Abstract
Mixed-solvent molecular dynamics (MixMD) is a hotspot-mapping technique that relies on molecular dynamics simulations of proteins in binary solvent mixtures. Previous work on MixMD has established the technique’s effectiveness in capturing binding sites of small organic compounds. In this work, we show that MixMD can identify both competitive and allosteric sites on proteins. The MixMD approach embraces full protein flexibility and allows competition between solvent probes and water. Sites preferentially mapped by probe molecules are more likely to be binding hotspots. There are two important requirements for the identification of ligand-binding hotspots: 1) hotspots must be mapped at very high signal-to-noise ratio and 2) the hotspots must be mapped by multiple probe types. We have developed our mapping protocol around acetonitrile, isopropanol, and pyrimidine as probe solvents because they allowed us to capture hydrophilic, hydrophobic, hydrogen-bonding, and aromatic interactions. Charged probes were needed for mapping one target, and we introduce them in this work. In order to demonstrate the robust nature and wide applicability of the technique, a combined total of 5 μs of MixMD was applied across several protein targets known to exhibit allosteric modulation. Most notably, all the protein crystal structures used to initiate our simulations had no allosteric ligands bound, so there was no pre-organization of the sites to predispose the simulations to find the allosteric hotspots. The protein test cases were ABL Kinase, Androgen Receptor, CHK1 Kinase, Glucokinase, PDK1 Kinase, Farnesyl Pyrophosphate Synthase and Protein-Tyrosine Phosphatase 1B. The success of the technique is demonstrated by the fact that the top-four sites solely map the competitive and allosteric sites. Lower-ranked sites consistently map other biologically relevant sites, multimerization interfaces, or crystal-packing interfaces. Lastly, we highlight the importance of including protein flexibility by demonstrating that MixMD can map allosteric sites that are not detected in half the systems using FTMap applied to the same crystal structures.
Graphical Abstract
Introduction
Traditional structure based drug discovery (SBDD) often relies on targeting an active site as a means of inhibiting protein function. However, such an approach may prove to be challenging in some protein targets. Allosteric sites on proteins allow an opportunity to circumvent such issues. Allostery has traditionally been defined as the modulation of function as a result of an effector binding at a site distant from the orthosteric site. Our evolving understanding of allosteric modulation has moved us from the sole view of an induced fit mechanism to include mechanisms dominated by population shift and conformational selection. Indeed, several studies have shown the existence of allostery in the absence of any notable change between the effector bound and unbound conformations, further strengthening the argument of a more dynamic view of allosteric mechanisms.1
Allostery is clearly important for drug design. It has a role in regulatory feedback mechanisms that control the activity of many enzymes, and they can provide an avenue to target disease.2 Furthermore, targeting allosteric sites can allow one to circumvent decreased effectiveness of inhibitors targeting the orthosteric/active site as a result of escape mutations. Moreover, it has been shown that targeting allosteric sites allows one to achieve selectivity when structural similarities in the orthosteric sites across multiple protein subtypes prevents one from achieving selectivity.3
Many discoveries of allosteric sites have been serendipitous outcomes of high throughput screens.4 Experimental approaches such as tethering thiol-containing small molecules to surface cysteine residues have also found success in identifying allosteric sites.5 There are several computational techniques that complement the detection of these allosteric sites. Computational methods for the detection of allosteric sites range from sequence-based analysis of evolutionarily conserved residues in the allosteric network6 to molecular dynamic (MD) simulations that detect an allosteric network through correlated motion of residues.7 These promising methods have only been applied to a handful of protein targets and further assessment needs to be done to evaluate their robustness.
To take full advantage of allosteric sites, it is essential to assess whether these sites are “druggable” and thereby amenable to drug discovery efforts. Common experimental approaches to assess the druggability of binding sites include NMR-based fragment screening8 and crystallography-based methods such as the multiple solvent crystal structures (MSCS) technique.9,10 Computational probe-mapping techniques, inspired by such experimental approaches, provide a cost-effective alternative. MixMD is one such probe-mapping technique that embraces the dynamic aspect of proteins. The MixMD method uses an MD simulation of the protein in a binary solvent of water and a miscible, organic solvent to determine the location where the solvent probes preferentially bind. Our earlier efforts in optimizing the MixMD technique have shown that full protein flexibility is required to map true hotspots with the method.11 Also, we have optimized the protocol to reduce the number of spurious minima identified on the protein surface.12 Spurious sites are a common drawback in similar methods. Most recently, we showed that a decrease in concentration of probes from 50% to 5% further improved the detection of hotspots over spurious sites.13
Probe-mapping techniques similar to MixMD have been presented by several groups. The first to be reported used MD simulations with isopropanol as a single probe at a concentration of 20%.14 A second probe-mapping technique termed SILCS utilized 1M benzene + 1M propane in water as the solvent mixture to carry out MD simulations.15,16 The third method used either isopropanol or a mixture of small fragments (acetic acid, acetamide, isopropylamine, and isopropanol) at a concentration of 20%.8 All probe-mapping techniques reported thus far rely on binning the probe locations onto a grid and identifying hotspots through some form of occupancy calculation. Each probe-mapping method has its merits and drawbacks. In our method, we have emphasized the use of water-miscible organic probes, simulated at low concentrations that are amenable to experimental verification.
Probe-mapping techniques such as MixMD take protein flexibility and competition of organic molecules with water into account. In principle, using drug-like fragments should facilitate the assessment of druggability for potential binding sites on the protein surface. Competition with water in MixMD allows one to explicitly assess if unfavorable solvation effects can impede binding. In this study, we extend MixMD in pursuit of allosteric sites and show that MixMD can map both active and allosteric sites on seven representative proteins. The most important factor in this study is that the crystal structures used to initiate the MixMD simulations had no allosteric ligands bound, so there was no preorganization of the allosteric site to bias the simulations.
Methods
Protocol for MixMD simulations
The crystal structures used as the starting conformations for the MixMD simulations are given in Table 1 by their PDB identification numbers (PDBid). The protein structures were stripped of water molecules and any cofactors or active-site ligands. This was followed by the addition of hydrogen atoms using Protonate 3D in MOE.17 The asparagine and glutamine residues were flipped as necessary to achieve optimal hydrogen bonding. Histidine tautomers were corrected when required. A sufficient number of sodium or chloride ions were added to neutralize each system using the tleap suite of AmberTools.18 A layer of probe molecules was added around the protein using tleap followed by the addition of a sufficient number of TIP3P19 water molecules as necessary to create a 5% v/v ratio of probe to water. The force field parameters for the probes acetonitrile, isopropanol, and pyrimidine were from our previous work.20 Methyl ammonium + acetate is a new probe set for MixMD, and the protocol and verification are given in the supplementary information (see Table S1 and Figures S1–S2). MD simulations were carried out in AMBER 1118 using the FF99SB21 force field. The SHAKE algorithm22 was used to restrain bonds to hydrogen atoms, and a time step of 2 fs was used. Particle Mesh Ewald as implemented for the GPUs (PMEMDCUDA23) was used. Non-bonded cutoff was 10 Å, and the Andersen Thermostat24 was used to maintain temperature at 300 K. Using this approach, three separate simulations with the probes acetonitrile, isopropanol, and pyrimidine were setup for each protein target. The systems were then subjected to an equilibration protocol to gradually increase the temperature and allow proper relaxation of all the atoms in the system as described previously.12 This was followed by a simulation of 20 ns. For each protein and probe pair, 10 independent simulations were carried out resulting in 200 ns of cumulative production simulation time.
