Summary
Cyclic AMP regulatory element binding protein and signal transducer and activator of transcription 3 (STAT3) may control inflammation by several mechanisms, one of the best characterized is the induction of the expression of the anti‐inflammatory cytokine interleukin‐10 (IL‐10). STAT3 also down‐regulates the production of pro‐inflammatory cytokines induced by immunoreceptor tyrosine‐based activation motif (ITAM)‐coupled receptors, a mechanism termed cross‐inhibition. Because signalling via ITAM‐dependent mechanisms is a hallmark of fungal pattern receptors, STAT3 activation might be involved in the cross‐inhibition associated with invasive fungal infections. The fungal surrogate zymosan produced the phosphorylation of Y705‐STAT3 and the expression of Ifnb1 and Socs3, but did not induce the interferon (IFN)‐signature cytokines Cxcl9 and Cxcl10 in bone marrow‐derived dendritic cells. Unlike lipopolysaccharide (LPS), zymosan induced IL‐10 and phosphorylated Y705‐STAT3 to a similar extent in Irf3 and Ifnar1 knockout and wild‐type mice. Human dendritic cells showed similar results, although the induction of IFNB1 was less prominent. These results indicate that LPS and zymosan activate STAT3 through different routes. Whereas type I IFN is the main effector of LPS effect, the mechanism involved in Y705‐STAT3 phosphorylation by zymosan is more complex, cannot be associated with type I IFN, IL‐6 or granulocyte–macrophage colony‐stimulating factor, and seems dependent on several factors given that it was partially inhibited by the platelet‐activating factor antagonist WEB2086 and high concentrations of COX inhibitors, p38 mitogen‐activate protein kinase inhibitors, and blockade of tumour necrosis factor‐α function. Altogether, these results indicate that fungal pattern receptors share with other ITAM‐coupled receptors the capacity to produce cross‐inhibition through a mechanism involving STAT3 and induction of SOCS3 and IL‐10, but that cannot be explained through type I IFN signalling.
Keywords: cytokines, dendritic cells, fungal infection, inflammation
Abbreviations
- BMDC
bone‐marrow‐derived dendritic cells
- CBP
CREB‐binding protein
- CRE
cAMP regulatory element
- CREB
CRE‐binding protein
- DC
human monocyte‐derived dendritic cells
- EP
E‐type prostanoid receptor
- GM‐CSF
granulocyte–macrophage colony‐stimulating factor
- IFN
interferon
- IFNAR1
IFN‐α and IFN‐β receptor subunit 1
- IL‐10
interleukin‐10
- IRF
IFN regulatory factor
- ITAM
immunoreceptor tyrosine‐based activation motif
- JAK
Janus kinase
- LPS
lipopolysaccharide
- MAPK
mitogen‐activated protein kinase
- MSK
mitogen‐ and stress‐activated kinases
- PAF
platelet‐activating factor
- PGE2
prostaglandin E2
- SOCS
suppressors of cytokine signalling
- STAT
signal transducer and activator of transcription
- TIR
Toll–interleukin receptor
- TLR
Toll‐like receptor
- TNF‐α
tumour necrosis factor‐α
- TRAM
TRIF‐related adaptor molecule
- TRIF
TIR‐domain‐containing adapter‐inducing IFN‐β
- WT
wild‐type
Introduction
The main function of interleukin‐10 (IL‐10) is its capacity to block cytokine production from T helper type 1 cells. Macrophages and dendritic cells (DC) are robust producers of IL‐10 and the balance among IL‐10, IL‐12 p70 and IL‐23 released from DC is a critical determinant of the polarization of the immune response. Renewed interest in the role of IL‐10 emerged from the study of the mechanisms of defence against invasive mycoses, a growing risk in patients with a compromised immune system. The commensal Candida is the most prevalent infectious fungus, but Cryptococcus, Aspergillus and Pneumocystis are also common opportunistic pathogens. The first line of defence against fungi is the innate immune system, which is mainly composed of phagocytic cells endowed with a set of pattern recognition receptors. A prominent production of IL‐10 accompanied by a low production of IL‐12 p70 characterizes the response to fungal patterns.1, 2 Given that an unbalanced production of IL‐10 may also contribute to the evasion by microbes of the immune response in other prevalent infections, e.g. Mycobacterium tuberculosis,3 the analysis of the mechanisms of IL‐10 regulation is of clinical relevance.
