Abstract
A SNP identified as rs548234, which is found in PRDM1, the gene that encodes BLIMP1, is a risk allele associated with systemic lupus erythematosus (SLE). BLIMP1 expression was reported to be decreased in women with the PRDM1 rs548234 risk allele compared with women with the nonrisk allele in monocyte-derived DCs (MO-DCs). In this study, we demonstrate that BLIMP1 expression is regulated by the binding of Kruppel-like factor 4 (KLF4) to the risk SNP. KLF4 is highly expressed in MO-DCs but undetectable in B cells, consistent with the lack of altered expression of BLIMP1 in B cells from risk SNP carriers. Female rs548234 risk allele carriers, but not nonrisk allele carriers, exhibited decreased levels of BLIMP1 in MO-DCs, showing that the regulatory function of KLF4 is influenced by the risk allele. In addition, KLF4 directly recruits histone deacetylases (HDAC4, HDAC6, and HDAC7), established negative regulators of gene expression. Finally, the knock down of KLF4 expression reversed the inhibitory effects of the risk SNP on promoter activity and BLIMP1 expression. Therefore, the binding of KLF4 and the subsequent recruitment of HDACs represent a mechanism for reduced BLIMP1 expression in MO-DCs bearing the SLE risk allele rs548234.
A risk allele associated with systemic lupus erythematosus generates a KLF4 binding site, resulting in repression of PRDM1 expression.
Introduction
Systemic lupus erythematosus (SLE) is a chronic inflammatory autoimmune disease of unknown etiology. It manifests as a production of multiple self-reactive antibodies targeting various organs in the body, generating a wide range of symptoms that contribute to disease pathogenesis (1, 2). Many studies suggest that genetic, immunologic, hormonal, and environmental factors contribute to lupus development. In lupus patients, there is a strong sex bias toward women, especially during their childbearing years (3, 4).
GWAS have assayed numerous SNPs in thousands of individuals and have identified hundreds of common genetic variants associated with over 80 diseases (http://www.genome.gov/gwastudies). Of these, 50 polymorphisms have been identified to predispose to SLE (reviewed in ref. 5) (6, 7). These risk alleles are found predominantly in genes that are associated with innate immunity: the interferon α signaling pathway and clearance pathways of apoptotic cells and immune complexes. Such genes include TLR7 (8, 9), interferon regulatory factor 5 (IRF5) (10), signal transducer and activator of transcription 4 (STAT4) (11, 12), interleukin-1 receptor-associated kinase 1 (IRAK1) (13, 14), tumor necrosis factor α–induced protein 3 (TNFAIP3) (15, 16), C1Q gene cluster (17), FCGR2A (18, 19), FCGR2B (20, 21), C-reactive protein (19, 22), and integrin α M (ITGAM) (23, 24). Polymorphisms in genes involved in lymphocyte signaling have also been identified. These may play a role in regulation of activation and suppression of lymphocytes, including PTPN22 (25), PDCD1 (26), LYN (27), and BLK (28).
Polymorphisms found in the intergenic region between positive regulatory domain I–binding factor 1 (PRDM1) and autophagy-related 5 (ATG5) have been identified as candidate factors for SLE in individuals of European (rs6568431, OR = 1.2, P = 7.12 × 10–10) (29) and Han Chinese (rs548234, OR = 1.25, P = 5.18 × 10–12) (30, 31) ancestries. BLIMP1, the protein encoded by PRDM1, is a transcriptional repressor expressed in various leukocytes. The regulation of BLIMP1 expression is important for maintaining a tolerogenic function in DCs, as demonstrated in DC-specific B lymphocyte–induced maturation protein 1 (Blimp1) knockout mice, in which a lupus-like phenotype developed in the female population. Due to increased IL-6 secretion from Blimp1-deficient DCs, these mice had an increased frequency of follicular T helper cells and germinal center B cells and contributing to disease pathogenesis (32). In healthy human monocyte-derived DCs (MO-DCs), the BLIMP1 expression level is lower in SLE SNP rs548234 C allele (risk) carriers compared with T allele (nonrisk) carriers. This difference is not observed in B lymphocytes, indicating a cell type–specific regulation of gene expression.
In this study, we investigated the role of polymorphism rs548234 and the molecular mechanism responsible for the regulation of BLIMP1 expression in MO-DCs. Our results revealed that a myeloid lineage transcription factor, Kruppel-like factor 4 (KLF4), binds to a sequence that is generated by the polymorphism and further recruits histone deacetylases 4, 6 and 7 (HDAC4, HDAC6 and HDAC7) to the region. We conclude that negative regulation by KLF4/HDACs in MO-DCs is responsible for the observed lineage-specific alteration in expression of BLIMP1 in risk allele carriers.
Results
Cell type–specific BLIMP1 mRNA levels in leukocytes from PRDM1 rs548234 carriers.
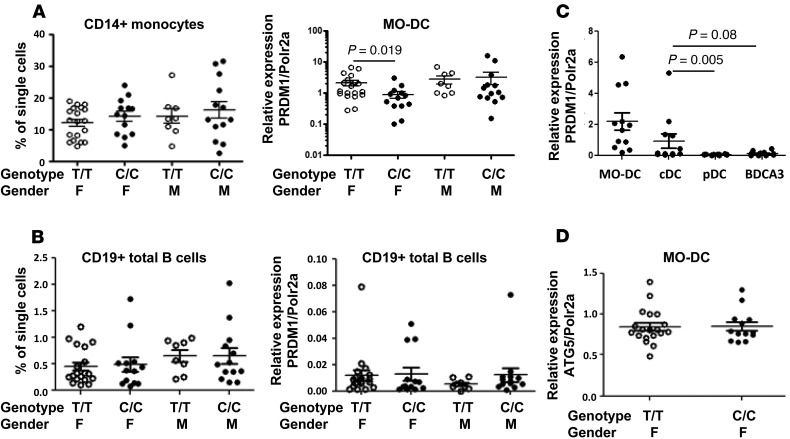
We previously reported that there is a decrease in the BLIMP1 level in MO-DCs, but not in total B cells, purified from rs548234 risk allele carriers compared with nonrisk controls (33). Here, we further confirm the previous observation with inclusion of both female and male individuals. In contrast to the female MO-DCs, there was no difference in BLIMP1 transcript in MO-DCs derived from male control allele (T/T) and male risk allele (C/C) carriers (Figure 1A). BLIMP1 expression in B cells was not different between control allele and risk allele carriers of both sexes (Figure 1B). There was no difference in frequency of CD14+ monocytes and total B cells in peripheral blood mononuclear cells (PBMCs) between control allele carriers and risk allele carriers (Figure 1, A and B). In addition, BLIMP1 expression in blood DCs was measured. Freshly isolated human conventional DCs (cDCs) expressed the highest level of BLIMP1, and this was comparable to the level expressed in MO-DCs (Figure 1C). These data suggest that BLIMP1 plays a role in blood cDCs as well as MO-DCs.
Figure 1. Cell type–dependent BLIMP1 expression.
Frequencies and the level of PRDM1 expression of blood CD14+ monocytes (A) and total B cells (B) from leukocytes prepared from female or male individuals with nonrisk (T/T) or risk (C/C) allele. CD14+ monocytes were cultured with granulocyte-macrophage colony-stimulating factor (GM-CSF) and IL-4 for 7 days for the generation of MO-DCs. MO-DCs and freshly isolated total B cells were further processed for total RNA preparation and qPCR for BLIMP1 expression. Relative expression of BLIMP1 was normalized to the level of housekeeping gene, POLR2A. Each dot represents an individual sample, and the bar represents the mean ± SEM (n = 5). (C) BLIMP1 (also known as PRDM1) expression was compared in DC subsets in human blood. Human blood DCs (cDC: Lin–CD11c+CD123–BDCA2–; pDC: Lin–CD11c–CD123hiBDCA2+; BDCA3 DC: Lin–CD11c+CD123–BDCA2–CD141+) and MO-DCs were purified and total RNAs were extracted. qPCR was performed, and the relative level of BLIMP1 was normalized to the level of the housekeeping gene. Each dot represents an individual sample, and the bar represents the mean ± SEM (n = 4). (D) The level of ATG5 was compared in MO-DCs from female nonrisk (T/T) or risk (C/C) carriers. Relative expression was normalized to the level of the housekeeping gene. Each dot represents an individual sample, and the bar represents the mean ± SEM (n = 5). The nonparametric, Mann-Whitney test was used for statistics.
SNP rs548234 resides in the intergenic region between PRDM1 and ATG5 on chromosome 6 (chr6: 106,120,159), 33,839 bp downstream and 64,324 bp upstream from the transcription initiation site of PRDM1 and ATG5, respectively. To determine whether the risk SNP regulates the level of ATG5 in MO-DCs, we measured ATG5 mRNA by qPCR. As shown in Figure 1D, there was no significant difference in ATG5 mRNA in MO-DCs from female controls or female risk SNP carriers.
The risk allele SNP generates a KLF4-binding site.
