ABSTRACT
Nuclear lamins are the major components of the nuclear lamina at the periphery of the nucleus, supporting the nuclear envelope and participating in many nuclear processes, including DNA replication, transcription and chromatin organization. A group of diseases, the laminopathies, is associated with mutations in lamin genes. One of the most striking cases is Hutchinson-Gilford progeria syndrome (HGPS) which is the consequence of a lamin A dominant negative mutant named progerin. Due to the abnormal presence of a permanent C-terminal farnesyl tail, progerin gradually accumulates on the nuclear membrane, perturbing a diversity of signalings and transcriptional events. The accumulation of progerin has led to the speculation that progerin possesses higher stability than the wild type lamin A protein. However, the low solubility of lamin proteins renders traditional immunoprecipitation-dependent methods such as pulse-chase analysis ineffective for comparing the relative stabilities of mutant and wild type lamins. Here, we employ a novel platform for inferring differences in lamin stability, which is based on normalization to a co-translated reporter protein following porcine teschovirus-1 2A peptide-mediated co-translational cleavage. The results obtained using this method support the notion that progerin is more stable than lamin A. Moreover, treatment of FTI reduces progerin relative stability to the level of wild type lamin A.
KEYWORDS: FTI, lamin A, lamin B1, progerin, protein relative stability, protein relative abundance
Introduction
The nuclear lamina, which is found at the interface between chromatin and the inner nuclear membrane, is an essential fibrillar network structure providing mechanical support to the nucleus in the cell.1,2 The major components of the lamina are nuclear lamin proteins. The A type lamins (lamin A, AΔ10, C, and C2 isoforms) are alternatively spliced products of the LMNA gene and are expressed primarily in differentiated cells.3-6 The B type lamin B1 is encoded by LMNB1, while lamin B2 and B3 are isoforms derived from LMNB2.3,7,8 Lamin proteins actively participate in various critical cellular functions including gene regulation, chromatin organization, nuclear envelope assembly, transcription, apoptosis, etc,2,9-11 and lamin mutations are associated with a wide range of human diseases known as laminopathies.12,13
One tragic example of human laminopathies is Hutchinson-Gilford progeria syndrome (HGPS), an extremely rare genetic disorder, affecting 1 out of 1 in 4–8 million persons, that manifests striking accelerated aging symptoms.12,14 Although this disease was first documented 130 y ago,15 the molecular mechanism that gives rise to progeria was not unraveled until 2003.16 The LMNA gene in HGPS patients carries a de novo nucleotide substitution from C to T at position 1824, which does not alter the amino acid sequence (G608G), but instead introduces a cryptic splice donor site that generates a 150-nucleotide deletion within the mRNA. The resulting protein product named progerin, carries a 50 amino acid in-frame deletion that prevents cleavage by ZMPSTE24, which normally removes the C-terminal farnesyl group as part of the processing of wild type lamin A.17-19 As a consequence, the permanently farnesylated progerin aberrantly accumulates on the nuclear envelope, eliciting numerous nuclear abnormalities in the cells of HGPS patients, including the hallmark phenotype – abnormal blebbed nuclei, as well as disrupted heterochromatin-lamin interactions and alterations in gene transcription.16,20-22 HGPS patients appear healthy at birth, but with a gradual accumulation of progerin in the nucleus, they start to display the traits of accelerated aging around 12 months, and unfortunately die of heart complications such as heart attacks or strokes in their early teens.12,23-26 Based on the observed buildup of progerin in the cells of HGPS patients, it has been speculated that progerin is less susceptible to proteolysis and more stable than the wild type lamin A.
The commonly used approach of measuring protein stability is pulse-chase analysis, which metabolically labels the protein of interest in the cells with a radioactive precursor for a short period, then chased with an excess of nonradioactive precursor molecules in the culture medium, followed by immunoprecipitation and SDS-PAGE to quantify the radiolabeled protein.27,28 However, successful deployment of this method, particularly the step of immunoprecipitation, largely depends on the solubility of the target protein. Although this method has been widely used to examine pre-lamin stability,29 the tendency of lamin proteins to polymerize into higher order insoluble structures in vitro at relatively low critical concentrations30 has a potential to interfere with the accurate assessment of lamin protein stability using this methodology. To overcome this limitation, we adapted the novel method originally employed by Rodriguez-Contreras and colleagues31 to demonstrate differential glucose transporter stability under various growth conditions in the protozoan parasite Leishmania mexicana. This simplified approach exploits the unique properties of viral 2A peptide sequences32 in a manner that does not require immunoprecipitation or radiolabelling of cells, and consequently avoids the complications arising from the treatment of cells with the translation inhibitor cycloheximide.
The 2A peptide was initially discovered and characterized in the foot and mouth disease virus (FMDV) which was shown to mediate the production of 2 polypeptides (i.e., 2A and 2B) from the virus' complex single open reading frame (ORF). Translation of the 19 amino acid 2A peptide coding sequence causes an intra-ribosomal “skipping” event between the final Gly residue of the 2A peptide and the first Pro residue of the next polypeptide, causing the release of the first polypeptide and reinitiating translation of the second polypeptide starting with Pro (Fig. 1A)(for simplicity, this process will be referred to as “cleavage”).32-35 Functional 2A peptide-like sequences have been discovered in several other viruses and retrotransposons, and various versions of the sequence have been exploited in molecular biology, gene therapy, and biotechnology applications because they enable the production of multiple polypeptides from single open reading frames.32 Because the 2 polypeptides resulting from a 2A-mediated co-translational cleavage event are inherently transcriptionally and translationally co-regulated, their relative abundance in the cell is determined solely by their post-translational stability. Differences in the abundance ratio of the polypeptides among cell types or under differential growth conditions will reflect alterations in the post-translational stability of one or both polypeptides. It has been reported that the 2A sequence from Porcine teschovirus-1 (P2A) has the highest cleavage efficiency among all 4 commonly used 2A sequences.36 Therefore we chose to use P2A in our study.
Figure 1.