At the transcriptional level, several scenarios of IL10 regulation have been disclosed. The one centred on the transcription factor cAMP regulatory element binding protein (CREB) and its co‐activators CREB‐binding protein (CBP) and CREB‐regulated transcription co‐activator is widely accepted in the case of the fungal mimic zymosan and in many cases lipopolysaccharide (LPS),4, 5, 6, 7 whereas other reports have focused on the role of signal transducer and activator of transcription 3 (STAT3) and an autocrine cycle dependent on interferon‐β (IFN‐β) and IFN regulatory factor 3 (IRF3) downstream of the Toll–interleukin receptor (TIR)‐domain‐containing adapter‐inducing IFN‐β (TRIF)/TRIF‐related adaptor molecule (TRAM) branch of the LPS/Toll‐like receptor 4 (TLR4) system.8, 9 The study of the response to fungal patterns has been facilitated by the use of the fungal surrogate zymosan, a cell‐wall extract from Saccharomyces cerevisiae that contains a β‐glucan component, recognized by the C‐type lectin receptor dectin‐1, and a mannose‐based component that mainly signals through TLR2 and the C‐type lectin receptor dectin‐2.10, 11, 12, 13 Although most views agreed on the lack of a significant induction of type I IFN upon fungal challenge, this issue should be revisited on the basis of recent findings. In fact, Ifnar1 −/− mice were reported to have an improved survival in response to Candida infection,14 and studies of humans with gain‐of‐function dominant mutations of STAT1 have shown an increased risk of invasive fungal infection.15 On the other hand, type I IFN was associated with host defence response against Candida 16 and another study unveiled a unique mechanism of induction of IFN‐β in mice by Candida through IRF5 activation downstream of dectin‐1 and the adaptor protein caspase recruitment domain‐containing protein 9,17 which might depend on IκB kinase‐β activity.18, 19 This mechanism was found in DC but not in macrophages, indicating that there may be cell‐dependency and, probably also species‐dependency. Another study has reported IFN‐β induction via tumour necrosis factor‐α (TNF‐α) and IRF1, so disclosing new mechanisms of regulation.20, 21 Given the strong induction of TNF‐α elicited by zymosan and the large list of secondary mediators that are released concomitantly and that can activate STAT3 – e.g. IL‐6, IL‐12, IL‐23, granulocyte–macrophage colony‐stimulating factor (GM‐CSF), chemokines, prostaglandin E2 (PGE2) and platelet‐activating factor (PAF)22, 23, 24, 25, 26 – it seems necessary to address the role of STAT3 in shaping the inflammatory response to fungal patterns. Our studies using mouse bone‐marrow‐derived DC (BMDC) and human monocyte‐derived DC have shown: (i) the activation of STAT3 by zymosan through a mechanism independent of type I IFN, (ii) the lack of involvement of the Janus kinase (JAK)/STAT3 system on the production of IL‐10 elicited by zymosan, and (iii) a cross‐inhibitory effect elicited by zymosan receptors on type I IFN responses.
Materials and methods
Reagents and cells
Zymosan, mannan from Saccharomyces cerevisiae, and the p38 mitogen‐activated protein kinase (MAPK) inhibitor SB203580 were obtained from Sigma Chemical Co. (St Louis, MO). The E‐type prostanoid receptor 2 (EP2) receptor antagonist PF‐04418948 and the EP4 receptor antagonist L‐161982 were from Cayman Chemical Industries (Ann Arbor, MI). Mannose‐depleted zymosan was obtained by boiling zymosan in 10 M NaOH. Interleukin‐10 was assayed with Biotrack ELISA systems from Amersham Biosciences (Chalfont St Giles, UK). The cell‐permeable STAT3 inhibitor peptide was from Millipore (Billerica, MA) (ref: 573096). Ruxolitinib and tofacitinib were from InvivoGen (San Diego, CA). Anti‐IL‐10 antibody (AF‐217‐NA) and anti‐IL‐6 (AF‐206‐NA) were from R&D Systems (Minneapolis, MN). The 12/15‐lipoxygenase inhibitor ethyl 3,4‐dihydroxybenzylidenecyanoacetate was from Santa Cruz Biotechnology, Inc. (Dallas, TX). The separation of monocytes and their differentiation into DC has been reported previously.27 Briefly, monocytes were cultured in the presence of GM‐CSF (800 U/ml) and IL‐4 (500 U/mL) for 5 days, in RPMI‐1640 medium supplemented with 4 mm l‐glutamine and 10% fetal bovine serum. Differentiation into DC was addressed by flow cytometry of CD40, CD80, CD83, CD86, CD11b and CD11c. Irf3 −/− mice were obtained by Dr Castrillo through a collaboration with Drs G. Cheng (University of California, Los Angeles) and T. Taniguchi (University of Tokyo).28 Ifnar1 −/− mice were kindly provided by Dr C. Ardavín (Centro Nacional de Biotecnología, Madrid, Spain) with permission from Dr U. Kalinke (Centre for Experimental and Clinical Infection Research, Hannover, Germany).29 Irf5 −/− mice were obtained from the Wellcome Trust Sanger Institute Mouse Genetics Project (Sanger MGP). Bone marrow from the femora and tibiae of C57BL/6 mice was used to generate BMDC by culture in the presence of murine GM‐CSF for 5 days. Purity of BMDC was confirmed by flow cytometry, which showed 85% cells to be positive for the integrin CD11c. Before carrying out the experiments, BMDC were maintained for 3 hr in the absence of GM‐CSF to avoid the activation of STAT3 elicited by GM‐CSF. The population of adherent cells, presumably composed of bone marrow macrophages and termed GM‐CSF macrophages, was used in some experiments. Candida albicans was provided by Dr Jesús Pla, from Universidad Complutense de Madrid.
Ethics
The study was approved by the Bioethical Committee of the Spanish Council of Research before starting, and so meets the standards of the Declaration of Helsinki in its revised version of 1975 and its amendments of 1983, 1989 and 1996. The written informed consent of all healthy donor participants was obtained at Centro de Hemoterapia y Hemodonación de Castilla y León Biobank. The researchers received the samples anonymously. The animal experiments were carried out with the permission of the local authority and conform to institutional standards.