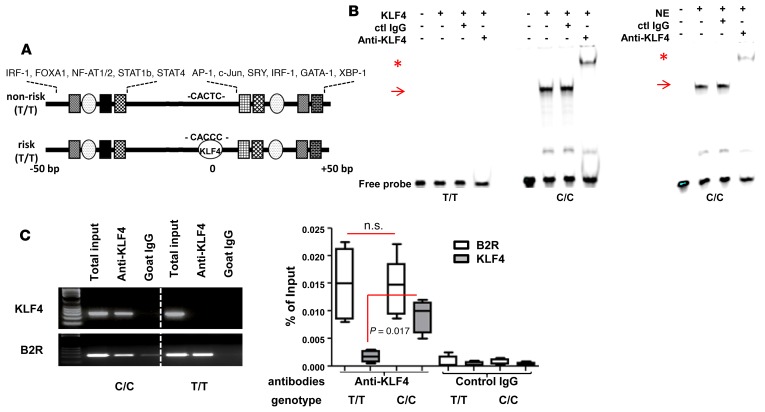
The majority of SNPs that have been identified by GWAS are located at an intergenic area and are largely unexplored. However, growing evidence suggests that many SNPs located in noncoding regions play an important role in regulating gene expression. They often generate enhancer binding motifs and alter chromatin structure (34, 35). In order to address this possibility, we investigated if there were binding motifs at the site in the risk C/C allele–containing DNA strand compared with the control T/T allele–containing DNA strand. The single nucleotide change from T to C generated a binding sequence for KLF4: CACCC (Figure 2A) (36). Therefore, we designed double-stranded (ds) oligonucleotides either the risk SNP or nonrisk SNP using the UCSC genome browser. Recombinant KLF4 protein and endogenous KLF4 showed specific binding to the ds oligonucleotide of the risk allele (C/C) but not to the ds oligonucleotide from the nonrisk allele (T/T) (Figure 2B). Next, we investigated whether KLF4 directly binds to the endogenous SNP-containing sequence by ChIP. KLF4 binding was detected in MO-DCs prepared from risk allele carriers but not in MO-DCs prepared from nonrisk allele carriers (Figure 2C). In order to confirm the expression and functionality of KLF4 in both risk allele and nonrisk allele individuals, we included the bradykinin B2 receptor (B2R) promoter in the ChIP assay, which is known to be bound by KLF4 (37). KLF4 binding to the B2R promoter was detectable, regardless of the donor genotype, proving that the ChIP assay for anti-KLF4 was unbiased in both SNP risk and nonrisk allele carriers. Hence, the differential binding of KLF4 to risk versus nonrisk SNP was due to sequence specificity in the genome. These data demonstrate that the risk allele generates a KLF4-binding site that is not present in the nonrisk allele.
Figure 2. KLF4 binds to the risk SNP.
(A) Schematic of genomic sequence around SNP rs548234. Putative binding sites of various transcription factors are found upstream and downstream of the SNP, and the KLF4-binding site is identified only in risk SNP by using Transfac and oPOSSUM 3.0 programs. (B) KLF binding with the risk allele probe (C/C) not with nonrisk allele probe (T/T) by EMSA. Recombinant KLF4 (left) or nuclear extract (NE) (5 μg) (right) was incubated with either T/T or C/C probe. To identify specificity of binding, either control or anti-KLF4 Ab was added. Arrows indicate KLF4 binding and asterisks indicate supershift. A representative image is shown from 3 independent experiments. (C) In vivo binding of KLF4 to the risk allele. To perform the ChIP assay, MO-DCs from either risk allele carriers (C/C) or nonrisk allele carriers (T/T) were prepared and incubated with either control goat IgG or anti-KLF Ab overnight. After precipitation, qPCR and PCR were performed with a primer set that amplifies the SNP region. The B2R gene was amplified as a positive control. Percentage of input was calculated relative level to the total input. In the box-and-whisker plot, horizontal bars indicate the medians, boxes indicate 25th to 75th percentiles, and whiskers indicate 10th and 90th percentiles (n = 4). The nonparametric, Mann-Whitney test was used for statistics.
Cell type– and sex-dependent KLF4 expression.
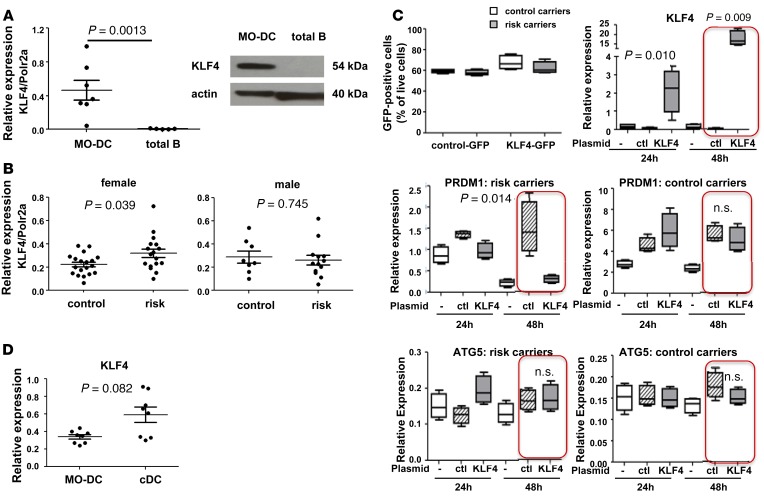
To test whether KLF4 is the regulatory factor that is responsible for cell type– and sex-specific regulation of BLIMP1 expression, KLF4 expression was measured in MO-DCs and in total B cells. Expression of KLF4 is important for myeloid lineage differentiation, and binding of KLF4 has been shown to either positively or negatively regulate gene expression depending on the interacting cofactors (36). There are several possibilities as to why there might be MO-DC–specific binding of KLF4 and reduction in BLIMP1: (a) KLF4 expression is limited to MO-DCs; (b) KLF4 expression is ubiquitous, but cofactor(s) of KLF4 for negative regulation are MO-DC specific; or (c) both KLF4 and its cofactor(s) are equally present in different cell types, but its accessibility to the SNP is regulated. We addressed the first hypothesis by measuring the mRNA for KLF4 in all cell types. KLF4 mRNA was present (at an average of half of the level of POLR2A mRNA) in MO-DCs but undetectable in B cells (~100-fold less than MO-DCs) (Figure 3A). A differential level of KLF4 protein in different cell types was also confirmed by Western blot (Figure 3A). We did not observe a detectable level of KLF4 mRNA and protein in total T cells, consistent with a MO-DC–specific regulatory mechanism (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/jci.insight.89569DS1). In order to understand why BLIMP1 levels were reduced in female risk allele carriers only, we compared the message level of KLF4 in MO-DCs prepared from female and male donors. KLF4 has been shown to be positively regulated by estrogen signals in breast cancer cells (38), suggesting that there might be a difference in KLF4 levels between females and males. There was no difference in the level of KLF4 mRNA analyzed from MO-DCs from all (including both risk and nonrisk allele carriers) female and male donors (data not shown); however, higher levels were observed in MO-DCs from female risk-allele carriers compared with female nonrisk carriers, but higher levels were not observed in MO-DCs from male risk carrier compared to male nonrisk allele carriers (Figure 3B).
Figure 3. Cell type–dependent expression of KLF4 and inverse correlation between BLIMP1 and KLF4.
(A) KLF4 mRNA (left) and KLF4 protein (right) were measured in MO-DCs and in total B cells. For mRNA, Each dot represents an individual sample, and the bar represents the mean ± SEM (n = 3). For protein, MO-DCs and total B cells were prepared, and 60 μg of total lysate was loaded for Western blotting. Actin is a loading control. A representative image is presented from 3 independent experiments. (B) The level of KLF4 mRNA was measured by qPCR and analyzed in risk allele carriers and control allele carriers: female (left) and male (right). Each dot represents an individual sample, and the bar represents the mean ± SEM (n = 10). (C) KLF4-expressing plasmid or control plasmid was transfected into MO-DCs prepared from nonrisk allele carriers or risk allele carriers. 48 hours after transfection, transfection efficiency was measured by GFP positivity and was around 60% in both control carriers and risk SNP carriers (top left). Total RNA was prepared, and mRNA levels of KLF4 (top right), BLIMP1 (middle row), and ATG5 (bottom row) were measured by qPCR. Relative expression was normalized to the level of HPRT1. In the box-and-whisker plot, horizontal bars indicate the medians, boxes indicate 25th to 75th percentiles, and whiskers indicate 10th and 90th percentiles (n = 3). (D) KLF4 mRNA was measured in MO-DCs and human blood cDCs. Each dot represents an individual sample, and the bar represents the mean ± SEM (n = 4). The nonparametric, Mann-Whitney test was used for statistics.
Next, we investigated whether KLF4 directly regulates BLIMP1 in MO-DCs. A KLF4-encoding plasmid was transfected into MO-DCs differentiated from risk allele carriers. In comparison to the BLIMP1 level in the control transfection, BLIMP1 was significantly decreased in KLF4 transfected MO-DCs (Figure 3C). Consistent with the observation of KLF4 binding to the risk allele only, overexpression of KLF4 did not affect the BLIMP1 level in nonrisk allele carrier MO-DCs. (Figure 3C). We also tested whether overexpression of KLF4 affects the level of ATG5 mRNA in MO-DCs from control and risk carriers; KLF4 did not alter ATG5 expression in either controls or risk carriers (Figure 3C).
