ABSTRACT
The packaging of genetic information in form of chromatin within the nucleus provides cells with the ability to store and protect massive amounts of information within a compact space. Storing information within chromatin allows selective access to specific DNA sequences by regulating the various levels of chromatin structure from nucleosomes, to chromatin fibers, loops and topological associating domains (TADs) using mechanisms that are being progressively unravelled. However, a relatively unexplored aspect is the energetic cost of changing the chromatin configuration to gain access to DNA information. Among the enzymes responsible for regulating chromatin access are the ATP-dependent chromatin remodellers that act on nucleosomes and use the energy of ATP hydrolysis to make chromatin DNA more accessible. It is assumed that the ATP used by these enzymes is provided by the mitochondria or by cytoplasmic glycolysis. We hypothesize that though this may be the case for cells in steady state, when gene expression has to be globally reprogramed in response to externals signals or stress conditions, the cell directs energy production to the cell nucleus, where rapid chromatin reorganization is needed for cell survival. We discovered that in response to hormones a nuclear ATP synthesis mechanism is activated that utilizing ADP-ribose and pyrophosphate as substrates.1 This extra view aims to put this process within its historical context, to describe the enzymatic steps in detail, to propose a possible structure of the ATP synthesising enzyme, and to shed light on how this may link to other reactions within the cell providing a perspective for future lines of investigation.
KEYWORDS: ATP, breast cancer, chromatin, chromatin remodelling, gene regulation, steroid hormones
Chromatin structure and mechanism of remodelling
The 4 m long DNA of our diploid cells needs to be compacted over 10,000 fold in chromatin in order to fit into the cell nucleus (5–10 × 10−6 m diameter) while still enabling dynamic access to the genetic information during the processes of transcription, repair and replication. DNA is first wrapped 1.6-fold around an octamer of the 4 core histones (H3, H4, H2A & H2B) to form the nucleosome core particle. The resulting beads-on-a-string chain of 11nm diameter is further compacted by the linker histones of the H1 family forming a nucleosome fiber of approximately 30 nm diameter. It is estimated that the nucleus contains around 30 million nucleosomes yielding in average one nucleosome every 200 base pairs of DNA. Originally nucleosomes were considered to be randomly positioned and to act as unspecific transcriptional repressors,2 but experiments on a few specific genomic regions and the recent availability of global nucleosome positioning information have revealed that nucleosomes can be specifically located relative to the DNA sequence and can even aid transcriptional regulation.3,4 It is becoming clear that not all nucleosomes are equal and that certain posttranslational modifications of the core or linker histones, as well the inclusion of histone variants, influence their dynamics and the accessibility of the underlying DNA sequences.5,6 The in depth description of this highly ordered first level of compaction is out of the reach of this review, but other excellent reviews have been recently published.7,8
The exact structure of the next level of chromatin compaction is still under debate.9 Initially, the 30 nm fiber was described as a solenoid with 6 nucleosomes per turn and the linker histones in the center,10 but several studies describing different topologies, including zigzag intermingling of 2 antiparallel strands have been also proposed, all of which share stabilization by nucleosome stacking,11,12 Recent cryo-EM images of nucleosomes arrays have provided one of the best models of the 30 nm fiber based on repeats of an intermingled tetra-nucleosome unit.13
Irrespective of the intermediate structure, chromatin fibers are further folded into chromatin loops, often connecting regulatory sequences such as promoters and enhancers, and these loops are compartmentalized into Topological Associating Domains (TADs), which are largely conserved between different cell types and insulated by ill-defined regions enriched in CTCF and cohesion binding sites.14 TADs are characterized by a high density of internal chromatin interactions, isolated from adjacent TADs. They have been shown to be epigenetic and functional domains exhibiting coordinated regulation of their coding and non-coding transcripts.15,16 Several TADs can aggregate in the nuclear periphery to form Lamina Associated Domains (LADs), usually heterochromatic and poorly transcribed. In some cases regulated genes can change their position in the nucleus, leaving the nuclear periphery when they are activated, or migrating to it when they are silenced (Review 17).