Immunoblots and chromatin immunoprecipitation assays
Proteins were separated by electrophoresis in SDS–PAGE and transferred to nitrocellulose membranes. The membranes were used for immunodetection of P‐Y705‐STAT3 (#9131), STAT3 (#9132), P‐Y701‐STAT1 (#9171S) and STAT1 (#9172) with reagents from Cell Signaling Technology (Beverly, MA). Anti‐COX‐2 antibody (sc‐1745) and anti‐IRF1 antibody (sc‐497 and sc‐13041) were from Santa Cruz Biotechnology, Inc. Anti‐IRF3 antibody (catalogue no. 39033) was from Active Motif (Carlsbad, CA). Anti‐IRF5 antibody (ab2932) was from abcam plc (Cambridge, UK). Quantification of the blots was carried out using biorad quantity one gel imaging software (Bio‐Rad, Hercules, CA). Chromatin immunoprecipitation assays were carried out as described previously in DC fixed with 1% formaldehyde.4 Chromatin sonication was carried out using a Bioruptor device. Cross‐links were reversed by heating at 67°C, and the DNA bound to the beads was isolated by extraction with phenol/chloroform/isoamylalcohol. PCR were carried out with primers designed from the IL10 promoter.
Real time RT‐PCR of IL10, IL12A, IL23A, SOCS3, CXCL9, CXCL10, CSF1, CSF2, CSF3, IFNB1 and GAPDH
Purified RNA was used for reverse transcription reactions. The resulting cDNA was amplified in a PTC‐200 apparatus equipped with a Chromo4 detector (Bio‐Rad) using SYBR Green I mix containing Hot Start polymerase (Thermo Fisher Scientific, Waltham, MA). The sets of primers for PCR are shown in the Supplementary material (Table S1). GAPDH was used as a housekeeping gene to assess the relative abundance of the different mRNA using the comparative CT (threshold cycle) method.
Results
Effect of Irf3 deletion on the production of IL‐10
Given that the production of IL‐10 in the LPS/TLR4 route depends on the induction of IFN‐β and the responses to the fungal mimic zymosan are elicited by C‐type lectin and TLR2 receptors, our first hypothesis was that IL‐10 production should display mechanistic differences in both cases given the central role of the TRIF/TRAM/IRF3/IFN‐β pathway in LPS/TLR4 signalling. In keeping with these views, a reduced production of IL‐10 was observed in BMDC of Irf3 −/− mice stimulated with LPS, whereas the response to 0·1 mg/ml zymosan was similar in both groups and significantly increased in Irf3 −/− mice stimulated with 1 mg/ml. Zymosan was a stronger stimulus than LPS because concentrations of IL‐10 as high as 7·5 ng/ml were observed, whereas LPS only induced concentrations of ~ 0·5 ng/ml (Fig. 1a). Il10 mRNA expression in response to LPS showed a significant decrease in Irf3 −/− mice compared with wild‐type (WT) animals. In contrast, the response to zymosan showed a tendency to be higher in the knockout mice, although it did not reach statistical significance (Fig. 1b,c). The WT mice showed a strong induction of Ifnb1 mRNA in response to 0·1 mg/ml zymosan, which was even higher than that elicited by LPS, as well as induction of Il23a mRNA (Fig. 1d). Deletion of Irf3 abrogated the expression of Ifnb1 in response to LPS, and reduced significantly the response to zymosan (Fig. 1d), which might be explained by a remaining IRF5‐dependent induction. The contribution of type I IFN signalling to the overall effect of zymosan was assessed by assaying Cxcl9 and Socs3 mRNA. Zymosan was a weak inducer of Cxcl9 but induced Socs3 expression at a level similar to that elicited by LPS (Fig. 1e), suggesting that Socs proteins could be involved in the cross‐inhibition of type I IFN signalling.30, 31, 32 Tyrosine phosphorylation of STAT1 and STAT3 was detected in both Irf3 −/− and WT mice (Fig. 1f, g). STAT1 phosphorylation was robust at 3 hr and decreased at 6 hr; it was difficult to disclose whether it was more robust in either the knockout or the WT animals after normalization of the blots (Fig. 1g). Taken together, these data indicate that IL‐10 protein production and Y705‐STAT3 phosphorylation in response to zymosan show a limited dependence on IRF3 and that zymosan may induce Ifnb1 mRNA expression in the absence of IRF3.
Figure 1.
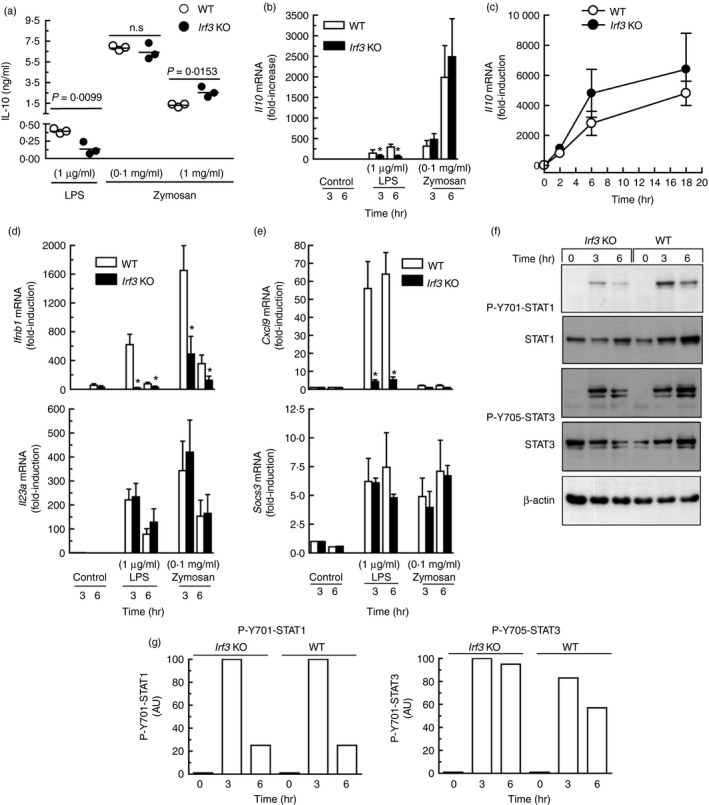
Effect of Irf3 deletion on cytokine production by mouse bone‐marrow‐derived dendritic cells (BMDC). (a) BMDC were incubated for 18 hr in the presence of either zymosan or lipopolysaccharide (LPS) at the concentrations indicated and the supernatants were collected for the assay of interleukin‐10 (IL‐10) protein. (b) Three independent pools of BMDC obtained from three wild‐type (WT) and three Irf3 −/− mice were stimulated for 3 and 6 hr and collected for the assay of Il10 mRNA. (c) Time–course of Il10 mRNA induction in response to 0·1 mg/ml zymosan in Irf3 −/− and WT mice. (d, e) Ifnb1, Il23a, Cxcl9 and Socs3 mRNA expression in response to LPS and zymosan in the same pools of animals studied in (b). (f) Effect of Irf3 deletion on Y701‐STAT1 and Y705‐STAT3 phosphorylation in BMDC stimulated with 0·1 mg/ml zymosan. (g) Densitometric scanning of the blots. This experiment was carried out using BMDC from an individual animal for each condition. Data in (a) and (c) represent mean ± SEM of three mice individually studied. n.s., not significant. AU, arbitrary units.