Table 1.
The PDBid codes and chain identities are given for the protein structures used in this MixMD study. Each chosen structure has been compared to all its allosteric-bound structures, and the range of all-atom RMSD is given for the residues within 4Å of the allosteric ligands.
| Protein | Starting conformation used for MixMD | Starting conformation differs from all allosteric-bound structures by a range of RMSD |
|---|---|---|
| ABL Kinase | 3KFA (Chain A)25 | 7.93 – 8.02 Å |
| Androgen Receptor | 2AM9 (Chain A)26 | 1.01 – 1.67 Å |
| PDK1 Kinase | 3RCJ (Chain A)27 | 1.07 – 2.15 Å |
| CHK1 Kinase | 1ZYS (Chain A)28 | 0.45 – 0.87 Å |
| Farnesyl Pyrophosphate Synthase | 4DEM (Chain F)29 | 1.26 – 2.14 Å |
| Glucokinase | 3IDH (Chain A)30 | 1.05 – 2.82 Å |
| Protein Tyrosine Phosphatase 1B | 2CMB (Chain A)31 | 2.02 – 2.12 Å |
Processing MixMD results
For each protein target, the location of all probe atoms from the last 5 ns of the ten runs were binned onto a grid of 0.5-Å spacing, using the ptraj module from AmberTools.18 The raw bin counts (x) in each of the grid points were converted to sigma values using the equation (x − μ)/σ where μ is the mean of all the binned grid data and σ is the standard deviation of all the binned grid data. This allows us to represent the location of the probes in a manner commonly implemented for electron density from X-ray crystallography. The resulting maps were contoured at various sigma values and examined in the presence of the average protein structure to identify locations of maximal occupancy. A higher sigma value for a particular location on the grid signifies a higher residence time for a probe molecule at that particular location across all ten MixMD simulation runs. The maps in this study have been color coded as orange for acetonitrile, blue for isopropanol, and magenta for pyrimidine to represent the respective MixMD simulations from which they have been derived. These maps were visualized in PyMOL.32
Results and Discussion
A recurring theme across all the protein targets was the consistent identification of the active and allosteric sites in the top-four hotspots identified by MixMD. The lower-ranked hotspots in each protein corresponded to cofactors or additives found in crystal structures and their protein-packing interfaces, which are in principle easier to desolvate. Below, we explain our choice of targets, our assessment of the cosolvent probes, and the results for each system. Lastly, we compare our method to FTMap33–36 and demonstrate that dynamic sampling and competition with water are needed for superior performance in mapping binding sites.
Choice of protein targets and conformations
The definition of allostery is broad and in general is used to imply anything that does not modulate a protein’s activity by interacting with the competitive site. Under such a definition, there are many protein targets to choose from to test the ability of MixMD to identify allosteric sites. In order to avoid misinterpretation of allosteric sites, we focused our attention on those targets for which experimental data clearly supported an allosteric mechanism; moreover, we limited our choice of protein targets to those that had a verified allosteric site confirmed through crystallography. This allowed a proper and fair comparison of our MixMD mapping results for competitive and allosteric molecules in crystal structures. In order to provide a robust analysis of the technique, we chose to start MixMD simulations from crystal structures with no allosteric ligand bound. Complex allosteric mechanisms exist where allosteric effectors modulate the quaternary relationship of a multimeric complex of a protein. Simulating a large, multimeric complex is computationally expensive. As an example, Bacterial L-lactate dehydrogenase exists in a tetrameric state that can be modulated by its allosteric effector, fructose 1,6-bisphosphate. In order to accurately map the allosteric sites, such a system would need to be simulated as a tetramer which is computationally expensive.37,38 These large systems were left out from the current analysis and will be the subject of a future study. Careful curation left us with seven protein targets that had crystal structures with a competitive ligand bound but no allosteric ligands: ABL Kinase, Androgen Receptor (AR), PDK1 Kinase, Farnesyl Pyrophosphate Synthase (FPPS), Glucokinase, CHK1 Kinase, and Protein Tyrosine Phosphatase 1B (PTP1B). It would have been best to start from an apo structure with no ligands in both sites, but these were not available for all the systems to allow an even comparison. We acknowledge that the set has several kinases, but this simply reflects the intense interest in allosteric control of kinases over the last several years.
Identifying and ranking hotspots on the protein surface
Assessing the relative importance of hotspots mapped on the protein surface is essential in establishing their significance. We assessed the mapped sites based on two main criteria. First and foremost, sites mapped at a high sigma value were given greater preference because they are maximally occupied. The occupancies are measured in units of σ, which is the standard deviation across the entire grid. The maps in our figures present “signals” that are 20–90 times the “noise” across the grid. This is one reason our method has fewer spurious minima than others. Second, hotspots must be mapped by more than one probe type, which implies “bindability” by diverse chemical functionalities. Indeed, such an approach to identifying hotspots has been highlighted by Vajda and co-workers in their FT-MAP technique where sites mapped by multiple probes were identified as hotspots.33 It is important to note that our use of binary solvent is essential when we require sites to be mapped by multiple probes. This is a condition that cannot necessarily be met in ternary solvent simulations that have been reported earlier.8,15,16 This gives MixMD a distinct advantage.
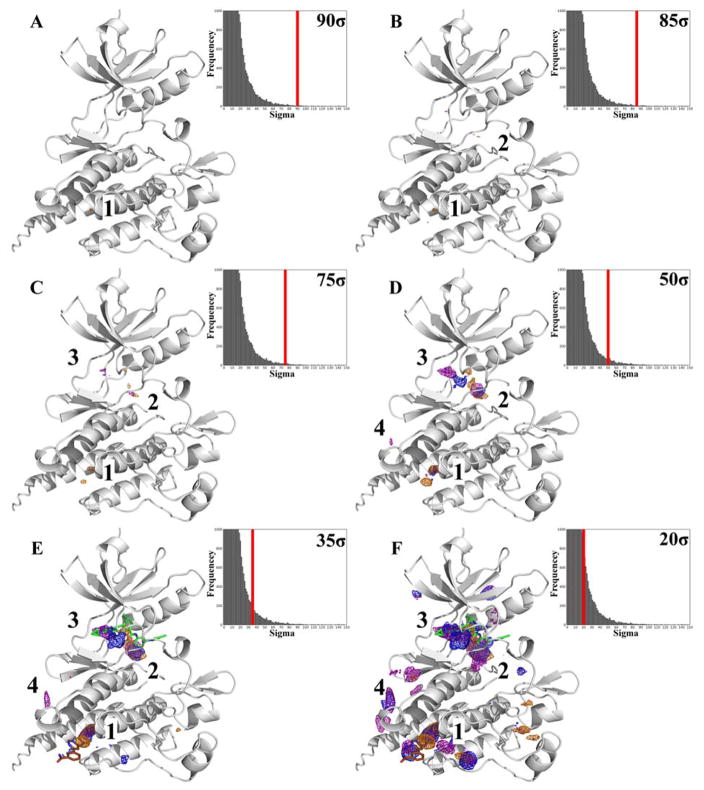
To illustrate the identification of hotspots with MixMD, Figure 1 shows ABL with probe occupancies contoured at varying sigma values. At 90σ in Figure 1A, the only hotspot mapped is in the allosteric site. However, upon decreasing it to 85σ (Figure 1B), we see that a second hotspot appears in the active site of ABL. As we continue decreasing the sigma value to 75σ (Figure 1C), a third hotspot appears which maps the hinge region of the active site in ABL. In lowering the sigma value further, we see that a fourth site appears on the protein surface at 50σ. Throughout this study, we have found it ideal to focus on the top-four hotspots. The supplemental information shows the same detailed process for ranking hotspots in all the other protein systems (see Figures S3–S11). In general, occupancy maps at 35σ clearly show the presence of the top-four sites before other spurious minima.
