Abstract
BCL6, a gene on chromosome 3, band q27, encodes a zinc finger transcriptional repressor that is needed for germinal center formation and has been implicated in the pathogenesis of some human lymphomas when it is mutated or involved in chromosomal rearrangements. To explore further the mechanisms of action of BCL6 in lymphomagenesis, we developed a transgenic mouse model mimicking a common translocation, the t(3, 14)(q27;q32), in human lymphomas. The transgenic mice develop normally and express the transgenic BCL6 protein constitutively in lymphocytes. A small fraction of the animals develop B and T cell lymphomas after a long latency period, but the incidence is dramatically enhanced following administration of N-ethyl-N-nitrosourea, a carcinogen that induces DNA mutations. The N-ethyl-N-nitrosourea-induced lymphomas spread widely, were exclusively T cell, expressed the BCL6 protein, and occurred only in the transgenic mice. Because BCL6 expression has been reported in a number of T cell tumors as well as in the more commonly occurring B cell lymphomas in humans, our transgenic mice provide a model for the study of human lymphomas.
BCL6, a gene on chromosome 3, band q27, was identified by us and others through its involvement in chromosomal rearrangements associated with a variety of non-Hodgkin lymphomas, especially diffuse large-cell B cell lymphomas (1, 2). The breakpoints cluster around the first (noncoding) exon of BCL6 (2), and it is thought that BCL6 expression becomes deregulated in ≈40% of diffuse large-cell B cell lymphomas by rearrangements in which normal BCL6 regulatory sequences are replaced by heterologous promoters (3). Additionally, in other lymphomas, mutations occur that affect BCL6 binding sites in the first (noncoding) exon and interfere with autoregulation (4). The BCL6 gene encodes a nuclear zinc finger protein, a transcriptional repressor, needed for germinal center formation (5). This protein is expressed at high levels in human lymphoid germinal center B cells, most cortical thymocytes, and some B and T cell lymphomas, but is not expressed in pre- or postgerminal center B cells or plasma cells (6).
The production of transgenic mice expressing human BCL6 has not been reported previously. It is postulated that the reason it is difficult to generate such mice by using conventional constructs is that they die during embryogenesis. We were able to generate BCL6 transgenic mice expressing human BCL6 specifically in lymphocytes in a two-mouse model that mimics a common translocation, the t(3,14)(q27;q32), seen in human lymphomas. We report the development of T cell lymphomas in a significant fraction of these mice after the administration of the carcinogen N-ethyl-N-nitrosourea (ENU).
Materials and Methods
BCL6 Transgenic Lines. We used an approach similar to that of Felsher and Bishop (7) to generate mice that express the human BCL6 transgene selectively in lymphocytes. This system requires two kinds of transgenic mice. The first, obtained from Felsher and Bishop, expresses the tetracycline-transactivating protein (tTA) under control of the inmmunoglobulin (IGH) enhancer and the SRα promoter (a fusion promoter containing the simian virus 40 early promoter and the R segment and part of the U5 sequence of the long-terminal repeat of human T cell leukemia virus type 1; ref. 8) (Eμ-tTA), permitting use of a version of the tet system in which tTA mediates the transcription of a transgene placed under the control of the tetracycline-responsive promoter (tet-o). In this system, the transgene is “on” in the absence of tetracycline or doxycycline and can be silenced by giving these drugs. We generated the second mouse by placing human BCL6 cDNA under the control of the tetracycline-responsive minimal promoter (tet-o-BCL6). We subcloned human full-length BCL6 cDNA, along with a 3′ construct containing an intron spanning 66 bp (gift of W. Müller, Gesellschaft für Biotechnologishe Forschung, Braunschweig, Germany; the construct was modified to include a part of the polylinker, the intron, and a 3′ end that was shortened by AseI digestion, blunting with Klenow polymerase, and ligation to the 3′ SwaI site) in the NotI site of the pBI-G Tet Vector (BD Biosciences/Clontech). This vector is a response plasmid that expresses a gene of interest as well as β-galactosidase (β-gal) from a bidirectional tet-responsive promoter. The construct was linearized with XmnI and AseI, purified, ultracentrifuged in a sucrose gradient, dialyzed against PBS, and microinjected into fertilized mouse ova that were implanted in pseudopregnant females. We identified multiple transgenic founder mice containing this BCL6 construct by Southern hybridization and blotting using DNA from tail clippings and 32P-labeled human BCL6 cDNA as a probe. Quantitation of band intensity relative to known standards was performed with imagequant software (Molecular Dynamics) to estimate transgene copy number in founders and F1 mice. Founders were derived in CD-1, then backcrossed to FVB/N before study. Offspring containing the BCL6 transgene were crossed with Eμ-tTA mice (FVB/N), and progeny containing both transgenes were identified by PCR of toe or tail DNA. These mice were tested for expression of the BCL6 transgene and used for additional studies. Mice were handled in accordance with institutional protocols.
