Abstract
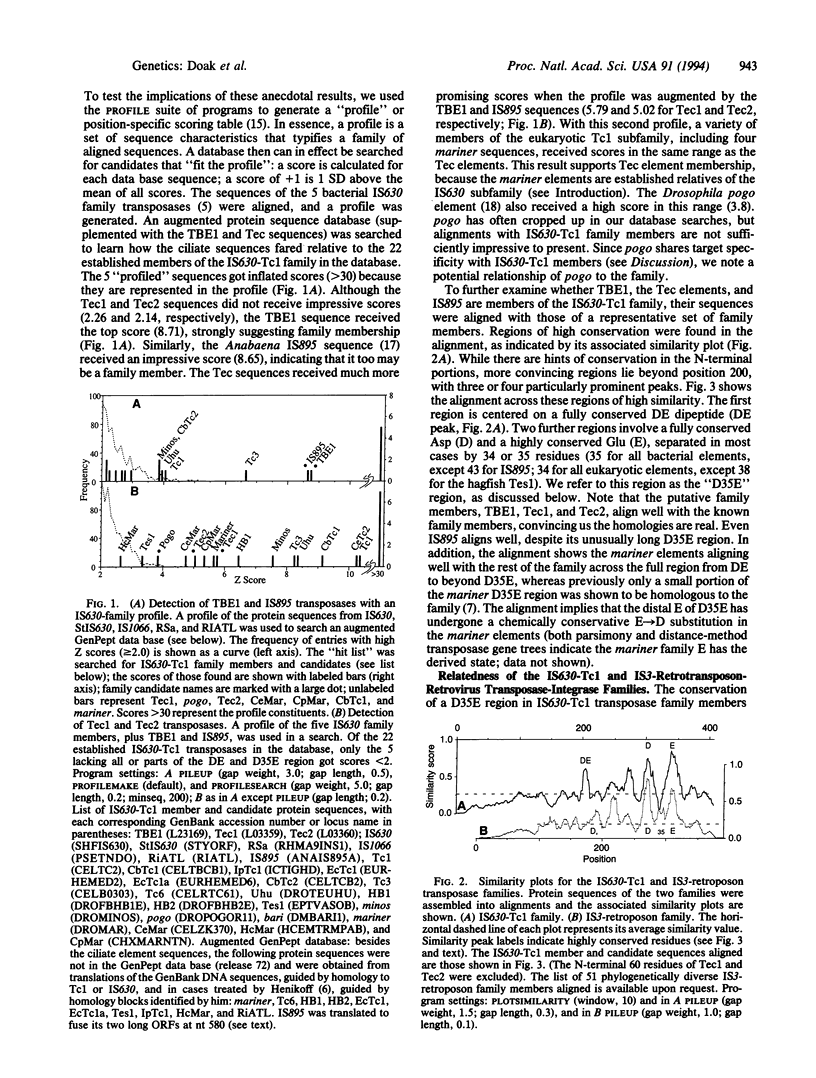
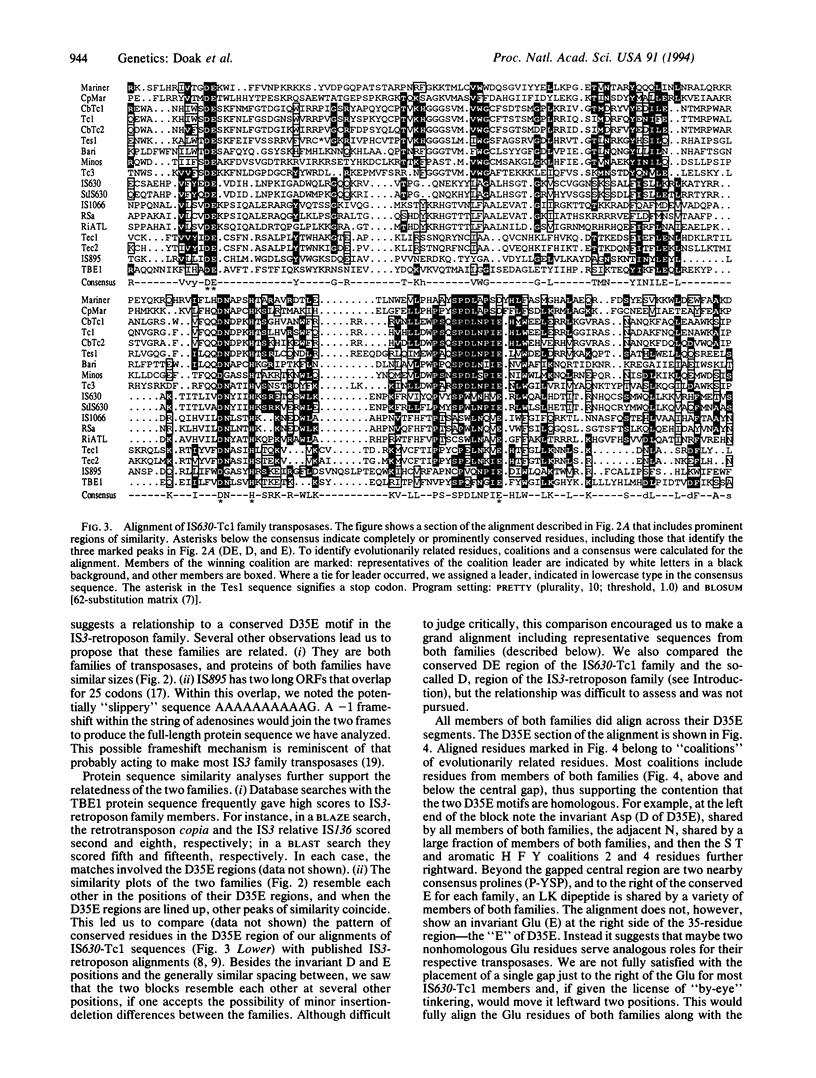
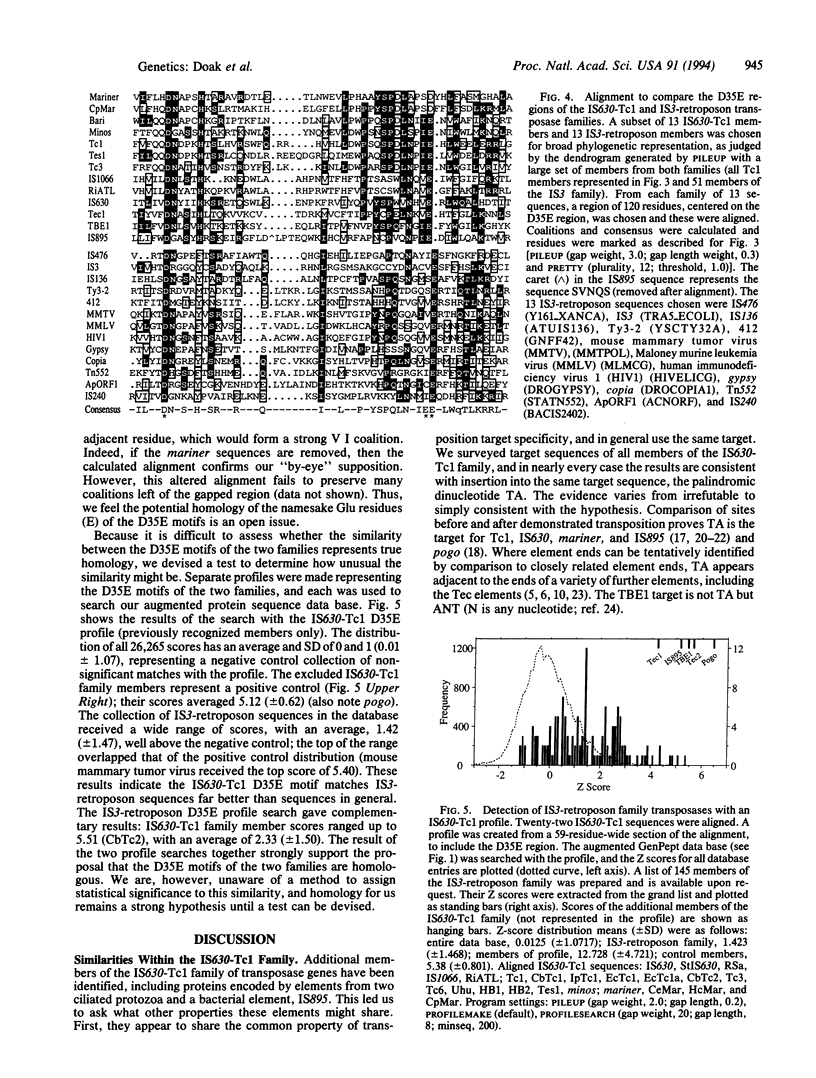
The transposon-like elements TBE1, Tec1, and Tec2 of hypotrichous ciliated protozoa appear to encode a protein that belongs to the IS630-Tc1 family of transposases. The Anabaena IS895 transposase also is placed in this family. We note that most family members transpose into the dinucleotide target, TA, and that members with eukaryotic hosts have a tendency for somatic excision that is carried to an extreme by the ciliate elements. Alignments including the additional members, and also mariner elements, show that transposases of this family share strongly conserved residues in a large C-terminal portion, including a fully conserved dipeptide, Asp-Glu (DE), and a block consisting of a fully conserved Asp and highly conserved Glu, separated by 34 or 35 residues (D35E). This D35E motif likely is homologous to the previously characterized D35E motif of the family of retroviral-retrotransposon integrases and IS3-like transposases. Because it is known that the IS3-retroposon D35E region is a critical portion of a domain capable of various in vitro transposition-related reactions, the results suggest that the two families share homologous catalytic transposase domains and that members of both families may share a common transposition mechanism.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alam J., Vrba J. M., Cai Y., Martin J. A., Weislo L. J., Curtis S. E. Characterization of the IS895 family of insertion sequences from the cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol. 1991 Sep;173(18):5778–5783. doi: 10.1128/jb.173.18.5778-5783.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bryan G., Garza D., Hartl D. Insertion and excision of the transposable element mariner in Drosophila. Genetics. 1990 May;125(1):103–114. doi: 10.1093/genetics/125.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman F. D., Engelman A., Palmer I., Wingfield P., Craigie R. Domains of the integrase protein of human immunodeficiency virus type 1 responsible for polynucleotidyl transfer and zinc binding. Proc Natl Acad Sci U S A. 1993 Apr 15;90(8):3428–3432. doi: 10.1073/pnas.90.8.