Figure 1.
The MixMD maps for ABL are contoured at varying sigma values from 90σ to 20σ to show the degree of molecular surface mapped by the probe atoms. The occupancy maps are color coded to represent MixMD results derived from different probes: acetonitrile (orange mesh), isopropanol (blue mesh), and pyrimidine (purple mesh). (A) At 90σ, the allosteric site shows the highest occupied points. (B) Maps contoured at 85σ, show a second hotspot in the active site. (C–F) When contouring the MixMD maps at successively lower sigma values, additional hotspots appear and are numbered based on their order of appearance. Unless sites are mapped by more than one probe type when contoured at 20σ they are ignored. The active site ligand (PDBid: 3KFA, green) and allosteric site ligand (PDBid: 3K5V, brown) are only shown for reference in E and F to orient the reader towards the location of the active and allosteric sites. We emphasize that no ligands were present in the MixMD simulations.
As the maps are contoured at lower sigma values successively in Figure 1D–F, the top-four sites increase in size and start to fuse, but smaller, less relevant sites start to appear. This fusion of mapped hotspots can be seen in the case of hotspot 2 and hotspot 3 that collectively map the entire competitive binding site (Figure 1F). Contouring our maps at 20σ allowed us to examine the full extent that the probes occupied the various binding sites. This was a common feature across all the protein targets in this study. Unless otherwise stated, all maps in our figures are contoured in two ways: one at 35σ to clearly show the presence of the top-four sites before other spurious minima and another at 20σ to show the full extent of mapping the binding sites. For clarity, those spurious sites are not shown in the “focused” 20σ figures. The 35σ maps are “raw” and show all sites in the occupancy grid.
Mapping active and allosteric sites with MixMD
ABL
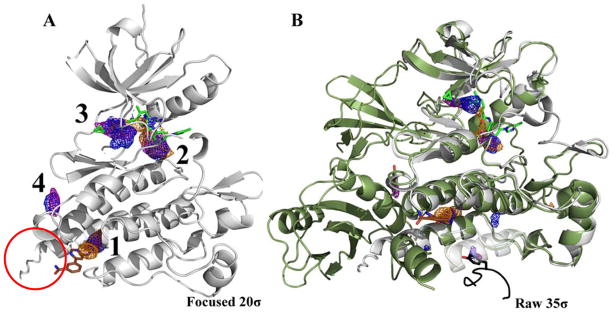
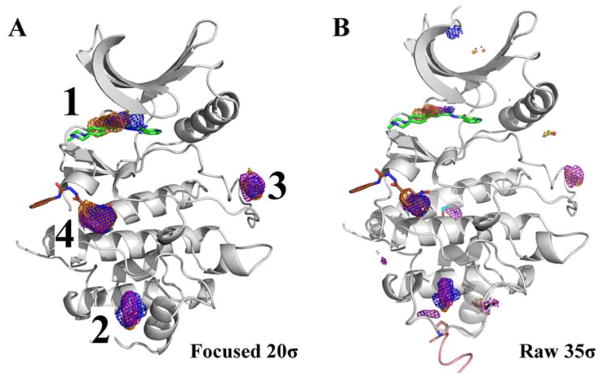
In analyzing the MixMD maps, we found that the active and allosteric sites are captured in the first-four hotspots for all systems examined. As mentioned earlier in the case of ABL, the top-three hotspots correspond to sub-sites in the active and allosteric sites. In ABL, the first hotspot lies in the allosteric site whereas the second and third hotspots map the entire active site (Figure 2A). However, it was interesting to find the fourth hotspot on the side of the protein. Upon checking the PDB for molecules that may complement this hotspot, we found that this location is part of the binding interface for the SH2 domain present in the full length protein. As shown in Figure 2B, the structure of the full length protein of ABL has a tyrosine residue from the SH2 domain occupying the location of the fourth hotspot. Clearly, this site has an important role in the functionality of ABL. One can envision that targeting such a site may likely disrupt the function of the kinase and thereby achieve allosteric modulation of ABL function. It is important to stress that while some sites mapped by MixMD may have no known allosteric regulatory role, these may be leveraged in the future to yield such a response.
Figure 2.
The first four hotspots from the ABL MixMD maps identify the active and allosteric sites: acetonitrile map (orange mesh), isopropanol map (blue mesh), and pyrimidine map (purple mesh). The active site ligand (PDBid: 3KFA, green) and the allosteric ligand (PDBid: 3K5V, brown) are shown for reference. (A) The four hotspots are shown contoured at 20σ with the spurious sites not shown. The red circle notes the αI′-helix that is referenced in a later section on conformational sampling. (B) MixMD maps of ABL contoured at 35σ with all sites shown. Superimposed on the structure are molecules from the PDB that occupy mapped hotspots on the protein surface. The crystal structure of the full length ABL protein (PDBid: 1OPK, dark green ribbon) was also superimposed to show the SH2 domain interface mapped by the fourth hotspot in MixMD. A different structure (PDBid: 1OPL) has a tyrosine residue (peptide in black) at the packing interface that occupies a small, lower-ranked hotspot at the bottom of the structure.
AR
In the case of the AR, the first-four hotspots map the active and allosteric sites (Figure 3A). As observed for ABL, the individual hotspots map sub-sites of the active and allosteric site which, when contoured at successively lower sigma values, fuse to map the entire binding site at 20σ. It is notable that for AR the active and the allosteric site are the only ones mapped in the first-four sites.
Figure 3.
(A) The location of just the top-four hotspots for the AR are shown contoured at 20σ: acetonitrile (orange), isopropanol (blue), and pyrimidine (purple). The active site ligand (PDBid: 2AM9, green) and the allosteric ligand (PDBid: 2PIX, brown) are shown for reference. A part of the α-helix obstructing the view has been hidden to provide a better view of the hotspots mapping the active site. (B) The MixMD maps are shown at 35σ that hotspots ranked five or lower correspond to crystal additives and packing contacts in other structures. Clearly, these are locations on the protein surface that can be desolvated and occupied. The different molecules are color coded as follows, Nuclear Receptor Co-Activator 2 in PDBid: 2QPY (black), glycerol in PDBid: 4HLW (yellow), and packing interface in PDBid: 2QPY (pink).
MixMD sites with lower sigma values could also provide relevant information for SBDD. The AR maps contoured at 35σ (Figure 3B) show that most lower-ranked hotspots mapped by multiple probes can be traced back to sites of biological and functional relevance (eg, locations of cofactors, substrates, or crystallographic additives). AR is activated by Nuclear Receptor Co-activator 2, and its binding location overlays with MixMD maps in Figure 3B. Several other crystal additives and protein-packing interfaces also coincide with the maps. This agreement between MixMD and the location of experimentally observed sites provides additional support that MixMD properly samples the protein surface for “easily desolvated” sites without getting stuck in irrelevant local minima. Similar results were observed for lower-ranked hotspots in other protein targets.