IL‐10 production in response to zymosan and Candida is independent of the IFN‐α/β receptor (IFNAR)
The BMDC of both Ifnar1 −/− and WT mice produced similar amounts of IL‐10 in response to zymosan (Fig. 2a). In contrast, the response to LPS was significantly decreased (Fig. 2b). An explanation for the lower levels of IL‐10 protein assayed in this set of experiments compared with the Irf3 −/− mice can be the different background of the animals, although in both cases mice were of the C57BL/6 strain. Mannose‐depleted zymosan induced a low amount of IL‐10 protein and heat‐killed Candida released IL‐10 to the same extent in both WT and knockout mice (Fig. 2c). Consistent with the results of the IL‐10 protein assays, the Ifnar1 −/− mice showed a reduced induction of Il10 mRNA in response to LPS, but no differences could be detected in response to zymosan (Fig. 2d). The expression of Ifnb1 mRNA showed similar values in both WT and Ifnar1 −/− mice (Fig. 2e), the levels at 2 hr being somewhat higher than at 4 hr in LPS‐treated BMDC, suggesting a rapid, and perhaps transient, trans‐activation of Ifnb1 consistent with a robust induction of IFN‐β protein (Fig. 2f).
Figure 2.
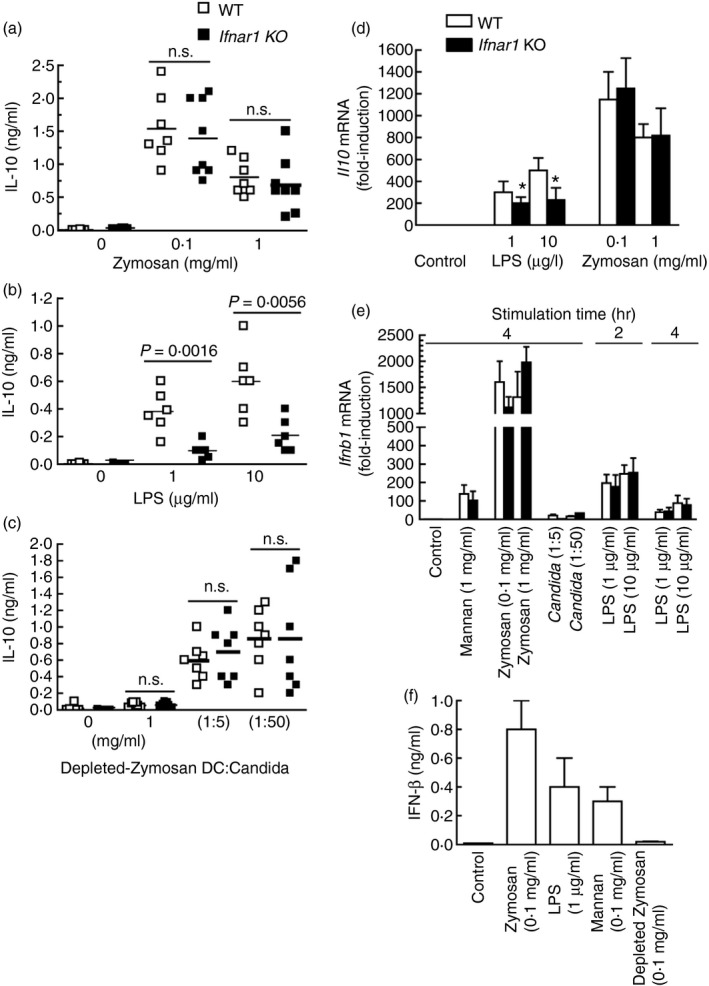
Production of interleukin‐10 (IL‐10) by wild‐type (WT) and Ifnar1 −/− mice. (a–c) Bone‐marrow‐derived dendritic cells (BMDC) were incubated in the presence of different stimuli and the IL‐10 protein was assayed in the supernatants after 18 hr of stimulation. Experiments with Candida albicans were carried out with heat‐killed Candida at two different ratios of DC : Candida as indicated. (d) Il10 mRNA induction in samples collected after 4 hr of stimulation. (e) Ifnb1 mRNA induction in samples collected at the times indicated. (f) Production of interferon‐β (IFN‐β) induced by different stimuli after 18 hr of incubation. Data in (d) represent mean ± SEM of six animals. Data in (e) represent mean ± SEM of three animals at 2 hr and nine animals at 4 hr. Data in (f) represent mean ± SEM of three mice.