To access nucleosomes the chromatin fiber has to be decondensed by poorly characterized mechanisms that often required the ATP-consuming activity of cohesins and condensins18 and modifications of linker histones by parylation and/or phosphorylation.19 The subsequent and continuous task of remodeling nucleosomes lies with multi-protein chromatin remodeling machines, all of which share an ATPase subunit that hydrolyses ATP.20 These chromatin remodelers use the energy of ATP hydrolysis to break and rebuild hundreds of interactions of core histones with DNA ultimately leading to core particle sliding, histone displacement or variant replacement. Posttranslational modifications of histone tails also contribute to chromatin remodeling. The combination of these multiple enzymatic activities results in a more open or closed chromatin structure, dictating the accessibility for the downstream effector proteins, but at very high energetic price. These energy requirements added to the energetic cost of maintaining transcription, RNA processing and export, DNA repair and replication during mitosis, make the nucleus a burdensome energy consuming organelle.
Hormone induced gene expression
We have been studying the mechanisms of gene regulation and chromatin remodelling events in the breast cancer cell line T47D that express progesterone receptor (PR) and estrogen receptor α (ERα). When T47D cells are deprived of mitogens for 16 hours prior to hormone exposure, growth slows down and cells accumulate in the G1 phase of the cell cycle reaching an almost quiescent state. This provides a synchronised cell population for studying the response to the synthetic progestin R5020 acting via PR.21 Before hormone exposure, most of PR is maintained as an inactive monomer complexed with the HSP90 and its associated chaperones and distributed between the cytoplasm and the cell nucleus. We assume that, energetically, in this low activity basal state gene expression and chromatin remodelling are sustained via diffusion of ATP from the mitochondria (Fig. 1).
Figure 1.
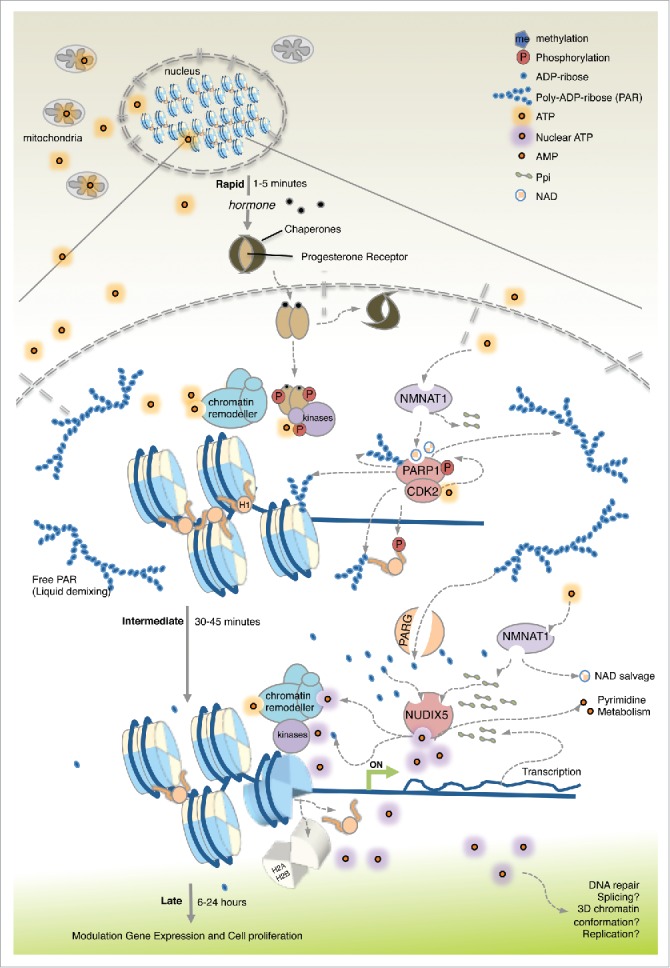
Energy focused events following hormone exposure of breast cancer cells. Hormone induced changes are shown separated into 3 phases. During the steady-state (before hormone) basic genome activity is maintained and the energy requirements are met by diffusion of ATP from the mitochondria. Rapidly upon hormone exposure (1–5 minutes) ligand-bound PR in complex with kinases is recruited to target genes. CDK2 phosphorylates PARP1 increasing its enzyme activity leading to local PARylation. Histone H1 phosphorylated by CDK2 and parylated is displaced. ATP consumed during this initial period is of mitochondrial origin and used by NMNAT1 to make NAD for PARP1 and PPi. After 30 min of hormone exposure (Intermediate) the increase in PAR triggers PARG, leading to accumulation of ADPR. NUDIX5 utilizes the increase in ADPR and PPi to generate ATP. ATP produced in the nucleus is used for stable displacement of H1 and H2A/H2B opening the chromatin for access of transcriptional regulators leading to gene activation/repression and ultimately to cell proliferation (late events).
