk share targets with the Drosophila JAK homolog, Hopscotch, which is a cytoplasmic tyrosine kinase. Stimulation of the JAK/STAT pathway by overexpressing hopscotchTum gives a strong lamellocyte phenotype. Another possibility is that Pvr and Alk activate the Jun-specific MAPK pathway. A role for this pathway in the lamellocyte response is suggested by the following: (i) a strong lamellocyte phenotype after overexpression of an activated form of Hemipterous, the Drosophila Jun kinase kinase; and (ii) a milder lamellocyte phenotype seen with wild-type Rac1, which is an activator of the Jun kinase pathway. A constitutively active form of Rac1 was found to be lethal when expressed with the Hemese-GAL4 driver and therefore could not be tested. Wild-type constructs of two other genes in the Jun kinase pathway, basket and misshapen, had no effect (Table 2), but it is uncertain whether the pathway is efficiently activated by the constructs used.
The effect of Aop is more difficult to fit into this context. Aop (also called Yan) is an ETS factor, which acts as a transcriptional repressor, in competition with the activating ETS factor Pointed. MAPK phosphorylation blocks this repression and leads to Aop being exported from the nucleus. The AopACT mutant protein has all possible MAPK phosphorylation sites mutated and acts as a constitutive repressor of Pointed. It therefore would be expected to have an antiproliferative effect, similar to that of dominant-negative Pvr. Instead, it gives a massive lamellocyte response and, surprisingly, a strong stimulation of hemocyte production. Even the crystal cells are affected. However, Pointed and Aop are known to respond both to Ras-MAPK and to Jun kinase signaling (48). It is possible that different MAPKs regulate a delicate balance between proliferation and lamellocyte activation by targeting different phosphorylation sites in the two ETS factors. Another problem is that no lamellocyte formation was observed when we overexpressed Imd (Table 2), although Imd is expected to activate Jun kinase signaling (49). Further experiments will be required to clarify the role of MAPKs in lamellocyte formation.
Finally, two negative regulators of the wingless pathway cause a lamellocyte phenotype, a dominant-negative form of Pangolin, the Drosophila homolog of the TCF transcription factor, and Shaggy, a negative regulator of the β-catenin homolog Armadillo. Although the effect of the Shaggy construct is weaker and more variable, both give rise to an increased number of lamel-locytes. This finding suggests that wingless signaling may be a negative regulator of the lamellocyte response.
Conclusions
It is clear that the lamellocyte response and the total hemocyte number are controlled independently. The latter may be regulated by receptor tyrosine kinases, by means of a single Ras-MAPK pathway. The lamellocyte response seems to be much more complex, responding to several different pathways. However, a high level of redundancy in this type of immune response cannot be excluded. Some of the complexity may be explained if the cellular immune response is controlled by means of cytokine signaling, involving different pathways in cytokine production and the reception of the signal. Another possibility is that some of the effects are artifacts of the overexpression system. When a kinase is highly overexpressed, it may act on targets that are not normally relevant (50). Finally, it is also possible that insects have sensors for a general imbalance in the cellular communication systems, as has been described for certain plant defense reactions (51). A perturbed balance is potentially a sign of pathogenic interference. When overexpressed, some genes could generate a similar imbalance, indirectly triggering an immune response. Whatever the case, the results described here provide a starting point to find out how an innate cellular immune defense can be activated.
Supplementary Material
Acknowledgments
We thank Ingrid Dacklin and the Umeå Fly and Worm Transgene Facility for assistance during the generation of Hemese-Gal transgenic lines, the Bloomington Stock Center for providing fly stocks, Anna-Karin Kronhamn for assistance with flies and crosses, and Shannon Albright for comments on the manuscript. UAS-Pvr.λ and UAS-Pvr.DN flies were kindly provided by P. Rørth (European Molecular Biology Laboratory, Heidelberg), UAS-Tum.l flies by D. Harrison (University of Kentucky, Lexington), UAS-dome and UAS-os (UAS-Upd) by J. Castelli-Gair Hombria (University of Cambridge, Cambridge, U.K.), Uas-Toll10b and UAS-imd by J.-M. Reichhart, UAS-dSmad2 by M. B. Connor, UAS-Med by S. Newfeld (Arizona State University, Tempe), and SaGa 49 E by A. Hofmann (Institut für Genetik der Freien Universität, Berlin). This research was supported by grants from the Swedish Research Council, the Swedish Cancer Society, and the Wallenberg Consortium North.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ALK, anaplastic lymphoma kinase; MAPK, mitogen-activated protein kinase; UAS, upstream activating sequence.