In order to determine the physiological relevance of KLF4 in blood DCs, the level of KLF4 mRNA was measured in blood cDCs and MO-DCs by qPCR. cDCs showed similar or higher levels of KLF4 compared with MO-DCs, although there was higher individual variation of KLF4 expression in the cDC subset (Figure 3D). Thus, KLF4 is likely to play a role in regulating BLIMP1 expression in blood cDCs as well as in MO-DCs.
These data suggest that the expression of KLF4 is monocyte/DC lineage specific. The level of KLF4 is inversely correlated with the level of BLIMP1, suggesting that it exerts a negative regulatory effect on BLIMP1 expression.
Enhancer activity of SNP genomic DNA and KLF4-dependent inhibitory effect on polymorphic allele.
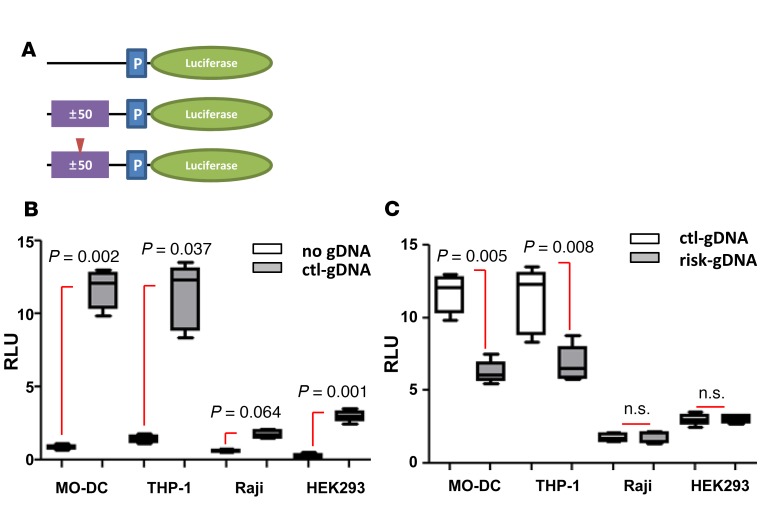
SNPs found in noncoding regions can generate enhancer binding motifs. This led us to explore whether genomic DNA (gDNA) containing SNP rs548234 regulates an enhancer effect on transcription. To determine this, small fragments (100 bp) of gDNA surrounding the SNP with either nonrisk allele (control-gDNA) or risk allele (risk-gDNA) were cloned into plasmids upstream of the Luciferase gene (Figure 4A). Plasmids with or without control-gDNA were transfected into various cell types, and their enhancing transcriptional regulatory activity was measured by luciferase activity. In all the tested cell types, control-gDNA plasmids were observed to have increased transcriptional activity compared with non-gDNA–containing plasmids, regardless of the level of KLF4 expression (~ 10-fold in myeloid cells and 2- to 5-fold in Raji and human embryonic kidney 293[HEK293] cell lines) (Figure 4B). Interestingly, MO-DCs or THP-1 cells that were transfected with risk-gDNA showed reduced enhancing activity compared with cells transfected with control-gDNA. Contrastingly, in cell types that express little to no KLF4, such as HEK293 and Raji B cells, risk-gDNA and control-gDNA shared similar enhancing activity (Figure 4C).
Figure 4. Gene regulatory function of SNP-containing genomic DNA.
(A) Diagram of reporter constructs containing genomic DNA (gDNA) (purple box). 100 bpgDNA (± 50 bp from SNP) from nonrisk allele (middle) or from risk allele (bottom; red arrowhead indicates risk allele) was inserted upstream of promoter region (P, blue box). (B and C) Each cell type was transfected with plasmid as indicated. Tk-Renilla with CMV promoter plasmid was cotransfected as a control. Cells were harvested 6 hours (MO-DC, THP-1, and Raji) or 48 hours (HEK293) after transfection, and luciferase level was measured. In the box-and-whisker plot, horizontal bars indicate the medians, boxes indicate 25th to 75th percentiles, and whiskers indicate 10th and 90th percentiles (n = 5). The nonparametric, Mann-Whitney test was used for statistics.
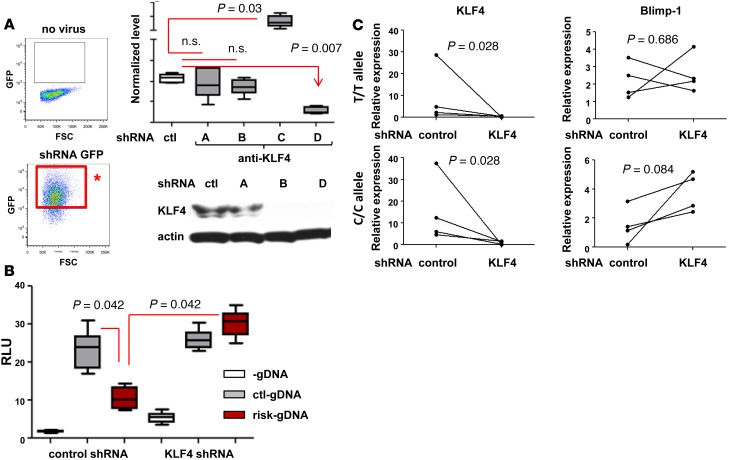
To confirm the role of KLF4 in regulating the cell-dependent enhancing activity observed in MO-DC and THP-1 cells, KLF4 expression was knocked down using KLF4 shRNA. Lentivirus expressing KLF4 shRNA and GFP was transduced into the cells (Figure 5A). As depicted in the diagram of Figure 5A (left column), GFP-positive shRNA expressing cells were purified for further analysis. One of the KLF4-targeting shRNAs successfully knocked down KLF4 mRNA and protein (Figure 5A, shRNA D). The enhancer activity of control-gDNA and risk-gDNA in KLF4-knocked down (KLFdl) THP-1 cell line was measured; if KLF4 was responsible for the reduction of enhancing activity in THP-1 cells, then similar activity levels would be expected between control-gDNA and risk-gDNA in the KLFdl THP-1 cell line. As seen in Figure 5B, risk-gDNA induced comparable transcriptional activity of the luciferase gene as control-gDNA in KLFdl THP-1 cells. We also investigated whether KLF4 deficiency can restore BLIMP1 expression in risk-carrier MO-DCs. MO-DCs differentiated from control or risk allele carriers were infected with either control shRNA or KLF4 shRNA, and GFP-positive MO-DCs were purified. Knockdown efficiency of KLF4 was confirmed by qPCR, and the level of BLIMP1 was measured. The level of BLIMP1 transcripts was increased in KLF4 shRNA–transfected MO-DCs. This reversion was observed in MO-DCs with risk allele carriers but not in MO-DCs with control carriers (Figure 5C).
Figure 5. Knockdown of KLF4 abolished the regulatory effect by risk allele.
(A) KLF4 knockdown by shRNA. Four different KLF4-targeting shRNA constructs (A, B, C, and C) and scrambled control shRNA lentivirus were infected into THP-1 cells. Four days after two rounds of infection, shRNA-positive cells were purified based on GFP expression (red box with asterisk). Cells with each construct were harvested, and mRNA and protein levels were measured by qPCR and Western blotting, respectively. Whisker plot represents the mean to max (n = 3). The Western blot image is representative of 3 experiments. (B) Promoter assay was performed in KLF4dl THP-1 cells. Each plasmid was transfected into either control shRNA or KLF4 shRNA (KLF4dl) as shown in figure, and luciferase activity was measured. In the box-and-whisker plot, horizontal bars indicate the medians, boxes indicate 25th to 75th percentiles, and whiskers indicate 10th and 90th percentiles (n = 3). (C) MO-DCs from control or risk allele carriers were infected with control shRNA or KLF4 shRNA lentivirus at day 5 and day 7 during differentiation. Four days after the second infection (day 9), GFP-positive MO-DCs were sorted and the level of KLF4 and BLIMP1 was measured by qPCR. Each dot represents an individual sample (n = 4). The nonparametric, Mann-Whitney test was used for statistics.
KLF4 interacts with HDACs to suppress BLIMP1 transcription.