Upon exposure to R5020, T47D cells rapidly respond by activating both non-genomic and genomic pathways.22,23 The small fraction of PR attached to the cell membrane activates the SRC/RAS/ERK and the CDK2 pathways, while the majority of soluble PR dissociates from chaperones and forms active homodimers. Activated ERK phosphorylates soluble PR at S294 and activates MSK1 forming a trimeric complex with PR that tethers the kinases to the its cognate Hormone Responsive Elements (HREs) in chromatin, where MSK1 modified histone H3 S1024 and CDK2 phosphorylates histone H1. The activated kinase CDK2 phosphorylates and activates the poly-ADP-ribose polymerase 1 (PARP1/ARDT1) resulting in an increase in local poly-ADP-ribose (PAR) levels.23 This leads to a rapid (1–5 min) displacement of histone H1 and of a repressive complex,25 opening chromatin. During a second phase (30 min) PR recruits histone modifiers and chromatin remodellers resulting in the displacement of histone H2A and H2B.26 Notably, not only activation of gene transcription but also active repression by hormone requires steroid receptors, kinases, chromatin remodellers and PARP1.27
PAR and the nuclear synthesis of ATP
PAR was originally described as the “third nucleic acid”28 and PARylation is a unique and abundant posttranslational protein modification. The formation of large branched charged chains of ADPR attached to proteins modifies protein-protein and protein-DNA interactions. Proteins containing a PAR-binding domain (PBD), such as the macro domain, can be modulated by PAR binding. More recently it has been shown that the formation of membrane-less organelles within the nucleus is dependent on PARylation, which provides a local environment for the concentration of chromatin factors and substrates (review 29).
We showed that rapid activation of PARP1 in response to hormone is essential for the cellular response in terms of chromatin remodelling, gene regulation and cell proliferation.23 The observation that the increase in PAR levels in response to hormone is transient peaking very early (5–10 min) and returning to basal levels within 60 min led us to investigate PAR degradation. PAR is hydrolysed by poly-ADP-ribose glycohydrolase (PARG) to ADPR, and we showed that specific inhibitors or siRNAs against PARG also compromised the hormonal gene regulation and subsequent cell proliferation. Thus, not only PAR synthesis but also its degradation are essential for hormone response, pointing to a possible role for accumulated ADPR.1
It is generally assumed that the energy required for nuclear functions in steady-state is provided by the mitochondria and in some cases by cytoplasmic glycolysis. We questioned whether this also applies to cells subjected to acute and global reprogramming of genome in response to external cues. There are indeed old findings suggesting the possibility of energy generation in the cell nucleus. Nearly 60 years ago Allfrey and Mirsky proposed that ATP could be generated in isolated nuclei.30 It was suggested that the substrate for nuclear oxidative phosphorylation originates in part from the ribose pathway.31 In 1989 an enzyme activity named ADP-ribose pyrophosphorylase was postulated in HeLa cell nuclei that would catalyze the formation of ATP and Ribose-5-phosphate (R-5-P) from ADP-ribose (ADPR) and PPi.32 Moreover, ATP generation via ADPR catabolism has been shown to be involved in DNA repair and replication.33 However, the enzymes catalyzing nuclear ATP synthesis were never isolated and the direct relationship to nuclear processes remained unproven. Given the relevance of PAR synthesis and degradation for hormone regulation we decided to address the potential synthesis of the nuclear ATP in the context of hormonal activation of quiescent breast cancer cells in culture, a process requiring changes in transcription activity of thousands of genes within a short period of time.4 Could the accumulation of ADPR be a source of local nuclear ATP?