BCL6 Transgene Expression. Total RNA was extracted from organs of transgenic and control mice, DNase-1-digested, reverse transcribed, and subjected to RT-PCR using β-actin primers that amplify 285 bp of cDNA (vs. 396 bp for genomic DNA) and primers that amplify a 289-bp fragment of the BCL6 transgene encompassing the 3′ end of the cDNA and the entire 66-bp intron placed beyond it. To determine expression in B and T cells, single-cell suspensions of mouse splenocytes were preincubated with anti-2.4G2 to prevent nonspecific binding of fluorochrome-conjugated antibodies, labeled with anti-CD3-PE and anti-CD19-FITC (BD Biosciences/Pharmingen), and sorted (DakoCytomation MoFlo-HTS) before RNA extraction and RT-PCR. The purity of the selected populations was determined by flow cytometry (DakoCytomation CyanLx). To ascertain inhibition of transgene expression, mice were given doxycycline hydrochloride (Sigma, 200 μg/ml) in drinking water changed once per week, and RNA expression was analyzed.
The bidirectional promoter in the construct permitted analysis of transgene expression at the protein level indirectly with the use of β-gal and directly by analysis of BCL6 protein expression. Cut tissue pieces of spleen and thymus were incubated in 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-gal) (9) for 18–20 h at room temperature. For β-gal visualization microscopically, tissues were fixed in cold 0.2% glutaraldehyde with 5 mM ethylenediamine tetraacetic acid (pH 8) and 2 mM magnesium chloride (MgCl2), placed in cold detergent solution (9) for 30 min, and incubated with 1 mg/ml X-gal for 18–19 h in the dark at 37°C. Triton X-100 (1%) was added to enhance penetration in spleen. Tissues for microscopy were rinsed and incubated for an additional day in PBS at 37°C (10).
To study BCL6 expression, spleen sections were deparaffinized, quenched in hydrogen peroxide (H2O2)/methanol, heated near boiling for 40 min in pH 10 antigen retrieval buffer (DAKO), stained with N-terminal BCL6 affinity-purified rabbit polyclonal IgG antibody (sc-858, Santa Cruz Biotechnology) overnight at 4°C, and incubated with peroxidase-conjugated affinity-purified donkey anti-rabbit IgG (H+L) (Jackson ImmunoResearch). Antigen–antibody binding was detected with diaminobenzidine (DAB) chromogen.
Immunization and ELISA. Transgenic animals and littermate controls aged 25 days to 18 months were immunized i.p. with sheep red blood cells (SRBCs) (Colorado Serum, Denver, 100 μl), keyhole limpet hemocyanin (KLH) (Sigma, 50–100 μg), or both, and killed 10–15 days later for analysis. ELISA for total IgG was performed in triplicate by using serially diluted serum samples from 9-month-old immunized mice on plates coated with SRBCs or KLH as appropriate.
Histology. Tissues were fixed in 10% buffered formalin, paraffinembedded, sectioned, and stained with hematoxylin/eosin. To minimize loss of β-gal stain during processing, tissues were paraffin-embedded by using an abbreviated protocol (TP1050, Leica), sectioned, and counterstained with nuclear fast red.
Flow Cytometry [Fluorescence-Activated Cell Sorting (FACS)]. Single cell suspensions were incubated for 15 min with anti-2.4G2 to prevent nonspecific binding and labeled with fluorochrome-conjugated antibodies (BD Biosciences/Pharmingen or Sigma) used according to the manufacturer's protocol. Analysis was performed on a BD FACScan flow cytometer.
Immunohistochemistry. BCL6 staining is described above. For T cell staining, tissues were deparaffinized, quenched, incubated in pH 10 antigen retrieval solution (DAKO) in a 98.6°C water bath for 40 min, and stained with CD3 rabbit polyclonal antibody (Cell Marque, Hot Springs, AR) for 60 min at room temperature. Antigen–antibody binding was detected by using the DAKO rabbit EnVision+ System (DAB). For B cell staining, tissues were incubated in near boiling pH 6 antigen retrieval solution for 12 min, stained with anti-IgM/IgG polyclonal rabbit antibodies (Lab Vision, Fremont, CA) overnight at 4°C, washed, and incubated with peroxidase-conjugated affinity-purified donkey anti-rabbit IgG (H+L). Antigen–antibody binding was detected with DAB chromogen. All sections were counterstained with dilute hematoxylin. Staining of spleen frozen sections with peanut agglutinin, anti-GL7, anti-B220, and anti-CD35 was performed similarly, except that antigen retrieval was not performed.