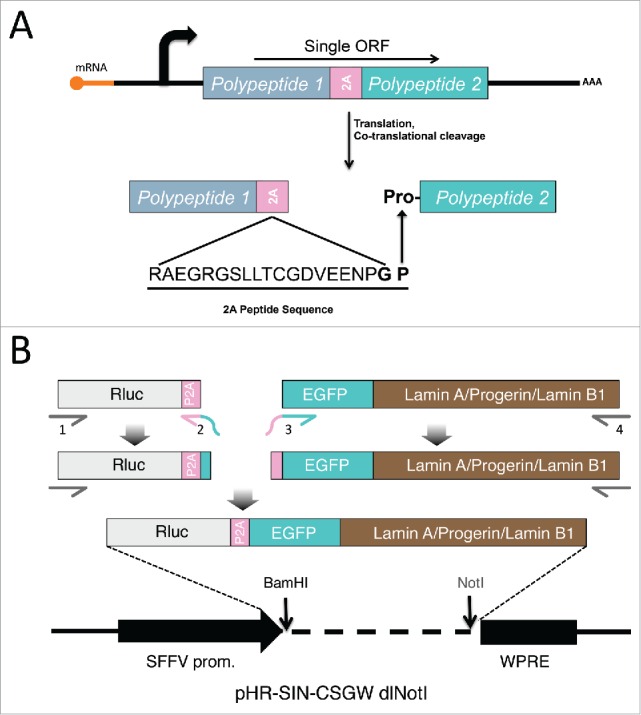
Schematic diagrams of processing and creation of 2A constructs. (A) Processing of 2A-linked constructs. DNA sequences of Polypeptide 1 and 2 are connected by a 2A motif and transcribed into a single ORF. The two polypeptides are then separated during translation by a co-translational, intraribosomal cleavage right before the proline at the end of the 2A sequence, resulting the proline at the N-terminus of polypeptide 2. (B) Generation of luciferase-P2A-lamin (lamin A, progerin, lamin B1) constructs. Segments of luciferase-P2A and EGFP tagged lamins were amplified and linked together via PCR. The subsequent long fragment was subcloned between the BamHI and NotI sites on the lentivector of pHR-SIN-CSGW dlNotI plasmid.
We reasoned that fusing Renilla luciferase (Rluc) to various EGFP-tagged lamin proteins (lamin A, progerin, and lamin B1) via a P2A peptide sequence (Fig. 1B) would allow the relative post-translational stabilities of the lamin proteins to be assessed by comparing the EGFP-lamin:Rluc ratios for each lamin type, since the stability of Rluc should be the same in all of the constructs. Different antibodies may present a discrepancy in protein detection efficiency, but fusing EGFP to each lamin eliminates this variability and allows uniform detection with an anti-EGFP antibody. Lamin stability was investigated in lamin A expressing fibroblasts and bone marrow mesenchymal stem cells (hBM-MSCs). Our results are consistent with the notion that progerin is more stable than wild type lamin A. Moreover, FTI treatment significantly reduced the post-translational stability of progerin to the level of wild type lamin A, which may provide new insights into future directions for the clinical therapy of HGPS.
Results
Progerin possesses higher post-translational stability than lamin A protein in primary fibroblasts and human bone marrow-derived mesenchymal stem cells (hBM-MSCs)
To connect Rluc and EGFP-tagged lamin proteins with P2A sequence, we applied a series of PCR reactions as illustrated in the schematic Fig. 1B. The subsequent Rluc-P2A-lamin constructs were then subcloned into the lentiviral expression vector for lentiviruses production in HEK293T cells as we previously described.37 To compare these lamins' relative stabilities using this P2A platform, we first transduced the lentiviruses in primary human fibroblasts (Fig. 2) and human bone marrow-derived mesenchymal stem cells (hBM-MSCs) (Fig. 3), both of which express comparable amounts of endogenous lamin A (Fig. S1). In both cell types, the majority of the EGFP-lamin proteins were successfully dissociated from Rluc protein under the effect of P2A motif, with a small fraction of uncleaved products (P2A-LA:8.8%; P2A-PG:10.7%; P2A-LB1: 25.3%) (Fig. S1). The expression of each EGFP-lamin was further validated using lamin-specific antibodies (Fig. 2A, Fig. 3A). The localization of each EGFP-lamin protein was identical to that of the corresponding endogeneous lamin (Fig. 2B, Fig. 3B), suggesting that these EGFP-lamins are properly integrated into the nuclear lamina network in fibroblasts and hBM-MSCs.
Figure 2.
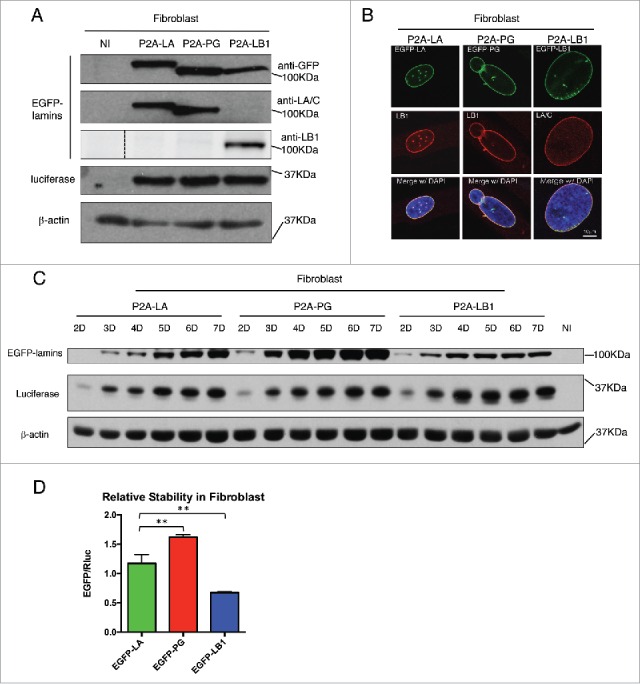
Comparing relative stability of lamin A, progerin and lamin B1 in human fibroblasts. (A) Western blotting analysis of viral infected human fibroblasts. Protein samples were immunoblotted with antibodies of GFP, lamin A/C, lamin B1, luciferase and β-actin. Non-infected fibroblast cells were used as a negative control. (B) Confocal fluorescence images. Infected fibroblasts expressing 2A-lamins (green) were fixed and stained with anti-lamin B1 (red) by immunofluorescence at 48 hours post infection. DNA was stained with DAPI (blue). A representative cell under each condition is shown. Bars, 10 μm. (C) Western blotting analysis on time course of viral infected human fibroblasts. Protein samples were immunoblotted with antibodies of GFP, luciferase and β-actin. (D) Quantification of lamins' relative stabilities in (C). The relative stability was calculated as the intensity ratio of EGFP/luciferase. Bar graph shows the average of day 5 to day 7 data. Results were generated from 3 biological replicates. *P < 0.05, **P < 0.01. P2A-LA, P2A-PG and P2A-LB1 refer to the constructs of luciferase-P2A-lamin A, luciferase-P2A-progerin and luciferase-P2A-lamin B1.