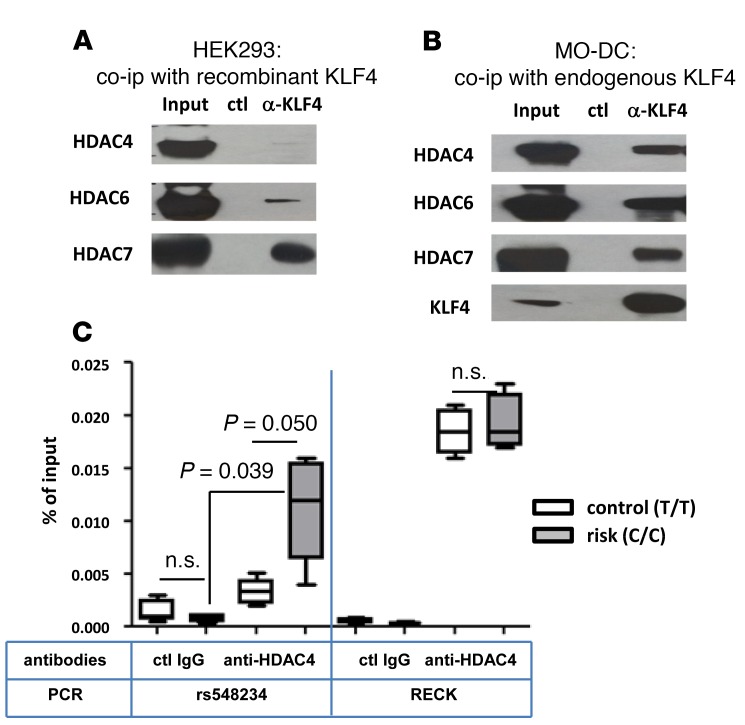
It has been well documented in various tumor cells that KLF family members, including KLF4, execute their gene regulatory function by recruitment of specific cofactors (reviewed in ref. 39), such as histone-modifying enzymes, like p300 (40) and HDAC7 (41). To investigate the molecular mechanism underlying the suppressive function of KLF4 in MO-DCs, a coimmunoprecipitation assay was performed. Immunoprecipitation of KLF4 coprecipitated HDAC6 and HDAC7 in KLF4-transfected HEK293 cells (Figure 6A). However, in MO-DCs, we found that KLF4 coprecipitated HDAC4 in addition to HDAC6 and HDAC7 (Figure 6B). Other HDACs (HDAC1, HDAC2, and HDAC3) and β-catenin showed no binding with KLF4 in MO-DCs (data not shown).
Figure 6. Direct interaction between KLF4 and HDACs and recruitment of HDAC4 to rs548234 risk allele.
Coimmunoprecipitation was performed with HEK293 cells (A) or MO-DCs (B) after KLF4 plasmid transfection. In both experiments, anti-KLF4 Ab or control Ab was incubated with total cell lysate overnight. Ab-protein complex was eluted, and Ab-bound proteins were separated by 4%–12% Bis-Tris protein gel. Untreated total input was loaded as a positive control. To detect HDACs, anti-HDAC antibodies were used. A representative image is shown from 3 independent experiments. (C) Recruitment of HDAC4 to the SNP region was measured by ChIP assay. Anti-HDAC4 Ab or control IgG was incubated with chromatin prepared from MO-DCs with the nonrisk control (T/T, white) or the risk (C/C, black) allele. Binding of HDAC4 to the SNP region was assessed by qPCR using SNP region-specific primers, and the percentage of input gDNA was calculated. Quantitation of the RECK promoter region was used as a positive control. In the box-and-whisker plot, horizontal bars indicate the medians, boxes indicate 25th to 75th percentiles, and whiskers indicate 10th and 90th percentiles (n = 4). The nonparametric, Mann-Whitney test was used for statistics.
Although the binding of KLF4 to HDAC cofactors HDAC4, HDAC6, and HDAC7 might explain its negative regulatory mechanism, we wanted direct evidence showing the recruitment of repressive HDACs to the risk SNP area. To test whether HDAC4 is recruited to the region around rs548234 in a genotype-dependent manner, we performed a ChIP assay using anti-HDAC4 Ab in MO-DCs derived from either or nonrisk SNP carriers. HDAC4 bound to the region around the risk SNP but not the control SNP (Figure 6C). HDAC4 bound to the reversion-inducing cysteine-rich protein with Kazal motifs (RECK) promoter region, which is known to be a target in ovarian cancer cells, in MO-DCs from both nonrisk SNP carriers in an equivalent manner (42). These data suggest that KLF4 could suppress transcription of BLIMP1 through the recruitment of HDACs.
Cell type–dependent KLF4 expression in SLE patients.
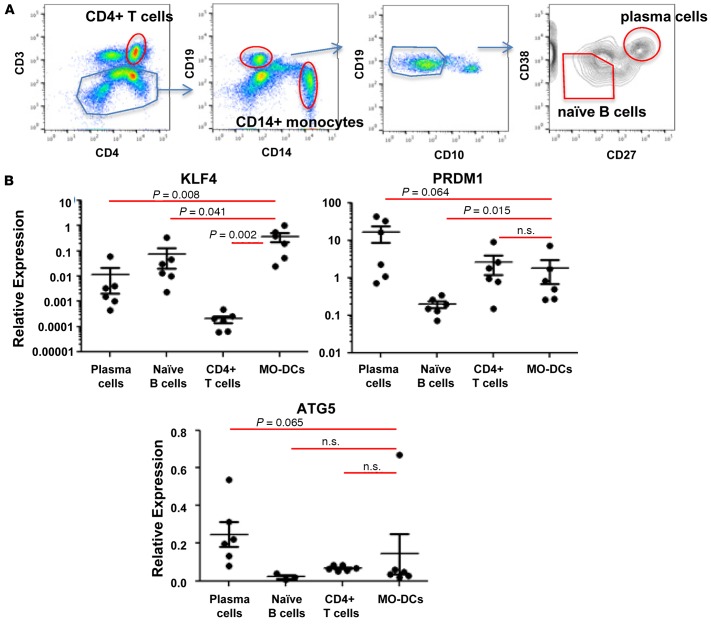
To assess the regulatory function of KLF4 in SLE patients, we investigated the expression of KLF4 and PRDM1 in various cell types from SLE patients. CD4+ T cells, naive B cells, and plasma cells were isolated from the blood of SLE patients (43), and MO-DCs were differentiated from CD14+ monocytes isolated from SLE patient’s blood (Figure 7). As shown in Figure 7B, KLF4 transcript was detected in MO-DCs, and the level of transcript was significantly higher than in naive B cells, plasma cells, and CD4+ T cells. PRDM1 expression was observed in all lymphocytes, although naive B cells exhibited a relatively low level compared with other cell types. In addition, we investigated whether increased KLF4 can suppress PRDM1 in MO-DCs from SLE patients with or without the risk allele. We observed a trend of low PRDM1 in KLF4-transfected MO-DCs harboring the risk allele but not the control allele (Supplemental Figure 2). These data support the view that the functional data we have observed in normal subjects are also likely to be relevant in patients with SLE.
Figure 7. Expression of KLF4, PRDM1, and ATG5 in leukocytes from SLE patients.
(A) Each leukocyte subset was purified from freshly prepared PBMCs, as indicated in the flow cytometry image. (B) Expression of KLF4, PRDM1, and ATG5 in different leukocyte cell types was measured by qPCR. Relative expression was normalized to the level of the housekeeping gene, HPRT1. Each dot represents an individual sample, and the bar represents the mean ± SEM (n = 4). The nonparametric, Mann-Whitney test was used for statistics.
Discussion
GWAS have been performed in large cohorts of control and SLE patients and have identified up to 1 million SNPs and about 50 genetic associations for SLE (28, 29, 44). SNP rs548234 is located between PRDM1 and ATG5, and its association with SLE was identified and confirmed by an independent study (31). In our study, we determined that there is a decreased level of BLIMP1 in MO-DCs carrying a risk allele compared with MO-DCs with a nonrisk allele, and this phenotype is cell type specific. We demonstrated that the polymorphism T → C generates a KLF4 direct binding sequence. The genomic region encompassing the SNP possesses an enhancer activity, thus increasing promoter activity. This enhancing activity of the risk allele DNA can be modulated depending on KLF4 expression. Expression of KLF4 can be detected in myeloid lineage, including monocytes and blood DCs, but not in CD4+ T cells and total B cells. KLF4 can interact with multiple HDACs in MO-DCs, implicating a negative regulatory role on promoter activity.
The PRDM1-ATG5 gene region was first reported to be associated with SLE in European and Chinese populations, a finding later confirmed by Zhou and colleagues who also recognized SNP rs548234 within this region (31). In addition, ATG5, a gene associated with SNP rs548234, was identified by Zhou as a candidate gene for SLE predisposition; the level of ATG5 was found to be increased in B lymphocytes, whereas BLIMP1 expression in B cells was not affected by the risk allele. We also observed that BLIMP1 expression is comparable regardless of allele in total B cells, confirming the data from the previous study. In fact, BLIMP1 levels are very low throughout B cell stages, except in plasmablasts or plasma cells, and this low expression profile may contribute to the minimal allele-specific differences of BLIMP1 expression. However, BLIMP1 expression is affected by the presence of the risk allele in MO-DCs. In contrast to B cells, MO-DCs express significant levels of BLIMP1 even without activation. cDCs (CD11c+ DCs in humans and CD11b+ cDCs in mice) express relatively higher levels of BLIMP1 compared with other subsets of DCs, suggesting that BLIMP1 may play an important role in these cell types.
It is not clear how immunological tolerance is broken in SLE. DCs are suggested to be critical players based on their immunological function and their altered phenotype in SLE patients. Multiple, but not all, studies have shown that circulating DCs (BDCA1+ DCs) from SLE patients have decreased T cell–activating capacity compared with DCs from healthy individuals (45, 46). In vitro–differentiated MO-DCs display substantial differences in phenotype and function between SLE patients and healthy individuals. MO-DCs differentiated from SLE patients express higher levels of costimulatory molecules and exhibit increased secretion of lymphocyte-activating cytokines, including IL-6 and B cell–activating factor (BAFF), thereby inducing enhanced proliferation of T cells (47, 48). Our previous study suggested that the level of BLIMP1 expression regulates the inflammatory function of MO-DCs. MO-DCs differentiated from healthy individuals with the risk allele have a low level of BLIMP1, a higher level of HLA– antigen D related (HLA-DR), and increased levels of proinflammatory cytokines following TLR stimulation, as observed in MO-DCs from SLE patients (33).