References
- 1.De Gregorio, E. & Lemaitre, B. (2002) Nature 419, 496–497. [DOI] [PubMed] [Google Scholar]
- 2.Hultmark, D. (2003) Curr. Opin. Immunol. 15, 12–19. [DOI] [PubMed] [Google Scholar]
- 3.Hoffmann, J. A. (2003) Nature 426, 33–38. [DOI] [PubMed] [Google Scholar]
- 4.Rizki, T. M. & Rizki, R. M. (1986) in Hemocytic and Humoral Immunity in Arthropods, ed. Gupta, A. P. (Wiley, New York), pp. 157–190.
- 5.Vass, E. & Nappi, A. J. (2000) J. Parasitol. 86, 1259–1270. [DOI] [PubMed] [Google Scholar]
- 6.Lavine, M. D. & Strand, M. R. (2002) Insect Biochem. Mol. Biol. 32, 1295–1309. [DOI] [PubMed] [Google Scholar]
- 7.Evans, C. J., Hartenstein, V. & Banerjee, U. (2003) Dev. Cell 5, 673–690. [DOI] [PubMed] [Google Scholar]
- 8.Carton, Y. & Nappi, A. J. (2001) Immunogenetics 52, 157–164. [DOI] [PubMed] [Google Scholar]
- 9.Russo, J., Brehelin, M. & Carton, Y. (2001) J. Insect Physiol. 47, 167–172. [DOI] [PubMed] [Google Scholar]
- 10.Lanot, R., Zachary, D., Holder, F. & Meister, M. (2001) Dev. Biol. 230, 243–257. [DOI] [PubMed] [Google Scholar]
- 11.Holz, A., Bossinger, B., Strasser, T., Janning, W. & Klapper, R. (2003) Development (Cambridge, U.K.) 130, 4955–4962. [DOI] [PubMed] [Google Scholar]
- 12.Rehorn, K. P., Thelen, H., Michelson, A. M. & Reuter, R. (1996) Development (Cambridge, U.K.) 122, 4023–4031. [DOI] [PubMed] [Google Scholar]
- 13.Munier, A. I., Doucet, D., Perrodou, E., Zachary, D., Meister, M., Hoffmann, J. A., Janeway, C. A., Jr., & Lagueux, M. (2002) EMBO Rep. 3, 1195–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernardoni, R., Vivancos, V. & Giangrande, A. (1997) Dev. Biol. 191, 118–130. [DOI] [PubMed] [Google Scholar]
- 15.Lebestky, T., Chang, T., Hartenstein, V. & Banerjee, U. (2000) Science 288, 146–149. [DOI] [PubMed] [Google Scholar]
- 16.Duvic, B., Hoffmann, J. A., Meister, M. & Royet, J. (2002) Curr. Biol. 12, 1923–1927. [DOI] [PubMed] [Google Scholar]
- 17.Lebestky, T., Jung, S. H. & Banerjee, U. (2003) Genes Dev. 17, 348–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watson, K. L., Justice, R. W. & Bryant, P. J. (1994) J. Cell Sci. 18, 19–33. [DOI] [PubMed] [Google Scholar]
- 19.Gateff, E. (1994) Int. J. Dev. Biol. 38, 565–590. [PubMed] [Google Scholar]
- 20.Govind, S. (1999) Oncogene 18, 6875–6887. [DOI] [PubMed] [Google Scholar]
- 21.Dearolf, C. R. (1998) Biochim. Biophys. Acta 1377, M13–M23. [DOI] [PubMed] [Google Scholar]
- 22.Mathey-Prevot, B. & Perrimon, N. (1998) Cell 92, 697–700. [DOI] [PubMed] [Google Scholar]
- 23.Sorrentino, R. P., Melk, J. P. & Govind, S. (2004) Genetics 166, 1343–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurucz, E., Zettervall, C.-J., Sinka, R., Vilmos, P., Pivarcsi, A., Ekengren, S., Hegedüs, Z., Ando, I. & Hultmark, D. (2003) Proc. Natl. Acad. Sci. USA 100, 2622–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brand, A. H. & Perrimon, N. (1993) Development (Cambridge, U.K.) 118, 401–415. [DOI] [PubMed] [Google Scholar]
- 26.Spradling, A. C. (1986) in Drosophila: A Practical Approach, ed. Roberts, D. B. (IRL, Oxford), pp. 175–197.