Figure 3.
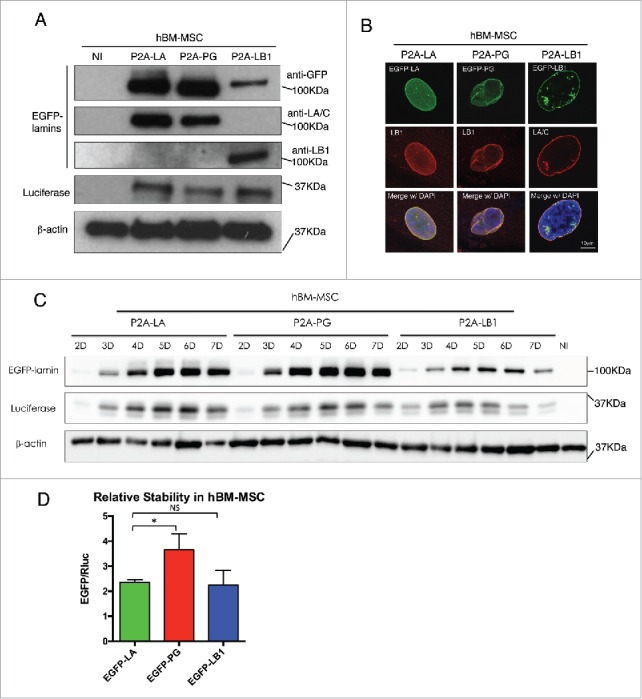
Comparing relative stability of lamin A, progerin and lamin B1 in hBM-MSCs. (A) Western blotting analysis of viral infected hBM-MSCs. Protein samples were immunoblotted with antibodies of GFP, lamin A/C, lamin B1, luciferase and β-actin. Non-infected hBM-MSCs cells were used as a negative control. (B) Confocal fluorescence images of infected hBM-MSCs expressing 2A-lamins (green) and stained with anti-lamin B1 (red) by immunofluorescence at 48 hours post infection. DNA was stained with DAPI (blue). A representative cell under each condition is shown. Bars, 10 μm. (C) Time course of viral infected hBM-MSCs were analyzed by Western blotting. Antibodies of GFP, luciferase and β-actin were utilized for immunoblotting. (D) Quantification of lamins' relative stabilities in (C) was calculated as the intensity ratio of EGFP/luciferase. Bar graph shows the average of day 5 to day 7 data. Results were generated from 3 biological replicates. *P < 0.05, **P < 0.01. P2A-LA, P2A-PG and P2A-LB1 refer to the constructs of luciferase-P2A-lamin A, luciferase-P2A-progerin and luciferase-P2A-lamin B1.
Next, time course experiments were applied to investigate the post-translational protein stability of the EGFP-lamins within a 7-day period after transduction. A gradual accumulation of the 3 EGFP-tagged lamins was noticeably observed in both fibroblasts and hBM-MSCs (Figs. 2C and 3C). Particularly in the fibroblasts, the proteins exhibited a rapid accumulation rate with greater slopes at earlier time points (day 2 - 4), and then reached the plateau by day 5 (EGFP-PG and EGFP-LB1) or 6 (EGFP-LA) (Fig. S2). A similar trend was also observed in hBM-MSCs.
To analyze the post-translational stability of these EGFP-tagged lamin variants, the EGFP signal was normalized to Rluc for quantification and the analysis presents an average of the plateau period from day 5 to day 7. The Rluc and the EGFP-tagged lamin/progerin are encoded within the same mRNA, and their translation initiates from the same ATG. Because the P2A-mediated “cleavage” that separates these 2 proteins occurs during translation, the post-translational stability of the Rluc and EGFP-lamin proteins is completely independent. Normalization to the co-translated Rluc control renders the contribution of all transcriptional and post-transcriptional regulatory mechanisms, except post-translational protein stability. Thus, we reason that the steady-state levels of EGFP-lamins (from day 5 to day 7) should directly reflect the post-translational stability of these proteins when normalized to Rluc. We further suggest that differences in the EGFP/Rluc ratio between the various EGFP-lamin/progerin fusions should reflect differences in relative post-translational stability. Based on this method, we found that among the 3 lamins, EGFP-progerin possessed the greatest relative stability in both cell types, followed by EGFP-lamin A and EGFP-LB1 (Figs. 2D and 3D). Interestingly, EGFP-lamin B1 was the least stable lamin variant in fibroblasts (Fig. 2D) and exhibited a stability similar to EGFP-LA in hBM-MSCs (Fig. 3D), despite the fact that it, like progerin, remains farnesylated.
Endogenous lamin A may not alter the post-translational stability of exogenously expressed A type lamin proteins
Because the 2A peptide-mediated reporter system relies on overexpression of exogenous EGFP-lamin proteins, which are expected to interact with endogenously encoded lamins, we wanted to determine if the presence of endogenous lamin A/C influenced the stability of the EGFP-lamin proteins. To address this issue, we used wild type mouse embryonic fibroblasts (MEF) and lamin A null MEF lines for lentiviral transduction. Like in fibroblasts and hBM-MSCs (Figs. 2 and 3), the fused proteins were successfully expressed and “cleaved” in both types of MEFs (Fig. 4A).