How does the noncoding risk SNP regulate expression of BLIMP1 in MO-DCs? Determining the functional role of SNPs found within noncoding intergenic or intronic regions can be challenging. Previous studies have determined that intergenic SNPs often participate in regulation of proximal gene expression either by the generation of a novel enhancer element or by interacting with a preexisting enhancer element (49, 50). Unlike promoters, distal enhancers are often cell type–specific, leading to tissue-specific risk SNP effects (51). The risk allele of rs548234 generates a KLF4-binding sequence that does not exist in the nonrisk SNP. KLF4 is a member of the family of KLF transcription factors, expressed in colon, skin, and brain. They regulate cell proliferation, differentiation and apoptosis (52). Due to its regulatory function in cell proliferation and differentiation, KLF4 has been studied in cancer most extensively. In contrast, studies on the function of KLF4 in autoimmunity are relatively sparse. A recent study from Tussiwand and colleagues found that KLF4 expression is required for a subset of cDCs to promote Th2 cell response in mice (53), highlighting a novel role of KLF4 in DC biology. However, it is not yet known whether such a KLF4-dependent subset of DCs exists in humans. Nonetheless, KLF4 expression is strictly limited to monocytes and myeloid cell lineage with the exception of CD8+ T cells (54) and memory B cells (55). Our study also confirmed the cell type–specific expression of KLF4 in blood leukocytes, permitting a cell type–specific regulation of the risk SNP of BLIMP1. Decreased BLIMP1 transcripts were observed only in MO-DCs of risk allele carrier, but not in B cells. In addition to MO-DCs, it might be interesting to investigate whether the BLIMP1 level is reduced in CD8+ T cells or in memory B cells from risk-SNP carriers as both cell types are reported to express KLF4.
How does KLF4 regulate BLIMP1 expression in MO-DCs? KLF4 (a molecule composed of 470 amino acids) can be divided into three functional domains: an activation domain (N-terminus), a repressive domain (central domain), and a DNA-binding domain (C-terminal) (52, 56). In myeloid cells, expression of KLF4 can be regulated by environmental stimuli, especially through inflammatory signaling (57, 58). KLF4 can regulate gene expression either positively or negatively depending on the coregulator. A positive regulatory function of KLF4 is mediated through an interaction with p300/CBP (52). Mechanisms for the repressive function of KLF4 include the recruitment of various HDACs (59, 60) or β-catenin/TCF-4 (61) or the direct competition with activating transcription factors (58). As seen in our coimmunoprecipitation study, KLF4 indeed interacted with three HDACs in MO-DCs. However, we were unable to detect any interactions with β-catenin or other HDACs (HDAC1/HDAC2/HDAC3), although high levels of β-catenin and HDAC1/HDAC2/HDAC3 were detected in MO-DCs (data not shown). The interaction of KLF4 with HDAC 4 found in MO-DCs but not in HEK293 cells, and interactions with HDAC6 and HDAC7 found in both cell types, led us to conclude that there is a cell type–dependent interaction profile of HDACs. Of these HDACs, we do not yet know which has a primary role in BLIMP1 regulation.
The negative regulatory activity of the risk-SNP was observed only in KLF4-expressing cell types, such as MO-DCs and THP-1 cells, and not in KLF4-negative cell types, like primary T cell (data not shown) and B cell lines. Further, knockdown of KLF4 expression in THP-1 cells removed the negative regulatory effect on the risk allele. We also observed that the genomic area encompassing the SNP rs548234 has a cell type–dependent enhancing activity. We consistently observed more than a 10-fold enhancing activity in myeloid cells (MO-DCs and THP-1) and approximately 3-fold in other cell types (HEK293 and B cell lines). These data imply that the SNP area might contain strong enhancing activity in myeloid lineage cells independent of the SNP, further suggesting that myeloid cells are the cell types most affected by the function of rs548234.
A similar level of KLF4 is found in male and female cells, but, interestingly, KLF4 transcripts are higher in MO-DCs from female risk-SNP carriers than non risk-SNP carriers. There are several possible explanations for this. First, the activated phenotype associated with the risk allele that we observed previously may contribute to this difference (33). Alternatively, common regulation on BLIMP1 expression through the sex hormone signaling pathways might be involved; both BLIMP1 and KLF4 have a putative estrogen responsive element (ERE) and a specificity protein 1–binding (Sp1-binding) sequence in their promoter. We have also considered the possibility that Blimp1 may exert a negative feedback on KLF4 expression. These possibilities will require further investigation.
In conclusion, we have identified a regulatory mechanism of the risk-SNP on BLIMP1 expression in human MO-DCs. The change from T to C generates a consensus binding sequence for KLF4, which is expressed in myeloid lineage cells but not in lymphoid lineage cells, resulting in lineage-specific regulation. This causes an inverse relationship between the expression levels of BLIMP1 and KLF4. An enhancer effect on transcription is also documented for the genomic sequence surrounding the SNP, and the sequence including the risk-SNP exhibits a reduced enhancer ability that is strictly dependent on the presence of KLF4. KLF4 suppresses gene transcription through the recruitment of HDACs in MO-DCs. This is the first functional evidence to our knowledge of how the SLE risk-SNP rs548234 regulates PRDM1 in human MO-DCs.
Methods
Preparations of PBMCs, blood DCs, and in vitro differentiation of MO-DCs.
Healthy PRDM1 rs548234 risk allele carriers and nonrisk allele controls were identified from the Genotype and Phenotype (GAP) registry at The Feinstein Institute for Medical Research. Both cohorts consisted of hormonally active females under 55 years old and were of various races and ethnicities. Participants consented for the study prior to their participation. Leukopacks were also purchased from the New York Blood Center. Total PBMCs were collected by Ficoll-Paque gradient centrifugation. Briefly, whole blood or leukopack was diluted with HBSS (Life Technologies) and layered on the Ficoll (GE healthcare life sciences). Cells were centrifuged at 750 g for 20 minutes without a break at room temperature. PBMCs were collected from the intermediate layer and washed 3 times with HBSS.
To generate MO-DCs, CD14+ monocytes were purified using the EasySep kit (Stem cell technologies) according to the manufacturer’s protocol. The purity of CD14+ cells was determined by flow cytometry LRSII (BD Biosciences). After purification, CD14+ monocytes were cultured with RPMI1640 supplemented with 10% heat-inactivated FBS, 1% penicillin-streptomycin (P/S), 1% L-glutamine, 100 ng/ml of recombinant human granulocyte-macrophage colony-stimulating factor (GM-CSF) (Peprotech), and 100 ng/ml of recombinant human IL-4 (Peprotech) for 7 days.
Blood DCs were prepared directly from PBMCs by sorting. Blood DC populations were identified after staining with the following Ab cocktail; nonmyeloid lineage markers (CD19, CD3, CD56), CD14, CD16, HLA-DR, CD11c, CD123, and CD141. From the lineage-negative and HLA-DR–positive population, CD14+CD16– monocytes were designated as a CD14+ monocyte, CD14–CD16+CD11c+ was designated as CD16+ DCs, CD14–CD16–CD11c–CD123+ was designated as pDCs, CD14–CD16–CD11c+CD123–CD141– was designated as cDCs, and CD14–CD16–CD11c+CD123–CD141+ was designated as BDCA3 DCs. Each sorted DC population was directly lysed with RLT buffer (QIAGEN) supplemented with 1% 2-ME and kept at –80 °C until use.
Lymphocyte isolation and MO-DC differentiation from SLE patients.
Whole blood samples from SLE patients were collected and genotyped by using Taqman SNP genotype systems (assay no. C_14436_10, ThermoFisher Scientific). PBMCs were prepared as described and monocytes, CD4+ T cells, plasma cells, and naive B cells were purified by FACSAria (BD Biosciences). Monocytes were cultured and differentiated into immature DCs in RPMI1640 medium supplemented with 100 ng/ml of GM-CSF and 100 ng/ml of IL-4 (both were purchased from Peprotech) for 7 days.
Cell lines.
The human monocytic cell line, THP-1, and the human B cell line, Raji, were maintained in RPMI 1640 supplemented with 10% FBS, 1% P/S, and 1% L-glutamine at 0.5 × 106 cells/ml. The HEK 293 cell line was purchased from ATCC (ATCC CRL-1573) and maintained in DMEM with 10% FBS, 1% P/S, and 1% L-glutamine.
RNA preparation and real-time PCR.
For each population, total RNA was extracted by either the RNeasy Mini- or Micro-kit (QIAGEN) or Direct-zol RNA Micro Prep (Zymo Research) based on the number of cells following the manufacturers’ protocol. A DNase digestion step was included in all the RNA preparation to exclude gDNA contamination. cDNA was generated using the iScript cDNA synthesis kit (Bio-Rad). Quantitative PCR was performed by real-time PCR analysis. Gene-specific primers were purchased from Taqman (Life Technologies), and quantitative PCR was performed using the Light cycler 480 II (Roche). POLR2A and ACTB were used as housekeeping genes, and relative expression of each gene was calculated by ΔΔCt.