PDK1 and CHK1
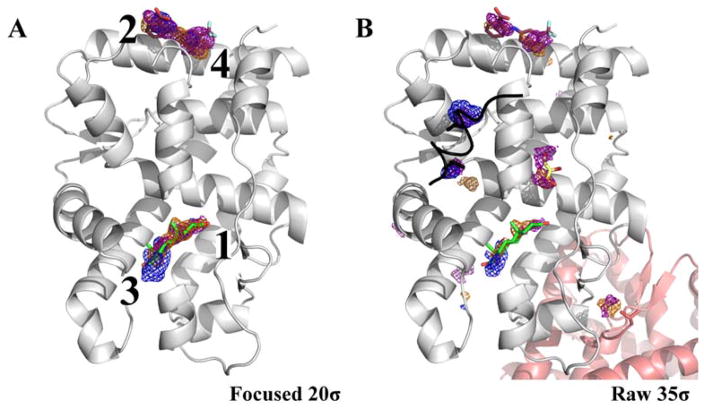
While such striking results were not achieved for PDK1, it is nonetheless important that allosteric sites and active sites were consistently captured in the first four hotspots (Figure 4). The active-site hinge region in PDK1 was mapped as the top hotspot whereas the allosteric site was mapped by the fourth hotspot. The second hotspot could be traced to a location occupied by a crystallographic additive in another PDB structure, and the third hotspot corresponded to the binding location of the proline ring of a peptide bound in the 3QC4 crystal structure of PDK1. These results suggest that MixMD identifies sites that could be easily desolvated, a prerequisite for druggable binding sites. Similar results were seen for CHK1 where the active site near the hinge region was mapped as the top hotspot (Figure 5). The allosteric site was ranked as the fourth hotspot and the hotspots ranked second and third denoted sub-sites for binding the peptide substrate of CHK1 on its surface.
Figure 4.
Just the top-four hotspots for PDK1 are shown contoured at 20σ, and raw occupancy maps are shown at 35σ. Maps are shown in mesh: acetonitrile (orange), isopropanol (blue), and pyrimidine (purple). The active site ligand (PDBid: 3RCJ, green) and the allosteric ligand from (PDBid: 4AW0, brown) are shown for reference. The first hotspot maps the hinge region of the active site. The second hotspot is located at the top of the protein. An additive bound at this site is overlaid on top from another PDK1 protein structure (PDBid: 3RWQ, pink). The third-ranked hotspot is known to bind a peptide (PDBid: 3QC4, yellow) and the fourth hotspot corresponds to the allosteric site. Lower-ranked hotspots in the 35σ frame overlap with protein contacts in PDBid: 3NAX (cyan) and PDBid: 3OTU (black).
Figure 5.
(A) CHK1 is shown with just the top-four hotspots contoured at 20σ. Maps are shown in mesh: acetonitrile (orange), isopropanol (blue), and pyrimidine (purple). The first and the fourth hotspots map the active (PDBid: 1ZYS, green) and allosteric site (PDBid: 3JVS, brown). The second and the third-ranked hotspots map sub-sites in the binding groove for the peptide substrate. (B) Maps contoured at 35σ show lower-ranked hotspots overlap with different CHK1 structures showing protein-packing interfaces (contact in structure 2YDK is shown in pink) and small crystal additives (structures 4FTO in black, 2YM8 in cyan, and 2XF0 in dark green).
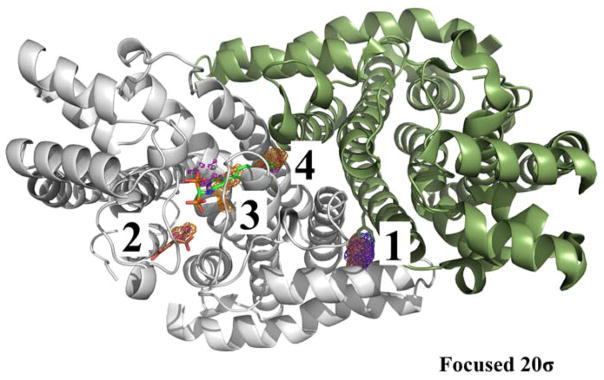
FPPS
MixMD simulations were performed on the proteins as monomers. This allowed the simulations to be completed in a reasonable amount of time. FPPS was interesting in this regard, since it functions as a dimer, but we simulated it as a monomer because the active and allosteric sites do not involve the second monomer (of course, second active and allosteric sites are contained in that second monomer). We assumed MixMD would identify part of the dimer interface. It is notable that the interface contains the first hotspot because dimerization must be highly favorable. A tyrosine residue from one of the monomers overlaps with the first hotspot as shown in Figure 6. This provides promising evidence in support of the use of MixMD as a technique to probe the location of biologically relevant binding partners of protein-protein interactions. The allosteric site was mapped by the second hotspot, and two sub-sites of the active site in FPPS were mapped by the third and fourth hotspots.
Figure 6.
The top-four hotspots for FPPS are contoured at 20σ. Maps are shown in mesh: acetonitrile (orange), isopropanol (blue), and pyrimidine (purple). The first hotspot maps the dimer interface. The second hotspot maps the allosteric site (PDBid: 3N5J, brown). The third- and fourth-ranked hotspots map two different sub-sites in the active site (PDBid: 4DEM, green). The protein was simulated as a monomer. However, the dimer is shown to illustrate that the top-ranked hotspot is located at the dimer interface. A tyrosine residue from one of the monomers colored dark green is shown to overlap with the first-ranked hotspot at the dimer interface.
Gluokinase reveals a small weakness of MixMD
We found that the active site of glukokinase was not well mapped by probes. In particular, the sub-site that binds sugar molecules in the active site was not mapped. Instead, the sugar-binding site was mapped by water molecules, which is likely reasonable given the structure of saccharides. Sugar-binding proteins are generally not considered druggable,39,40 so this result supports our proposal that MixMD preferentially identifies druggable binding sites. Part of the ATP cofactor’s site was mapped with the fourth hotspot, but charged probes would be needed to map the phosphate tail of ATP. Notably, the allosteric site was extensively mapped by the first hotspot when contoured at 20σ (Figure 7). However, the second- and third-ranked hotspots could not be traced back to examples from the PDB of molecules that could bind at these locations.
Figure 7.
Glucokinase is shown with just the top-four hotspots contoured at 20σ. Maps are shown in mesh: acetonitrile (orange), isopropanol (blue), and pyrimidine (purple). The first hotspot extensively maps the allosteric site (PDBid: 3H1V, brown). The fourth-ranked hotspot maps part of the ATP binding site on glucokinase (PDBid: 3FGU, grey). B) In addition to the top-four hotspots, a few very small sites are present at 35σ. They are clearly less occupied and lower ranked. A protein packing interface from PDB structure 3QIC (cyan) overlaps one of these small sites.