Figure 4.

Examining lamin relative stabilities in both LA+/+ and LA-/- MEFs. (A) Western blotting analysis of viral infected MEF cells. Proteins were probed with antibodies of GFP, human lamin A/C (mAB3211, which only recognizes human lamin A/C), lamin A/C (N-18), luciferase and β-actin. Non-infected cells were used as a negative control. (B) Time course of viral infected hBM-MSCs were analyzed by Western blotting. Antibodies of GFP, luciferase, human lamin A/C and β-actin were utilized for immunoblotting. (C) The quantification was calculated as the intensity ratio of EGFP/luciferase. Bar graph shows the average of day 4 to day 6 data. Results were generated from 3 biological replicates. P2A-LA, P2A-PG and P2A-LB1 refer to the constructs of luciferase-P2A-lamin A, luciferase-P2A-progerin and luciferase-P2A-lamin B1.
Next, the post-translational stability of EGFP-LA and EGFP-PG was determined by time course experiments from 4 d to 6 d post-transduction when the steady-state expression was achieved (Fig. 4B). The quantification analysis was done by averaging all the EGFP/Rluc ratio of each lamin protein (Fig. 4C), as in Figs. 2 and 3. We found that the relative protein levels of EGFP-LA and EGFP-PG in wild type and LA null MEFs were essentially identical, suggesting the post-translational degradation of exogenously expressed human A type lamins was not impacted by the presence of endogenous mouse lamin A.
It should be noted that these wild type or lamin A null MEF cells were derived from mouse embryos. This species-mismatched cell line might explain why we did not observe a significant increase in the relative amount of human progerin compare with human lamin A. While the results from this experiment implies that the presence of endogenous lamin A/C does not affect the post-translational stability of the exogenously expressed EGFP-lamins, MEFs lines are not the optimal system and further validation using a human LA/C null fibroblast or MSC lines are desired.
FTI treatment reduces progerin stability in fibroblasts
Farnesyltransferase inhibitors (FTI) block farnesylation of progerin, relocalize the protein away from the nuclear envelope, and alleviate the prominent nuclear phenotypes in human progeria fibroblasts.38-41 The past studies on FTI did not elaborate how inhibition of farnesylation by FTI affects post-translational lamin protein stability, specifically in the lamins with farnesyl tail such as progerin and lamin B1. To study this, the P2A-EGFP-lamin system was applied in fibroblast cells treated with FTI for 6 d (Fig. 5A). During the treatment, nucleoplasmic aggregates of EGFP-lamin A, EGFP-progerin or EGFP-lamin B1were observed (Fig. S3). The quantification was presented as averaged data of the EGFP/Rluc ratio from day 4 to day 6, when the protein steady-state was achieved (Fig. 5B). Interestingly, we found that FTI significantly reduced progerin's stability to the level of wild type lamin A, whereas lamin A had no significant changes in stability after FTI treatment (Fig. 5B).
Figure 5.

Effects of FTI on lamins relative stabilities in human fibroblasts. (A) Western blotting analysis of viral infected fibroblasts upon the treatment of FTI. DMSO treated cells were mock control. (B) Quantification of the relative stability in (A) is presented as EGFP/luciferase ratios. Bar graph shows the average of day 4 to day 6 data. Results were generated from 3 biological replicates. *P < 0.05, **P < 0.01. P2A-LA, P2A-PG and P2A-LB1 refer to the constructs of luciferase-P2A-lamin A, luciferase-P2A-progerin and luciferase-P2A-lamin B1.
To our surprise, the other farnesylated lamin, EGFP-LB1, displayed an opposite response to FTI treatment, where its post-translational stability was largely increased by farnesylation inhibition. Yet, Adam and his colleagues have previously reported reduced endogenous lamin B1 expression in fibroblasts treated with FTI.42 To address this potential discrepancy, we compared endogenous LB1 and exogenous LB1 protein levels in FTI treated and untreated fibroblast cells (Fig. S4). In agreement with Adam et al., the endogenous LB1 protein decreased with FTI treatment in both non-infected and P2A-LB1 lentiviral-infected fibroblasts (Fig. S4 A and B). Consistently, we observed an increased level of exogenously expressed EGFP-LB upon FTI treatment (Fig. S4C). As suggested by Adam et al., the decreased endogenous lamin B1 level was likely due to the downregulated lamin B1 mRNA by FTI treatment. Whereas for EGFP-lamin B1, its transcription is driven by a spleen focus-forming virus (SFFV) promoter, which may be independent of the influence of FTI. Therefore, the accumulation of EGFP-lamin B1 in the FTI treatment, as revealed by the P2A method, indeed reflects only the post-translational protein stability of EGFP-lamin B1.
Discussion
In this study, we employed a viral P2A-sequence based comparison system to demonstrate that progerin is post-translationally more stable than wild type lamin A in fibroblasts and hBM-MSCs. Our results are in agreement with the previous observation that progerin protein accumulates during cellular aging.16,21,44 FTI significantly reduced progerin's post-translational stability to the level of wild type lamin A, which provides additional evidence to support the beneficial effects of FTI in HGPS cells, animal models and patient clinical trials.39,41,45 Interestingly, our study showed that EGFP-lamin B1s post-translational stability was increased upon the treatment of FTI. A previous study reported a reduction in endogenous lamin B1 protein in FTI-treated cells, which was likely due to the downregulation of lamin B1 mRNA level by FTI.43 In our experiment, the transcription of the exogenous EGFP-lamin B1 was driven by an SFFV promoter which does not show a noticeable response to FTI treatment (Fig. S4). Furthermore, in the P2A system, the normalization of EGFP-lamins to the Rluc control accounts for any differences in mRNA abundance or translation rate. Thus, only the post-translational stability of EGFP-lamin B1 was assessed and compared across samples. Our study suggests that normally farnesylated LB1 is less stable than the non-farnesylated LB1. Previous findings have shown that disrupted farnesylation by mutations in the CAAX motif of LB1 mislocalize the protein to the nucleoplasm.46,47 A recent study has reported that lamin B1 degradation involves nucleus-to-cytoplasm vesicular transport that delivers lamin B1-LC3 to the lysosomes.49 Based on these data, a possible explanation is that the removal of the farnesyl tail from lamin B1 may disassociate the protein away from the nuclear lamina, which disrupts the LC3-mediated exporting vesicle formation, causing an increase in the stability of lamin B1.