Monitoring Development of Spontaneous Lymphomas. A cohort of aging mice backcrossed four times to FVB/N (transgenics and littermate controls) was followed until death or 3 years, whichever came first. Mice aged 6 months or more were autopsied.
Monitoring Lymphoma Development After ENU Administration. A subgroup of mice backcrossed five times to FVB/N (transgenics and littermate controls, usually positive for either the Eμ-tTA or BCL6 transgene, but occasionally wild type) were injected once i.p. with ENU [Sigma; 100 mg/kg dissolved in PBS (pH 6)] to promote carcinogenesis or with PBS only. Littermate controls received injections at the same ages as the transgenic mice. Mice with incurable conditions were killed. Each mouse that died was autopsied. Tissue sections were evaluated by a hematopathologist blinded to the mouse line and treatment and two anatomic pathologists (one blinded).
Statistical Analysis. The incidence of lymphoma and overall mortality in the groups by 433 days after injection were compared with use of the log-rank test. In the analysis of lymphoma incidence, 4 of 27 ENU-injected transgenics (group 1), 1 of 25 PBS-injected transgenics (group 2), 1 of 24 ENU-injected controls (group 3), and 2 of 28 PBS-injected controls (group 4) had to be excluded because of severe organ autolysis at death.
Results
BCL6 Transgenic Lines. Of 52 mice derived from microinjected zygotes, 12 transgenic founders were identified by Southern analysis. Estimates of transgene copy number by Southern blotting ranged from 4 to >30. No increased embryonal lethality was evident. Transgenic mice developed normally and had healthy litters of average size (8–11 pups). Healthy transgenic mice and littermate controls killed at varying ages, from 3 weeks to adulthood, showed no consistent differences in weight. Similarly, weight and histologic examination of spleen, thymus, and other organs showed no consistent differences in healthy immunized or nonimmunized transgenic animals as compared to controls. Furthermore, ELISA for total IgG levels in serum from immunized mice, immunohistochemistry on frozen sections of spleen from immunized mice using markers for follicular dendritic cells (FDC), germinal center cells (GCC), and B cells, and FACS performed on splenocytes and thymocytes of immunized and nonimmunized transgenic animals and littermate controls at various ages revealed no significant differences. The markers included anti-CD19 and -B220 for B cells; anti-CD3, -CD4, and -CD8 for T cells; anti-CD35 for FDC; peanut agglutinin and anti-GL7 for GCC; anti-CD69 for activated leukocytes; anti-IgM and -IgD for B cell subsets; and anti-CD138 for plasma cell-associated.
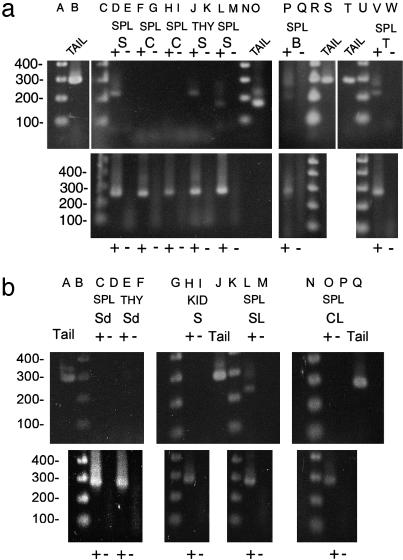
BCL6 Transgene Expression. RT-PCR on total RNA (Fig. 1) showed expression of transgene RNA in spleen and thymus of animals transgenic for tet-o-BCL6 and Eμ-tTA but, as expected, not in organs containing few, if any, lymphocytes (e.g., kidney) or in control mice. Two mouse lines, one containing four copies of tet-o-BCL6 and another 20 copies, gave similar data and were used for further studies. B and T cells (populations >99% pure) both expressed transgene RNA. As expected, doxycycline inhibited transgene expression.
Fig. 1.
RT-PCR of RNA. Reverse transcription lanes are marked +; – lanes indicate no reverse transcription using same primers as in preceding lane. (a Upper) Lanes A, 1 kb plus DNA ladder; B, RT-PCR with construct primers that amplify 289 bp from tail (genomic) DNA; C, 1 kb plus ladder; D, RT-PCR, cDNA from spleen (SPL) of study (S) mouse (transgenic) reveals the 223-bp band expected when the 66-bp intron is spliced out; F–I, SPL cDNA of control (C) mice containing only BCL6 (lane F) or Eμ-tTA transgene (lane H), as expected, no band is found; J, thymus (THY), S mouse, shows expected splice; L, different primer pair in transgene 5′ to intron reveals 170-bp band and, as expected, is the same size in genomic DNA (lane O); P and V, cDNA from flow-sorted (>99% pure) B cells (lane P) and T cells (lane V) from spleen reveals partial splicing with original primer pair. (b Upper) Construct primers spanning intron. Lanes C–F, RT-PCR, cDNA of SPL and THY from 2 S mice on doxycycline (Sd) show down-regulation of transgene expression; H, kidney, S mouse, as expected, no transgene expression was found; L, cDNA from SPL, S mouse with B cell lymphoma (SL), shows splice; lane O, SPL of C mouse with lymphoma (CL), as expected, no band was found. (a Lower and b Lower) RT-PCR with β-actin primers shows cDNA (285-bp band) in each lane (+) where reverse transcriptase was added.
