Abstract
Approximately 50% of high-grade serous ovarian cancers (HGSOCs) have defects in genes involved in homologous recombination (HR) (i.e., BRCA1/2). Preclinical models to optimize therapeutic strategies for HR-deficient (HRD) HGSOC are lacking. We developed a preclinical platform for HRD HGSOCs that includes primary tumor cultures, patient-derived xenografts (PDXs), and molecular imaging. Models were characterized by immunohistochemistry, targeted sequencing, and reverse-phase protein array analysis. We also tested PDX tumor response to PARP, CHK1, and ATR inhibitors. Fourteen orthotopic HGSOC PDX models with BRCA mutations (BRCAMUT) were established with a 93% success rate. The orthotopic PDX model emulates the natural progression of HGSOC, including development of a primary ovarian tumor and metastasis to abdominal viscera. PDX response to standard chemotherapy correlated to that demonstrated in the patient. Pathogenic mutations and HGSOC markers were preserved after multiple mouse passages, indicating retention of underlying molecular mechanisms of carcinogenesis. A BRCA2MUT PDX with high p-CHK1 demonstrated a similar delay of tumor growth in response to PARP, CHK1, and ATR inhibitors. A poly (ADP-ribose) polymerase (PARP) inhibitor radiotracer correlated with PARP1 activity and showed response to PARP inhibition in the BRCA2MUT PDX model. In summary, the orthotopic HGSOC PDX represents a robust and reliable model to optimize therapeutic strategies for BRCAMUT HGSOC.
An orthotopic ovarian patient-derived-xenograft model recapitulates the natural progression of ovarian cancer.
Introduction
High-grade serous ovarian cancer (HGSOC) remains the fifth leading cause of cancer death in women in the U.S., with over 14,000 deaths estimated for 2016 (1). More than 80% of women present in advanced stages and 80% will develop recurrent disease despite aggressive surgical removal of tumor and initial high response rates to standard-of-care frontline platinum/taxane chemotherapy (2–4). Therefore, a critical need remains to develop new effective therapeutic strategies that exploit the unique biology and genetics of HGSOC. There has been a lack of molecularly characterized preclinical models maintaining fidelity of the original tumor in homologous recombination–deficient (HRD) HGSOC (5).
There are limitations to traditional experimental models used for HGSOC research. Xenograft studies to determine which drugs warrant testing in clinical trials have traditionally been performed using established cell lines. However, established cell lines change after multiple passages, are selected for their ability to grow in tissue culture, and often do not genomically resemble HGSOC (5–7). In addition, the most frequently used subcutaneous xenograft model does not recapitulate the native ovarian microenvironment (8). Indeed, recent studies suggest that most, if not all, HGSOCs arise in the fallopian tube fimbria, with the ovarian and peritoneal implants representing metastasis from the original site (9–11).
Patient-derived xenografts (PDXs) maintain the characteristics of the patients’ original tumor including histology, mutational status, gene expression, and clinical behavior while remaining biologically stable through multiple passages in mice (12–14). They represent an evolution of the cell line xenograft model in which fresh tumor tissue is obtained directly from patients and subsequently implanted orthotopically into immunodeficient mice (15, 16). PDXs allow for harvesting and re-implantation in successive mice generations serving as a biorepository for each patient (17). Given that PDXs maintain the principle characteristics of the patients’ original tumor (16), they serve as reliable models to study therapeutic response and resistance.
Poly (ADP-ribose) polymerase (PARP) inhibitors block the repair of single-strand DNA breaks. This leads to double-strand DNA breaks which cannot be repaired effectively in BRCA-mutant (BRCAMUT) cancers, capitalizing on synthetic lethality (18, 19). Given that ~50% of HGSOCs have defects in homologous recombination (HR) (i.e., BRCA1/2MUT) (20), PARP inhibitors are rational therapies for this subset of HGSOC. One PARP inhibitor (PARPi), olaparib (AZD2281), resulted in an overall response rate of ~35% in heavily pretreated patients, which led to FDA approval for recurrent HGSOC germline BRCAMUT carriers (21). Unfortunately, clinical responses are limited and nondurable; therefore, alternative therapeutic strategies are urgently needed.
Cell cycle checkpoint signaling inhibitors are currently being explored in HGSOC. Ataxia telangiectasia and Rad3-related (ATR) and its downstream kinase checkpoint kinase 1 (CHK1), activated by DNA replication stress and DNA double-strand breaks to prevent cell cycle progression, permit the completion of DNA replication and DNA repair processes (22, 23). Cells with increased levels of oncogenic stress (i.e., cyclin E and Myc overexpression, RAS mutation), p53 deficiency, and loss of specific DNA repair proteins, are especially sensitive to ATR inhibition (24–29). Drugs targeting ATR (i.e., AZD6738 and VX-970) and CHK1 (i.e., MK8776, SCH 900776, LY2606368, and CCT245737) are in early phase I/II clinical trial development (https://clinicaltrials.gov/).
Herein, we present a robust and reliable BRCA-deficient platform for optimization of therapeutics for HRD HGSOC. Our models represent true orthotopic PDX cancers whereby tumor tissue (undisrupted) is transplanted onto the mouse fallopian tube fimbria/ovary (representing the most likely site of origin of HGSOC. The transplanted tumor tissue then develops into a primary ovarian tumor in ~95% of cases with subsequent metastasis to abdominal viscera, mimicking the natural progression of HGSOC. Response to standard chemotherapy, histology and morphologic patterns, and genomic signatures are maintained in the PDX tumors from the parent tumor and remain stable when passaged in mice. Finally, we illustrate how such a model can be used as an experimental platform to investigate targeting the ATR-CHK1 axis as an alternative approach to capitalize on synthetic lethality in BRCAMUT HGSOC.
Results
A potentially novel BRCAMUT orthotopic ovarian PDX platform mimics the natural progression of HGSOC.
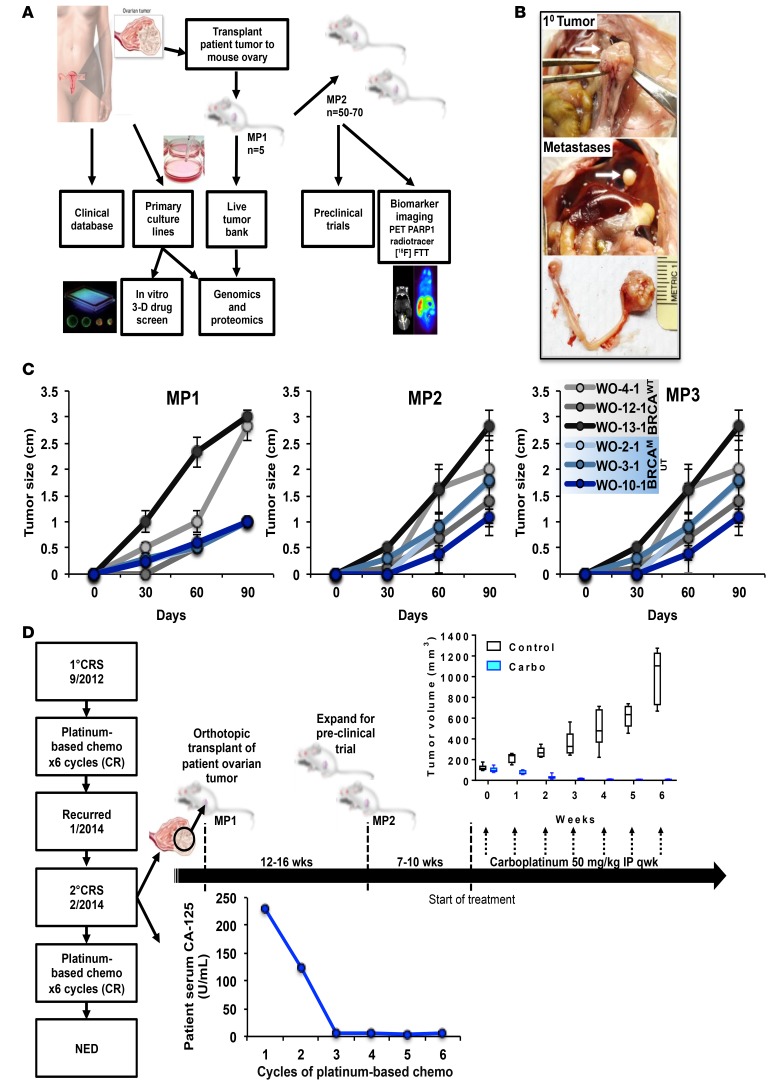
The schema for our preclinical platform is illustrated in Figure 1A. Patient tumor at the time of tumor debulking surgery (removal of all possible visible tumor) was removed to develop both primary cell cultures and PDX models for that patient. Fresh, undigested tumor chunks (~2 × 2 × 2 mm) were transplanted onto the fallopian tube fimbria/ovary of 5 NSG mice (Supplemental Figure 1A; supplemental material available online with this article; doi:10.1172/jci.insight.89760DS1). PDX tumors (mouse passage 1 [MP1]) were then passaged and expanded to perform preclinical trials or banked for future studies. Drug studies used ultrasound to provide an accurate assessment of tumor volume over time (Supplemental Figure 1B). Studies evaluating PARP inhibitors were accompanied by molecular imaging using the PARPi analogue tracer [18F]FluorThanatrace ([18F]FTT), which binds to PARP1, serving as a functional biomarker (30, 31). At the same time tumors can be grown in vitro in 2D and 3D cultures and used for drug screens (not shown). PDXs and primary tumor cultures underwent histological, genomic, and proteomic analyses to identify potential therapeutic targets and streamline models for future studies. Cell and PDX models were analyzed using a custom 154 ovarian cancer gene panel (comprised of ovarian cancer and HR-related and actionable genes) using targeted sequencing of full genes or all exons (Supplemental Figure 2). Reverse phase protein array analysis (RPPA) evaluating 308 phosphorylated and total proteins was also used to correlate alterations at the gene and protein level (32) (Figure 1A).
Figure 1. A potentially novel BRCAMUT ovarian patient-derived-xenograft (PDX) platform.