We show that the P2A sequence efficiently mediated the disassociation of Rluc and EGFP-tagged lamin proteins in different cell lines, including fibroblasts, hBM-MSCs and MEFs, suggesting the extensive applicability of this method. The normalization of steady-state levels of EGFP-tagged lamins to those of the cotranslationally separated Rluc protein controlled for differences in transcription, mRNA stability, and translation rates between samples, and allowed differences in post-translational stability between the various lamins to be inferred. The main advantages of the P2A peptide-mediated post-translational reporter system are that it is much simple to implement, it avoids potentially confounding pleiotropic effects from cycloheximide inhibition of translation, and it provides a means to look at the relative stability of insoluble proteins. One of the main drawbacks of the technique in its current configuration is that it cannot provide a direct measurement of protein half-life, and only allows relative changes in protein stability to be inferred for a protein under different growth conditions,31 or between protein variants as presented here for the lamins. Rodriguez-Contreras and colleagues used a variation of the P2A-peptide technique to examine changes in stability of the LmxGT1 glucose transporter in response to glucose starvation, and demonstrated that the fold-change in LmxGT1 stability determined via this technique was essentially identical to the fold-change in half-life determined via the cycloheximide block technique.31 This serves as a validation of the underlying concepts of the technique, and emphasizes the direct relationship between protein half-life and steady-state protein abundance. We have noticed that the dynamics of normalized protein accumulation (lamin/luc) over time were highly reproducible and specific for each lamin type (Fig. S2). Since the rate of increase in protein abundance should be directly proportional to the half-life of the protein, it may be possible to use this rate to calculate protein half-life in a manner similar to the “approach to steady-state labeling” method described previously.50 In that method the cells were labeled with a continuous supply of [3H] uridine and the rate of specific mRNA that accumulated at a steady-state level was measured. Then the half-life of the mRNA was calculated based on the time required to reach its steady-state.50 We intend to explore this possibility in the future.
Materials and methods
Cell culture and cell treatment
Human primary skin fibroblasts were obtained from the Progeria Research Foundation and cultured in MEM (Life Technologies) supplemented with 20% FBS (Gemini Bio-Products) and 2 mm l-glutamine (Life Technologies) at 37 °C supplied with 5% CO2. Human bone marrow mesenchymal stem cells (hBM-MSCs) purchased from Rooster Bio were maintained in αMEM (Corning) supplemented with 10% heat-inactivated FBS (Seradigrn), 2 mm l-glutamine and 1% MEM non-essential amino acid (NEAA) (Life Technologies) in 5% CO2 at 37 °C. Control and lamin A null mouse embryonic fibroblasts were kindly provided by Dr. Jan Lammerding and grown in DMEM (Lonza) supplemented with 10% FBS. In the FTI treatment experiment, FTI (J&J) at a final concentration of 2 μM was added to culture media at the time of viral infection. Medium was changed every other day with Lonafarnib supplementation.
Plasmid construction
Plasmids of pEGFP-C1-LA, pEGFP-C1-PG and pEGFP-C1-LB1 were constructed based on the pEGFP-C1 vector (Clontech). The lentiviral vector pHR-SIN-CSGW dlNotI was obtained as previously described.37 Briefly, Rluc-P2A in pRP-M-Rluc-P2A-GFP plasmid and EGFP-lamins in above mentioned lamin plasmids were amplified by PCR using primer sets P1 (5′-GGTCCAGCGGATCCATGGCTTCCAAGGTG-3′) and P2 (5′-GCCCTTGCTCACCATCGGACCTGGGTTCTC-3′), targeting Rluc-P2A, as well as P3 (5′-GAGAACCCAGGTCCGATGGTGAGCAAGGGC-3′) and P4 (for LA/PG: 5′-GGTAGCCTGCGGCCGCAGATTACATGATGCTGCAGTTCTGG-3′, for LB1: 5′-GGTAGCCTGCGGCCGCTTACATAATTGCACAGCTTCTATTGG-3′), targeting EGFP-lamins. The primer P2 completely overlapped with P3, which allowed the fragments of Rluc-P2A and EGFP-lamins to automatically ligate together in a second round of PCR reaction using P1 and P4. The ligated large fragments were subsequently sub-cloned into the BamHI and NotI sites of pHR-SIN-CSGW dlNotI.
Virus generation and viral infection
HEK293T cells were co-transfected with lentiviral plasmids and 2 virus packaging vectors, pHR-CMV-8.2ΔR and pCMV-VSVG, utilizing Fugene 6 (Promega). Culture supernatants were collected on 48 hrs and 72 hrs post-transfection, and filtered through 0.45-μm filters to remove any nonadherent 293T cells, followed by concentration at 25k RPM for 2 hours in 4°C by Optima™ L-100K Ultracentrifuge (Beckman Coulter). The virus pellets were re-suspended in 1 ml of cold DMEM/F12 (Lonza), then stored at −80 °C. Next, fibroblasts, hBM-MSCs, MEFs or iPSCs were infected by lentiviruses in media supplemented with Polybrene (Santa Cruze Biotechnology) with the final concentration of 8 μg/ml. The medium was changed every other day post-infection until the cells were harvested.
Western blotting
Western blotting analysis was performed as previously described.51 Briefly, infected cells were lysed in Laemmli Sample Buffer containing 5% β-mercaptoethanol (Bio-Rad) to obtain whole cell lysates. Proteins were separated on 10% SDS-PAGE gels, followed by transferring onto nitrocellulose membranes (Bio-Rad) for antibody detection. Bands were imagined by enhanced chemiluminescence (Clarity™ Western ECL Substrate, Bio-Rad). Quantification was executed by Image Lab™ Software (Bio-Rad).