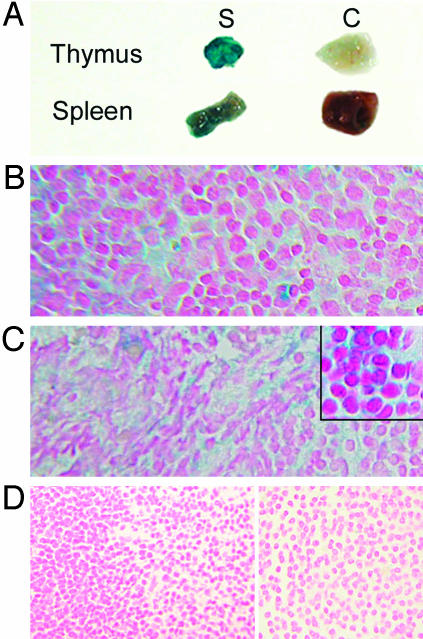
Because the construct in which the transgene was cloned permits use of β-gal analysis to study expression, we stained thymus and spleen tissues with X-gal. Thymus of transgenic animals, but not controls, revealed strongly positive blue-green cytoplasmic staining, extending from the cortex into the medulla, and β-gal expression also was positive in spleens of transgenic mice but not controls (Fig. 2). Spleens of normal mice contain few cells expressing BCL6 (11). Thus, in nonimmunized wild-type mice, which have few, if any, germinal centers, BCL6 staining is minimal. In contrast, spleens from our nonimmunized transgenic mice, which likewise have few germinal centers, when incubated with BCL6 antibody, revealed a strongly diffusely positive BCL6 (nuclear) expression pattern (Fig. 3).
Fig. 2.
β-Gal staining of mouse thymus and spleen. (A) Representative cut tissue pieces of thymus (Upper) and spleen (Lower) from study (S) mice (transgenic) stain deeply blue-green as compared with minimal background staining in controls (C). (B–D) Microsopic analysis of thymus (B) and spleen (C) from transgenic mice reveal the blue-green cytoplasmic staining expected with X-gal, whereas tissues from control mice do not stain (D). Sections are counterstained with nuclear fast red.
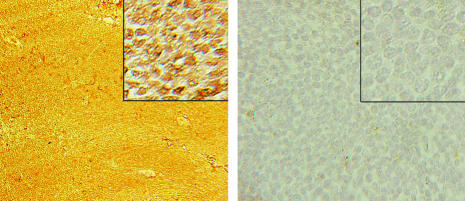
Fig. 3.
Immunohistochemistry. Paraffin-embedded sections of spleen stained with rabbit polyclonal antibodies to N-terminal BCL6 are shown. The nuclei in the lymphocytes of the transgenic mouse spleen (Left) stain brown, indicating that they are positive for BCL6 (23-day-old spleen containing mostly white pulp), whereas the cells in the wild-type mouse spleen (Right) do not recognize anti-BCL6.
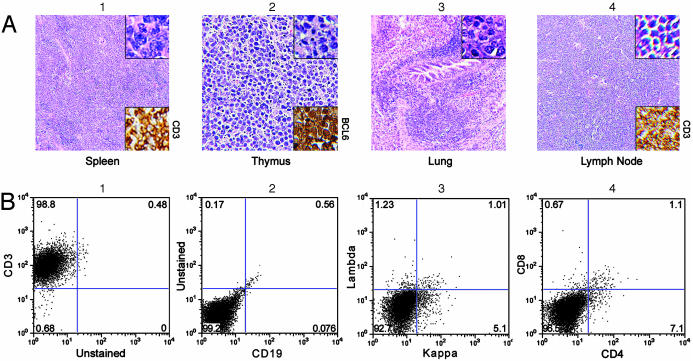
Development of Spontaneous Lymphomas. Two of 15 noninjected transgenic mice (all dead), but none of 17 littermate controls (one still living at 3 years) followed for at least 2 years developed BCL6-positive splenic B cell lymphomas. Both tumors showed a diffuse histologic pattern with a mixture of small, intermediate, and large cells. Cleaved nuclei were not a feature. These tumors do not fit precisely into the proposed classification for murine lymphoid neoplasms (12). RT-PCR performed on one tumor detected the splice site in our construct, indicating that RNA from our transgene was present (Fig. 1b, lane L). In the younger of these mice (727 days at death), the lymphoma effaced the normal splenic architecture but did not spread to other organs. However, in the older animal (963 days), tumor spread was noted in lung, bone marrow (paratrabecular nodules), liver, and lymph nodes (Fig. 4A). FACS of this tumor (spleen) showed 100% kappa staining and IgG plus IgD positivity but no IgM (Fig. 4B). T cells were present in reduced numbers, but the ratio of CD4+/CD8+ cells was normal. Additionally, one transgenic mouse (not a littermate of the original 15) developed BCL6-positive T cell lymphoma in spleen and bone marrow spontaneously at 435 days.