(A) Patient tumors removed during surgery are transplanted onto the fallopian tube fimbria/ovaries of NSG mice for PDX development and grown in tissue culture. Mouse tumors are then used to establish a live tumor bank or expanded for preclinical trials. Models can also be used for functional biomarker studies. Targeted genomic sequencing and reverse phase protein array (RPPA) analysis is performed on all patient and PDX samples. Primary tumor cultures may be used for in vitro 2D and 3D drug screening. (B) Representative BRCAMUT PDX (c.8945delAA; WO-2-1) 10 weeks after transplantation showing primary tumor replacing host ovary (upper panel), diaphragmatic metastasis (middle panel), and primary ovarian tumor and normal mouse ovary (lower panel). (C) PDX tumor size by palpation over time for select PDX models comparing BRCAMUT (WO-2-1, WO-3-1, WO-10-1) with BRCAWT (WO-4-1, WO-12-1, WO-13-1) HGSOC over 3 mouse passages (MPs). BRCAMUT PDXs demonstrated slower growth rates compared with BRCAWT at MP1 (0.11 mm/day vs. 0.29 mm/day; P < 0.001). However, in subsequent passages MP2 and MP3, growth rates were similar (0.19 mm/day for BRCAMUT vs. 0.25 mm/day for BRCAWT; P = 0.08 for MP2, 0.177 mm/day for BRCAMUT vs. 0.18 mm/day for BRCAWT; P = 0.9 for MP3). Dots represent mean of determinations with SEM. A linear mixed-effects model was used to compare growth rates/day between groups. Individual mouse data shown in dot plots in Supplemental Figure 4. (D) Example of preclinical platform using a BRCA2MUT (c.8945delAA) PDX model (WO-2). The patient was diagnosed with ovarian cancer in 2012 and had primary cytoreductive surgery (1° CRS) followed by standard chemotherapy (carboplatin and paclitaxel) and had a complete remission (CR). Tumor obtained at secondary CRS for recurrent disease in January 2014 was used for PDX generation. PDX tumors grew in 12 to 16 weeks to approximately 1,000 mm3. Tumors were harvested and expanded for a preclinical trial. Mice were randomized to control or carboplatin 50 mg/kg weekly and tumor growth as measured by weekly ultrasound. All mice showed CR. Similarly, the patient’s tumor also responded to a platinum-based chemotherapy regimen as illustrated in the graph of her serum CA-125. The box-and-whisker plots show the median, with boxes extending from the 25th to 75th percentile and the whiskers extending from minimum to maximum values of the dataset. Control, n = 9 mice; carboplatin, n = 6 mice.
The orthotopic transplant approach, whereby undigested tumor chunks are transplanted directly onto the mouse fallopian tube fimbria/ovary, ultimately recapitulates the natural progression of ovarian cancer with development of a primary ovarian tumor (Figure 1B and Supplemental Figure 1A) and metastasis to the diaphragm (Figure 1B and Supplemental Figure 3). Growth rates for BRCAMUT PDXs (WO-2, -3, and -10) were then compared with wild-type (BRCAWT) PDXs (WO-4, -12, and -13) over 3 passages in mice. BRCAMUT PDXs demonstrated slower take rates compared with BRCAWT when first passaged in mice (patient tumor transplanted to mice ovary and tumor develops = MP1). However, subsequent passages (MP2, tumor from MP1 transplanted to mice and tumor forms; and MP3, tumor that develops from MP2) demonstrated similar growth rates (Figure 1C and Supplemental Figure 4).
Response to standard-of-care chemotherapy in the PDX was compared with that of the patient from which the PDX was derived (Figure 1D). A patient harboring a BRCA2 mutation (c.8945delAA) was initially diagnosed with HGSOC in 2012. She had primary cytoreductive surgery (1° CRS) followed by standard chemotherapy with a platinum-based regimen resulting in complete remission (CR). Her cancer recurred in January 2014 and at the time of her secondary tumor debulking surgery, tumor tissue was obtained for our preclinical platform (WO-2-1). Fresh, undigested tumor was transplanted and grew over the following 12–16 weeks, reaching maximum size. The tumor was then harvested and expanded for a preclinical trial. Mice were randomized into 2 arms (drug and control). One arm was treated with weekly i.p. carboplatin over 6 weeks, resulting in 100% complete response rate, which mimicked the patient’s response to a platinum-based regimen as illustrated by normalization of her serum CA-125 by cycle 3 of chemotherapy. The control group received vehicle only.
Patient tumor histology, morphology, and ovarian cancer markers are preserved in PDXs.
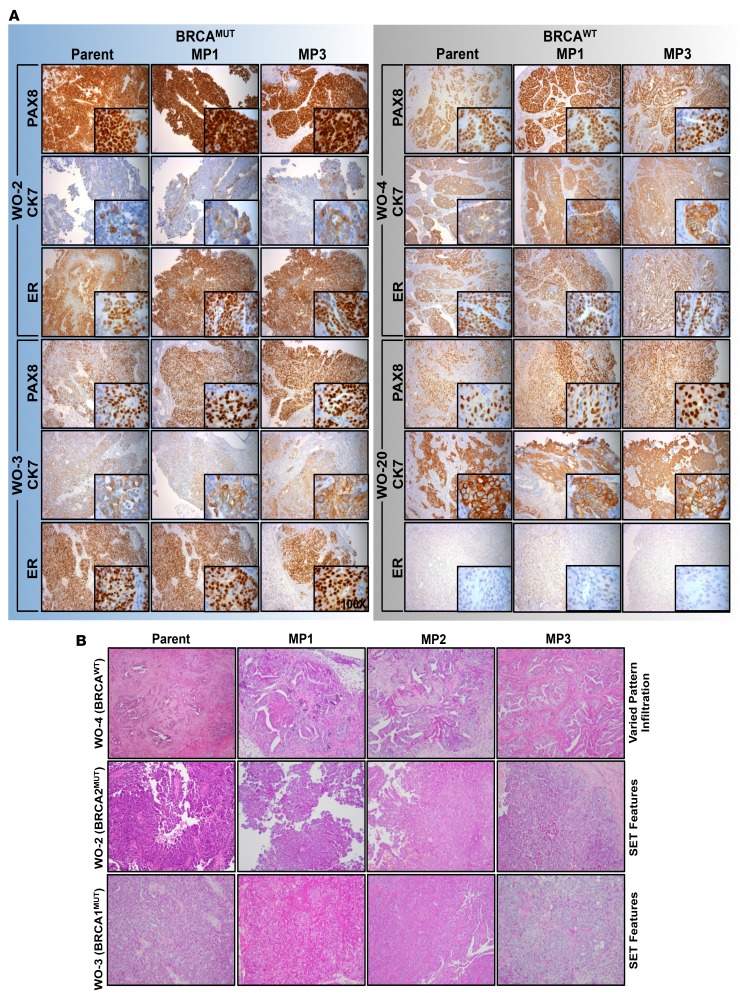
Epithelial HGSOC markers, PAX8, a nuclear stain, and CK7, a plasma membrane stain, were positive in all patient tumors (PAX8, n = 11 of 11; CK7, n = 10 of 10) and preserved over multiple mouse passages (MP1 through MP3) in all PDXs evaluated. PAX8 and CK7 immunohistological staining are illustrated for a BRCA1MUT (WO-3), BRCA2MUT (WO-2), and 2 BRCAWT (WO-4 and WO-20) patient and corresponding PDX models (Figure 2A and Supplemental Figure 5). Estrogen receptor (ER), a nuclear stain demonstrated in approximately 76% of HGSOC (33), was similarly expressed in 72.7% (n = 8 of 11) of patient tumors. ER positivity was defined as greater than 10% of cells demonstrating nuclear staining per field. All BRCAMUT patient tumors were ER+ (n = 6 of 6) and 40% (n = 2 of 5) BRCAWT patient tumors were ER+. ER status was preserved over MP1 to MP3 in 92% (n = 23 of 25) of samples assessed. BRCA1 protein expression was also evaluated and BRCA1WT tumors stained positive (n = 5), although staining varied from strong (WO-4, WO-19, and WO-20) to low (WO-13 and WO-24). BRCA1 protein expression was absent (WO-10 and WO-16) or very low (WO-3, WO-8, and WO-21) in BRCA1MUT patient samples and their matched PDXs (Supplemental Figure 6).
Figure 2. Ovarian cancer epithelial markers and morphologic characteristics in parent tumors are preserved over multiple mouse passages.
(A) BRCAMUT patient-derived-xenograft (PDX) models (WO-2, WO-3) and 2 BRCAWT PDX models (WO-4, WO-20) were evaluated by H&E and immunohistochemistry for epithelial ovarian cancer markers. PAX8 (paired box 8, nuclear stain), CK7 (cytokeratin 7, cytoplasmic stain), and ER (estrogen receptor, nuclear stain) in parent tumor and matched PDXs up to mouse passage 3 (MP1–MP3) are shown. Magnification is ×10 for large panels and ×100 for inserts. (B) Five matched BRCAMUT patient/PDXs and 5 matched BRCAWT patient /PDXs were reviewed for SET (solid, pseudoendometrioid, and translational cell carcinoma-like) morphology in a blinded fashion. WO-4 (BRCAWT), WO-2-1 (BRCA2MUT), and WO-3-1 (BRCA1MUT) representative parent and PDX tumors of MP1, MP2, and MP3 H&E sections shown demonstrating SET criteria in BRCAMUT and not in BRCAWT tumors. Parent WO-2-1 tumor showed micropapillary features, while the MP1–MP3 tumors showed both micropapillary and solid architecture. Magnification is ×10 for all panels.
Morphological characteristics that predict BRCAMUT status were then evaluated. BRCA1/2MUT HGSOCs have more frequent solid, pseudoendometrioid, and transitional cell carcinoma (SET features) (34) (Figure 2B). SET patterns were present in 60% (n = 3 of 5) of BRCAMUT parent tumors evaluated. Sixty-seven percent of (n = 2 of 3) of tumors having SET features were preserved from parent through MP3 (1 patient tumor did not have SET but MP1–3 did). No BRCAWT parent tumors or their PDXs had SET features.
Finally, we evaluated PDX tumors for human stroma content. Parent tumors were compared with PDX tumors at MP1 using a human-specific ALU probe labeled with fluorescein isothiocyanate (green nuclei) that detects Alu repeat sequences of human DNA (35). Colocalization of Alu and DAPI was demonstrated in the PDX stroma, suggesting the presence of human cells, although the majority of tumor stroma was composed of mouse cells (Supplemental Figure 7).
Genomic characterization of BRCAMUT PDXs.