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caizzi R., Caggese C., Pimpinelli S. Bari-1, a new transposon-like family in Drosophila melanogaster with a unique heterochromatic organization. Genetics. 1993 Feb;133(2):335–345. doi: 10.1093/genetics/133.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler M., Fayet O. Translational frameshifting in the control of transposition in bacteria. Mol Microbiol. 1993 Feb;7(4):497–503. doi: 10.1111/j.1365-2958.1993.tb01140.x. [DOI] [PubMed] [Google Scholar]
- Dreyfus D. H. Evidence suggesting an evolutionary relationship between transposable elements and immune system recombination sequences. Mol Immunol. 1992 Jun;29(6):807–810. doi: 10.1016/0161-5890(92)90191-y. [DOI] [PubMed] [Google Scholar]
- Engelman A., Craigie R. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J Virol. 1992 Nov;66(11):6361–6369. doi: 10.1128/jvi.66.11.6361-6369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayet O., Ramond P., Polard P., Prère M. F., Chandler M. Functional similarities between retroviruses and the IS3 family of bacterial insertion sequences? Mol Microbiol. 1990 Oct;4(10):1771–1777. doi: 10.1111/j.1365-2958.1990.tb00555.x. [DOI] [PubMed] [Google Scholar]
- Gribskov M., Lüthy R., Eisenberg D. Profile analysis. Methods Enzymol. 1990;183:146–159. doi: 10.1016/0076-6879(90)83011-w. [DOI] [PubMed] [Google Scholar]
- Harris L. J., Baillie D. L., Rose A. M. Sequence identity between an inverted repeat family of transposable elements in Drosophila and Caenorhabditis. Nucleic Acids Res. 1988 Jul 11;16(13):5991–5998. doi: 10.1093/nar/16.13.5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Detection of Caenorhabditis transposon homologs in diverse organisms. New Biol. 1992 Apr;4(4):382–388. [PubMed] [Google Scholar]
- Henikoff S., Henikoff J. G. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrick G., Cartinhour S., Dawson D., Ang D., Sheets R., Lee A., Williams K. Mobile elements bounded by C4A4 telomeric repeats in Oxytricha fallax. Cell. 1985 Dec;43(3 Pt 2):759–768. doi: 10.1016/0092-8674(85)90249-1. [DOI] [PubMed] [Google Scholar]