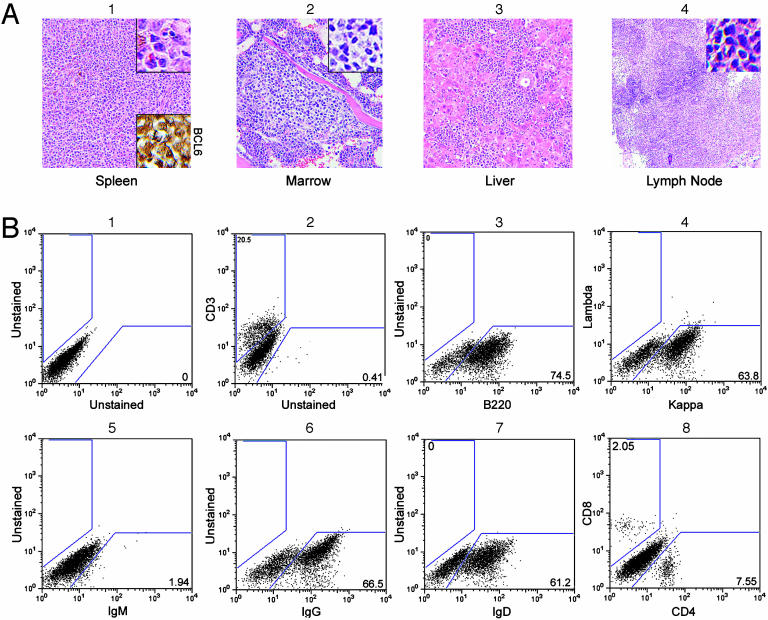
Fig. 4.
B cell lymphoma. (A)(1) Hematoxylin/eosin stains reveal that the structure of the spleen is effaced with a mixture of small, medium, and large tumor cells that stain for BCL6 (Lower Inset). (2) Paratrabecular tumor nodules are present in the bone marrow. (3 and 4) Tumor cells infiltrate the liver (3) and partially efface a mesenteric lymph node (4). (Original magnifications, ×100 in A and ×300 in Insets.) (B) FACS shows the tumor is B cell, because most of the cells stain with anti-B220 (3). They are kappa positive (4). Staining is positive for IgG (6) and IgD (7) but not IgM (5). T cells are present in reduced numbers (2), but the CD4/CD8 ratio is normal (8).
In spleens from two transgenic animals, but not in any controls, the B cells surrounding the periarteriolar lymphoid sheath were irregular in size and shape and seemed focally to encroach upon the adjacent red pulp. We considered this disorganized B cell expansion to be a possible precursor to lymphomatous transformation.
One control died of BCL6-negative large-cell T cell lymphoma with spread in multiple organs at 564 days. No B cell lymphomas or BCL6-positive T cell tumors occurred in controls.
Lymphoma Development After ENU Administration. Seven T cell lymphomas developed in 23 analyzable transgenics (30%, or 26% of the total group of 27 mice) injected with ENU (group 1), but not in any of the other injected groups (P ≤ 0.005; see description of groups 1–4 in Materials and Methods). Five lymphomas occurred in 12 mice (42%) containing 20 copies of the BCL6 transgene, and two occurred in 11 mice (18%) with four copies. Six mice received the injection between 66 and 95 days, but one was injected at 254 days. Lymphomas developed 163–299 days after injection, caused massive enlargement of the thymus, commonly invaded perithymic fat, muscle, liver, and lung, effaced the architecture of bone marrow, lymph nodes, and spleen (Fig. 5A), sometimes invaded other organs (e.g., kidney and uterus), and were strongly BCL6-positive. Histology showed sheets of small- to medium-sized pleomorphic tumor cells with a vesicular nucleus and sometimes a prominent nucleolus. Often there was evidence of high cell turnover (“starry sky” pattern). The mice succumbed rapidly to these tumors, but it was possible to do FACS on two; both were CD3+ but CD4– and CD8– (Fig. 5B). In addition, B220 FACS (not shown) performed on one tumor was positive.
Fig. 5.