These results show a limited role of type I IFN signalling on the production of IL‐10 elicited by zymosan and Candida, agree with earlier reports showing a limited effect of mannose‐depleted zymosan on IL‐10 production, and point to the mannose‐based component of zymosan as the main pathogen‐associated molecular patterns involved in the production of IL‐10.
Functional consequences of the phosphorylation of Y705‐STAT3 by zymosan components and comparison with the human system
Given that STAT3 activity seemed independent of type I IFN signalling, its possible role in the development of cross‐inhibition came into prominence. Y705‐STAT3 phosphorylation by LPS was clearly observed at 3 hr and reached a maximum between 6 and 18 hr. Zymosan showed a similar effect, although it was not detected at 3 hr (Fig. 3a). This time–course was consistent with the involvement of STAT3 in the induction of Il10 by LPS, as Y705‐STAT3 phosphorylation preceded the expression of Il10 mRNA (Fig. 3b). In keeping with previous reports, zymosan was a weak stimulus of Il12a mRNA expression33, 34 and a strong inducer of Il23a and Ifnb1 mRNA (Fig. 3b). P‐Y705‐STAT3 was observed to a similar extent in both WT and Ifnar1 −/− mice in response to zymosan and Candida (Fig. 3c). Human DC showed a time–course of Y705‐STAT3 phosphorylation (Fig. 4a) and a pattern of cytokine induction similar to those observed in mouse BMDC (Fig. 4b), with the exception of a tendency of IL10 mRNA to decrease at 18 hr and a lower induction of IFNB1 mRNA. In contrast, human macrophages showed a somewhat higher induction (Fig. 4c). To address the mechanism underlying STAT3 activation in human DC, the effects of the different components of zymosan and some cytokines were assayed. Zymosan, mannan and high concentrations of IL‐10 were robust inducers, whereas mannose‐depleted zymosan was a weak stimulus (Fig. 5a). Only mannan activated the phosphorylation of Y701‐STAT1 robustly (Fig. 5b). Zymosan was more active than mannan and depleted‐zymosan to induce IL10 and IL23A mRNA (Fig. 5c), whereas the IFN‐β‐inducible genes CXCL9 and CXCL10 showed reduced expression compared with the effect of IFN‐β (Fig. 5d). Interleukin‐10 induced its own mRNA to a low extent at a concentration of 10 ng/ml, which seems to be close to the maximum concentration that could be reached under physiological conditions. The selectivity of this effect being confirmed by the inhibition with anti‐IL‐10 antibody (Fig. 5e). Consistent with the lack of expression of its cognate receptor, IL‐23, which signals through a receptor coupled to the JAK/STAT system in some types of lymphoid cells,35 did not induce the expression of IL10 mRNA. These findings show limited evidence for type I IFN responses in the phosphorylation of Y705‐STAT3 and agree with an actual inhibition of type I IFN responses by the ITAM‐dependent receptors dectin‐1 and dectin‐2. Overall, the results observed in mice can be translated into humans with only a few cautions.
Figure 3.
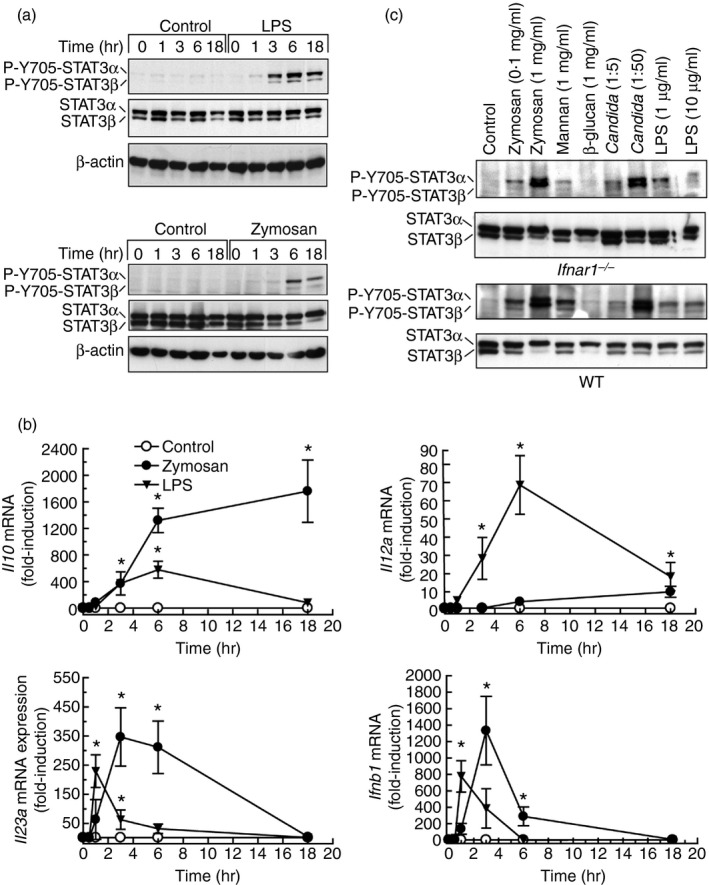
Y705‐STAT3 phosphorylation and cytokine expression in response to different stimuli in bone‐marrow‐derived dendritic cells (BMDC). (a) Time–course of Y705‐STAT3 phosphorylation in BMDC of mice stimulated with 10 μg/ml lipopolysaccharide (LPS) and 0·1 mg/ml zymosan. (b) Induction of Il10, Il12a, Il23a and Ifnb1 mRNA in response to zymosan and LPS. (c) Effect of different stimuli on Y705‐STAT3 phosphorylation in cells from Ifnar1 −/− and wild‐type (WT) mice stimulated for 6 hr. Data in (b) represent mean ± SEM of three different experiments. *P < 0·05.