- Jahn C. L., Doktor S. Z., Frels J. S., Jaraczewski J. W., Krikau M. F. Structures of the Euplotes crassus Tec1 and Tec2 elements: identification of putative transposase coding regions. Gene. 1993 Oct 29;133(1):71–78. doi: 10.1016/0378-1119(93)90226-s. [DOI] [PubMed] [Google Scholar]
- Khan E., Mack J. P., Katz R. A., Kulkosky J., Skalka A. M. Retroviral integrase domains: DNA binding and the recognition of LTR sequences. Nucleic Acids Res. 1991 Feb 25;19(4):851–860. doi: 10.1093/nar/19.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klobutcher L. A., Jahn C. L. Developmentally controlled genomic rearrangements in ciliated protozoa. Curr Opin Genet Dev. 1991 Oct;1(3):397–403. doi: 10.1016/s0959-437x(05)80306-5. [DOI] [PubMed] [Google Scholar]
- Kulkosky J., Jones K. S., Katz R. A., Mack J. P., Skalka A. M. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol Cell Biol. 1992 May;12(5):2331–2338. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuuchi K. Transpositional recombination: mechanistic insights from studies of mu and other elements. Annu Rev Biochem. 1992;61:1011–1051. doi: 10.1146/annurev.bi.61.070192.005051. [DOI] [PubMed] [Google Scholar]
- Prère M. F., Chandler M., Fayet O. Transposition in Shigella dysenteriae: isolation and analysis of IS911, a new member of the IS3 group of insertion sequences. J Bacteriol. 1990 Jul;172(7):4090–4099. doi: 10.1128/jb.172.7.4090-4099.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson H. M. The mariner transposable element is widespread in insects. Nature. 1993 Mar 18;362(6417):241–245. doi: 10.1038/362241a0. [DOI] [PubMed] [Google Scholar]
- Rosenzweig B., Liao L. W., Hirsh D. Target sequences for the C. elegans transposable element Tc1. Nucleic Acids Res. 1983 Oct 25;11(20):7137–7140. doi: 10.1093/nar/11.20.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schukkink R. F., Plasterk R. H. TcA, the putative transposase of the C. elegans Tc1 transposon, has an N-terminal DNA binding domain. Nucleic Acids Res. 1990 Feb 25;18(4):895–900. doi: 10.1093/nar/18.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenzen T., Matsutani S., Ohtsubo E. Site-specific transposition of insertion sequence IS630. J Bacteriol. 