In order to measure ATP levels in T47D cells following hormone we used 2 methods; a FRET-based measurement of the ATP/ADP ratio in cytoplasmic, nuclear or mitochondrial compartments in individual living cells, and a luciferase based method that allows high-throughput population measurements. We found that nuclear ATP levels increase rapidly (after 5 min) in cells exposed to hormone reaching a maximum at 15–20 min and returning to basal levels after 60–80 min.1 The increase of nuclear ATP was dependent on an initial pulse of ATP of mitochondrial origin, as inhibition of mitochondrial ATP prior to hormone exposure prevented nuclear ATP formation. However, inhibition of mitochondrial ATP synthesis, shortly after hormone addition (10 min) did not affect the increase in ATP, indicating that subsequent nuclear ATP synthesis is independent of mitochondria.
A mass spec screen of PAR interacting proteins identified a novel PAR target involved in ADPR metabolism, namely Nucleotide Diphosphate linked to moiety X 5 (NUDIX5/NUDT5). We found that NUDIX5 interacts with both PARG and PAR by co-immunoprecipitation following hormone.1 In addition, using siRNA combined with both global gene expression arrays and quantitative PCR we were able to demonstrate that the presence of NUDIX5 is essential for hormone induced gene expression changes and cell proliferation in both T47D and MCF7 breast cancer cells. Prior to hormone exposure NUDIX5 hydrolyses ADPR to AMP and R-5-P, however following hormone, due to the local increase in ADPR and PPi, NUDIX5 catalyzes not only the formation of AMP but also the synthesis of ATP within the cell nucleus. Recombinant NUDIX5 can catalyze these 2 reactions in vitro, though hydrolysis to AMP is the dominant reaction.
Using both FRET-based and luciferase-based sensors we were able to show that nuclear ATP generation is dependent on the combined actions of PARP1, PARG and NUDIX5. As discussed earlier, during the very early events, (Fig. 1), PAR accumulates and participates in general chromatin opening, perhaps generating a microenvironment by liquid demixing that favors the concentration of effector proteins and substrates.34 Such membrane-less organelles are transient in nature, like PAR accumulation. Indeed, the elevated nuclear ATP levels observed after 10–15 min inhibit the activity of PARP1, ensuring that the nucleus can return to a basal state.
In contrast chromatin remodelling measured by H1 and H2A displacement at later stages after PAR accumulation and degradation (Fig. 1), is dependent on PARP1, PARG and NUDIX5, and affects similarly activated and repressed genes. We hypothesize that this mechanism of nuclear energy generation is a “safety measure,” to avoid cell death by parthanato's characterized by detrimental decrease in both NAD+ and ATP levels.35 Transient nuclear ATP generated via the action of NUDIX5 ensures the coverage of the energetic cost associated with massive rapid global changes in chromatin induced by hormone.
Substrate sources and product accumulation
One of the open intriguing questions posed from this work is the source of PPi. It is possible that PPi concentrations are sufficiently abundant within the cell nucleus due to the basal reactions taking place, or perhaps is locally generated during the process of transcription, facilitating local ATP generation by NUDIX5. This suggest that there may be no specific mechanism in place to generate PPi at the chromatin. However, nuclear ATP generation is not only dependent on the combined actions of PARP, PARG and NUDIX, but also on the generation of NAD+ by the nuclear enzyme nicotinamide mono nucleotide adenylyl transferase 1 (NMNAT1).
NMNAT1 uses NMN and ATP to generate NAD+ and PPi within the nucleus and is the nuclear source of NAD+ for PARP1. We hypothesis that NMNAT1 could also be a local source of PPi for NUDIX5 at the regions of chromatin requiring extensive chromatin remodelling. NMNAT1 interacts with PARP1 and NUDIX5 and NMNAT1, PARP1 and PARG regulate similar sets of genes and co-localize on specific regulated regions of the genome.1,36 Measurement of total cellular PPi levels is possible with commercial kits or mass spec and the local measurement of nanomolar concentration of PPi within cells was recently described.37 This new methodology may provide the required information to understand the nuclear source of PPi following hormone exposure.
Irrespective of the source it is likely that the unfavourable energetics of the reaction (ADPR + PPi > ATP + R-5-P) in vitro my improve due to the local increase in both ADPR and PPi substrates or to alternative conformations of the interacting enzymes in the cell nucleus.