Western blotting.
Cells were lysed and proteins were extracted in a radio immunoprecipitation assay buffer (RIPA buffer) (25 mM Tris-HCl [pH 7.6], 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS; Pierce) and complete protease inhibitor (Roche) and phosphatase inhibitor cocktail (Sigma-Aldrich). The lysates were obtained after centrifugation, and protein concentration was measured using the BCA protein assay kit (Pierce, Thermo Scientific). The lysates were separated by 4%–12% Bis-Tris PAGE. After transfer to PVDF membrane (Hybond-C; GE Amersham), the membrane was stained with ponceau S (Sigma-Aldrich) to confirm protein transfer. The PVDF membrane was then destained with 5% acetic acid and washing several times with 0.1% Tween-20 in Tris-buffered saline (TBS-T) (pH 7.6) and blocked for 1 hour at room temperature with 5% non-fat dry milk in TBS-T buffer. The blocked membranes were then incubated with the primary antibodies for overnight at 4°C. HDAC1 mouse monoclonal Ab (mAb) (10E2), HDAC2 mouse mAb (3F3), HDAC3 mouse mAb (7G6C5), HDAC4 rabbit mAb (D15C3), HDAC6 rabbit mAb (D2E5), and KLF4 rabbit mAb (D1F2) were purchased from cell signaling technology. HDAC7 rabbit polyclonal antibodies (pAbs) and KLF4 rabbit mAb (EPR3550) were purchased from Abcam. The membrane was washed 4 times (15 minutes each) with TBS-T buffer and then incubated with secondary antibodies conjugated with HRP (1:20,000) for 1 hour at room temperature. The immunoreactive proteins were visualized with the ECL detection reagents (Thermo).
Plasmids and transient transfections.
An expression vector for human KLF4 (EX-Z5703-M61) and control ORF expression vector (EX-NEG-M68) were purchased from Genecopoeia and luciferase vector pGL4.25 was purchased from Promega. For transient transfection studies, Lonza Nucleofector system and Lipofectamine (Invitrogen) were used for THP-1 cells and MO-DCs or for HEK293 cells, respectively. Briefly, day 5 cultured MO-DCs were harvested and 1 μg or 5–10 μg of expression vector was mixed with 1 × 106 cells in 100 μl of Nucleofector solution. The cells and plasmid mixture were transferred to cuvettes, and electroporation was performed using a preselected program for each cell type. Immediately after electroporation, 500 μl of culture medium was added to the cells and plated for recovery. For Lipofectamine, HEK293 cells were seeded in 6-well plates and grown to approximately 75% confluence at the time of transfection, following the manufacture’s protocol.
ChIP.
ChIP was performed as previously described with minor modifications (33). For KLF4, 10 μg of anti-KLF4 goat pAbs (AF3640) or control antibodies (AB-108-C) (both purchased from R&D Systems) was incubated with 100 μl of protein-G magnetic beads (Novex, Thermo Fisher Scientific) at 4°C for overnight with gentle rotating. For HDAC4 ChIP, 3 μg of anti-HDAC4 (clone D15C3) rabbit mAb or normal rabbit IgG (both purchased from Cell Signaling Technologies) was bound to protein-G magnetic beads. The next day, THP-1 cells or MO-DCs were fixed with 1% formaldehyde for 10 minutes at room temperature and quenched with125 mM glycine. The cells were rinsed with ice-cold PBS and RIPA buffer (Thermo Scientific) containing protease inhibitors (Roche). The cell lysate was sonicated 15 times for 30 seconds (Misonixsonicator 3000) and was allowed cool on ice for 1 minute between pulses. Magnetic bead–conjugated antibodies were incubated with the cell lysate at 4°C overnight with rotation. The next day, unbound lysates were washed with wash buffer (300 mM LiCl, 50 mM HEPES [pH 7.6], 1 mM EDTA, 0.7% DOC, and 1% NP-40) 10 times. Anti-KLF4–bound protein/DNA complexes were eluted with elution buffer (50 mM Tris [pH 8.0], 10 mM EDTA, and 1% SDS), and the protein was separated from the DNA complex. Eluted DNA was cleaned using a DNA purification kit (QIAGEN), and PCR was performed. Primers to amplify the B2R (a positive control of KLF4 ChIP) were 5′-GCAGAGCGGAGAGCGAAGG-3′ and 5′-GCCTGATGTCCCCACCGTC-3′. The PCR conditions were as follows: 94°C for 5 minutes; 94°C for 15 seconds, 60°C for 15 seconds, and 72°C for 15 seconds for 35 cycles; and 72°C for 10 minutes. Primers to amplify RECK (positive control for HDAC4 ChIP) were 5′-CATAACAAAGAGCCCTGGTACG-3′ and 5′-CTGCTCCTTCTGCTGGCC-3′(42). The qPCR conditions were 94°C for 10 minutes; 94°C for 15 seconds, 60°C for 15 seconds, and 72°C for 15 seconds for 55 cycles; and 72°C for 10 minutes. Primers to amplify the SNP area were 5′-CAAAGCTTCCAGGCTTTTACA-3′ and 5′-TGAACCAAAGAAGGAAAAGTCAA-3′. The PCR condition was 94°C for 5 minutes; 94°C for 15 seconds, 53°C for 15 seconds, and 72°C for 15 seconds for 40 cycles; and 72°C for 10 minutes.
shRNA clones and lentivirus infection.
shRNA plasmids for KLF4 or scrambled control plasmids were purchased from Origene Tech (pGFP-C-shLenti; TL316853). Lentiviruses were produced with cotransfection of pLP1, pLP2, pLP/vsvg lentiviral packaging DNAs (Invitrogen). shRNA lentiviral plasmid (with a molecular ratio of plasmid of 1.5:2.5:2:4) was introduced into HEK293 cells by calcium-phosphate transfection, following the manufacture’s protocol (Invitrogen). Transduction was performed by spin infection protocol. Briefly, virus titer was determined, and MO-DCs or THP-1 cells were resuspended into virus-containing supernatant (1 million cells/ml of supernatant) in the presence of 4 μg/ml of polybrene (Santa Cruz Biotech.) and centrifuged at 950 g for 90 minutes at room temperature. After infection, cells were incubated for 6 hours in a CO2 incubator. Medium was changed with fresh culture medium and cells were maintained for 4 days.
Coimmunoprecipitation.
KLF4 rabbit mAb (D1F2, 5 μg) or control IgG mAb was incubated with protein G-magnetic beads (1.5 mg, DynaBead, Thermo) overnight with continuous rotation at 4°C. The next day, KLF4-overexpressing HEK293 cells or MO-DCs were lysed with RIPA buffer (Pierce, Thermo) with protease inhibitor (Roche), phosphatase inhibitor (Sigma-Aldrich), and 1% Triton X-100. Protein measured with a BCA protein assay kit (Pierce, Thermo) and 300 μg of cell lysates were mixed with KLF4 mAb–bead complex for 10 minutes at room temperature with rotation. The Ab-bead-lysate complex was washed 4 times and magnetically isolated (DynaMag, Thermo). After the last wash, the Ab-bead-Ag complex was transferred to a new tube. Proteins were eluted with 30 μl elution buffer (from the kit) and 10 μl LDS sample buffer/Reducing agent mix (NuPAGE, Thermo). After incubation at 90°C for 10 minutes, the supernatant was transferred to a new tube. Immunoblotting was performed with different antibodies to detect proteins in the complex.
Luciferase assay.
100 bp of normal and risk allele genomic sequences were cloned into pGL4.25 Luciferase vector. Optimal conditions for transfection, plasmid concentration, and incubation time after transfection were determined. HEK293 cells were transfected with 5 μg of Luc plasmid and 1 μg of Tk-Renilla by Lipofectamine and harvested after 48 hours transfection; 2 × 106 THP-1 cells and MO-DCs were transfected with 10 μg of Luc plasmid and 1 μg of Tk-Renilla plasmid by Nucleofector and harvested at 6 hours after transfection; and 5 × 106 Raji B cells were transfected with 20 μg of Luc plasmid and 2 μg of Tk-Renilla plasmid by Nucleofector and harvested at 6 hours transfection. Transfected cells were lysed and measured with the Dual-Glo Luciferase assay kit (Promega), following the manufacture’s protocol. Relative luciferase units were normalized by the luciferase value of Tk-Renilla of each sample.
EMSA.
Oligonucleotides were synthesized and labeled with IRD700 at the 5′-end (Integrated DNA technologies), and ds oligonucleotide was prepared in annealing buffer at a concentration of 20 pmol. Gel preparation and electrophoresis were performed as described previously (56, 62). Briefly, recombinant KLF4 (10 ng, Origene) or nuclear extract was incubated in binding buffer (10 mM Tris, 50 mM KCl, 3.5 mM DTT) supplemented with 2 μg poly (dI-dC), 5 mM MgCl2, 20mM ZnCl2, 1% DTT/Tween, and sonicated salmon sperm DNA for 5 minutes, and the probe was added for additional 15 minutes incubation at room temperature. 1 μg of anti–KLF4 antibodies (D1F2) or unlabeled oligos (2 nmol) was added to the binding mixture to test specificity of the binding. Protein-DNA complexes were resolved on a prerun 5% Tris/Borate/EDTA (TBE) polyacrylamide gel at 100 V for 1.5~2.0 hours in 0.5× TBE buffer. The image was scanned by Odyssey (LI-COR Biosciences). The oligonucleotide sequences for the probe are as follows: nonrisk allele, 5′-TGTCTTCTCTCACATTGTCTTGACTT-3′, and risk allele, 5′-TGTCTTCTCTCACCCTTGTCTTGACT-3′.