PTP1B reveals a clear need for charged probes in MixMD
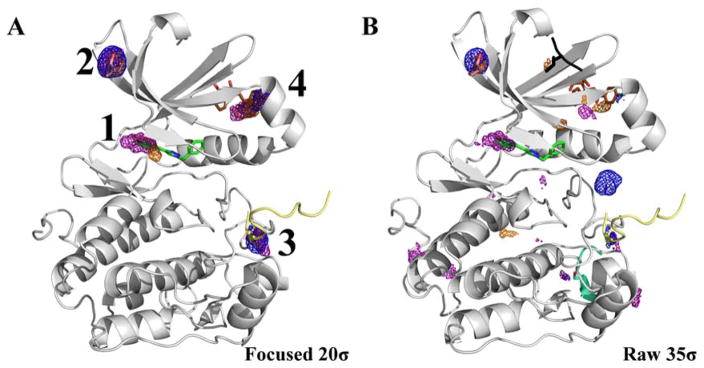
In PTP1B, our standard probes mapped the allosteric site as expected (second hotspot in Figure 8A). The first, third, and fourth hotspots captured the locations of crystal additives and protein-interaction sites as shown in Figure 8A. However, the active site was not identified.
Figure 8.
(A) The location of the top-4 hotspots is shown for PTP1B. Maps for the organic probes are shown in mesh: acetonitrile (orange), isopropanol (blue), and pyrimidine (purple). The first hotspot maps the location of an additive seen in a crystal structure (PDBid: 3RWQ, pink but difficult to see in this view). The second hotspot maps the allosteric site (PDBid: 1T49, brown). Hotspot 3 is located near the packing interface PDBid: 2CMC (cyan). Hotspot 4 is located near the packing interface of PDBid: 4GRY. The raw 35σ structure is in the supplemental information. The active site ligand (PDBid: 2CMB, green) is shown for reference. (B) A MixMD simulation of acetate + methyl ammonium was carried out, and the top-four hotspots are shown contoured at 20σ. Acetate hotspots are in red mesh and methyl ammonium hotspots are in blue. A fragment of the substrate is overlaid on top of the protein (PDBid: 1G1F, black). The top-two ranked hotspots are from the acetate ion mapping both sites known to bind phosphorylated tyrosines.
It would have been fulfilling to map the binding site of ATP’s phosphates in glucokinase, but it was not a compelling case for developing charged probes. PTP1B was the example that proved we needed them. The active site of PTP1B is charged and known to bind phosphorylated residues. Our MixMD probes did not map the binding site, which is known to accommodate inhibitors. Similar results were obtained by Bakan et al. who had initially carried out a simulation of PTP1B with isopropanol probes and found no mapping of the active site.8 However, they showed that acetate probes could map the active site. Indeed, we found that a MixMD simulations of acetate + methyl ammonium ions (a ternary solution balanced for system neutrality) mapped the active site well, specifically identifying the sites known to bind phosphorylated tyrosines as hotspots 1 and 2 (Figure 8B). PTP1B acts as a negative regulator of insulin signaling by dephosphorylating key residues of the insulin receptor which inactivates it.41
We have developed MixMD to identify druggable sites, and our probes were chosen because they are similar to drug-like molecules. Clearly, the MixMD technique can be adapted to identify other functionally relevant sites by tailoring the set of probes. However, using maps from charged probes has the hazard of pulling the search for leads out of drug-like space. Targeting the charged binding site of PTP1B in the context of drug discovery has proven difficult as multiple iterations of medicinal chemistry efforts have been met with limited success in replacing the charged site on inhibitors.42,43 Several reviews and druggability detection methods on the subject have expressed the view of PTP1B as an undruggable target due to the difficult and slow progress in optimizing compounds.43–45 In fact, this is one of the primary reasons drug discovery efforts targeting phosphatases have been ignored in favor of kinases, even though kinases and phosphatases are known to work in tandem to regulate major disease-related pathways.43 The discovery of an allosteric site on PTP1B has renewed interest in alternative strategies of targeting PTB1B.46
Comparison of MixMD to FTMap
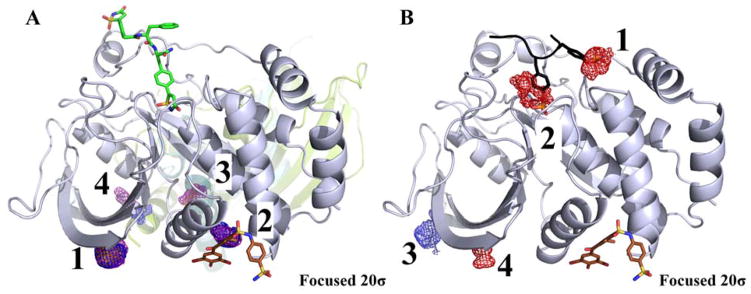
MixMD simulations take a long time; do such costly simulations have an advantage over quick methods that map proteins using static crystal structures. Table 1 shows that the conformational change between competitive- and allosteric-bound states is large for ABL, but the rest of the systems have RMSD < 3Å. Those conformational changes are small enough that some methods might be able to predict the allosteric site using only the competitive-bound crystal structures we used to initiate the MixMD simulations. If other methods cannot identify the allosteric sites from the crystal structures, it would highlight the importance of including protein flexibility and explicit competition with water through MD simulations.
A reviewer specifically asked that we add a comparison of MixMD against the widely used FTMap method.33–36 FTMap identifies hotspots by minimizing millions of small probe molecules on a static protein structure. Druggable sites are identified by clusters of different, overlapping probe types.33 FTMap uses 16 types of probe molecules, and at least two kinds of probes must have minima that overlap on the protein surface. The crystal structures we used for MixMD (see Table 1) were submitted to the FTMap server,36 and the results are summarized in Figure 9. In four of the seven systems (ABL, AR, CHK1, and PTP1B), the allosteric sites were not identified by FTMap. Furthermore, many potentially spurious sites outside the competitive and allosteric sites were identified (black molecules in Figure 9), which makes it difficult to know which sites to target in prospective applications. Lastly, we should note that FTMap had small difficulties with FPPS. Sites 1 and 2 mapped the competitive site well and occupied the same space as known ligands, but only the far edges of sites 3 and 6 had small overlap with the a few atoms in ligands. For the allosteric site, FTMap’s site 4 was the same sort of small overlap at the edges. All other matching sites in all other systems in Figure 9 had excellent overlap with the whole probes occupying the same volumes as the known ligands.
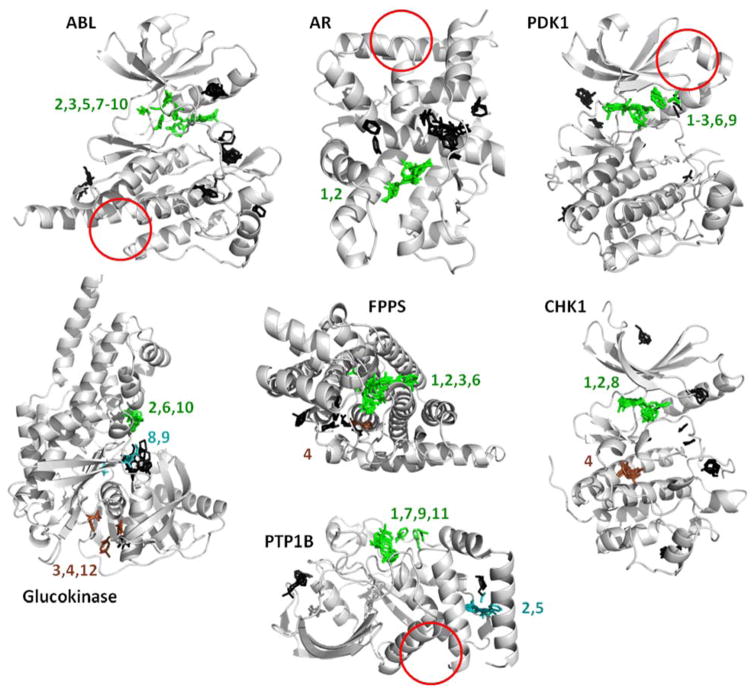
Figure 9.