Figure 4.
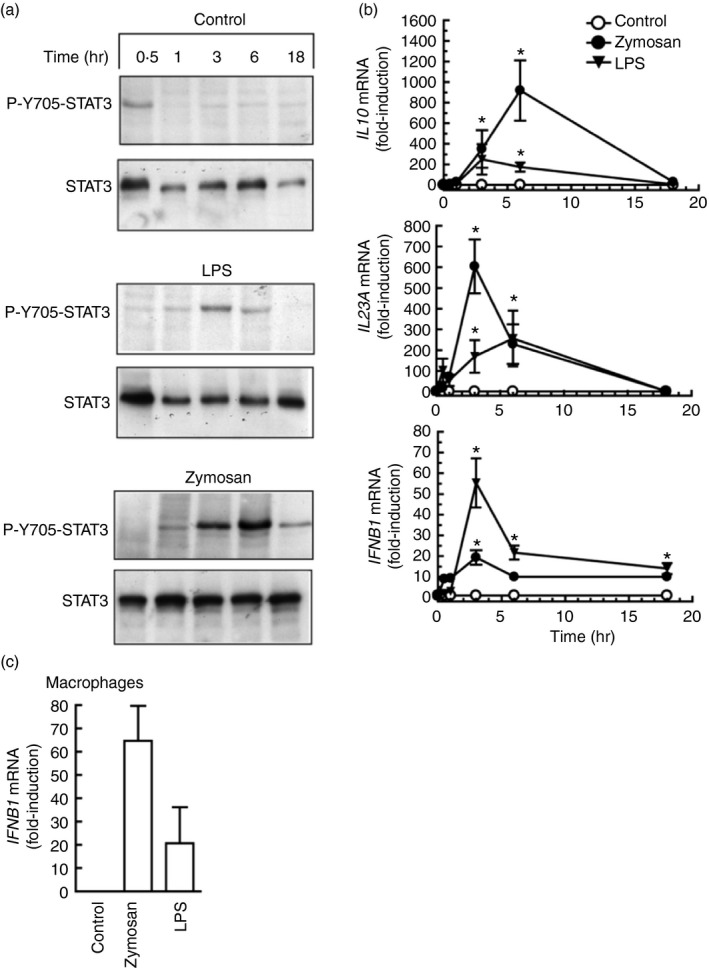
Y705‐STAT3 phosphorylation and cytokine expression induction in human dendritic cells (DC). (a) Time–course of Y705‐STAT3 phosphorylation. (b) Induction of IL10,IL23A, and IFNB1 mRNA in response to 0·1 mg/ml zymosan and 10 μg/ml lipopolysaccharide (LPS). (c) Effect of zymosan and LPS on IFNB1 mRNA induction in human macrophages stimulated for 6 hr with 1 mg/ml zymosan and 10 μg/ml LPS. Data in (b) and (c) represent mean ± SEM of three different experiments. *P < 0·05 compared with controls.
Figure 5.
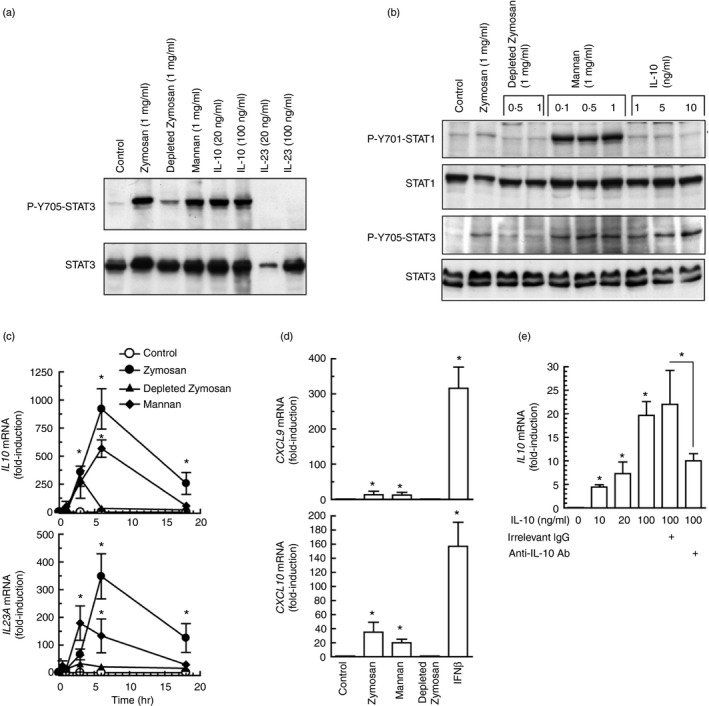
Effect of the different components of zymosan on Y705‐STAT3 and Y701‐STAT1 phosphorylation, and on cytokine induction. (a, b) Human dendritic cells (DC) were stimulated for 6 hr with different stimuli and cell lysates were used for the assay of Y705‐STAT3 and Y701‐STAT1 phosphorylation. (c, d) Effect of zymosan, mannan and mannose‐depleted zymosan at the concentration of 1 mg/ml on the induction of the mRNA of IL10,IL23A,CXCL9 and CXCL10. Interferon‐β (IFN‐β) was used as a positive control at the concentration of 1000 U/ml. (e) Effect of interleukin‐10 (IL‐10) and control irrelevant antibody on the effect of exogenous IL‐10 on IL10 mRNA induction expression. Antibodies were used at the concentration of 10 μg/ml. Data represent mean ± SEM of four different experiments. *P < 0·05 as compared to controls.