A potential model for the ATP synthesising machinery: NMNAT1/NUDIX5
A crystal structure of human NUDIX5/NUDT5 homodimer bound with one ADPR on each of the 2 active sites has been reported.38 This structure illustrates the hydrolysis of ADPR to AMP and R-5-P but is not easily compatible with PPi binding. We postulated that the homodimer is stabilized in the cell by the phosphorylation of T45 that interacts with L28 of the opposite monomer.1 Upon hormone exposure T45 is dephosphorylated leading to destabilization of the homodimer.1 We hypothesize that recombinant NUDIX5 homodimer exists in equilibrium between the dominant ADPR hydrolysing conformation and a minor ATP synthesising conformation, in which the relative orientation of the monomer flips yielding more open active sites that can accommodate the presence of PPi required for ATP synthesis (Fig. 2). We also have experimental evidence for the formation of a hexameric complex with recombinant NUDIX5. Therefore, we tried to model a NUDIX5 hexamer, and this was only possible with the flipped monomer conformation and not with the published conformation of the dimer (Fig. 2). Phosphorylation of T45 is predicted to prevent the flipping of NUDIX5 monomers, hindering the formation of the hexamer. Thus, dephosphorylation of NUDIX5 may trigger the switch to a conformational change that activates the energy generating molecular machine.
Figure 2.
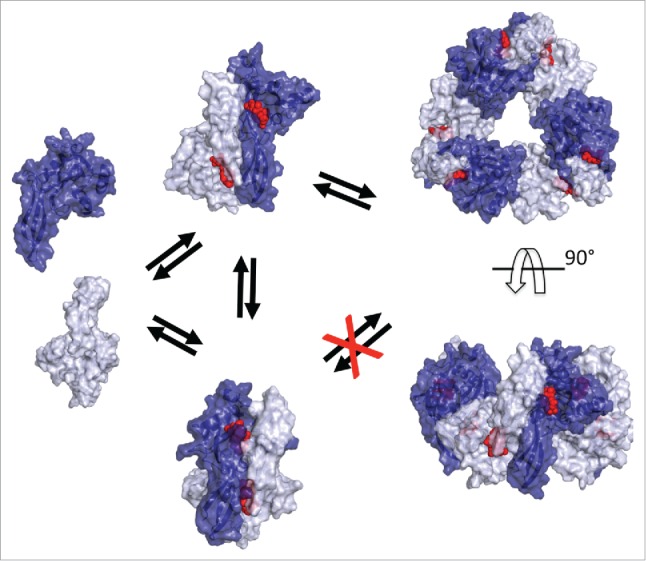
Model of NUDIX5 dimers, and hexamer. From left to right: 1) Surface representation of NUDIX5 monomers (light and dark blue); 2) Two versions of the NUDIX5 homodimer: the published one (PDB code 2DSC) at the bottom cannot bind PPi as required for making ATP; the “flipped” conformation at the top facilitated by T45 dephosphorylation could bind PP1; 3) in silico predicted NUDIX5 homo-hexameric structure based on the “flipped” homodimer, top view (top) and side view (bottom). The model of NUDIX5 hexamer was obtained using MODELLER and refined with ROSETTA. Structures shown in the figure were rendered using PyMOL (http://www.pymol.org). ADPR is shown in red.
Open questions
It has been recently proposed that the cell-specific global 3D structure of eukaryotic genomes is generated and maintained by the mechanism of “loop extrusion” that requires the ATP dependent activity of cohesins in interphase and condensins in mitosis.39 We are now exploring whether this essential process for gene regulation also depends on the nuclear generation of ATP.
Given the relevance of nuclear ATP synthesis for chromatin remodelling and the importance of chromatin reorganization in the DNA damage response, stress response, cell differentiation during embryogenesis, cell reprogramming to iPSC and their differentiation and possibly DNA replication, we expect these processes to require the pathway PARP1/PARG/NUDIX5, or other pathways for nuclear ATP synthesis waiting to be discovered. The possible existence of alternative pathways comes from the wide conservation of NUDIX5 in plants and animals with the exception of diptera.