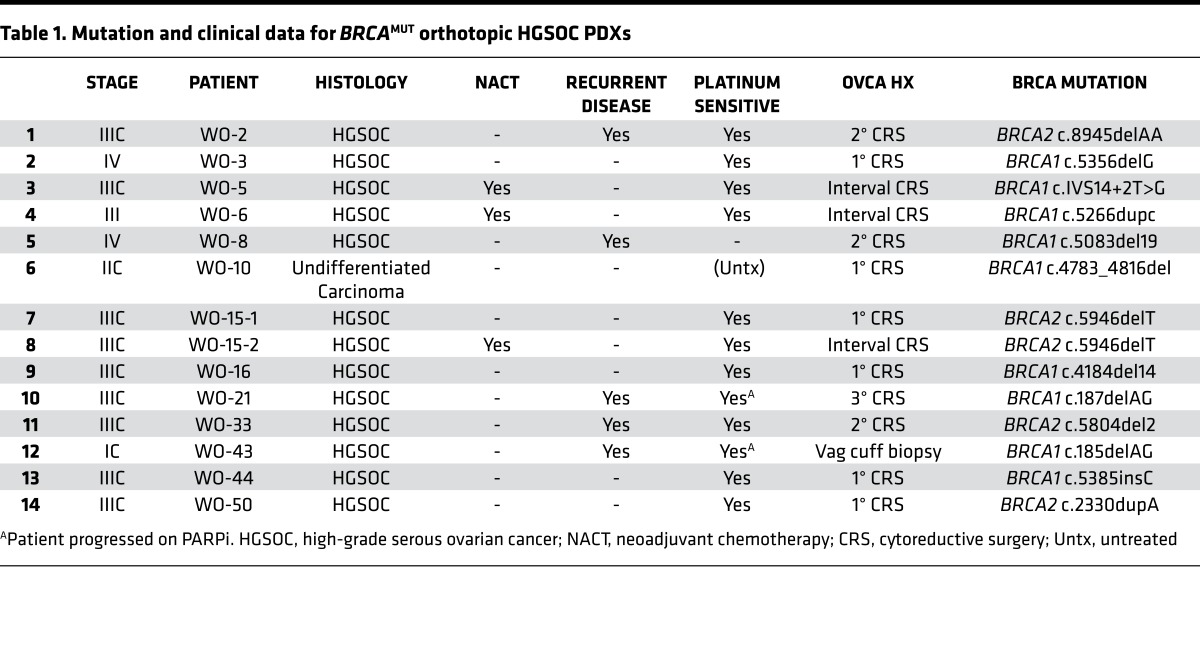
We have developed over 40 PDX models using an orthotopic transplant approach with a 93% success rate (n = 37 of 40), 100% for MP2 (15 of 15), and 100% for MP3 (7 of 7). Of these, 14 HGSOC PDXs have been generated from BRCA1/2 mutation carriers (Table 1 and Figure 3) with no substantive differences in success rate (n = 14 of 15). Palpable tumors developed within 4 to 6 weeks of transplant.
Table 1. Mutation and clinical data for BRCAMUT orthotopic HGSOC PDXs.
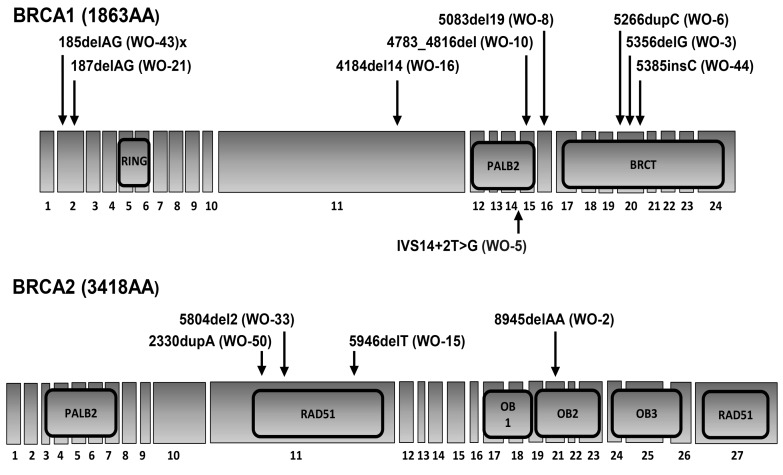
Figure 3. Illustration of the BRCA1 and BRCA2 genes, including exons, introns, and functional domains noting location of BRCA mutations in the patient-derived-xenograft (PDX) models.
RING, really interesting new gene; PALB2, partner and localizer of BRCA2; BRCT, BRCA1 C-terminal; OB, oligonucleotide/oligosaccharide-binding domain.
Of the 14 germline BRCAMUT HGSOC PDX models, 64% (n = 9 of 14) harbor a BRCA1 mutation and the remaining contain a BRCA2 mutation (Table 1). The majority of patients, 86% (n = 12 of 14), had advanced disease (stage IIIC–IV). Of the 64% (n = 9 of 14) of patients who presented with an initial disease occurrence, 33% (n = 3 of 9) received neoadjuvant chemotherapy (NACT) followed by interval CRS. Of the 36% (n = 5 of 14) of patients who presented with recurrent disease (3 for 2° CRS, 1 for 3° CRS, 1 for vaginal cuff biopsy), 1 was platinum resistant and 2 progressed on PARPi. One patient (WO-15) was first sampled at the time of her planned 1° CRS (WO-15-1); however, because of unresectable disease, she received NACT and was subsequently sampled again at the time of her interval CRS (WO-15-2) (Table 1). BRCA mutation gene locations on exons and introns and functional domains for each PDX are demonstrated (Figure 3).
Genomic pathways are preserved in original tumor compared with tumor passaged in PDXs.
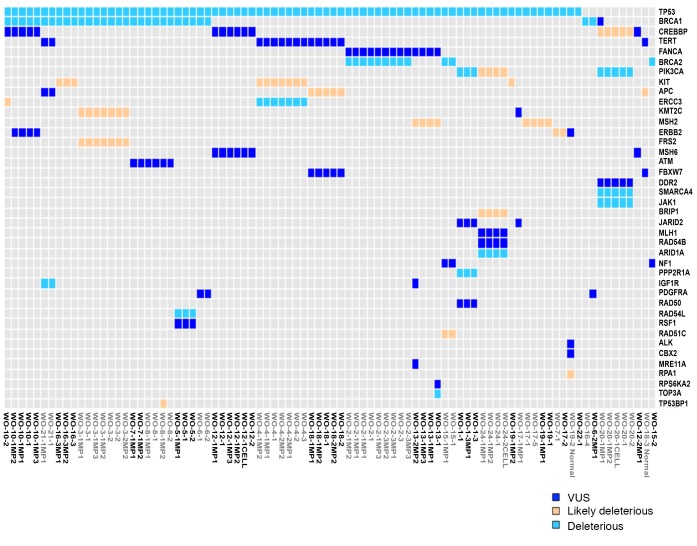
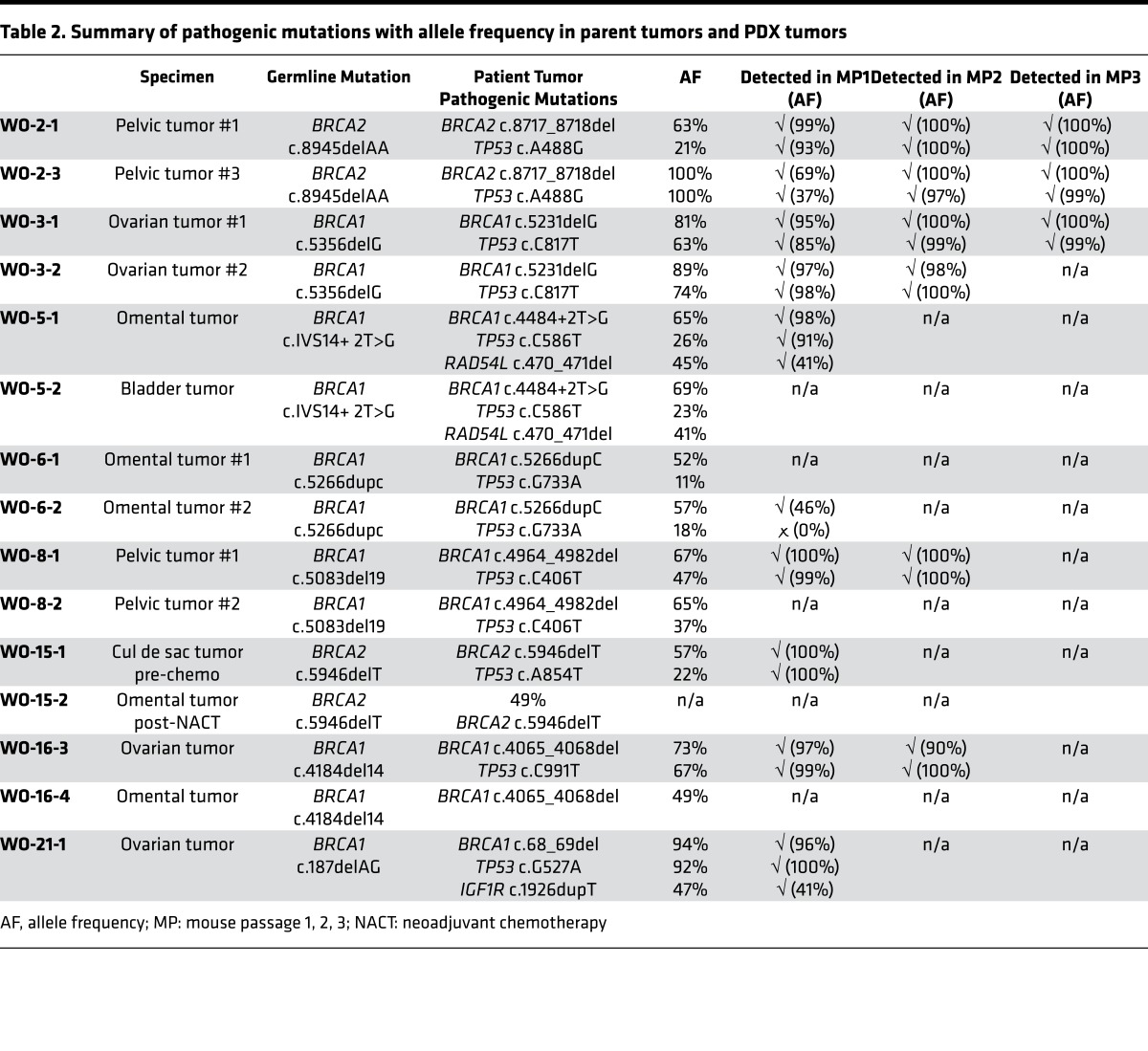
To find potential targetable gene mutations we developed a 157 ovarian cancer gene panel. This panel is comprised of all genes relevant to HGSOC including HR-related genes and actionable targets (Supplemental Figure 2). Twenty-one PDX models, matched with their parent tumor, underwent sequencing using the ovarian cancer panel (Figure 4). Few deleterious mutations were encountered overall, but the most common were TP53 and BRCA1/2. Other deleterious mutations included PIK3CA, ERCC3, SMARCA4, JAK1, ARID1A, PPP2R1A, IGF1R, RAD54L, and TOP3A. Likely deleterious mutations included CREBBP, PIK3CA, KIT, APC, KMT2C, MSH2, ERBB2, FRS2, BRIP1, RAD51C, RPA1, and TP53BP1. Variants of undetermined significance (VUS) are also shown (Figure 4). Deleterious mutations were preserved in the PDX model and in the original patient tumor in 94% of cases. Likely deleterious and VUS were preserved in 82% and 74% of cases, respectively. In particular, BRCAMUT PDX models up to MP3 were then compared with the parent tumor from which they were derived. Multiple tumor locations (ovary, omentum, and pelvic tumors) were analyzed per PDX model. Deleterious mutations present in the parent tumor were overall preserved through mouse passages (TP53, 92%; BRCA1, 100%; BRCA2, 100%, RAD54L, 100%; IGF1R, 100%) but with increasing allele frequency (Table 2).