T cell lymphomas. (A) Hematoxylin/eosin-stained sections show that the architecture of the spleen (1), thymus (2), and mesenteric lymph node (4) are effaced with pleomorphic small to medium-sized tumor cells that also infiltrate the lung (3). Images in 1 and 3 are from the same mouse, whereas images in 2 and 4 are from a different mouse. The tumors stain with anti-CD3 (Lower Insets in 1 and 4) and are BCL6-positive (Lower Inset in 2). They often show a starry sky pattern (4). (Original magnifications, ×75 in 1, 3, and 4, ×200 in 2, ×500 in Upper Inset of 1, ×300 in Upper Insets of 2-4.) (B) FACS confirms the tumors consist of T cells, because they stain with anti-CD3 (1) but not with anti-CD19 (2) or antibodies to kappa/lambda (3). They are CD4–,CD8– (4).
No B cell lymphomas developed in animals injected with ENU. Various other neoplasms occurred in the ENU-injected animals, most often pulmonary adenocarcinoma with a papillary pattern. The incidence of these tumors was similar in transgenics and controls.
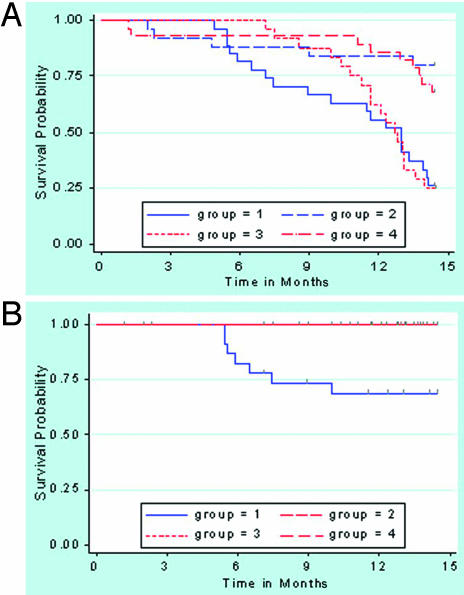
Time to Event Analysis. Although the overall survival probability was not significantly different between the two ENU-injected groups (groups 1 and 3, Fig. 6A), death probability from lymphoma in group 1 (transgenic-ENU) was significantly higher than in the other groups (P = 0.005, 0.004, and 0.003, respectively; Fig. 6B).
Fig. 6.
Kaplan–Meier curves. (A) Curve of overall survival shows that the survival probability was not significantly different between the mice in the ENU-treated groups (transgenics, group 1; controls, group 3) or between those in the PBS-treated groups (transgenics, group 2; controls, group 4). The survival probability was lower in the ENU groups compared with the PBS-treated groups (all P ≤ 0.001). (B) Curve of death from lymphomas, which occurred only in ENU-treated transgenics (group 1), and was significantly higher than in any other group (all P≤ 0.005).
Discussion
It has long been thought that BCL6 becomes an oncogene important in the pathogenesis of some human lymphomas when it is mutated or involved in a variety of translocations that result in deregulation of its normal function. The events involved in the oncogenic process are not well understood. In an effort to create an animal model that might permit further exploration of the mechanism of action of human BCL6 in lymphomagenesis, we generated a BCL6 transgenic mouse model that mimics a common translocation found in human lymphomas (1), the t(3,14)(q27;q32), in which the normal BCL6 regulatory sequences are lost, and, instead, IGH sequences are juxtaposed upstream of BCL6 coding sequences. The transgenic animals reported here express BCL6 from the transgene constitutively specifically in lymphocytes, both B and T, and demonstrated no developmental or growth abnormalities. Although we anticipated that we might have to silence the transgene by giving doxycycline during development to prevent toxicity, this was not necessary. However, because transgenic mice may show adaptation to genetic lesions during development, it is possible that if BCL6 expression were suppressed by treating pregnant mice with doxycycline, the effects of reexpressing the protein in newborn or adult mice might be different.
The overall incidence of spontaneous malignancies of the hematopoietic system in mice is estimated to be 1–2%, but this varies depending on the strain and environmental conditions (13). In 71 FVB/N mice followed for 2 years, no lymphomas had occurred by age 14 months, but, by 24 months, four animals (5.6%) had developed lymphomas (14). Some B cell lymphomas in genetically engineered mouse models have been BCL6 positive (11), but we have been unable to find reports of the BCL6 status of spontaneous B cell lymphomas in wild-type mice. The histologic subtypes of B cell lymphomas in mice are multiple and include follicular and diffuse large cell lymphomas (11, 12, 14, 15). We describe two B cell lymphomas that occurred in unmanipulated older BCL6 transgenic mice (13% of mice that reached 2 years), suggesting that a long latency period during which additional genetic events may occur might be required before the development of lymphomas. A factor that may have influenced the time frame of lymphoma development is the housing of our mice in specific pathogen-free facilities (SPF), a requirement in our institution. The time course for lymphoma development in NFS.V+ mice raised in SPF was 5 months longer than in those raised in conventional facilities (16). Although the B cell lymphomas in our mice did not have a follicular histologic pattern seen commonly in human tumors, the paratrabecular nature of the bone marrow foci (often seen in human follicular lymphoma) in our oldest mouse suggests that this lymphoma may have evolved from a germinal center cell.