PARP1, PARG and aberrant NAD+ metabolism have received significant attention as drug targets for the treatment of breast cancer and other types of cancers not limited to breast.40,41 NUDIX5 is the only NUDIX family overexpressed in breast cancer patient samples, in correlation with elevated levels of PARP1.1 Given that stratifying patients based on NUDIX5 expression levels reveals a poorer outcome for patients with high NUDIX5 levels. We believe that the unique structure of the NUDIX5 may provide a target for selective inhibition of ADPR-derived ATP synthesis opening new avenues for combinatorial drug discovery.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- [1].Wright RH, Lioutas A, Le Dily F, Soronellas D, Pohl A, Bonet J, Nacht AS, Samino S, Font-Mateu J, Vicent GP, et al.. ADP-ribose-derived nuclear ATP synthesis by NUDIX5 is required for chromatin remodeling. Science 2016; 352:1221-5; PMID:27257257; http://dx.doi.org/ 10.1126/science.aad9335 [DOI] [PubMed] [Google Scholar]
- [2].Han M, Grunstein M. Nucleosome loss activates yeast downstream promoters in vivo. Cell 1988; 55:1137-45; PMID:2849508; http://dx.doi.org/ 10.1016/0092-8674(88)90258-9 [DOI] [PubMed] [Google Scholar]
- [3].Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell 2008; 132:887-98; PMID:18329373; http://dx.doi.org/ 10.1016/j.cell.2008.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ballare C, Castellano G, Gaveglia L, Althammer S, Gonzalez-Vallinas J, Eyras E, Le Dily F, Zaurin R, Soronellas D, Vicent GP, et al.. Nucleosome-driven transcription factor binding and gene regulation. Mol Cell 2013; 49:67-79; PMID:23177737; http://dx.doi.org/ 10.1016/j.molcel.2012.10.019 [DOI] [PubMed] [Google Scholar]
- [5].Bowman GD, Poirier MG. Post-translational modifications of histones that influence nucleosome dynamics. Chem Rev 2015; 115:2274-95; PMID:25424540; http://dx.doi.org/ 10.1021/cr500350x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Talbert PB, Henikoff S. Histone variants-ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol 2010; 11:264-75; PMID:20197778 [DOI] [PubMed] [Google Scholar]
- [7].Venkatesh S, Workman JL. Histone exchange, chromatin structure and the regulation of transcription. Nat Rev Mol Cell Biol 2015; 16:178-89; PMID:25650798 [DOI] [PubMed] [Google Scholar]
- [8].Luger K, Dechassa ML, Tremethick DJ. New insights into nucleosome and chromatin structure: an ordered state or a disordered affair?. Nature Rev Mol Cell Biol 2012; 13:436-47; PMID:22722606; http://dx.doi.org/ 10.1038/nrm3382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Maeshima K, Hihara S, Eltsov M. Chromatin structure: does the 30-nm fibre exist in vivo? Curr Opin Cell Biol 2010; 22:291-7; PMID:20346642; http://dx.doi.org/ 10.1016/j.ceb.2010.03.001 [DOI] [PubMed] [Google Scholar]
- [10].Finch JT, Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci U S A 1976; 73:1897-901; PMID:1064861; http://dx.doi.org/ 10.1073/pnas.73.6.1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wu C, Bassett A, Travers A. A variable topology for the 30-nm chromatin fibre. EMBO Reports 2007; 8:1129-34; PMID:18059311; http://dx.doi.org/ 10.1038/sj.embor.7401115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Travers A. Structural biology. The 30-nm fiber redux. Science 2014; 344:370-2; PMID:24763580; http://dx.doi.org/ 10.1126/science.1253852 [DOI] [PubMed] [Google Scholar]
- [13].Song F, Chen P, Sun D, Wang M, Dong L, Liang D, Xu RM, Zhu P, Li G. Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science 2014; 344:376-80; PMID:24763583; http://dx.doi.org/ 10.1126/science.1251413 [DOI] [PubMed] [Google Scholar]