Figure 4. Genomic profiles are preserved from parent tumors to PDX tumors.
Targeted sequencing of full genes or all exons was performed using a 157-targeted-gene panel on both BRCAMUT and BRCAWT patient tumors with matched patient-derived xenografts (PDXs) highlighting deleterious, likely deleterious, and variants of unknown significance (VUS).
Table 2. Summary of pathogenic mutations with allele frequency in parent tumors and PDX tumors.
Targeting the ATR/CHK1 pathway in BRCAMUT HGSOC PDX models.
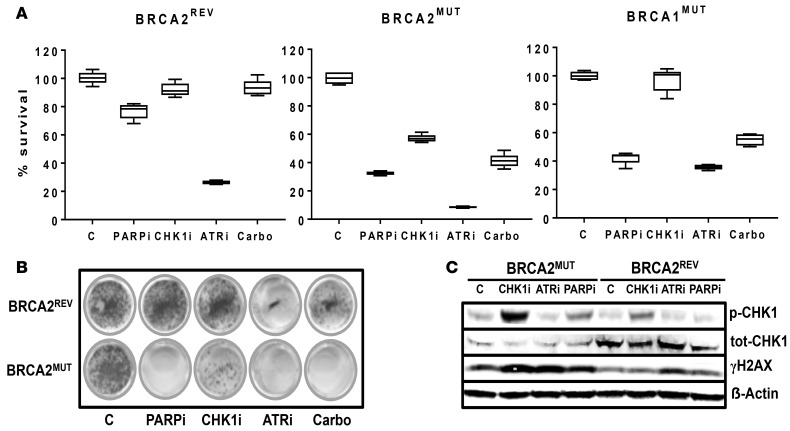
Blocking cell cycle checkpoints is an alternative approach for inducing synthetic lethality in HRD HGSOCs. We first evaluated ATR/CHK1 inhibition in comparison with PAPRi in vitro. As expected, PARPi (AZD2281) at 1 μM and carboplatin at 30 μM were significantly more cytotoxic to BRCAMUT (PEO1, JHOS4) cells than BRCAREV (PEO4 known platinum resistant) cells. CHK1i (MK8776) was significantly more cytotoxic in the BRCA2MUT cells than BRCAWT; however, there was minimal effect in JHOS4 BRCA1MUT cells. The ATR inhibitor (ATRi) (AZD6738) was cytotoxic in both BRCAMUT and BRCAWT cells (Figure 5A). In vitro, PARPi and ATRi/CHK1i were both as cytotoxic as chemotherapy in BRCAMUT cells. Colony forming assays confirmed viability assays (Figure 5B). In BRCA2MUT cells (PEO1) after 24 hours of treatment with PARPi (AZD2281), CHK1i (MK8776), and ATRi (AZD6738) there was an increase in the phosphorylation of γH2AX relative to control, indicative of accumulation of DNA damage. With ATRi, there was an increase in γH2AX in BRCAWT cells (PEO4) relative to control, but not with other agents tested. In the BRCA2MUT cells (PEO1), CHK1i increased CHK1 (Ser345) phosphorylation as expected, given inhibition of enzyme activity and a likely feedback loop, confirming the drug target as shown in prior reports (36) (Figure 5C).
Figure 5. Targeting the ATR-CHK1 axis in BRCA1/2MUT cell models.
(A) Viability of established ovarian cancer cells after treatment with 1 μM PARPi (AZD2281), 1 μM CHK1i (MK8776), 1 μM ATRi (AZD6738), and 30 μM carboplatin was assessed with an MTT assay. Cell models included PEO1 (BRCA2MUT), JHOS4 (BRCA1MUT), and the homologous recombination–proficient cells, PEO4 (BRCA2REV). Cells were incubated in their respective drug concentrations for 5 days. PARPi was significantly more cytotoxic in BRCAMUT relative to BRCA2REV cells (32.9% ± 0.7% cell viability for BRCA2MUT cells and 42.3% ± 2.0% for BRCA1MUT cells vs. 76.7% ± 2.0% for BRCA2REV cells; P < 0.00001 and P = 0.0002 for BRCA2 vs. BRCAREV and BRCA1 vs. BRCAREV, respectively). Carboplatin treatment appeared to have a similar effect to that of PARPi in all lines: BRCA2MUT (41.2% ± 3.9% cell viability; P = 0.07 carboplatin compared with PARPi); BRCA1MUT (55.0% ± 5.2% viability; P = 0.0003 carboplatin vs. PARPi); and BRCA2REV (96.3% ± 4.6% viability; P = 0.01 carboplatin vs. PARPi). Only the BRCA2MUT cell model was sensitive to 1 μM CHK1i (56.6% ± 1.1% cell viability). Both BRCAMUT and BRCA2REV cells were sensitive to ATRi monotherapy (8.6% ± 0.2%, 38.9% ± 1.4%, and 26.3% ± 0.5% cell viability for BRCA2MUT, BRCA1MUT, and BRCA2REV cells, respectively; P < 0.001 for all lines ATRi vs. carboplatin treatment). The box-and-whisker plots show the median, with boxes extending from the 25th to 75th percentile and the whiskers extending from minimum and maximum values of the dataset (n = 5 per group). Two-way ANOVA was performed before the Tukey’s honestly significant difference test to determine whether there was an overall difference between the groups. The data shown are a single representative experiment with 5 determinations. Three independent experiments were performed. (B) Colony forming assay was performed with 1 μM PARPi, 1 μM CHK1i, 1 μM ATRi, and 30 μM carboplatin treatment in BRCA2MUT (PEO1) and BRCA2REV (PEO4) cell models. Cells were incubated in their respective drug concentrations for 13 days. Colony formation was decreased in BRCA2MUT compared with BRCA2REV cells with PARPi (3.3% ± 1.3% vs. 116.8% ± 5.2%; P < 0.00001), CHK1i (28.6% ± 3.1% vs. 94.1% ± 6.0%; P = 0.0001), and carboplatin (3.9% ± 1.2% vs. 43.3% ± 2.8%; P < 0.00001). All values are percentage colony formation relative to control. Both cell models showed decreased colony formation with ATRi (6.4% ± 1.6% for BRCA2MUT vs. 10.9% ± 1.4% for BRCA2REV; P = 0.08). (C) BRCA2MUT (PEO1) and BRCA2REV (PEO4) cells were treated with 1 μM PARPi, 1 μM CHK1i, and 1 μM ATRi and lysates were collected after 24 hours of treatment. Western blot for the indicated total and phosphoproteins is shown.
To examine and confirm direct effects of drugs on the ATR/CHK1 pathway, cells were treated with aphidicolin (APH). APH inhibits DNA polymerase activity and causes the accumulation of single-strand DNA, which potently activates the ATR kinase. As expected, CHK1 (Ser345) phosphorylation increased following 4 hours of treatment with APH, and PARPi did not further increase CHK1 (Ser345) phosphorylation in the context of replication stalling. However, CHK1 (Ser345) phosphorylation was significantly decreased by ATRi, confirming sufficient dosing with the ATR inhibitor (Supplemental Figure 8).
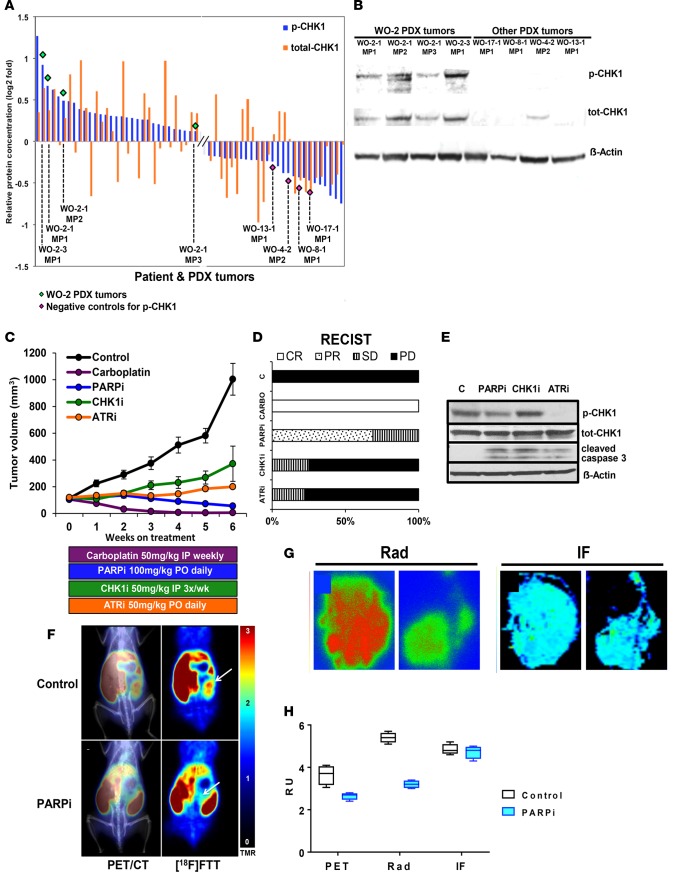
RPPA of 308 phosphorylated and total proteins was used to select a BRCAMUT PDX model for preclinical studies to evaluate targeting the ATR/CHK1 pathway with CHK1i and ATRi in comparison with PARPi. WO-2-1 (BRCA2 c.8945delAA) PDX tumors demonstrated elevated p-CHK1 (phospho-Ser345) relative to other PDX tumors by RPPA and was confirmed with Western blot (Figure 6, A and B).
Figure 6. Targeting the ATR-CHK1 axis in a BRCA2MUT PDX model.