Immunofluorescence staining and microscopy
Viral infected fibroblasts, hBM-MSCs and MEFs were fixed in 4% paraformaldehyde/phosphate buffered saline (PBS) for 20 min at room temperature (RT). They were then permeabilized with 0.5% Triton X-100 in PBS for 5 min at RT. After blocking in 4% BSA/TBS at RT for 1 h, and probing with the primary antibodies overnight at 4 °C, cells were incubated with secondary antibody at RT for 1 h in the dark. Secondary antibodies used were Alexa Fluor® 594 donkey anti-goat IgG (Invitrogen) and Alexa Fluor® 594 donkey anti-mouse IgG (Invitrogen). Lastly, the cells were stained by VECTASHIELD® Mounting Medium with DAPI (H-1200, VECTOR). Immunofluorescence microscopy was performed on a Leica SP5 X Confocal Microscope (Leica Microsystems, Inc.). For iPSCs, fluorescence images were taken by a Zeiss Axio Observer Microscope (Carl Zeiss, Inc.).
Antibodies
The antibodies used in western blotting and immunofluorescence were: mouse-anti-human Lamin A/C (MAB3211, Millipore), goat-anti-mouse Lamin A/C (N-18, Santa Cruz Biotechnology), goat-anti-Lamin B1 (sc-6217, Santa Cruz Biotechnology), mouse anti-β-Actin peroxidase conjugated (A3854, Sigma), rabbit-anti-GFP antibody (ab290, Abcam), mouse-anti-Renilla Luciferase Antibody (MAB4410, Millipore).
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- [1].Fawcett DW. On the occurrence of a fibrous lamina on the inner aspect of the nuclear envelope in certain cells of vertebrates. Am J Anat (1966); 119:129-45; PMID:6007824; http://dx.doi.org/ 10.1002/aja.1001190108 [DOI] [PubMed] [Google Scholar]
- [2].Goldman RD, Gruenbaum Y, Moir RD, Shumaker DK, Spann TP. Nuclear lamins: building blocks of nuclear architecture. Genes Dev (2002); 16:533-47; PMID:11877373; http://dx.doi.org/ 10.1101/gad.960502 [DOI] [PubMed] [Google Scholar]
- [3].Krohne G, Benavente R, Scheer U, Dabauvalle MC. The nuclear lamina in Heidelberg and W??rzburg: A personal view. Eur J Cell Biol (2005); 84:163-179; PMID:15819398; http://dx.doi.org/ 10.1016/j.ejcb.2004.12.005 [DOI] [PubMed] [Google Scholar]
- [4].McKeon FD, Kirschner MW, Caput D. Homologies in both primary and secondary structure between nuclear envelope and intermediate filament proteins. Nature (1986); 319:463-8; PMID:3453101; http://dx.doi.org/ 10.1038/319463a0 [DOI] [PubMed] [Google Scholar]
- [5].Furukawa K, Inagaki H, Hotta Y. Identification and cloning of an mRNA coding for a germ cell-specific A-type lamin in mice. Exp Cell Res (1994); 212:426-30; PMID:8187835; http://dx.doi.org/ 10.1006/excr.1994.1164 [DOI] [PubMed] [Google Scholar]
- [6].Machiels BM, Zorenc AH, Endert JM, Kuijpers HJ, van Eys GJ, Ramaekers FC, Broers JL. An alternative splicing product of the lamin A/C gene lacks exon 10. J Biol Chem (1996); 271:9249-53; PMID:8621584; http://dx.doi.org/ 10.1074/jbc.271.16.9249 [DOI] [PubMed] [Google Scholar]
- [7].Pollard KM, Chan EK, Grant BJ, Sullivan KF, Tan EM, Glass CA. In vitro posttranslational modification of lamin B cloned from a human T-cell line. Mol Cell Biol (1990); 10:2164-75; PMID:2325650; http://dx.doi.org/ 10.1128/MCB.10.5.2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Furukawa K, Hotta Y. cDNA cloning of a germ cell specific lamin B3 from mouse spermatocytes and analysis of its function by ectopic expression in somatic cells. EMBO J (1993); 12:97-106; PMID:8094052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Spann TP, Goldman AE, Wang C, Huang S, Goldman RD. Alteration of nuclear lamin organization inhibits RNA polymerase II-dependent transcription. J Cell Biol (2002); 156:603-8; PMID:11854306; http://dx.doi.org/ 10.1083/jcb.200112047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bridger JM, Foeger N, Kill IR, Herrmann H. The nuclear lamina. Both a structural framework and a platform for genome organization. FEBS J (2007); 274:1354-61 [DOI] [PubMed] [Google Scholar]
- [11].Gruenbaum Y, Foisner R. Lamins: Nuclear Intermediate Filament Proteins with Fundamental Functions in Nuclear Mechanics and Genome Regulation. Annu Rev Biochem (2014); 84:150306093657004. [DOI] [PubMed] [Google Scholar]
- [12].Capell BC, Collins FS. Human laminopathies: nuclei gone genetically awry. Nat Rev Genet (2006); 7:940-52; PMID:17139325; http://dx.doi.org/ 10.1038/nrg1906 [DOI] [PubMed] [Google Scholar]
- [13].Cao K, Park C. Cellular Basis of Laminopathies (2012); 1-7 [Google Scholar]
- [14].Sarkar PK, Shinton RA. Hutchinson-Guilford progeria syndrome. Postgrad Med J (2001); 77:312-7; PMID:11320273; http://dx.doi.org/ 10.1136/pmj.77.907.312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hutchinson J. Congenital absence of hair and mammary glands with atrophic condition of the skin and its appendages, in a boy whose mother had been almost wholly bald from alopecia areata from the age of Six. Med Chir Trans (1886); 69:473-7; PMID:20896687; http://dx.doi.org/ 10.1177/095952878606900127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Eriksson M, Brown WT, Gordon LB, Glynn MW, Singer J, Scott L, Erdos MR, Robbins CM, Moses TY, Berglund P, et al.. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature (2003); 423:293-8; PMID:12714972; http://dx.doi.org/ 10.1038/nature01629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sinensky M, Fantle K, Trujillo M, McLain T, Kupfer A, Dalton M. The processing pathway of prelamin A. J Cell Sci (1994); 107:( Pt 1, 61-7; PMID:8175923 [DOI] [PubMed] [Google Scholar]