- [14].Dekker J, Marti-Renom MA, Mirny LA. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat Rev Genetics 2013; 14:390-403; PMID:23657480; http://dx.doi.org/ 10.1038/nrg3454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, Lee AY, Yen CA, Schmitt AD, Espinoza CA, Ren B. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature 2013; 503:290-4; PMID:24141950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Le Dily F, Bau D, Pohl A, Vicent GP, Serra F, Soronellas D, Castellano G, Wright RH, Ballare C, Filion G, et al.. Distinct structural transitions of chromatin topological domains correlate with coordinated hormone-induced gene regulation. Genes Dev 2014; 28:2151-62; PMID:25274727; http://dx.doi.org/ 10.1101/gad.241422.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gonzalez-Sandoval A, Gasser SM. On TADs and LADs: spatial control over gene expression. Trends Genetics 2016; 32:485-95; PMID:27312344; http://dx.doi.org/ 10.1016/j.tig.2016.05.004 [DOI] [PubMed] [Google Scholar]
- [18].Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA. Formation of chromosomal domains by loop extrusion. Cell Reports 2016; 15:2038-49; PMID:27210764; http://dx.doi.org/ 10.1016/j.celrep.2016.04.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hottiger MO. Nuclear ADP-ribosylation and its role in chromatin plasticity, cell differentiation, and epigenetics. Annual Rev Biochem 2015; 84:227-63; PMID:25747399; http://dx.doi.org/ 10.1146/annurev-biochem-060614-034506 [DOI] [PubMed] [Google Scholar]
- [20].Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annual Rev Biochem 2009; 78:273-304; PMID:19355820; http://dx.doi.org/ 10.1146/annurev.biochem.77.062706.153223 [DOI] [PubMed] [Google Scholar]
- [21].Truss M, Bartsch J, Schelbert A, Hache RJ, Beato M. Hormone induces binding of receptors and transcription factors to a rearranged nucleosome on the MMTV promoter in vivo. EMBO J 1995; 14:1737-51; PMID:7737125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ballare C, Uhrig M, Bechtold T, Sancho E, Di Domenico M, Migliaccio A, Auricchio F, Beato M. Two domains of the progesterone receptor interact with the estrogen receptor and are required for progesterone activation of the c-Src/Erk pathway in mammalian cells. Mol Cell Biol 2003; 23:1994-2008; PMID:12612073; http://dx.doi.org/ 10.1128/MCB.23.6.1994-2008.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wright RH, Castellano G, Bonet J, Le Dily F, Font-Mateu J, Ballare C, Nacht AS, Soronellas D, Oliva B, Beato M. CDK2-dependent activation of PARP-1 is required for hormonal gene regulation in breast cancer cells. Genes Dev 2012; 26:1972-83; PMID:22948662; http://dx.doi.org/ 10.1101/gad.193193.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Vicent GP, Ballare C, Nacht AS, Clausell J, Subtil-Rodriguez A, Quiles I, Jordan A, Beato M. Induction of progesterone target genes requires activation of Erk and Msk kinases and phosphorylation of histone H3. Mol Cell 2006; 24:367-81; PMID:17081988; http://dx.doi.org/ 10.1016/j.molcel.2006.10.011 [DOI] [PubMed] [Google Scholar]
- [25].Vicent GP, Nacht AS, Font-Mateu J, Castellano G, Gaveglia L, Ballare C, Beato M. Four enzymes cooperate to displace histone H1 during the first minute of hormonal gene activation. Genes Dev 2011; 25:845-62; PMID:21447625; http://dx.doi.org/ 10.1101/gad.621811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Beato M, Vicent GP. Impact of chromatin structure and dynamics on PR signaling. The initial steps in hormonal gene regulation. Mol Cell Endocrinol 2012; 357:37-42; PMID:21945605; http://dx.doi.org/ 10.1016/j.mce.2011.09.004 [DOI] [PubMed] [Google Scholar]