Statistics.
Statistical significance was determined with a nonparametric, Mann-Whitney test, and P ≤ 0.05 was considered significant.
Study approval.
The protocol for study of human samples was approved by the IRB of The Feinstein Institute for Medical Research (approval 19-081A for GaP healthy volunteers and HS15-0652 for SLE patients). All the participants were informed by consent form prior to their participation.
Author contributions
All contributing authors have agreed to the submission of this manuscript for publication. SJK conceived and designed the study and analyzed and interpreted the data. SHJ performed the experiments and helped interpret data, HC performed gene expression studies, PKG provided the study samples, and BD conceived the study and helped interpret the data. SJK and SHJ wrote the manuscript, and BD and PKG participated in critical review of the manuscript.
Supplementary Material
Acknowledgments
We are especially thankful to M. Keogh, M. DeFranco, and G. Klein from the Genotype and Phenotype Registry at the Feinstein Institute for recruiting PRDM1 genotyped subjects. We also thank C. Aranow and M. Mackay at the Feinstein Institute for recruiting SLE patients. This work was supported by grants from the US National Institutes of Health (R01 AR065209 to SJK and SHJ) and Alliance of Lupus Research (to BD).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:JCI Insight. 2017;2(1):e89569. doi:10.1172/jci.insight.89569.
Contributor Information
Su Hwa Jang, Email: Sjang1@northwell.edu.
Helen Chen, Email: Hchen5@northwell.edu.
Betty Diamond, Email: bdiamond@nshs.edu.
Sun Jung Kim, Email: sjkim@nshs.edu.
References
- 1.Bootsma H, et al. The predictive value of fluctuations in IgM and IgG class anti-dsDNA antibodies for relapses in systemic lupus erythematosus. A prospective long-term observation. Ann Rheum Dis. 1997;56(11):661–666. doi: 10.1136/ard.56.11.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hahn BH. Antibodies to DNA. N Engl J Med. 1998;338(19):1359–1368. doi: 10.1056/NEJM199805073381906. [DOI] [PubMed] [Google Scholar]
- 3.Chakravarty EF, Bush TM, Manzi S, Clarke AE, Ward MM. Prevalence of adult systemic lupus erythematosus in California and Pennsylvania in 2000: estimates obtained using hospitalization data. Arthritis Rheum. 2007;56(6):2092–2094. doi: 10.1002/art.22641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lahita RG. The role of sex hormones in systemic lupus erythematosus. Curr Opin Rheumatol. 1999;11(5):352–356. doi: 10.1097/00002281-199909000-00005. [DOI] [PubMed] [Google Scholar]
- 5.Deng Y, Tsao BP. Genetic susceptibility to systemic lupus erythematosus in the genomic era. Nat Rev Rheumatol. 2010;6(12):683–692. doi: 10.1038/nrrheum.2010.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barcellos LF, et al. High-density SNP screening of the major histocompatibility complex in systemic lupus erythematosus demonstrates strong evidence for independent susceptibility regions. PLoS Genet. 2009;5(10):e1000696. doi: 10.1371/journal.pgen.1000696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.International MHC Autoimmunity Genetics Network, et al. Mapping of multiple susceptibility variants within the MHC region for 7 immune-mediated diseases. Proc Natl Acad Sci USA. 2009;106(44):18680–18685. doi: 10.1073/pnas.0909307106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawasaki A, et al. TLR7 single-nucleotide polymorphisms in the 3’ untranslated region and intron 2 independently contribute to systemic lupus erythematosus in Japanese women: a case-control association study. Arthritis Res Ther. 2011;13(2):R41. doi: 10.1186/ar3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.dos Santos BP, et al. TLR7/8/9 polymorphisms and their associations in systemic lupus erythematosus patients from southern Brazil. Lupus. 2012;21(3):302–309. doi: 10.1177/0961203311425522. [DOI] [PubMed] [Google Scholar]
- 10.Niewold TB, Kelly JA, Flesch MH, Espinoza LR, Harley JB, Crow MK. Association of the IRF5 risk haplotype with high serum interferon-alpha activity in systemic lupus erythematosus patients. Arthritis Rheum. 2008;58(8):2481–2487. doi: 10.1002/art.23613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abelson AK, et al. STAT4 associates with systemic lupus erythematosus through two independent effects that correlate with gene expression and act additively with IRF5 to increase risk. Ann Rheum Dis. 2009;68(11):1746–1753. doi: 10.1136/ard.2008.097642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kariuki SN, Kirou KA, MacDermott EJ, Barillas-Arias L, Crow MK, Niewold TB. Cutting edge: autoimmune disease risk variant of STAT4 confers increased sensitivity to IFN-alpha in lupus patients in vivo. J Immunol. 2009;182(1):34–38. doi: 10.4049/jimmunol.182.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacob CO, et al. Identification of IRAK1 as a risk gene with critical role in the pathogenesis of systemic lupus erythematosus. Proc Natl Acad Sci USA. 2009;106(15):6256–6261. doi: 10.1073/pnas.0901181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhai Y, et al. Association of interleukin-1 receptor-associated kinase (IRAK1) gene polymorphisms (rs3027898, rs1059702) with systemic lupus erythematosus in a Chinese Han population. Inflamm Res. 2013;62(6):555–560. doi: 10.1007/s00011-013-0607-2. [DOI] [PubMed] [Google Scholar]
- 15.Adrianto I, et al. Association of a functional variant downstream of TNFAIP3 with systemic lupus erythematosus. Nat Genet. 2011;43(3):253–258. doi: 10.1038/ng.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Musone SL, et al. Multiple polymorphisms in the TNFAIP3 region are independently associated with systemic lupus erythematosus. Nat Genet. 2008;40(9):1062–1064. doi: 10.1038/ng.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Radanova M, Vasilev V, Dimitrov T, Deliyska B, Ikonomov V, Ivanova D. Association of rs172378 C1q gene cluster polymorphism with lupus nephritis in Bulgarian patients. Lupus. 2015;24(3):280–289. doi: 10.1177/0961203314555173. [DOI] [PubMed] [Google Scholar]
- 18.Karassa FB, Trikalinos TA, Ioannidis JP, FcgammaRIIa-SLE Meta-Analysis Investigators Role of the Fcgamma receptor IIa polymorphism in susceptibility to systemic lupus erythematosus and lupus nephritis: a meta-analysis. Arthritis Rheum. 2002;46(6):1563–1571. doi: 10.1002/art.10306. [DOI] [PubMed] [Google Scholar]
- 19.Jönsen A, et al. Association between SLE nephritis and polymorphic variants of the CRP and FcgammaRIIIa genes. Rheumatology (Oxford) 2007;46(9):1417–1421. doi: 10.1093/rheumatology/kem167. [DOI] [PubMed] [Google Scholar]
- 20.Li X, et al. A novel polymorphism in the Fcgamma receptor IIB (CD32B) transmembrane region alters receptor signaling. Arthritis Rheum. 2003;48(11):3242–3252. doi: 10.1002/art.11313. [DOI] [PubMed] [Google Scholar]
- 21.Chen JY, et al. Association of a transmembrane polymorphism of Fcgamma receptor IIb (FCGR2B) with systemic lupus erythematosus in Taiwanese patients. Arthritis Rheum. 2006;54(12):3908–3917. doi: 10.1002/art.22220. [DOI] [PubMed] [Google Scholar]
- 22.Russell AI, et al. Polymorphism at the C-reactive protein locus influences gene expression and predisposes to systemic lupus erythematosus. Hum Mol Genet. 2004;13(1):137–147. doi: 10.1093/hmg/ddh021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan Y, Li LH, Pan HF, Tao JH, Sun ZQ, Ye DQ. Association of ITGAM polymorphism with systemic lupus erythematosus: a meta-analysis. J Eur Acad Dermatol Venereol. 2011;25(3):271–275. doi: 10.1111/j.1468-3083.2010.03776.x. [DOI] [PubMed] [Google Scholar]
- 24.Toller-Kawahisa JE, et al. The variant of CD11b, rs1143679 within ITGAM, is associated with systemic lupus erythematosus and clinical manifestations in Brazilian patients. Hum Immunol. 2014;75(2):119–123. doi: 10.1016/j.humimm.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 25.Wu H, et al. Association analysis of the R620W polymorphism of protein tyrosine phosphatase PTPN22 in systemic lupus erythematosus families: increased T allele frequency in systemic lupus erythematosus patients with autoimmune thyroid disease. Arthritis Rheum. 2005;52(8):2396–2402. doi: 10.1002/art.21223. [DOI] [PubMed] [Google Scholar]
- 26.Liu JL, et al. Association between the PD1.3A/G polymorphism of the PDCD1 gene and systemic lupus erythematosus in European populations: a meta-analysis. J Eur Acad Dermatol Venereol. 2009;23(4):425–432. doi: 10.1111/j.1468-3083.2009.03087.x. [DOI] [PubMed] [Google Scholar]