Mechanism of STAT3 activation in human DC
Several secondary mediators induced by zymosan may phosphorylate STAT3 at Y705 by JAK‐ and Src‐dependent routes.22, 36 In keeping with the limited effect of IL‐10 on STAT3 activation, a neutralizing anti‐IL‐10 antibody reduced Y705‐STAT3 phosphorylation only slightly, even when combined with an anti‐IL‐6 neutralizing antibody (Fig. 6a). The JAK inhibitors ruxolitinib and tofacitinib blocked the effect of zymosan. Ruxolitinib showing a robust effect at a lower concentration (Fig. 6b). STAT3 showed a mobility shift in response to zymosan that was observed in the absence of Y705‐STAT phosphorylation and could be explained by other post‐translational modifications, e.g. acetylation of lysine residues.37, 38 Notably, the JAK inhibitors did not affect the expression of IL10 mRNA in response to zymosan, whereas they inhibited the response to LPS and enhanced the IL23A mRNA induction in response to both stimuli (Fig. 6c, d), suggesting that the JAK‐activity‐dependent down‐regulation of IL23A expression could be explained by a direct blockade of IL‐10 signalling, given that the expression of IL10 mRNA was not affected. The possible involvement of PGE2 was assessed because it is an autocrine effector of IL‐10 production and may influence CREB4 and STAT324 activation. Although a direct effect of PGE2 on P‐Y705‐STAT3 was not observed, PGE2 receptor antagonists showed a slight inhibitory effect on zymosan and mannan responses (Fig. 7a). Given the prominent role of COX‐2 in the autocrine production of PGE2 and in STAT3 activation,39 its expression was addressed. Zymosan and mannan were robust inducers, whereas mannose‐depleted zymosan was inactive (Fig. 7b). These results disclose some commonality of the mechanisms involved in the induction of both PTGS2/COX2 and IL10. Stimulation with PAF did not induce Y705‐STAT3 phosphorylation, even when combined with PGE2, but similar to the effect of COX inhibitors, the PAF receptor antagonist showed some inhibitory effect that was enhanced in some cases by the combined addition of the COX inhibitors sc‐560 and sc‐236 (Fig. 7c–e). This effect is consistent with the expression of functional PAF receptors in DC and its cooperation with the signalling routes elicited by other agonists.40, 41, 42 Densitometric analysis of the inhibition of P‐Y705‐STAT3 phosphorylation in three independent experiments showed a 38% inhibition (Fig. 7e). Given that the transcriptome induced by fungal patterns disclosed that genes displaying growth factor activity are induced to a maximal extent43 and GM‐CSF (encoded by the gene CSF2) is a strong activator of STAT3, we addressed the mRNA expression of CSF1, CSF2 and CSF3. As shown in Fig. 7f, CSF2 mRNA showed maximal increase at 4 hr, making it likely its involvement as an autocrine activator of Y705‐STAT3 phosphorylation. However, stimulation with zymosan in the presence of a blocking antibody of GM‐CSF did not influence the expression of IL10 mRNA (Fig. 7d). The effect of GM‐CSF could also be ruled out by the absence of Y694‐STAT5 phosphorylation, a definite marker of GM‐CSF signalling (not shown). Given that a portion of the cross‐inhibition elicited by ITAM‐containing receptors has been associated with a signalling cascade dependent on p38 MAPK and mitogen‐ and stress‐activated kinases 1/2 (MSK1/2), which induces Y705‐STAT3 phosphorylation and Il10 expression,30, 31 the effect of the p38 MAPK inhibitor SB203580 was addressed. SB203580 showed 30% inhibition of Y705‐STAT3 phosphorylation in a densitometric scanning (Fig. 8a), whereas it inhibited by ~ 80% IL10 and IL23A mRNA expression, suggesting that its overall effect might depend on several targets (Fig. 8b). The inhibitor of Src kinases PP2 showed a tendency to enhance IL10 mRNA expression and did not influence Y705‐STAT3 phosphorylation (Fig. 8b, c). The Millipore STAT3 inhibitor did not elicit any significant effect (Fig. 8b). Together, these results show the involvement of JAK in the activation of STAT3 elicited by zymosan. This activation has a minimal effect on IL10 expression, but may contribute to diminish the pro‐inflammatory effects associated with IL23A induction expression. In addition, our data rule out the involvement of some purported effectors on the induction of Y705‐STAT3 phosphorylation and suggest the contribution of several mechanisms involving the lipid mediators PAF and PGE2, as well as p38 MAPK.
Figure 6.