- [27].Nacht AS, Pohl A, Zaurin R, Soronellas D, Quilez J, Sharma P, Wright RH, Beato M, Vicent GP. Hormone-induced repression of genes requires BRG1-mediated H1.2 deposition at target promoters. EMBO J 2016; 35:1822-43; PMID:27390128; http://dx.doi.org/ 10.15252/embj.201593260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Chambon P, Weill JD, Mandel P. Nicotinamide mononucleotide activation of new DNA-dependent polyadenylic acid synthesizing nuclear enzyme. Biochem Biophys Res Commun 1963; 11:39-43; PMID:14019961; http://dx.doi.org/ 10.1016/0006-291X(63)90024-X [DOI] [PubMed] [Google Scholar]
- [29].Morales J, Li L, Fattah FJ, Dong Y, Bey EA, Patel M, Gao J, Boothman DA. Review of poly (ADP-ribose) polymerase (PARP) mechanisms of action and rationale for targeting in cancer and other diseases. Critical Rev Eukaryotic Gene Expression 2014; 24:15-28; PMID:24579667; http://dx.doi.org/ 10.1615/CritRevEukaryotGeneExpr.2013006875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Allfrey VG, Mirsky AE. The role of deoxyribonucleic acid and other polynucleotides in Atp synthesis by isolated cell nuclei. Proc Natl Acad Sci US A 1957; 43:589-98; PMID:16590060; http://dx.doi.org/ 10.1073/pnas.43.7.589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Betel I. The endogenous substrate for nuclear oxidative phosphorylation. Arch Biochem Biophys 1969; 134:271-4; PMID:5354764; http://dx.doi.org/ 10.1016/0003-9861(69)90283-5 [DOI] [PubMed] [Google Scholar]
- [32].Tanuma S. Evidence for a novel metabolic pathway of (ADP-ribose)n: pyrophosphorolysis of ADP-ribose in HeLa S3 cell nuclei. Biochem Biophys Res Communications 1989; 163:1047-55; PMID:2551267; http://dx.doi.org/ 10.1016/0006-291X(89)92327-9 [DOI] [PubMed] [Google Scholar]
- [33].Oei SL, Ziegler M. ATP for the DNA ligation step in base excision repair is generated from poly(ADP-ribose). J Biol Chem 2000; 275:23234-9; PMID:10930429 [DOI] [PubMed] [Google Scholar]
- [34].Altmeyer M, Neelsen KJ, Teloni F, Pozdnyakova I, Pellegrino S, Grofte M, Rask MB, Streicher W, Jungmichel S, Nielsen ML, et al.. Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose). Nat Communications 2015; 6:8088; PMID:26286827; http://dx.doi.org/ 10.1038/ncomms9088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].David KK, Andrabi SA, Dawson TM, Dawson VL. Parthanatos, a messenger of death. Front Biosci (Landmark Ed) 2009; 14:1116-28; PMID:19273119; http://dx.doi.org/ 10.2741/3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zhang T, Berrocal JG, Yao J, DuMond ME, Krishnakumar R, Ruhl DD, Ryu KW, Gamble MJ, Kraus WL. Regulation of poly(ADP-ribose) polymerase-1-dependent gene expression through promoter-directed recruitment of a nuclear NAD+ synthase. J Biol Chem 2012; 287:12405-16; PMID:22334709; http://dx.doi.org/ 10.1074/jbc.M111.304469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Chao D, Ni S. Nanomolar pyrophosphate detection and nucleus staining in living cells with simple terpyridine-Zn(II) complexes. Scientific Reports 2016; 6:26477; PMID:27198968; http://dx.doi.org/ 10.1038/srep26477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zha M, Guo Q, Zhang Y, Yu B, Ou Y, Zhong C, Ding J. Molecular mechanism of ADP-ribose hydrolysis by human NUDT5 from structural and kinetic studies. J Mol Biol 2008; 379:568-78; PMID:18462755; http://dx.doi.org/ 10.1016/j.jmb.2008.04.006 [DOI] [PubMed] [Google Scholar]
- [39].Dekker J, Mirny L. The 3D Genome as moderator of chromosomal communication. Cell 2016; 164:1110-21; PMID:26967279; http://dx.doi.org/ 10.1016/j.cell.2016.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Fauzee NJ, Pan J, Wang YL. PARP and PARG inhibitors–new therapeutic targets in cancer treatment. Pathology Oncol Res 2010; 16:469-78; PMID:20383759; http://dx.doi.org/ 10.1007/s12253-010-9266-6 [DOI] [PubMed] [Google Scholar]
- [41].Dang CV. Links between metabolism and cancer. Genes Dev 2012; 26:877-90; PMID:22549953; http://dx.doi.org/ 10.1101/gad.189365.112 [DOI] [PMC free article] [PubMed] [Google Scholar]