(A) Reverse phase protein array (RPPA) analysis for 308 total and phosphoproteins in patient tumors and their corresponding PDXs were analyzed. Two proteins (p-CHK1 on Ser345 and total CHK1) were selected and plotted on a log2-fold scale. WO-2-1 (BRCA2MUT c.8945delAA) demonstrated elevated p-CHK1 relative to other tumors. (B) PDX tumors with high and low p-CHK1 by RPPA were compared by Western blot for p-CHK1 and total CHK1. (C and D) WO-2-1 PDXs were randomized into the following groups: vehicle control; PARPi (AZD2281) 100 mg/kg by oral gavage daily; CHK1i (MK8776) 50 mg/kg i.p. every 3 days; ATRi (AZD6738) 50 mg/kg daily; and carboplatin 50 mg/kg weekly. Tumor volume was measured by weekly ultrasound. There was a significant decrease in all treatment groups relative to control (P < 0.0001) and a significant difference relative to carboplatin treatment (P = 0.0001 for PARPi, P = 0.0072 for CHK1i, and P < 0.00001 for ATRi). ANOVA analysis was conducted to evaluate differences among means. Tukey’s honestly significant difference (HSD) test was used for all pairwise mean comparisons. PARPi treatment decreased average tumor volume by 35% over treatment duration with 68.8% (n = 11 of 16) and 31.2% (n = 5 of 16) showing partial remission (PR) and stable disease (SD), respectively, by RECIST 1.1 (37). CHK1i treatment resulted in a 25% (n = 2 of 8) SD rate and 75% (n = 6 of 8) with PD (progression of disease). ATRi treatment resulted in a 22.2% (n = 2 of 9) SD rate, and 77.8% (n = 7 of 9) with PD in average tumor volume, respectively. Carboplatin treatment resulted in 100% CR. Each symbol represents mean of determinations with SEM. Control, n = 7; PARPi n = 16; carboplatin, n = 5; ATRi, n = 9; CHK1i, n = 6 (see Supplemental Figure 9 for individual mouse response). (E) Lysates from PDX tumors after 2 weeks of treatment were immunoblotted for the indicated proteins. (F) PET imaging of PARP-1 with [18F]FTT and PET/CT fused images are shown in untreated and PARPi-treated mouse. WO-2-1 (BRCA2MUT c.8945delAA) PDX received oral doses of PARPi (AZD2281) 50 mg/kg every day for 1 month and on the day of PET imaging prior to the imaging study. White arrows point to tumors. (G) Digital autoradiography (left panel) of tumor sections and PARP-1 immunofluorescence (IF; right panel) of adjacent tumor sections comparing control with PARPi-treated tumor sections (n = 2) is shown. Representation of 1 of 2 independent experiments is shown. (H) Correlative data shown for PET, autoradiography (Rad), and IF modalities for determining PARP-1 expression. Graphical representation of 3 independent experiments and associated results are shown. Four regions of interest were drawn in each dataset. The box-and-whisker plot shows the median, with boxes extending from the 25th to 75th percentile and the whiskers extending from minimum to maximum values of the dataset (n = 4).
We then compared ATRi/CHK1i and PARPi with standard-of-care chemotherapy in a BRCAMUT PDX model WO-2-1 (BRCA2 c.8945delAA). After PDX tumors were established, mice were randomized to 5 treatment arms. There was a statistically significant decrease in tumor volume in all treatment groups as measured by weekly ultrasound (for representative image see Supplemental Figure 1) relative to control (P < 0.001 for all groups) (Figure 6C). Carboplatin treatment resulted in a 100% complete response by week 3. PARPi and ATRi/CHK1i all resulted in tumor suppression but not tumor regression, as monotherapy and were all inferior to carboplatin chemotherapy. There were no complete responses with PARPi alone even with prolonged treatment (22 weeks, data not shown) measured by RECIST 1.1 (37) (Figure 6D). Individual PDX responses to treatment are illustrated in Supplemental Figure 9. Mouse weights were stable in all groups over treatment duration, suggesting acceptable drug toxicity (Supplemental Figure 10). PDX tumors were dissociated and lysates evaluated for drug effects. There was an increase in p-CHK1 in the CHK1i treatment group relative to control at 1 week after treatment initiation (Figure 6E). ATRi decreased the p-CHK1 protein, demonstrating target engagement. Cleaved caspase 3, an apoptosis marker, was increased in all treatment groups relative to control (Figure 6E). Another PDX model that was generated from a BRCA1 mutation carrier who was platinum sensitive but recently progressed on PARPi (olaparib) after 16 months of treatment was tested. This patient’s tumor was harvested and sequenced after progression on PARPi and demonstrated persistence of the BRCA1 mutation without reversion (Table 2). ATRi/CHK1i and PARPi treatment was compared with vehicle control and similarly, ATRi and CHK1i each suppressed PDX growth compared with control (Supplemental Figure 11). PARPi treatment suppressed PDX growth initially with early signs of drug resistance, but data were limited due to the small sample size. This patient subsequently received another PARPi, rucapaib (1 year after prior PARPi), and similar to her PDX, responded for a brief time (only 3 months) before progression.
[18F]FTT uptake on PET/CT correlates with PARP-1 activity level in vivo and response to olaparib in a BRCA2-mutant OVCA PDX model.
PET imaging of PARP-1 with the PARPi analogue tracer [18F]FTT was then performed in a HGSOC PDX model before and after PARPi treatment. PET imaging studies revealed differences in radiotracer uptake in the control versus PARPi-treated animal (Figure 6F) (30, 38). This result suggests that PARPi competitively inhibits the [18F]FTT from binding to PARP-1, as demonstrated by a decrease in radiotracer uptake (Figure 6F).
To validate PET imaging findings, we performed ex vivo digital autoradiography for PARP-1 with [125I]KX1, an iodinated analogue of [18F]FTT, using tumor sections from imaged mice (31). This allowed us to evaluate specific radiotracer signal from the tumor ex vivo and to compare the control versus PARPi-treated animal. We found that there was indeed a difference in radiotracer binding in tumor sections that followed a similar trend to what was measured in the PET imaging study (Figure 6G). These data confirm the results from our PET imaging study. This suggests that the difference in radiotracer signal is caused either by intratumoral PARPi concentrations or decreased expression of PARP-1.
To evaluate whether PARP-1 expression may have decreased over the course of 1 month with PARPi treatment, we performed PARP-1 immunofluorescence on tumor sections adjacent to those used in autoradiography experiments. We found no difference in PARP-1 expression measured in the control versus PARPi-treated tumors (Figure 6G). While the control tumor had grown large over the course of the therapy study, the PARPi-treated tumor was difficult to visualize by PET alone; however, with the assistance of CT imaging, regions of interest were drawn around the tumor for comparison. This provides strong evidence that both PET and autoradiography studies revealed differences in radiotracer uptake due to competitive inhibition, despite similar protein intratumoral PARP-1 concentrations (Figure 6H).
Discussion
There is currently a lack of preclinical platforms that allow rapid translation of scientifically rational laboratory discoveries into clinical trials (39). Relevant experimental animal models are particularly lacking in this regard. Here, we have developed a preclinical pipeline to study HRD HGSOC using an orthotopic ovarian cancer transplant PDX model. This model recapitulates the heterogeneity of the parent tumor and remains stable when passaged in mice. Our model mimics the natural progression of this disease and response to standard chemotherapy as well as maintains the morphologic patterns, biomarkers, and genomic signatures of the original tumor. We also include molecular functional imaging (i.e., PARPi analogue) for biomarker discovery in our platform. Further, our PDX platform represents a living tissue bank that can readily be used to test novel targeted therapies.
The promise of in vitro and in vivo studies for targeted therapies in ovarian cancer has not translated into success in the clinical setting until now (39). PARPi treatment, for example, in BRCAMUT ovarian cancer cell line models has been shown to decrease cell viability, increase apoptosis, and completely inhibit tumor growth in BRCAMUT in vivo models (18, 19). However, clinical trials using PARPi treatment have shown modest response rates of ~40%. Furthermore, these responses are primarily partial and rarely complete; in the few tumors that do respond, resistance ultimately develops (21). Similarly, our models showed tumor suppression but not complete tumor regression with PARPi, simulating what is seen in the clinic.
Reasons for such a discrepancy between preclinical and clinical findings can be related to both the in vitro and in vivo models used for study. Even with the evolution of ovarian cancer cell lines, approximately 70% of published ovarian cancer studies use established cell lines that do not genomically resemble HGSOCs (5). Further, there are limitations to the different types of in vivo HGSOC models. Subcutaneous xenografts, while preserving mutational heterogeneity and oncogene expression profiles (40), are not positioned in the right anatomic location or microenvironment and do not model the natural progression of ovarian cancer, as they do not typically metastasize (8). Intraperitoneal xenografts, while they may mimic aspects of tumor metastasis, may miss early transforming events in metastasis (8, 41). Practically, there may be a low engraftment rate (40) and failure of discrete mass formation (12, 40), making assessing treatment response and passaging in mice difficult. Subrenal capsule xenografts involve an invasive, meticulous engrafting procedure (42). Finally, there is the complex nature and heterogeneity of ovarian cancer itself (43). Different metastatic sites in a single patient may demonstrate differences in genomic profiles and copy number variations. Over the course of different treatment regimens and recurrences, as is common in ovarian cancer, there may be shifts in the clonal subpopulations and selection and emergence of resistant clones (43, 44). Finally, the stroma, extracellular matrix, and immune cells likely also play a role in both the progression and response to different treatment modalities (45, 46).
In this study we have demonstrated that our orthotopic PDX model is a feasible and more clinically relevant approach to the development of a preclinical platform for the study of BRCA1/2MUT HGSOC when compared with historic cell line models. Our model allows the study of intertumoral and interpatient heterogeneity by creating PDX models for different regions of disease from one patient and different patients, respectively. Fresh tumor tissue is obtained directly from patients and subsequently implanted into immunodeficient mice. Thus, unlike cell lines and cell suspension made from patients’ tumors, using undigested tumor chunks will preserve some of these non–cell-autonomous factors, especially when assessed in low passage in vivo (12, 47). These non–cell-autonomous factors, like human stroma, which our model demonstrates at least in early passages, can be drivers of disease progression, thereby representing potential therapeutic targets (41, 47). Furthermore, unlike the initial in vivo studies with PARPi that showed complete inhibition in BRCAMUT models, our model correlated with what is typically seen in the clinical setting; that is, we observed partial responses and rare complete responses with PARPi treatment. In fact, with prolonged treatment, resistance to PARPi developed by week 21 (data not shown).
Other strengths of our model include our focus on a subset of HGSOC, specifically those deficient in HR. We have established a repository of PDXs with a variety of BRCA1/2 mutations that will permit us to study responses to various therapies as well as characterize resistance mechanisms, depending on the particular mutation profile (48). In addition, the study provides evidence that our orthotopic HGSOC PDX model can be used for evaluation of functional biomarkers using [18F]FTT uptake on PET/CT for PARPi therapy studies. Future studies evaluating the role of PARP-1 expression as a predictive biomarker for PARPi therapy will employ this technology. Studies in small cell lung cancer have already suggested that PARP-1 levels are potent predictors for response to PARPi, independent of BRCA1/2 mutations (49, 50).