- [18].De Sandre-Giovannoli A, Bernard R, Cau P, Navarro C, Amiel J, Boccaccio I, Lyonnet S, Stewart CL, Munnich A, Le Merrer M, et al.. Lamin a truncation in Hutchinson-Gilford progeria. Science (2003); 300:2055; PMID:12702809; http://dx.doi.org/ 10.1126/science.1084125 [DOI] [PubMed] [Google Scholar]
- [19].D'Apice MR, Tenconi R, Mammi I, van den Ende J, Novelli G. Paternal origin of LMNA mutations in Hutchinson-Gilford progeria. Clin Genet (2004); 65:52-4; http://dx.doi.org/ 10.1111/j..2004.00181.x [DOI] [PubMed] [Google Scholar]
- [20].Cao K, Capell BC, Erdos MR, Djabali K, Collins FS. A lamin A protein isoform overexpressed in Hutchinson-Gilford progeria syndrome interferes with mitosis in progeria and normal cells. Proc Natl Acad Sci U S A (2007); 104:4949-54; PMID:17360355; http://dx.doi.org/ 10.1073/pnas.0611640104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Goldman RD, Shumaker DK, Erdos MR, Eriksson M, Goldman AE, Gordon LB, Gruenbaum Y, Khuon S, Mendez M, Varga R, et al.. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci U S A (2004); 101:8963-8; PMID:15184648; http://dx.doi.org/ 10.1073/pnas.0402943101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].McCord RP, Nazario-Toole A, Zhang H, Chines PS, Zhan Y, Erdos MR, Collins FS, Dekker J, Cao K. Correlated alterations in genome organization, histone methylation, and DNA-lamin A/C interactions in Hutchinson-Gilford progeria syndrome. Genome Res (2013); 23:260-9; PMID:23152449; http://dx.doi.org/ 10.1101/gr.138032.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].DeBusk FL. The Hutchinson-Gilford progeria syndrome. Report of 4 cases and review of the literature. J Pediatr (1972); 80:697-724; PMID:4552697; http://dx.doi.org/ 10.1016/S0022-3476(72)80229-4 [DOI] [PubMed] [Google Scholar]
- [24].Baker PB, Baba N, Boesel CP. Cardiovascular abnormalities in progeria. Case report and review of the literature. Arch Pathol Lab Med (1981); 105:384-6; PMID:6894691 [PubMed] [Google Scholar]
- [25].Gordon LB, Massaro J, Sr D'Agostino RB, Campbell SE, Brazier J, Brown WT, Kleinman ME, Kieran MW; Collaborative Progeria Clinical Trials. Impact of farnesylation inhibitors on survival in Hutchinson-Gilford progeria syndrome. Circulation (2014); 130:27-34; PMID:24795390; http://dx.doi.org/ 10.1161/CIRCULATIONAHA.113.008285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ullrich NJ, Gordon LB. Hutchinson-Gilford progeria syndrome. Handb Clin Neurol (2015); 132:249-64; PMID:26564085; http://dx.doi.org/ 10.1016/B978-0-444-62702-5.00018-4 [DOI] [PubMed] [Google Scholar]
- [27].Zhou P. Determining protein half-lives. Methods Mol Biol (2004); 284:67-77; PMID:15173609 [DOI] [PubMed] [Google Scholar]
- [28].Fritzsche S, Springer S. Pulse-chase analysis for studying protein synthesis and maturation. Curr Protoc Protein Sci (2014); 78:30.3.1-23 [DOI] [PubMed] [Google Scholar]
- [29].Bertacchini J, Beretti F, Cenni V, Guida M, Gibellini F, Mediani L, Marin O, Maraldi NM, de Pol A, Lattanzi G, et al.. The protein kinase Akt/PKB regulates both prelamin A degradation and Lmna gene expression. FASEB J (2013); 27:2145-55; PMID:23430973; http://dx.doi.org/ 10.1096/fj.12-218214 [DOI] [PubMed] [Google Scholar]
- [30].Dechat T, Adam SA, Taimen P, Shimi T, Goldman RD. Nuclear lamins. Cold Spring Harb Perspect Biol (2010); 2:a000547; PMID:20826548; http://dx.doi.org/ 10.1101/cshperspect.a000547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Rodriguez-Contreras D, Aslan H, Feng X, Tran K, Yates PA, Kamhawi S, Landfear SM. Regulation and biological function of a flagellar glucose transporter in Leishmania mexicana: a potential glucose sensor. FASEB J (2015); 29:11-24; PMID:25300620; http://dx.doi.org/ 10.1096/fj.14-251991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].De Felipe P, Luke GA, Hughes LE, Gani D, Halpin C, Ryan MD. E unum pluribus: Multiple proteins from a self-processing polyprotein. Trends Biotechnol (2006); 24:68-75; PMID:16380176; http://dx.doi.org/ 10.1016/j.tibtech.2005.12.006 [DOI] [PubMed] [Google Scholar]
- [33].Ryan MD, King AM, Thomas GP. Cleavage of foot-and-mouth disease virus polyprotein is mediated by residues located within a 19 amino acid sequence. J Gen Virol (1991); 72:(Pt 11, 2727-32; PMID:1658199; http://dx.doi.org/ 10.1099/0022-1317-72-11-2727 [DOI] [PubMed] [Google Scholar]
- [34].Donnelly ML, Luke G, Mehrotra A, Li X, Hughes LE, Gani D, Ryan MD. Analysis of the aphthovirus 2A/2B polyprotein ‘cleavage’ mechanism indicates not a proteolytic reaction, but a novel translational effect: a putative ribosomal ‘skip’. J Gen Virol (2001); 82:1013-25; PMID:11297676; http://dx.doi.org/ 10.1099/0022-1317-82-5-1013 [DOI] [PubMed] [Google Scholar]