1990 Jul;172(7):3830–3836. doi: 10.1128/jb.172.7.3830-3836.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tudor M., Lobocka M., Goodell M., Pettitt J., O'Hare K. The pogo transposable element family of Drosophila melanogaster. Mol Gen Genet. 1992 Mar;232(1):126–134. doi: 10.1007/BF00299145. [DOI] [PubMed] [Google Scholar]
- Vincent K. A., Ellison V., Chow S. A., Brown P. O. Characterization of human immunodeficiency virus type 1 integrase expressed in Escherichia coli and analysis of variants with amino-terminal mutations. J Virol. 1993 Jan;67(1):425–437. doi: 10.1128/jvi.67.1.425-437.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink C., Oude Groeneger A. M., Plasterk R. H. Identification of the catalytic and DNA-binding region of the human immunodeficiency virus type I integrase protein. Nucleic Acids Res. 1993 Mar 25;21(6):1419–1425. doi: 10.1093/nar/21.6.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K. R., Herrick G. Expression of the gene encoded by a family of macronuclear chromosomes generated by alternative DNA processing in Oxytricha fallax. Nucleic Acids Res. 1991 Sep 11;19(17):4717–4724. doi: 10.1093/nar/19.17.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K., Doak T. G., Herrick G. Developmental precise excision of Oxytricha trifallax telomere-bearing elements and formation of circles closed by a copy of the flanking target duplication. EMBO J. 1993 Dec;12(12):4593–4601. doi: 10.1002/j.1460-2075.1993.tb06148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gent D. C., Groeneger A. A., Plasterk R. H. Mutational analysis of the integrase protein of human immunodeficiency virus type 2. Proc Natl Acad Sci U S A. 1992 Oct 15;89(20):9598–9602. doi: 10.1073/pnas.89.20.9598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer J. R., Zehnder A. J., de Vos W. M. Identification of a novel composite transposable element, Tn5280, carrying chlorobenzene dioxygenase genes of Pseudomonas sp. strain P51. J Bacteriol. 1991 Nov;173(22):7077–7083. doi: 10.1128/jb.173.22.7077-7083.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]