We also demonstrate that our preclinical platform can be used to investigate alternative approaches to modulate DNA repair activity, capitalizing on synthetic lethality in BRCAMUT HGSOC. RPPA identified which existing PDX models may be suitable models, based on p-CHK1 expression, for a preclinical study comparing PARPi with ATRi/CHK1i and standard chemotherapy. We found that ATRi and CHK1i have modest activity, but similar to PARPi, are inferior to standard chemotherapy as single agents in models with high p-CHK1. PARPi, along with cell cycle checkpoint inhibitors, will likely be best applied as combination therapies. While cancer cells harboring some degree of replicative stress upregulate ATR and CHK1 activity to survive (26, 27, 29, 51), inhibiting either one of these proteins alone may not be sufficient to cause cell death, at least when using the drugs and recommended regimens applied herein.
There is emerging evidence that ATRi may be more effective than CHK1i in killing cancer cells. ATR may be able to stabilize replication forks independently of CHK1 (52), so while CHK1i may increase origin firing and thus replication stress, ATR is still able to stabilize replication forks and allow cell survival (53). Additionally, ATR can suppress origin firing and the intra-S checkpoint independently of CHK1 (54, 55). In contrast, CHK1 may be activated independently of ATR (56) and may prevent replication catastrophe in S-phase cells, even in the presence of an ATRi (57). Such a network of alternative DNA repair pathways may contribute to our preclinical findings and suggests that targeting more than one pathway may be necessary to achieve complete tumor regression. Additional research is required to examine the optimal clinical benefit from ATRi and CHK1i inhibition, which may depend on the genetics of the tumor as well as the drug and treatment regimen used. The PDX model described herein may be of particular utility for these comparative studies.
Importantly, our model is lacking the human immune system, a major limitation. We are working to humanize these models to allow evaluation of drug effects on the tumor immune microenvironment and vice versa. Clinical trials are both costly and time consuming. Thus, being able to better predict which treatment regimens will work for a select group of patients is critical to increase the success rate and to get effective drugs to patients who are most likely to benefit.
In conclusion, strategies to optimize approaches capitalizing on synthetic lethality with HRD are needed. Orthotopic BRCAMUT PDXs serve as excellent experimental models for therapeutic studies. Genomic and proteomic characterization of PDXs can help strategize therapeutic options for evaluation in preclinical PDX trials. ATRi and CHK1i are active in BRCAMUT HGSOC preclinical models and may serve as alternative approaches for the treatment of this disease and warrant further investigation.
Methods
Cell lines and reagents.
PEO1 and PEO4 ovarian epithelial tumor cell lines were grown in RPMI media with 10% FBS and penicillin/streptomycin (gift from Andrew Godwin, University of Kansas, Lawrence, Kansas, USA). JHOS4 (gift from Ronny Drapkin, University of Pennsylvania) ovarian epithelial tumor cell lines were grown in DMEM/F12 media with 10% FBS, and penicillin/streptomycin. Mutation profiles for all cell lines were confirmed with targeted sequencing of full genes or all exons using our 157-targeted-gene panel. PEO1 harbors a BRCA2 (c.C4965G), TP53 (c.G731A), and CREBBP (c.G3712T) mutation. JHOS4 has a BRCA1 (c.5278-1G>A) and TP53 (c.T440G) mutation. PEO4 was derived from the same patient as PEO1 and has a BRCA2 reversion mutation as well as the same TP53 mutation. Authenticity of cell lines was confirmed by short tandem repeat (STR) analysis, and mycoplasma testing was negative.
Establishment of PDXs.
NOD-SCID IL2Rγ−/− (NSG) mice were purchased from the Jackson Laboratory. All mice were housed according to the policies of the IACUC of the Wistar Institute. PDXs were generated by sectioning of fresh tumor tissue obtained from the Hospital of the University of Pennsylvania and engrafting tumor chunks (~2 mm3) pieces orthotopically to the fallopian tube fimbria/ovary of 5- to 8-week-old female mice. Briefly, after anesthetized mice were shaved, a small 1- to 2-cm vertical incision was made through the skin over the left flank. The peritoneal cavity was then entered through a 1-cm vertical incision. The left ovarian bursa was elevated and exteriorized. Once the left ovary was identified on the anterior side of the bursa, two to three 2 × 2 × 2 mm tumor chunks were sutured to the ovary/fallopian tube fimbria using 5.0 PDS suture. The tumor transplant was then placed back inside the peritoneal cavity, and ~20 μl of Matrigel was placed over the transplant. The peritoneal cavity and then the skin were closed (Supplemental Figure 1A). Once the MP1 PDX tumor reached approximately 700–1,000 mm3, it was harvested and retransplanted for expansion in serial generations (MP2), which were used for in vivo drug response studies. Fresh patient tumor was minced and cryopreserved in 5% DMSO plus FBS for later thawing and transplantation. Samples were also snap frozen for DNA, RNA, and protein analysis. Authenticity of PDX tumors was confirmed by STR analysis. Tumors were also digested using collagenase for development of primary tumor cultures (58).
Preclinical trial and tissue preparation.
For tumor transplantation, banked cryovials of tumor chunks were thawed, washed twice with HBSS, and transplanted to the fallopian tube fimbria/ovary of 5- to 8-week-old NSG female mice. Once tumors were palpated, tumor length (l) and width (w, smaller of 2 measurements) were measured by ultrasound (M-TURBO Ultrasound, SonoSite) with an HFL38x 13-6MHz linear transducer on each mouse and tumor volume (V = [l × w2]/2) was calculated. Once tumor volume reached 70–100 mm3, animals (n = 50) were randomized using Tumor Manager (Biopticon) into 5 treatment groups: carboplatin (50 mg/kg i.p. injection weekly; Hospira); MK8776 (CHK1i, 50 mg/kg i.p. every 3 days; Selleckchem); AZD2281 (PARPi, 100 mg/kg oral gavage [OG] daily); AZD6738 (ATRi, 50 mg/kg OG daily; AstraZeneca); and vehicle. Tumor volume and body weight was measured every week. When tumor volume was greater than or equal to 1,000 mm3, animals were euthanized according to IACUC guidelines. Three animals were sacrificed after 2 weeks of treatment to evaluate drug-target effects. Tumors were collected and snap frozen for DNA, RNA, and protein analysis. Tumors were also fixed in 10% formalin for morphological and immunohistochemical analysis. Treatment response in individual mice was also assessed using RECIST 1.1 (37).
Immunohistochemistry.
Samples were fixed in 10% formalin. Tissues were dehydrated in graded ethanol solutions, cleared in xylene, and embedded in paraffin. Paraffin blocks were cut into 4- to 6-μm sections and placed onto glass slides. After deparaffinization and rehydration, antigen retrieval was done via pressure cooker. Slides were incubated in 1× target retrieval solution at 120°C for 4 minutes at 18 to 20 psi. Endogenous hydrogen peroxidase activity was blocked with hydrogen peroxide for 10 minutes followed by rinsing with wash buffer. Slides were individually incubated with cytokeratin 7 (1:2,000 dilution, catalog M7018, DAKO), PAX8 (1:800 dilution, catalog 10336-1-AP, Proteintech Group), ER (1:200 dilution, clone EP1, DAKO), BRCA1 N-terminal (1:500 dilution, clone MS110, Calbiochem), and BRCA1 C-terminal (1:10,000 dilution, catalog 07-434, EMD/Millipore) antibodies in wash buffer for 60 minutes. Alternatively, slides were incubated with appropriate isotype controls and diluted similarly. Slides were washed and incubated with anti-rabbit horseradish peroxidase polymer for 30 minutes, followed by a further wash. Slides were developed using 3,3′-diaminobenzidine (DAB) chromogen for 10 minutes and then washed with water. After staining, slides were counterstained, dehydrated, and cleared using a Leica autostainer. Finally, slides were coverslipped with mounting reagent. In situ hybridization was performed using a human-specific Alu DNA probe (PR026-100, BioGenex) and staining was performed as described (35). DAPI (4′,6-diamidino-2-phenylindole) was used to label nuclear DNA.
Slides were also reviewed and SET (solid, pseudoendometriod, and transitional cell carcinoma-like morphology) features (34) were assessed by a gynecologic oncology pathologist (RS) in a blinded manner using light microscopy.
DNA sequencing and mutation analysis.
Somatic mutation screening was performed using a custom designed targeted massively parallel sequencing protocol. For each patient tumor and PDX, 500 ng genomic DNA was sheared randomly into 200-bp fragments with the Covaris LE220 Focused-UltraSonicator. Sheared DNA was A-tailed and ligated with adaptor-embedded indexes using the NEBNext Ultra DNA Library Prep Kit for Illumina (New England BioLabs). DNA quality, fragment size, and concentration of library preps were measured using Agilent’s DNA 1000 chips in conjunction with the 2100 Bioanalyzer (Agilent Technologies). High-quality samples were pooled and hybridized to a custom capture library using Agilent SureSelect kits. DNA quality, size, and concentration were assessed using the Qubit 2.0 fluorometer (Life Technologies) and the Bioanalyzer. Library captures were sequenced using on the HiSeq 2500 (Illumina) at the University of Pennsylvania Next Generation Sequencing Core.
Our custom capture panel included 157 genes, primarily tumor suppressor genes, involved in pathways implicated in ovarian and/or breast cancer susceptibility and tumorigenesis, such as HR, mismatch repair, or checkpoint inhibition (Supplemental Figure 2).
Alignment to human reference genome NCBI Build 37 was performed with the Burrows-Wheeler Aligner (59) and reads were analyzed using the Genome Analysis Toolkit for variant calling (60). Disambiguation of sequencing read origin was performed by mapping reads to both the mouse and human genome. Reads that mapped with high accuracy to the mouse genome were removed. ANNOVAR was used for variant annotation (61, 62). Samples were removed due to poor sequencing quality if more than 10% of targets had 0% coverage or if at least 10× coverage was achieved in less than 50% of targets. Variant identification and classification were performed using a strict filtering and analysis pipeline that has previously been described and validated (63, 64). The locus-specific databases ClinVar, dbSNP, and COSMIC were utilized to help identify suspected deleterious variants, and any missense variant calls that could not be confirmed in the literature were considered VUS and excluded from analysis. Pindel (65) was utilized to detect large rearrangements or indels. All PDX and parent tumors were authenticated using STR (Wistar Genomics Facility).
RPPA analysis.
RPPA was performed by the MD Anderson Center RPPA core facility as previously described (66) and data reported as Normalized Linear and Normalized Log2. Three hundred eight phosphorylated and total proteins were evaluated (https://www.mdanderson.org/education-and-research/resources-for-professionals/scientific-resources/core-facilities-and-services/functional-proteomics-rppa-core/index.html). Unsupervised hierarchical clustering was performed on RBN log2 median-centered protein values using Cluster 3.0 software (http://rana.lbl.gov/EisenSoftware.htm). Results were visualized using Treeview software (http://rana.lbl.gov/EisenSoftware.htm).
Western blot.