- 27.Jiang, G., den Hertog, J., Su, J., Noel, J., Sap, J. & Hunter, T. (1999) Nature 401, 606–610. [DOI] [PubMed] [Google Scholar]
- 28.Finley, K. D., Edeen, P. T., Foss, M., Gross, E., Ghbeish, N., Palmer, R. H., Taylor, B. J. & McKeown, M. (1998) Neuron 21, 1363–1374. [DOI] [PubMed] [Google Scholar]
- 29.Andres, A. J. & Thummel, C. S. (1994) in Drosophila melanogaster: Practical Uses in Cell and Molecular Biology, Methods in Cell Biology, eds. Goldstein, L. S. B. & Fyrberg, E. A. (Academic, San Diego), Vol. 44, pp. 565–573. [Google Scholar]
- 30.Rizki, T. M. & Rizki, R. M. (1980) Experientia 36, 1223–1226. [Google Scholar]
- 31.Ashburner, M. (1989) Drosophila: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 32.Duffy, J. B. (2002) Genesis 34, 1–15. [DOI] [PubMed] [Google Scholar]
- 33.Gerlitz, O., Nellen, D., Ottiger, M. & Basler, K. (2002) Int. J. Dev. Biol. 46, 173–176. [PubMed] [Google Scholar]
- 34.Goto, A., Kadowaki, T. & Kitagawa, Y. (2003) Dev. Biol. 264, 582–591. [DOI] [PubMed] [Google Scholar]
- 35.Asha, H., Nagy, I., Kovacs, G., Stetson, D., Ando, I. & Dearolf, C. R. (2003) Genetics 163, 203–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rebay, I. (2002) Dev. Biol. 251, 1–17. [DOI] [PubMed] [Google Scholar]
- 37.Duchek, P., Somogyi, K., Jekely, G., Beccari, S. & Rørth, P. (2001) Cell 107, 17–26. [DOI] [PubMed] [Google Scholar]
- 38.Heino, T. I., Karpanen, T., Wahlström, G., Pulkkinen, M., Eriksson, U., Alitalo, K. & Roos, C. (2001) Mech. Dev. 109, 69–77. [DOI] [PubMed] [Google Scholar]
- 39.Cho, N. K., Keyes, L., Johnson, E., Heller, J., Ryner, L., Karim, F. & Krasnow, M. A. (2002) Cell 108, 865–876. [DOI] [PubMed] [Google Scholar]
- 40.Lorén, C. E., Scully, A., Grabbe, C., Edeen, P. T., Thomas, J., McKeown, M., Hunter, T. & Palmer, R. H. (2001) Genes Cells 6, 531–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morris, S. W., Kirstein, M. N., Valentine, M. B., Dittmer, K. G., Shapiro, D. N., Saltman, D. L. & Look, A. T. (1994) Science 263, 1281–1284. [DOI] [PubMed] [Google Scholar]
- 42.Fujimoto, J., Shiota, M., Iwahara, T., Seki, N., Satoh, H., Mori, S. & Yamamoto, T. (1996) Proc. Natl. Acad. Sci. USA 93, 4181–4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuefer, M. U., Look, A. T., Pulford, K., Behm, F. G., Pattengale, P. K., Mason, D. Y. & Morris, S. W. (1997) Blood 90, 2901–2910. [PubMed] [Google Scholar]
- 44.Lorén, C. E., Englund, C., Grabbe, C., Hallberg, B., Hunter, T. & Palmer, R. H. (2003) EMBO Rep. 4, 781–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Voas, M. G. & Rebay, I. (2004) Dev. Dyn. 229, 162–175. [DOI] [PubMed] [Google Scholar]
- 46.Brunner, D., Oellers, N., Szabad, J., Biggs, W. H., III, Zipursky, S. L. & Hafen, E. (1994) Cell 76, 875–888. [DOI] [PubMed] [Google Scholar]
- 47.Biggs, W. H., III, Zavitz, K. H., Dickson, B., van der Straten, A., Brunner, D., Hafen, E. & Zipursky, S. L. (1994) EMBO J. 13, 1628–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hsu, T. & Schulz, R. A. (2000) Oncogene 19, 6409–6416. [DOI] [PubMed] [Google Scholar]
- 49.Boutros, M., Agaisse, H. & Perrimon, N. (2002) Dev. Cell 3, 711–722. [DOI] [PubMed] [Google Scholar]
- 50.Li, J. & Li, W. X. (2003) Genetics 164, 247–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schneider, D. S. (2002) Cell 109, 537–540. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.