- [35].de Felipe P, Luke GA, Brown JD, Ryan MD. Inhibition of 2A-mediated ‘cleavage’ of certain artificial polyproteins bearing N-terminal signal sequences. Biotechnol J (2010); 5:213-23; PMID:19946875; http://dx.doi.org/ 10.1002/biot.200900134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kim JH, Lee SR, Li LH, Park HJ, Park JH, Lee KY, Kim MK, Shin BA, Choi SY. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One (2011); 6:1-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Xiong Z-M, Choi JY, Wang K, Zhang H, Tariq Z, Wu D, Ko E, LaDana C, Sesaki H, Cao K. Methylene blue alleviates nuclear and mitochondrial abnormalities in progeria. Aging Cell (2015); 15:279-90; PMID:26663466; http://dx.doi.org/ 10.1111/acel.12434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Toth JI, Yang SH, Qiao X, Beigneux AP, Gelb MH, Moulson CL, Miner JH, Young SG, Fong LG. Blocking protein farnesyltransferase improves nuclear shape in fibroblasts from humans with progeroid syndromes. Proc Natl Acad Sci U S A (2005); 102:12873-8; PMID:16129834; http://dx.doi.org/ 10.1073/pnas.0505767102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Yang SH, Meta M, Qiao X, Frost D, Bauch J, Coffinier C, Majumdar S, Bergo MO, Young SG, Fong LG. A farnesyltransferase inhibitor improves disease phenotypes in mice with a Hutchinson-Gilford progeria syndrome mutation. J Clin Invest (2006); 116:2115-21; PMID:16862216; http://dx.doi.org/ 10.1172/JCI28968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Capell BC, Erdos MR, Madigan JP, Fiordalisi JJ, Varga R, Conneely KN, Gordon LB, Der CJ, Cox AD, Collins FS. Inhibiting farnesylation of progerin prevents the characteristic nuclear blebbing of Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci U S A (2005); 102:12879-84; PMID:16129833; http://dx.doi.org/ 10.1073/pnas.0506001102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Gordon LB, Kleinman ME, Miller DT, Neuberg DS, Giobbie-Hurder A, Gerhard-Herman M, Smoot LB, Gordon CM, Cleveland R, Snyder BD, et al.. Clinical trial of a farnesyltransferase inhibitor in children with Hutchinson – Gilford progeria syndrome 2012); 2-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Adam SA, Butin-Israeli V, Cleland MM, Shimi T, Goldman RD. Disruption of lamin B1 and lamin B2 processing and localization by farnesyltransferase inhibitors. Nucleus (2013); 4:142-50; PMID:23475125; http://dx.doi.org/ 10.4161/nucl.24089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Adam SA, Butin-Israeli V, Cleland MM, Shimi T, Goldman RD. Disruption of lamin B1 and lamin B2 processing and localization by farnesyltransferase inhibitors. Nucleus (2013); 4:142-50; PMID:23475125; http://dx.doi.org/ 10.4161/nucl.24089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Cao K, Blair CD, Faddah DA, Kieckhaefer JE, Olive M, Erdos MR, Nabel EG, Collins FS. Progerin and telomere dysfunction collaborate to trigger cellular senescence in normal human fibroblasts (2011); 121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Capell BC, Olive M, Erdos MR, Cao K, Faddah DA, Tavarez UL, Conneely KN, Qu X, San H, Ganesh SK, et al.. A farnesyltransferase inhibitor prevents both the onset and late progression of cardiovascular disease in a progeria mouse model. Proc Natl Acad Sci U S A (2008); 105:15902-7; PMID:18838683; http://dx.doi.org/ 10.1073/pnas.0807840105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Verstraeten VL, Peckham LA, Olive M, Capell BC, Collins FS, Nabel EG, Young SG, Fong LG, Lammerding J. Protein farnesylation inhibitors cause donut-shaped cell nuclei attributable to a centrosome separation defect. Proc Natl Acad Sci (2011); 108:4997-5002; PMID:21383178; http://dx.doi.org/ 10.1073/pnas.1019532108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Maske CP, Hollinshead MS, Higbee NC, Bergo MO, Young SG, Vaux DJ. A carboxyl-terminal interaction of lamin B1 is dependent on the CAAX endoprotease Rce1 and carboxymethylation. J Cell Biol (2003); 162:1223-32; PMID:14504265; http://dx.doi.org/ 10.1083/jcb.200303113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Jung H-J, Nobumori C, Goulbourne CN, Tu Y, Lee JM, Tatar A, Wu D, Yoshinaga Y, de Jong PJ, Coffinier C, et al.. Farnesylation of lamin B1 is important for retention of nuclear chromatin during neuronal migration. Proc Natl Acad Sci U S A (2013); 110, E1923-32; PMID:23650370; http://dx.doi.org/ 10.1073/pnas.1303916110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Dou Z, Xu C, Donahue G, Shimi T, Pan JA, Zhu J, Ivanov A, Capell BC, Drake AM, Shah PP, et al.. Autophagy mediates degradation of nuclear lamina. Nature (2015); 527:1-17; ; http://dx.doi.org/ 10.1038/nature15548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].GREENBERG JR. High Stability of Messenger RNA in Growing Cultured Cells. Nature (1972); 240:102-4; PMID:4564814; http://dx.doi.org/ 10.1038/240102a0 [DOI] [PubMed] [Google Scholar]
- [51].Wu D, Flannery AR, Cai H, Ko E, Cao K. Nuclear localization signal deletion mutants of lamin A and progerin reveal insights into lamin A processing and emerin targeting. Nucleus (2014); 5:66-74; PMID:24637396; http://dx.doi.org/ 10.4161/nucl.28068 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