Cells and tissues were harvested and lysed in RIPA buffer (33) containing a cocktail of protease and phosphatase inhibitors (Calbiochem). Following protein concentration determination (Bio-Rad), cell lysates were resolved in reducing SDS-PAGE gels and immunoblotted with antisera specific for p-CHK1 (Ser345, catalog 2348), γH2AX (catalog 9718) (both Cell Signaling Technology), and CHK1 (catalog sc-8408, Santa Cruz Biotechnology). The species-appropriate horseradish peroxidase–conjugated secondary antibody was used, followed by detection with an Odyssey Quantitative Fluorescent Imaging System (LI-COR Biotechnology). Anti–β-actin (catalog sc-69879, Santa Cruz Biotechnology) was used as an internal control.
Cytotoxicity assays.
Cells (5 × 103) were seeded on 96-well plates and treated with the indicated doses of PARPi (AZD2281), CHK1i (MK8776), ATRi (AZD6738), and carboplatin for 5 days. At the end of the treatment period, the viable cell number was determined by MTT colorimetric assay. Cells were incubated with 10 μl MTT at 5 mg/ml (Sigma-Aldrich) for 2 hours at 37°C. DMSO was added and the absorbance was measured in a microplate reader at a wavelength of 570 nm. IC50s were calculated using GraphPad Prism.
Colony formation assay.
Cells (5 × 103) were plated onto 12-well plates and incubated at 37°C. After seeding, cells were treated with drugs for 10 to 14 days. Media and drugs were refreshed every 5 days. When the cells grew to visible colonies, the colonies were washed once with PBS and fixed with 4% paraformaldehyde for 10 minutes. The cells were then stained with 0.2% crystal violet. Images of entire wells were scanned and the colony forming area was quantified using ImageJ (NIH). For each sample, the results from 3 replicates were averaged (67).
PET imaging of PARP-1 with [18F]FTT in ovarian cancer PDX model.
PET imaging was performed in 2 BRCA2MUT PDXs. Mice were previously enrolled in PARPi (AZD2281) therapy experiments and at the end of the study a control and an PARPi-treated mouse underwent PET imaging. The PARPi-treated mouse received oral doses of 50 mg/kg PARPi (AZD2281) every day for 1 month and on the day of PET imaging prior to the imaging study. Mice were anesthetized via nose cone at 2% to 3% isoflurane, 1 l/min oxygen, and body temperature was maintained by heating pad. Intravenous tail vein injections of [18F]FTT (200 microcuries) were administered immediately before the start of imaging and mice were scanned for 60 minutes on a Phillips Mosaic small animal PET scanner. Regions of interest were drawn manually over the tumors as well as a muscle region (background) to produce tumor to muscle ratios for each animal. CT was performed for each mouse on a Phillips mini CT scanner for anatomical reference. At the conclusion of the study the mice were sacrificed and tumors were harvested for correlative assays.
PARP-1 autoradiography with [125I]KX1.
Digital autoradiography was performed on tumor sections derived from each mouse used in PET imaging studies. Briefly, tumors were flash frozen in liquid nitrogen and embedded in OCT. Tumors were mounted in a Leica CM1950 cryostat and were sectioned at 5 μm. Tissue sections were placed on slides and stored at –80°C. On the day of autoradiography experiments, slides were removed from storage and allowed to equilibrate to room temperature for 30 minutes. Slides were then washed in PBS for 3 minutes. Next, 200 μl PBS containing [125I]KX1 was added to the slide and allowed to equilibrate for 30 minutes. Sections were then washed 3 times in PBS and exposed for 30 minutes to reusable phosphor films. Lastly, the film was read on a GE Typhoon FLA 7000 digital imager. Regions of interest were drawn around tissue sections and average intensity was calculated using GE image analysis software.
PARP-1 immunofluorescence.
Tumor sections were fixed in cold acetone chilled on dry ice for 10 minutes. Next, sections were washed with PBS 3 times for 5 minutes each. Sections were then blocked with 5% BSA in PBS for 1 hour. A hydrophobic ring was drawn around the tumor section. Primary PARP-1 antibody (clone 46D11, Cell Signaling Technology) was diluted in PBS (1:200) and incubated overnight at 4°C. Sections were washed 3 times with PBS, 5 minutes per wash. Fluorescent secondary antibody diluted in PBS (1:500) was added to the section and incubated at room temperature in a dark box for 1 hour. Finally, sections were imaged on a LiCor OdYSSEY XL (LI-COR Biotechnology). Five regions of interest were drawn in each tumor section and used to quantify relative PARP-1 expression in the control and PARPi-treated tumor.
Statistics.
MTT, colony formation assays, and Western blot assays were done at least thrice and mean ± SEM displayed in bar graphs. One- or two-way ANOVA was conducted to assess differences among means. Following a significant ANOVA result (P ≤ 0.05) rejecting the null hypothesis that means are the same across the treatment groups, the Tukey’s honestly significant difference (HSD) test was used for all pairwise mean comparisons. This multiple comparison procedure ensures actual family-wise error rates no greater than prespecified 5%. A linear mixed-effects model was used to estimate growth rate over time. All statistical analyses were completed using Stata MP Version 14.0 (StataCorp).
Study approval.
Animals utilized in these studies were housed and cared for by the University Laboratory Animal Resources under the direction of a veterinarian. Ordering, housing, and caring for mice is centralized and carried out according to NIH and USDA standards for animal care. This facility is fully accredited by AAALAC, demonstrating compliance with NIH guidelines. The protocol for animal use was approved by the University IUCUC (approval number 112635). Written consent was obtained from all human participants who provided samples for PDXs. An IRB approved protocol was obtained to collect patient samples for research (number 702679).
Author contributions
FS designed research studies, conducted experiments, acquired and analyzed data, provided reagents, and wrote the manuscript. EG, HK, BW, and KN conducted experiments, acquired and analyzed data, and wrote the manuscript. CK and MH provided reagents and analyzed data. PB, RZ, and XZ analyzed data. SJ conducted experiments and acquired and analyzed data. YL and RS acquired and analyzed data. GM and Mehran M acquired and analyzed data, provided reagents, and wrote the manuscript. RHM, YZ, GC, RD, and NJ acquired and analyzed data and provided reagents. EB analyzed data and wrote the manuscript. JT acquired data and provided reagents. Mark M analyzed data, provided reagents, and wrote the manuscript.
Supplementary Material
Acknowledgments
The authors thank Mei Zheng for IHC preparation and the Herlyn and Drapkin laboratory group for helpful discussions. We also would like to thank the Sand family for their support for this project.
This work was supported by NIH grants 1K08CA151892-01A1 (to FS) and P01 CA114046 (to MH), Basser Team Science grant (to FS, EB, and RZ) from the University of Pennsylvania, and the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation grant (to MH and RD).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:JCI Insight. 2017;2(1):e89760. doi:10.1172/jci.insight.89760.
Contributor Information
Erin George, Email: emg2106@gmail.com.
Hyoung Kim, Email: hyoungk@mail.med.upenn.edu.
Clemens Krepler, Email: ckrepler@Wistar.org.
Brandon Wenz, Email: bwenz@mail.med.upenn.edu.
Mehran Makvandi, Email: makvandi@mail.med.upenn.edu.
Janos L. Tanyi, Email: Janos.Tanyi@uphs.upenn.edu.
Rugang Zhang, Email: rzhang@wistar.org.
Patricia Brafford, Email: brafford@Wistar.org.
Stephanie Jean, Email: stephanie.jean@uphs.upenn.edu.
Yiling Lu, Email: yilinglu@mdanderson.org.
Gordon B. Mills, Email: gmills@mdanderson.org.
Meenhard Herlyn, Email: herlynm@wistar.org.
Mark Morgan, Email: mark.morgan@uphs.upenn.edu.
Xiaochen Zhang, Email: xzhang@phs.psu.edu.
Robert Soslow, Email: soslowr@mskcc.org.
Ronny Drapkin, Email: rdrapkin@mail.med.upenn.edu.
Neil Johnson, Email: neil.johnson@fccc.edu.
Ying Zheng, Email: yingzhen@mail.med.upenn.edu.
George Cotsarelis, Email: cotsarel@mail.med.upenn.edu.