T cell lymphomas may occur in wild-type mice (13), and high-grade T cell tumors have been noted in NFS.V+ mice, but the incidence of BCL6 positivity was only 20%, with staining at low levels (11). Only one of our transgenic mice spontaneously developed BCL6-positive T cell lymphoma, but with exposure to ENU, a DNA alkylating agent that causes single-base mutations, the incidence of T cell lymphoma was enhanced (30% of analyzable transgenics with death at an average of 306 days). The observed tumors related to ENU expressed BCL6 and were exclusively T cell (Fig. 5A). The CD4– CD8– B220+ phenotype we noted has been reported in expanded clones of alpha–beta T cells (17) and gamma–delta T cells (18) in certain mutant mice.
The Felsher and Bishop model for the MYC gene driven by the same EμSR control element (7) also resulted in T cell lymphomas, although, unlike our tumors, they were CD4+ and CD8+ and occurred without ENU administration. These authors attribute the propensity for T cell neoplasia to the high levels of gene expression produced by the SRα promoter in T cells. The fact that our tumors occurred only in transgenic animals and expressed abundant BCL6 supports the notion that the BCL6 protein in these lymphomas is derived from the transgene rather than from endogenous murine BCL6. Expression of the BCL6 protein has been reported in a number of human T cell neoplasms, e.g., CD30+ T cell anaplastic large cell lymphomas, focally in angioimmunoblastic T cell lymphomas, and in precursor T cell lymphoblastic lymphomas, including one, like our tumors, with a “precursor thymocyte phenotype” (CD4–, CD8–) (6), suggesting that deregulation of the BCL6 protein may contribute to the pathogenesis of these kinds of tumors as well as the more commonly occurring B cell lymphomas in humans. Thus, our model may have relevance for the study of both B and T human lymphomas.
It has been found that BCL6 mutations may accumulate during the transformation of human follicular lymphoma to aggressive diffuse large B cell lymphoma (19). Similarly, our BCL6 transgenic mice develop an increased incidence of lymphomas after a “second hit” by a chemical carcinogen that induces mutations, a phenomenon that also has been reported in other mouse models of lymphoma or leukemia (20, 21). Thus, this study demonstrates the importance of BCL6 and the relevance of cooperating mutations in the development of lymphomas.
Acknowledgments
We thank L. Degenstein for microinjections of mouse ova, initial mouse matings, and cryopreservation of mouse embryos; Anatomic Pathology for tissue processing; Hematopathology for CD3 staining methodology; D. Wiler for skilled assistance with illustrations; R. Duggan for expert help with FACS; Drs. I. Aifantis and U. Storb (University of Chicago, Chicago) for selected antibodies; the Mouse Facilities and Animal Resources Center staff for superb mouse care; and Dr. W. Müller for the pepBsplice construct. We are grateful to Drs. M. Utset, H. M. Shen, U. Storb, I. Aifantis, H. Schreiber, Y. Li, G. Langan, and M. Baron for helpful discussions. We acknowledge Drs. Manuel Diaz and Giuseppina Nucifora for their careful reviews and thoughtful comments. This study was supported by National Institutes of Health Grant CA63365 (to B.W.B.), the Training and Research Support Program of the University of Chicago Hospitals Laboratories (B.W.B. and J.M.B.), an award to the University of Chicago's Division of Biological Sciences under the Research Resources Program for Medical Schools of the Howard Hughes Medical Institute (to B.W.B.), the University of Chicago Department of Pathology (B.W.B.), Hematology Research Funds at the University of Chicago donated by S. Samsky and E. Lanzl (to J.M.B.), and University of Chicago Cancer Center Support Grant P30 CA144599.
Abbreviations: ENU, N-ethyl-N-nitrosourea; tTA, tetracycline-transactivating protein; β-gal, β-galactosidase; X-gal, 5-bromo-4-chloro-3-indolyl β-d-galactoside; FACS, fluorescence-activated cell sorting.