- 27.Lu R, et al. Genetic associations of LYN with systemic lupus erythematosus. Genes Immun. 2009;10(5):397–403. doi: 10.1038/gene.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hom G, et al. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N Engl J Med. 2008;358(9):900–909. doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- 29.Gateva V, et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat Genet. 2009;41(11):1228–1233. doi: 10.1038/ng.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han JW, et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet. 2009;41(11):1234–1237. doi: 10.1038/ng.472. [DOI] [PubMed] [Google Scholar]
- 31.Zhou XJ, et al. Genetic association of PRDM1-ATG5 intergenic region and autophagy with systemic lupus erythematosus in a Chinese population. Ann Rheum Dis. 2011;70(7):1330–1337. doi: 10.1136/ard.2010.140111. [DOI] [PubMed] [Google Scholar]
- 32.Kim SJ, Zou YR, Goldstein J, Reizis B, Diamond B. Tolerogenic function of Blimp-1 in dendritic cells. J Exp Med. 2011;208(11):2193–2199. doi: 10.1084/jem.20110658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim SJ, Gregersen PK, Diamond B. Regulation of dendritic cell activation by microRNA let-7c and BLIMP1. J Clin Invest. 2013;123(2):823–833. doi: 10.1172/JCI64712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen CY, Chang IS, Hsiung CA, Wasserman WW. On the identification of potential regulatory variants within genome wide association candidate SNP sets. BMC Med Genomics. 2014;7:34. doi: 10.1186/1755-8794-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li G, Pan T, Guo D, Li LC. Regulatory Variants and Disease: The E-Cadherin -160C/A SNP as an Example. Mol Biol Int. 2014;2014:967565. doi: 10.1155/2014/967565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feinberg MW, et al. The Kruppel-like factor KLF4 is a critical regulator of monocyte differentiation. EMBO J. 2007;26(18):4138–4148. doi: 10.1038/sj.emboj.7601824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saifudeen Z, Dipp S, Fan H, El-Dahr SS. Combinatorial control of the bradykinin B2 receptor promoter by p53, CREB, KLF-4, and CBP: implications for terminal nephron differentiation. Am J Physiol Renal Physiol. 2005;288(5):F899–F909. doi: 10.1152/ajprenal.00370.2004. [DOI] [PubMed] [Google Scholar]
- 38.Hu D, Zhou Z, Davidson NE, Huang Y, Wan Y. Novel insight into KLF4 proteolytic regulation in estrogen receptor signaling and breast carcinogenesis. J Biol Chem. 2012;287(17):13584–13597. doi: 10.1074/jbc.M112.343566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tetreault MP, Yang Y, Katz JP. Krüppel-like factors in cancer. Nat Rev Cancer. 2013;13(10):701–713. doi: 10.1038/nrc3582. [DOI] [PubMed] [Google Scholar]
- 40.Evans PM, Zhang W, Chen X, Yang J, Bhakat KK, Liu C. Kruppel-like factor 4 is acetylated by p300 and regulates gene transcription via modulation of histone acetylation. J Biol Chem. 2007;282(47):33994–34002. doi: 10.1074/jbc.M701847200. [DOI] [PubMed] [Google Scholar]
- 41.Ai W, Zheng H, Yang X, Liu Y, Wang TC. Tip60 functions as a potential corepressor of KLF4 in regulation of HDC promoter activity. Nucleic Acids Res. 2007;35(18):6137–6149. doi: 10.1093/nar/gkm656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahn MY, et al. Histone deacetylase inhibitor, apicidin, inhibits human ovarian cancer cell migration via class II histone deacetylase 4 silencing. Cancer Lett. 2012;325(2):189–199. doi: 10.1016/j.canlet.2012.06.017. [DOI] [PubMed] [Google Scholar]
- 43.Caraux A, et al. Circulating human B and plasma cells. Age-associated changes in counts and detailed characterization of circulating normal CD138- and CD138+ plasma cells. Haematologica. 2010;95(6):1016–1020. doi: 10.3324/haematol.2009.018689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.International Consortium for Systemic Lupus Erythematosus Genetics (SLEGEN) et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40(2):204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scheinecker C, Zwölfer B, Köller M, Männer G, Smolen JS. Alterations of dendritic cells in systemic lupus erythematosus: phenotypic and functional deficiencies. Arthritis Rheum. 2001;44(4):856–865. doi: 10.1002/1529-0131(200104)44:4<856::AID-ANR142>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 46.Jin O, et al. Systemic lupus erythematosus patients have increased number of circulating plasmacytoid dendritic cells, but decreased myeloid dendritic cells with deficient CD83 expression. Lupus. 2008;17(7):654–662. doi: 10.1177/0961203308089410. [DOI] [PubMed] [Google Scholar]
- 47.Ding D, Mehta H, McCune WJ, Kaplan MJ. Aberrant phenotype and function of myeloid dendritic cells in systemic lupus erythematosus. J Immunol. 2006;177(9):5878–5889. doi: 10.4049/jimmunol.177.9.5878. [DOI] [PubMed] [Google Scholar]
- 48.Decker P, Kötter I, Klein R, Berner B, Rammensee HG. Monocyte-derived dendritic cells over-express CD86 in patients with systemic lupus erythematosus. Rheumatology (Oxford) 2006;45(9):1087–1095. doi: 10.1093/rheumatology/kel061. [DOI] [PubMed] [Google Scholar]
- 49.Yao L, Tak YG, Berman BP, Farnham PJ. Functional annotation of colon cancer risk SNPs. Nat Commun. 2014;5:5114. doi: 10.1038/ncomms6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Turner AW, et al. Functional Analysis of a Novel Genome-Wide Association Study Signal in SMAD3 That Confers Protection From Coronary Artery Disease. Arterioscler Thromb Vasc Biol. 2016;36(5):972–983. doi: 10.1161/ATVBAHA.116.307294. [DOI] [PubMed] [Google Scholar]
- 51.Heintzman ND, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459(7243):108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Geiman DE, Ton-That H, Johnson JM, Yang VW. Transactivation and growth suppression by the gut-enriched Krüppel-like factor (Krüppel-like factor 4) are dependent on acidic amino acid residues and protein-protein interaction. Nucleic Acids Res. 2000;28(5):1106–1113. doi: 10.1093/nar/28.5.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tussiwand R, et al. Klf4 expression in conventional dendritic cells is required for T helper 2 cell responses. Immunity. 2015;42(5):916–928. doi: 10.1016/j.immuni.2015.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weinreich MA, Takada K, Skon C, Reiner SL, Jameson SC, Hogquist KA. KLF2 transcription-factor deficiency in T cells results in unrestrained cytokine production and upregulation of bystander chemokine receptors. Immunity. 2009;31(1):122–130. doi: 10.1016/j.immuni.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klaewsongkram J, Yang Y, Golech S, Katz J, Kaestner KH, Weng NP. Krüppel-like factor 4 regulates B cell number and activation-induced B cell proliferation. J Immunol. 2007;179(7):4679–4684. doi: 10.4049/jimmunol.179.7.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yet SF, et al. Human EZF, a Krüppel-like zinc finger protein, is expressed in vascular endothelial cells and contains transcriptional activation and repression domains. J Biol Chem. 1998;273(2):1026–1031. doi: 10.1074/jbc.273.2.1026. [DOI] [PubMed] [Google Scholar]
- 57.Alder JK, et al. Kruppel-like factor 4 is essential for inflammatory monocyte differentiation in vivo. J Immunol. 2008;180(8):5645–5652. doi: 10.4049/jimmunol.180.8.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Feinberg MW, Cao Z, Wara AK, Lebedeva MA, Senbanerjee S, Jain MK. Kruppel-like factor 4 is a mediator of proinflammatory signaling in macrophages. J Biol Chem. 2005;280(46):38247–38258. doi: 10.1074/jbc.M509378200. [DOI] [PubMed] [Google Scholar]
- 59.Noti JD, Johnson AK, Dillon JD. The leukocyte integrin gene CD11d is repressed by gut-enriched Kruppel-like factor 4 in myeloid cells. J Biol Chem. 2005;280(5):3449–3457. doi: 10.1074/jbc.M412627200. [DOI] [PubMed] [Google Scholar]
- 60.Wei X, Xu H, Kufe D. Human mucin 1 oncoprotein represses transcription of the p53 tumor suppressor gene. Cancer Res. 2007;67(4):1853–1858. doi: 10.1158/0008-5472.CAN-06-3063. [DOI] [PubMed] [Google Scholar]
- 61.Zhang W, et al. Novel cross talk of Kruppel-like factor 4 and beta-catenin regulates normal intestinal homeostasis and tumor repression. Mol Cell Biol. 2006;26(6):2055–2064. doi: 10.1128/MCB.26.6.2055-2064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim S, Elkon KB, Ma X. Transcriptional suppression of interleukin-12 gene expression following phagocytosis of apoptotic cells. Immunity. 2004;21(5):643–653. doi: 10.1016/j.immuni.2004.09.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.