Fiona Simpkins, Email: fiona.simpkins@uphs.upenn.edu.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Salani R, et al. Posttreatment surveillance and diagnosis of recurrence in women with gynecologic malignancies: Society of Gynecologic Oncologists recommendations. Am J Obstet Gynecol. 2011;204(6):466–478. doi: 10.1016/j.ajog.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Zivanovic O, Aldini A, Carlson JW, Chi DS. Advanced cytoreductive surgery: American perspective. Gynecol Oncol. 2009;114(2 Suppl):S3–S9. doi: 10.1016/j.ygyno.2008.11.033. [DOI] [PubMed] [Google Scholar]
- 4.Armstrong DK, et al. Intraperitoneal cisplatin and paclitaxel in ovarian cancer. N Engl J Med. 2006;354(1):34–43. doi: 10.1056/NEJMoa052985. [DOI] [PubMed] [Google Scholar]
- 5.Domcke S, Sinha R, Levine DA, Sander C, Schultz N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat Commun. 2013;4:2126. doi: 10.1038/ncomms3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wenger SL, Senft JR, Sargent LM, Bamezai R, Bairwa N, Grant SG. Comparison of established cell lines at different passages by karyotype and comparative genomic hybridization. Biosci Rep. 2004;24(6):631–639. doi: 10.1007/s10540-005-2797-5. [DOI] [PubMed] [Google Scholar]
- 7.Hausser HJ, Brenner RE. Phenotypic instability of Saos-2 cells in long-term culture. Biochem Biophys Res Commun. 2005;333(1):216–222. doi: 10.1016/j.bbrc.2005.05.097. [DOI] [PubMed] [Google Scholar]
- 8.Bobbs AS, Cole JM, Cowden Dahl KD. Emerging and evolving ovarian cancer animal models. Cancer Growth Metastasis. 2015;8(Suppl 1):29–36. doi: 10.4137/CGM.S21221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crum CP, Drapkin R, Kindelberger D, Medeiros F, Miron A, Lee Y. Lessons from BRCA: the tubal fimbria emerges as an origin for pelvic serous cancer. Clin Med Res. 2007;5(1):35–44. doi: 10.3121/cmr.2007.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piek JM, van Diest PJ, Verheijen RH, Kenemans P. Cell cycle-related proteins p21 and bcl-2: markers of differentiation in the human fallopian tube. Histopathology. 2001;38(5):481–482. doi: 10.1046/j.1365-2559.2001.1163c.x. [DOI] [PubMed] [Google Scholar]
- 11.Perets R, Drapkin R. It’s totally tubular....riding the new wave of ovarian cancer research. Cancer Res. 2016;76(1):10–17. doi: 10.1158/0008-5472.CAN-15-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weroha SJ, et al. Tumorgrafts as in vivo surrogates for women with ovarian cancer. Clin Cancer Res. 2014;20(5):1288–1297. doi: 10.1158/1078-0432.CCR-13-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. doi: 10.14694/EdBook_AM.2014.34.e258. Scott CL, Mackay HJ, Haluska P, Jr. Patient-derived xenograft models in gynecologic malignancies. Am Soc Clin Oncol Educ Book. 2014:e258–e266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu JF, et al. Establishment of patient-derived tumor xenograft models of epithelial ovarian cancer for pre-clinical evaluation of novel therapeutics [published online ahead of print August 29, 2016] Clin Cancer Res. doi: 10.1158/1078-0432.CCR-16-1237. doi: 10.1158/1078-0432.CCR-16-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hidalgo M, et al. Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer Discov. 2014;4(9):998–1013. doi: 10.1158/2159-8290.CD-14-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tentler JJ, et al. Patient-derived tumour xenografts as models for oncology drug development. Nat Rev Clin Oncol. 2012;9(6):338–350. doi: 10.1038/nrclinonc.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morton CL, Houghton PJ. Establishment of human tumor xenografts in immunodeficient mice. Nat Protoc. 2007;2(2):247–250. doi: 10.1038/nprot.2007.25. [DOI] [PubMed] [Google Scholar]
- 18.Bryant HE, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly (ADP-ribose) polymerase. Nature. 2005;434(7035):913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 19.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434(7035):917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 20.Cancer Genome Atlas Research Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaufman B, et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J Clin Oncol. 2015;33(3):244–250. doi: 10.1200/JCO.2014.56.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huntoon CJ, et al. ATR inhibition broadly sensitizes ovarian cancer cells to chemotherapy independent of BRCA status. Cancer Res. 2013;73(12):3683–3691. doi: 10.1158/0008-5472.CAN-13-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karnitz LM, Zou L. Molecular pathways: targeting ATR in cancer therapy. Clin Cancer Res. 2015;21(21):4780–4785. doi: 10.1158/1078-0432.CCR-15-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peasland A, et al. Identification and evaluation of a potent novel ATR inhibitor, NU6027, in breast and ovarian cancer cell lines. Br J Cancer. 2011;105(3):372–381. doi: 10.1038/bjc.2011.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reaper PM, et al. Selective killing of ATM- or p53-deficient cancer cells through inhibition of ATR. Nat Chem Biol. 2011;7(7):428–430. doi: 10.1038/nchembio.573. [DOI] [PubMed] [Google Scholar]
- 26.Gilad O, et al. Combining ATR suppression with oncogenic Ras synergistically increases genomic instability, causing synthetic lethality or tumorigenesis in a dosage-dependent manner. Cancer Res. 2010;70(23):9693–9702. doi: 10.1158/0008-5472.CAN-10-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toledo LI, et al. A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nat Struct Mol Biol. 2011;18(6):721–727. doi: 10.1038/nsmb.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruzankina Y, Schoppy DW, Asare A, Clark CE, Vonderheide RH, Brown EJ. Tissue regenerative delays and synthetic lethality in adult mice after combined deletion of Atr and Trp53. Nat Genet. 2009;41(10):1144–1149. doi: 10.1038/ng.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schoppy DW, et al. Oncogenic stress sensitizes murine cancers to hypomorphic suppression of ATR. J Clin Invest. 2012;122(1):241–252. doi: 10.1172/JCI58928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edmonds CE, et al. [(18)F]FluorThanatrace uptake as a marker of PARP1 expression and activity in breast cancer. Am J Nucl Med Mol Imaging. 2016;6(1):94–101. [PMC free article] [PubMed] [Google Scholar]
- 31.Makvandi M, et al. A radiotracer strategy to quantify PARP-1 expression in vivo provides a biomarker that can enable patient selection for PARP inhibitor therapy. Cancer Res. 2016;76(15):4516–4524. doi: 10.1158/0008-5472.CAN-16-0416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tibes R, et al. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Mol Cancer Ther. 2006;5(10):2512–2521. doi: 10.1158/1535-7163.MCT-06-0334. [DOI] [PubMed] [Google Scholar]
- 33.Simpkins F, et al. Src Inhibition with saracatinib reverses fulvestrant resistance in ER-positive ovarian cancer models in vitro and in vivo. Clin Cancer Res. 2012;18(21):5911–5923. doi: 10.1158/1078-0432.CCR-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soslow RA, et al. Morphologic patterns associated with BRCA1 and BRCA2 genotype in ovarian carcinoma. Mod Pathol. 2012;25(4):625–636. doi: 10.1038/modpathol.2011.183. [DOI] [PubMed] [Google Scholar]
- 35.Yang R, et al. Direct conversion of mouse and human fibroblasts to functional melanocytes by defined factors. Nat Commun. 2014;5:5807. doi: 10.1038/ncomms6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Syljuåsen RG, et al. Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol Cell Biol. 2005;25(9):3553–3562. doi: 10.1128/MCB.25.9.3553-3562.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eisenhauer EA, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45(2):228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 38.Zhou D, et al. Synthesis, [¹8F] radiolabeling, and evaluation of poly (ADP-ribose) polymerase-1 (PARP-1) inhibitors for in vivo imaging of PARP-1 using positron emission tomography. Bioorg Med Chem. 2014;22(5):1700–1707. doi: 10.1016/j.bmc.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hasan N, Ohman AW, Dinulescu DM. The promise and challenge of ovarian cancer models. Transl Cancer Res. 2015;4(1):14–28. doi: 10.3978/j.issn.2218-676X.2015.01.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dobbin ZC, et al. Using heterogeneity of the patient-derived xenograft model to identify the chemoresistant population in ovarian cancer. Oncotarget. 2014;5(18):8750–8764. doi: 10.18632/oncotarget.2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lengyel E, et al. Epithelial ovarian cancer experimental models. Oncogene. 2014;33(28):3619–3633. doi: 10.1038/onc.2013.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee CH, et al. Establishment of subrenal capsule xenografts of primary human ovarian tumors in SCID mice: potential models. Gynecol Oncol. 2005;96(1):48–55. doi: 10.1016/j.ygyno.2004.09.025. [DOI] [PubMed] [Google Scholar]
- 43.Blagden SP. Harnessing pandemonium: the clinical implications of tumor heterogeneity in ovarian cancer. Front Oncol. 2015;5:149. doi: 10.3389/fonc.2015.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bashashati A, et al. Distinct evolutionary trajectories of primary high-grade serous ovarian cancers revealed through spatial mutational profiling. J Pathol. 2013;231(1):21–34. doi: 10.1002/path.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liotta LA, Kohn EC. The microenvironment of the tumour-host interface. Nature. 2001;411(6835):375–379. doi: 10.1038/35077241. [DOI] [PubMed] [Google Scholar]
- 46.Zhang L, et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N Engl J Med. 2003;348(3):203–213. doi: 10.1056/NEJMoa020177. [DOI] [PubMed] [Google Scholar]
- 47.Cassidy JW, Caldas C, Bruna A. Maintaining tumor heterogeneity in patient-derived tumor xenografts. Cancer Res. 2015;75(15):2963–2968. doi: 10.1158/0008-5472.CAN-15-0727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bouwman P, Jonkers J. Molecular pathways: how can BRCA-mutated tumors become resistant to PARP inhibitors? Clin Cancer Res. 2014;20(3):540–547. doi: 10.1158/1078-0432.CCR-13-0225. [DOI] [PubMed] [Google Scholar]
- 49.Byers LA, et al. Proteomic profiling identifies dysregulated pathways in small cell lung cancer and novel therapeutic targets including PARP1. Cancer Discov. 2012;2(9):798–811. doi: 10.1158/2159-8290.CD-12-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cardnell RJ, et al. Proteomic markers of DNA repair and PI3K pathway activation predict response to the PARP inhibitor BMN 673 in small cell lung cancer. Clin Cancer Res. 2013;19(22):6322–6328. doi: 10.1158/1078-0432.CCR-13-1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Toledo LI, Murga M, Fernandez-Capetillo O. Targeting ATR and Chk1 kinases for cancer treatment: a new model for new (and old) drugs. Mol Oncol. 2011;5(4):368–373. doi: 10.1016/j.molonc.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Elvers I, et al. CHK1 activity is required for continuous replication fork elongation but not stabilization of post-replicative gaps after UV irradiation. Nucleic Acids Res. 2012;40(17):8440–8448. doi: 10.1093/nar/gks646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Toledo LI, et al. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell. 2013;155(5):1088–1103. doi: 10.1016/j.cell.2013.10.043. [DOI] [PubMed] [Google Scholar]
- 54.Couch FB, et al. ATR phosphorylates SMARCAL1 to prevent replication fork collapse. Genes Dev. 2013;27(14):1610–1623. doi: 10.1101/gad.214080.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luciani MG, Oehlmann M, Blow JJ. Characterization of a novel ATR-dependent, Chk1-independent, intra-S-phase checkpoint that suppresses initiation of replication in Xenopus. J Cell Sci. 2004;117(Pt 25):6019–6030. doi: 10.1242/jcs.01400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang XH, Shiotani B, Classon M, Zou L. Chk1 and Claspin potentiate PCNA ubiquitination. Genes Dev. 2008;22(9):1147–1152. doi: 10.1101/gad.1632808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Buisson R, Boisvert JL, Benes CH, Zou L. Distinct but concerted roles of ATR, DNA-PK, and Chk1 in countering replication stress during S phase. Mol Cell. 2015;59(6):1011–1024. doi: 10.1016/j.molcel.2015.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ince TA, et al. Characterization of twenty-five ovarian tumour cell lines that phenocopy primary tumours. Nat Commun. 2015;6:7419. doi: 10.1038/ncomms8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chang X, Wang K. wANNOVAR: annotating genetic variants for personal genomes via the web. J Med Genet. 2012;49(7):433–436. doi: 10.1136/jmedgenet-2012-100918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31(13):3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maxwell KN, et al. Prevalence of mutations in a panel of breast cancer susceptibility genes in BRCA1/2-negative patients with early-onset breast cancer. Genet Med. 2015;17(8):630–638. doi: 10.1038/gim.2014.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fishbein L, et al. Whole-exome sequencing identifies somatic ATRX mutations in pheochromocytomas and paragangliomas. Nat Commun. 2015;6:6140. doi: 10.1038/ncomms7140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25(21):2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hew KE, et al. MAPK activation predicts poor outcome and the MEK inhibitor, Selumetinib, reverses antiestrogen resistance in ER-positive high-grade serous ovarian cancer. Clin Cancer Res. 2016;22(4):935–947. doi: 10.1158/1078-0432.CCR-15-0534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guzmán C, Bagga M, Kaur A, Westermarck J, Abankwa D. ColonyArea: an ImageJ plugin to automatically quantify colony formation in clonogenic assays. PLoS ONE. 2014;9(3):e92444. doi: 10.1371/journal.pone.0092444. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.