These are the hotspots provided by the FTMap server, run on the same crystal structures we used to initiate the MixMD simulations (see Table 1). FTMap does well identifing all the active sites, but it does not detect the allosteric sites in half the systems (red circles). The view of the proteins is the same as the previous figures to allow the reader to compare the results. Hotspots are determined from clusters of probe molecules and ranked according to their average energy (cluster numbers are shown). Each cluster above is colored based on whether it overlaps with known ligands. Green indicates a cluster that overlaps with competitive ligands; brown clusters overlap allosteric ligands; black clusters do not overlap either binding site and their rank numbers are not shown. Cyan clusters in glucokinase overlap correct positions that were not mapped by MixMD. Cyan clusters in PTP1B map the site of a large crystallographic additive, which also was not mapped by MixMD. Please note that numbering in FTMap starts at zero, and we have shifted the numbers to start at one.
FTMap has a strong track record, and it came as no surprise that it identified the active sites in every one of the proteins. Some pre-organization of the binding sites was possible from the competitive ligands bound in the crystal structures, but that was an advantage to both FTMap and MixMD. We were pleased that MixMD simulations achieved superior results using only three cosolvents (plus charged probes for PTP1B) in comparison to the sixteen diverse probes used in FTMap. However, there were a few instances where that probe diversity may have given FTMap an advantage over MixMD. FTMap actually identifies the sugar site in glucokinase that we missed. It also identifies the binding site of the phosphate tail of ATP in glukokinase. After our PTP1B simulations, we did not go back and add charged probes in our simulations of glucokinase, so we do not know if MixMD would have identified the site of the phosphate tail using the more representative probes. FTMap also has interesting results for PTP1B that are different from our own. Sites 2 and 5 from FTMap overlay the site of an octylglucoside that was present in the crystal structure; octylglucoside is a non-ionic detergent additive. We are not aware of any biological significance for the molecule, but it creates a large cavity where the His-tag on the N-terminus is packed against the C-terminus of PTP1B in the crystal structure. The cavity closes in the MixMD simulations, but it is completely reasonable that FTMap identified this cavity.
Limits of conformational sampling with MixMD
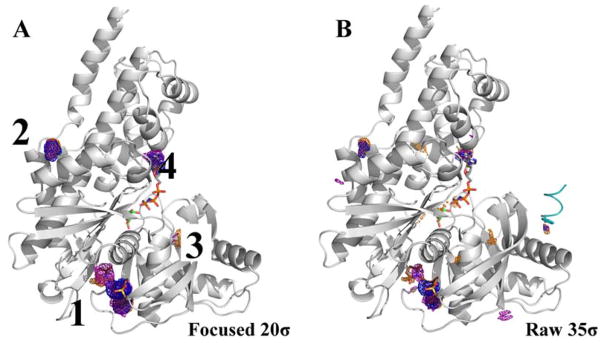
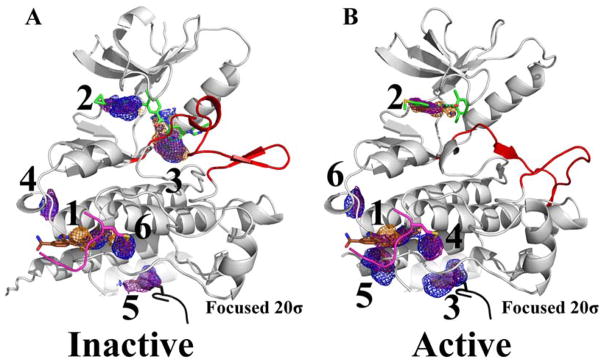
Proteins exist in an ensemble of functionally relevant conformations, and studying the effect of starting conformation on MixMD performance is especially important in the context of allosteric control. As noted in the previous section, Table 1 shows that ABL is the member of our set with the largest conformational change possible. If the reader compares the ABL structure in Figure 2A (average structure from the MixMD simulations) to the ABL structure in Figure 9 (crystal structure used to initiate MixMD and FTMap), it is clear that there is a conformational change in the αI′-helix (inside the red circle in Figure 2A, but the αI’-helix is to the left of the red circle around the allosteric site of ABL in Figure 9). MixMD started with helix in the extended conformation, but the structures spontaneously adopted the bent conformation by the end of all the simulations. The kinked αI′-helix is seen in the autoinhibitory state when a myristate tail binds in the adjacent allosteric site. It is known that molecules can bind to the allosteric site with the helix in either state, but inhibition only happens when the helix is induced to bend.47 This raises an important point that ligands may bind to the sites identified by MixMD and result in allosteric regulation – either inhibition or activation – but they may bind without any alteration of activity.
Another issue for ABL is the active (DFG-in) and inactive (DFG-out) conformation of the activation loop. The simulations shown in Figure 2 have the DFG-out conformation, so we ran an additional set of MixMD simulations using the DFG-in, active conformation from PDB structure 1M52 to examine the effect of starting conformation. We found there was no inter-conversion between the two conformational states of the DFG loop during either set of the MixMD simulations. However, it is important to note that the competitive and allosteric sites were consistently mapped as the top-two hotspots starting from either conformation.
Interestingly, the third-ranked hotspot in MixMD of the inactive form (Figure 10A) was missing in MixMD results from the active form (Figure 10B). This is because the activation loop in the active form occupies the hotspot present in the inactive form and precludes the mapping of this site by probe molecules. Also, we saw different rankings of hotspots between the two conformations. For instance, the fourth-ranked hotspot in the inactive form of ABL (Figure 10A) is ranked sixth in the active form (Figure 10B). In its place, a hotspot in the peptide substrate binding site takes precedence in the active form of ABL. This is in perfect agreement from the standpoint of catalytic activity, as the DFG-in form of ABL binds peptide substrates to phosphorylate them. These subtle changes in MixMD rankings starting from different conformations opens up exciting prospects for the use of MixMD in understanding the functional relevance of different conformations of proteins.
Figure 10.
MixMD simulations were performed using the (A) DFG-out inactive (PDBid: 3KFA) and (B) DFG-in active (PDBid: 1M52) forms of ABL. The activation loop is colored red to show the difference in this region between the two protein conformations. Maps for the organic probes are shown in mesh: acetonitrile (orange), isopropanol (blue), and pyrimidine (purple). The top-six hotspots are shown for both conformations to illustrate the rearrangement of the hotspot rankings. For both conformations, the allosteric and active sites were ranked first and second, respectively. The third-ranked hotspot in the inactive conformation is now occupied by the activation loop in the active form of ABL, this leads to a rearrangement in raking of several sites on the protein. Two protein packing interfaces are shown colored black (PDBid: 1OPL) and pink (PDBid: 3QRK). The active site ligand (PDBid: 3KFA, green) and allosteric site ligand (PDBid: 3K5V, brown) are shown for reference.