References
- 1.Baron, B. W., Nucifora, G., McCabe, N., Espinosa, R., III, Le Beau, M. M. & McKeithan, T. W. (1993) Proc. Natl. Acad. Sci. USA 90, 5262–5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ye, B. H., Lista, F., LoCoco, F., Knowles, D. M., Offit, K., Chaganti, R. S. K. & Dalla-Favera, R. (1993) Science 262, 747–750. [DOI] [PubMed] [Google Scholar]
- 3.Ye, B. H., Chaganti, S., Chang, C.-C., Niu, H., Corradini, P., Chaganti, R. S. K. & Dalla-Favera, R. (1995) EMBO J. 14, 6209–6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pasqualucci, L., Migliazza, A., Basso, K., Houldsworth, J., Chaganti, R. S. K. & Dalla-Favera, R. (2003) Blood 101, 2914–2923. [DOI] [PubMed] [Google Scholar]
- 5.Dent, A. L., Shaffer, A. L., Yu, X., Allman, D. & Staudt, L. M. (1997) Science 276, 589–592. [DOI] [PubMed] [Google Scholar]
- 6.Hyjek, E., Chadburn, A., Liu, Y. F., Cesarman, E. & Knowles, D. M. (2001) Blood 97, 270–276. [DOI] [PubMed] [Google Scholar]
- 7.Felsher, D. W & Bishop, J. M. (1999) Mol. Cell 4, 199–207. [DOI] [PubMed] [Google Scholar]
- 8.Bodrug, S. E., Warner, B. J., Bath, M. L., Lindeman, G. J., Harris, A. W. & Adams, J. M. (1994) EMBO J. 13, 2124–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hogan, B., Beddington, R., Costantini, F. & Lacy, E. (1994) Manipulating the Mouse Embryo (Cold Spring Harbor Lab. Press, Plainview, NY), 2nd Ed., pp. 373–374.
- 10.Sanes, J. R., Johnson, Y. R., Kotzbauer, P. T., Mudd, J., Hanley, T., Martinou, J. C. & Merlie, J. P. (1991) Development (Cambridge, U.K.) 113, 1181–1191. [DOI] [PubMed] [Google Scholar]
- 11.Qi, C.-F., Hori, M., Coleman, A. E., Torrey, T. A., Taddesse-Heath, L., Ye, B. H., Chattopadhyay, S. K., Hartley, J. W. & Morse, H. C., III (2000) Leukemia Res. 24, 719–732. [DOI] [PubMed] [Google Scholar]
- 12.Morse, H. C., III, Anver, M. R., Fredrickson, T. N., Haines, D. C., Harris, A. W., Harris, N. L., Jaffe, E. S., Kogan, S. C., MacLennan, I. C. M., Pattengale, P. K., et al. (2002) Blood 100, 246–258. [DOI] [PubMed] [Google Scholar]
- 13.Percy, D. H. & Barthold, S. W. (1993) in Pathology of Laboratory Rodents and Rabbits (Iowa State Univ. Press, Ames), p. 61.
- 14.Ward, J. M., Anver, M. R., Mahler, J. F. & Devor-Henneman, D. E. (2000) in Pathology of Genetically Engineered Mice, eds. Ward, J. M., Mahler, J. F., Maronpot, R. R. & Sundberg, J. P. (Iowa State Univ. Press, Ames), p. 176.
- 15.Hori, M., Xiang, S., Qi, C.-F., Chattopadhyay, S. K., Fredrickson, T. N., Hartley, J. W., Kovalchuk, A. L., Bornkamm, G. W., Janz, S., Copeland, N. G., et al. (2001) Blood Cells Mol. Dis. 27, 217–222. [DOI] [PubMed] [Google Scholar]
- 16.Morse, H. C., III, McCarty, T., Qi, C.-F., Torrey, T. A., Naghashfar, Z., Chattopadhyay, S. K., Fredrickson, T. N. & Hartley, J. W. (2003) Adv. Immunol. 81, 97–121. [DOI] [PubMed] [Google Scholar]
- 17.Hamad, A. R., Mohamood, A. S., Trujillo, C. J., Huang, C. T., Yuan, E. & Schneck, J. P. (2003) J. Immunol. 171, 2421–2426. [DOI] [PubMed] [Google Scholar]
- 18.Hughes, D. P., Hayday, A., Craft, J. E., Owen, M. J. & Crispe, I. N. (1995) J. Exp. Med. 182, 233–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lossos, I. S., Warnke, R. & Levy, R. (2002) Leukemia 16, 1857–1862. [DOI] [PubMed] [Google Scholar]
- 20.Castilla, L. H., Garrett, L., Adya, N., Orlic, D., Dutra, A., Anderson, S., Owens, J., Eckhaus, M., Bodine, D. & Liu, P. P. (1999) Nat. Genet. 23, 144–146. [DOI] [PubMed] [Google Scholar]
- 21.Higuchi, M., O'Brien, D., Kumaravelu, P., Lenny, N., Yeoh, E.-J. & Downing, J. R. (2002) Cancer Cell 1, 63–74. [DOI] [PubMed] [Google Scholar]