Conclusion
MixMD simulations map hotspots on protein surfaces while allowing for protein flexibility and competition with water. In this study, we have successfully demonstrated the application of MixMD to several allosteric systems, identifying the active and allosteric sites within the top-four sites. Also, sites that do not correspond to active and allosteric sites represent locations of cofactor binding sites and protein multimerization/packing interfaces. While our choice of probes reflects the need to map druggable and easy-to-desolvate binding sites, we have proven that one can easily extend the technique by employing a different set of probes to map charged binding sites if needed. Furthermore, we highlight the advantage of MixMD simulations to identify allosteric sites compared to the hotspot-mapping technique FTMap. We have also explored the role of protein starting conformation on MixMD, using ABL as a test case. The subtle changes in MixMD rankings between the active and inactive conformations of ABL were found to reflect the underlying differences in the functional relevance of these protein conformations. In future studies, we intend to explore the relationship between protein starting conformation and MixMD results in further detail in order to establish the significance of these findings.
Acknowledgments
We thank Dr. Charles L. Brooks III for providing access to the Gollum clusters at the University of Michigan, and the IBM Matching Grants Program for granting the GPU units for high-performance MD simulations. We greatly appreciate the generous donation of the MOE software from Chemical Computing Group. This work has been supported by the National Institutes of Health (GM65372).
Footnotes
Supporting Information
Force field parameters and protocols used for the acetate + methyl ammonium MixMD, a detailed depiction of the MixMD hotspot-ranking process for the protein systems studied, and agreement of MixMD maps with experimental crystallographic data from the PDB are presented.
References
- 1.Tsai CJ, del Sol A, Nussinov R. Allostery: Absence of a Change in Shape Does Not Imply That Allostery Is Not at Play. J Mol Biol. 2008;378:1–11. doi: 10.1016/j.jmb.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nussinov R, Tsai CJ. Allostery in Disease and in Drug Discovery. Cell. 2013;153:293–305. doi: 10.1016/j.cell.2013.03.034. [DOI] [PubMed] [Google Scholar]
- 3.Ma L, Seager MA, Wittmann M, Jacobson M, Bickel D, Burno M, Jones K, Graufelds VK, Xu G, Pearson M, et al. Selective Activation of the M1 Muscarinic Acetylcholine Receptor Achieved by Allosteric Potentiation. Proc Natl Acad Sci. 2009;106:15950–15955. doi: 10.1073/pnas.0900903106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hardy JA, Wells JA. Searching for New Allosteric Sites in Enzymes. Curr Opin Struct Biol. 2004;14:706–715. doi: 10.1016/j.sbi.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 5.Hardy JA, Lam J, Nguyen JT, O’Brien T, Wells JA. Discovery of an Allosteric Site in the Caspases. Proc Natl Acad Sci U S A. 2004;101:12461–12466. doi: 10.1073/pnas.0404781101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reynolds KA, McLaughlin RN, Ranganathan R. Hot Spots for Allosteric Regulation on Protein Surfaces. Cell. 2011;147:1564–1575. doi: 10.1016/j.cell.2011.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McClendon CL, Friedland G, Mobley DL, Amirkhani H, Jacobson MP. Quantifying Correlations Between Allosteric Sites in Thermodynamic Ensembles. J Chem Theory Comput. 2009;5:2486–2502. doi: 10.1021/ct9001812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bakan A, Nevins N, Lakdawala AS, Bahar I. Druggability Assessment of Allosteric Proteins by Dynamics Simulations in the Presence of Probe Molecules. J Chem Theory Comput. 2012;8:2435–2447. doi: 10.1021/ct300117j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allen KN, Bellamacina CR, Ding X, Jeffery CJ, Mattos C, Petsko GA, Ringe D. An Experimental Approach to Mapping the Binding Surfaces of Crystalline Proteins†. J Phys Chem. 1996;100:2605–2611. [Google Scholar]
- 10.Mattos C, Ringe D. Locating and Characterizing Binding Sites on Proteins. Nat Biotechnol. 1996;14:595–599. doi: 10.1038/nbt0596-595. [DOI] [PubMed] [Google Scholar]
- 11.Lexa KW, Carlson HA. Full Protein Flexibility Is Essential for Proper Hot-Spot Mapping. J Am Chem Soc. 2011;133:200–202. doi: 10.1021/ja1079332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lexa KW, Carlson HA. Improving Protocols for Protein Mapping through Proper Comparison to Crystallography Data. J Chem Inf Model. 2013;53:391–402. doi: 10.1021/ci300430v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ung PMU, Ghanakota P, Graham SE, Lexa KW, Carlson HA. Identifying Binding Hot Spots on Protein Surfaces by Mixed-Solvent Molecular Dynamics: HIV-1 Protease as a Test Case. Biopolymers. 2016;105:21–34. doi: 10.1002/bip.22742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seco J, Luque FJ, Barril X. Binding Site Detection and Druggability Index from First Principles. J Med Chem. 2009;52:2363–2371. doi: 10.1021/jm801385d. [DOI] [PubMed] [Google Scholar]
- 15.Guvench O, MacKerell AD. Computational Fragment-Based Binding Site Identification by Ligand Competitive Saturation. PLoS Comput Biol. 2009;5:e1000435. doi: 10.1371/journal.pcbi.1000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raman EP, Yu W, Guvench O, MacKerell AD. Reproducing Crystal Binding Modes of Ligand Functional Groups Using Site-Identification by Ligand Competitive Saturation (SILCS) Simulations. J Chem Inf Model. 2011;51:877–896. doi: 10.1021/ci100462t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Molecular Operating Environment. Chemical Computing Group Inc; Montreal, Canada: 2010. [Google Scholar]
- 18.Case DA, Darden T, Cheatham T, Simmerling CL, Wang J, Duke RE, Luo R, Walker R, Zhang W, Merz K, et al. Amber 11. University of California; 2010. [Google Scholar]
- 19.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of Simple Potential Functions for Simulating Liquid Water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 20.Lexa KW, Goh GB, Carlson HA. Parameter Choice Matters: Validating Probe Parameters for Use in Mixed-Solvent Simulations. J Chem Inf Model. 2014;54:2190–2199. doi: 10.1021/ci400741u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of Multiple Amber Force Fields and Development of Improved Protein Backbone Parameters. Proteins Struct Funct Bioinforma. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ryckaert JP, Ciccotti G, Berendsen HJ. Numerical Integration of the Cartesian Equations of Motion of a System with Constraints: Molecular Dynamics of N-Alkanes. J Comput Phys. 1977;23:327–341. [Google Scholar]
- 23.Götz AW, Williamson MJ, Xu D, Poole D, Le Grand S, Walker RC. Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 1. Generalized Born. J Chem Theory Comput. 2012;8:1542–1555. doi: 10.1021/ct200909j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andersen HC. Molecular Dynamics Simulations at Constant Pressure And/or Temperature. J Chem Phys. 1980;72:2384–2393. [Google Scholar]
- 25.Zhou T, Commodore L, Huang WS, Wang Y, Sawyer TK, Shakespeare WC, Clackson T, Zhu X, Dalgarno DC. Structural Analysis of DFG-in and DFG-out Dual Src-Abl Inhibitors Sharing a Common Vinyl Purine Template. Chem Biol Drug Des. 2010;75:18–28. doi: 10.1111/j.1747-0285.2009.00905.x. [DOI] [PubMed] [Google Scholar]
- 26.Pereira de Jésus-Tran K, Côté PL, Cantin L, Blanchet J, Labrie F, Breton R. Comparison of Crystal Structures of Human Androgen Receptor Ligand-Binding Domain Complexed with Various Agonists Reveals Molecular Determinants Responsible for Binding Affinity. Protein Sci. 2006;15:987–999. doi: 10.1110/ps.051905906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merkul E, Klukas F, Dorsch D, Grädler U, Greiner HE, Müller TJJ. Rapid Preparation of Triazolyl Substituted NH-Heterocyclic Kinase Inhibitors via One-Pot Sonogashira coupling–TMS-deprotection–CuAAC Sequence. Org Biomol Chem. 2011;9:5129–5136. doi: 10.1039/c1ob05586k. [DOI] [PubMed] [Google Scholar]
- 28.The citation for structure 1ZYS at the PDB is listed as “To be published”. Stavenger RA, Zhao B, Zhou B-BS, Brown MJ, Lee D, Holt DA. [accessed May 27, 2016];Pyrrolo[2,3-B]pyridines Inhibit the Checkpoint Kinase Chk1. http://www.rcsb.org/pdb/explore/explore.do?structureId=1zys.
- 29.Lin YS, Park J, De Schutter JW, Huang XF, Berghuis AM, Sebag M, Tsantrizos YS. Design and Synthesis of Active Site Inhibitors of the Human Farnesyl Pyrophosphate Synthase: Apoptosis and Inhibition of ERK Phosphorylation in Multiple Myeloma Cells. J Med Chem. 2012;55:3201–3215. doi: 10.1021/jm201657x. [DOI] [PubMed] [Google Scholar]
- 30.Petit P, Antoine M, Ferry G, Boutin JA, Lagarde A, Gluais L, Vincentelli R, Vuillard L. The Active Conformation of Human Glucokinase Is Not Altered by Allosteric Activators. Acta Crystallogr D Biol Crystallogr. 2011;67:929–935. doi: 10.1107/S0907444911036729. [DOI] [PubMed] [Google Scholar]
- 31.Ala PJ, Gonneville L, Hillman MC, Becker-Pasha M, Wei M, Reid BG, Klabe R, Yue EW, Wayland B, Douty B, et al. Structural Basis for Inhibition of Protein-Tyrosine Phosphatase 1B by Isothiazolidinone Heterocyclic Phosphonate Mimetics. J Biol Chem. 2006;281:32784–32795. doi: 10.1074/jbc.M606873200. [DOI] [PubMed] [Google Scholar]
- 32.The PyMOL Molecular Graphics System. Schrödinger, LLC; [Google Scholar]
- 33.Brenke R, Kozakov D, Chuang GY, Beglov D, Hall D, Landon MR, Mattos C, Vajda S. Fragment-Based Identification of Druggable “hot spots” of Proteins Using Fourier Domain Correlation Techniques. Bioinformatics. 2009;25:621–627. doi: 10.1093/bioinformatics/btp036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kozakov D, Hall DR, Chuang GY, Cencic R, Brenke R, Grove LE, Beglov D, Pelletier J, Whitty A, Vajda S. Structural Conservation of Druggable Hot Spots in Protein–protein Interfaces. Proc Natl Acad Sci. 2011;108:13528–13533. doi: 10.1073/pnas.1101835108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bohnuud T, Beglov D, Ngan CH, Zerbe B, Hall DR, Brenke R, Vajda S, Frank-Kamenetskii MD, Kozakov D. Computational Mapping Reveals Dramatic Effect of Hoogsteen Breathing on Duplex DNA Reactivity with Formaldehyde. Nucleic Acids Res. 2012;40:7644–7652. doi: 10.1093/nar/gks519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kozakov D, Grove LE, Hall DR, Bohnuud T, Mottarella SE, Luo L, Xia B, Beglov D, Vajda S. The FTMap Family of Web Servers for Determining and Characterizing Ligand-Binding Hot Spots of Proteins. Nat Protoc. 2015;10:733–755. doi: 10.1038/nprot.2015.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iwata S, Kamata K, Yoshida S, Minowa T, Ohta T. T and R States in the Crystals of Bacterial L–lactate Dehydrogenase Reveal the Mechanism for Allosteric Control. Nat Struct Mol Biol. 1994;1:176–185. doi: 10.1038/nsb0394-176. [DOI] [PubMed] [Google Scholar]
- 38.Ohta T, Yokota K, Minowa T, Iwata S. Mechanism of Allosteric Transition of Bacterial L-Lactate Dehydrogenase. Faraday Discuss. 1992;93:153–162. doi: 10.1039/fd9929300153. [DOI] [PubMed] [Google Scholar]
- 39.Hopkins AL, Groom CR. The Druggable Genome. Nat Rev Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 40.Aretz J, Wamhoff E-C, Hanske J, Haymann D, Rademacher C. Computational and Experimental Prediction of Human C-Type Lectin Receptor Druggability. Front Immunol. 2014;5 doi: 10.3389/fimmu.2014.00323. article 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tonks NK. PTP1B: From the Sidelines to the Front Lines! FEBS Lett. 2003;546(1):140–148. doi: 10.1016/s0014-5793(03)00603-3. [DOI] [PubMed] [Google Scholar]
- 42.Barr AJ. Protein Tyrosine Phosphatases as Drug Targets: Strategies and Challenges of Inhibitor Development. Future Med Chem. 2010;2:1563–1576. doi: 10.4155/fmc.10.241. [DOI] [PubMed] [Google Scholar]
- 43.De Munter S, Köhn M, Bollen M. Challenges and Opportunities in the Development of Protein Phosphatase-Directed Therapeutics. ACS Chem Biol. 2013;8:36–45. doi: 10.1021/cb300597g. [DOI] [PubMed] [Google Scholar]
- 44.Blaskovich MAT. Drug Discovery and Protein Tyrosine Phosphatases. Curr Med Chem. 2009;16:2095–2176. doi: 10.2174/092986709788612693. [DOI] [PubMed] [Google Scholar]
- 45.Cheng AC, Coleman RG, Smyth KT, Cao Q, Soulard P, Caffrey DR, Salzberg AC, Huang ES. Structure-Based Maximal Affinity Model Predicts Small-Molecule Druggability. Nat Biotechnol. 2007;25:71–75. doi: 10.1038/nbt1273. [DOI] [PubMed] [Google Scholar]
- 46.Hansen SK, Cancilla MT, Shiau TP, Kung J, Chen T, Erlanson DA. Allosteric Inhibition of PTP1B Activity by Selective Modification of a Non-Active Site Cysteine Residue†. Biochemistry (Mosc) 2005;44:7704–7712. doi: 10.1021/bi047417s. [DOI] [PubMed] [Google Scholar]
- 47.Jahnke W, Grotzfeld RM, Pellé X, Strauss A, Fendrich G, Cowan-Jacob SW, Cotesta S, Fabbro D, Furet P, Mestan J, et al. Binding or Bending: Distinction of Allosteric Abl Kinase Agonists from Antagonists by an NMR-Based Conformational Assay. J Am Chem Soc. 2010;132:7043–7048. doi: 10.1021/ja101837n. [DOI] [PubMed] [Google Scholar]