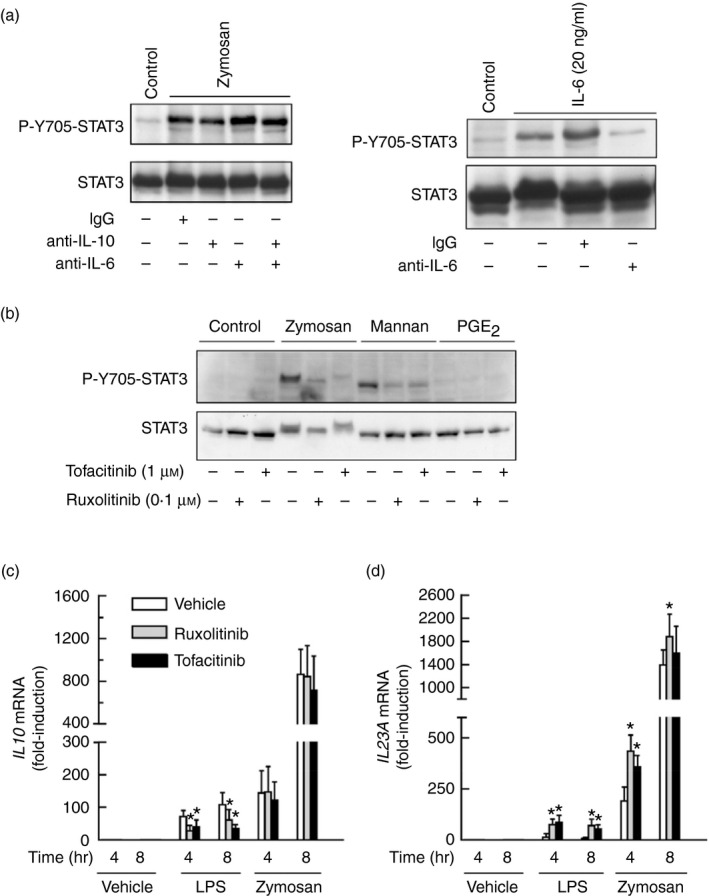
Effect of anti‐interleukin‐10 (IL‐10) and anti‐IL‐6 antibody on Y705‐STAT3 phosphorylation in response to zymosan. (a) Human dendritic cells (DC) were incubated for 30 min with 10 μg/ml of the different antibodies before the addition of 1 mg/ml zymosan. Cell lysates were collected at 6 hr to assay Y705‐STAT3 phosphorylation. To confirm the blocking activity of anti‐IL‐6 antibody, DC were stimulated with 20 ng/ml of IL‐6 for 1 hr and after this time, lysates were collected to assay Y705‐STAT3 (right panel). (b) Effect of ruxolitinib and tofacitinib on Y705‐STAT3 phosphorylation elicited by 1 mg/ml zymosan and mannan, and 1 μm prostaglandin E2 (PGE 2). (c, d) Effect of ruxolitinib and tofacitinib on IL10 and IL23A mRNA expression. Data represent mean ± SEM of three independent experiments. *P < 0·05.
Figure 7.
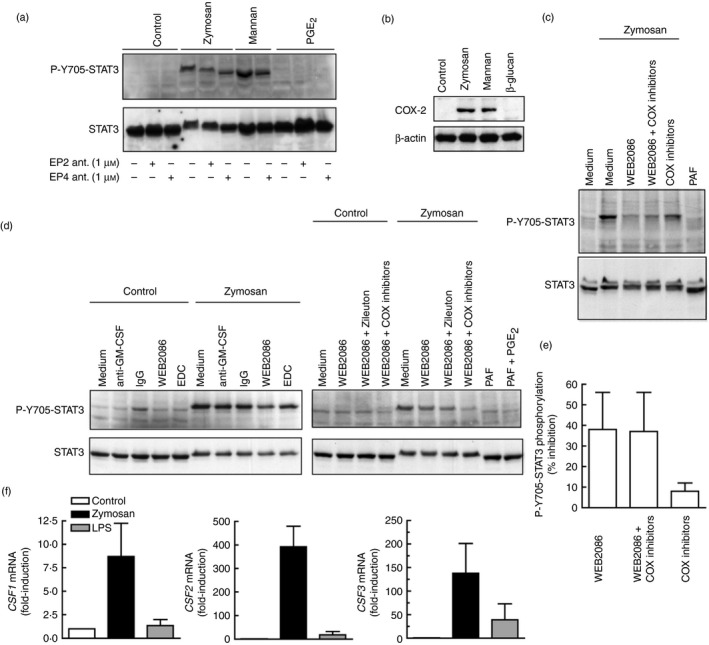
Analysis of the effect of lipid mediators and granulocyte–macrophage colony‐stimulating factor (GM‐CSF) on Y705‐STAT3 phosphorylation in human dendritic cells (DC). (a) Effect of the antagonists of E prostanoid receptor 2, PF‐04418948 and EP4, L‐161982, and exogenous prostaglandin E2 (PGE 2) (1 μm) on Y705‐STAT3 phosphorylation. (b) Effect of the different components of zymosan on COX2 induction. (c–e) The effect of the blockade of endogenous mediators on Y705‐STAT3 was assessed with anti‐GM‐CSF and IgG control antibody (10 μg/ml), the platelet‐activating factor (PAF) receptor antagonist WEB2086 (30 μm), the COX‐1 inhibitor sc‐560 (50 nm), the COX‐2 inhibitor sc‐236 (50 nm), the 5‐lipoxygenase inhibitor zileuton (10 μm), and the 12/15‐lipoxygenase inhibitor ethyl 3,4‐dihydroxybenzylidenecyanoacetate (3 μm). PAF was used at a concentration of 1 μm. A typical experiment of Y705‐STAT3 phosphorylation inhibition by WEB2086 is shown in (c), together with the densitometric analysis of a set of three independent experiments in (e). (f) Induction of CSF1,CSF2 and CSF3 mRNA in response to zymosan and lipopolysaccharide (LPS). Results show mean ± SEM of three independent experiments. *P < 0·05 as compared to controls.
Figure 8.
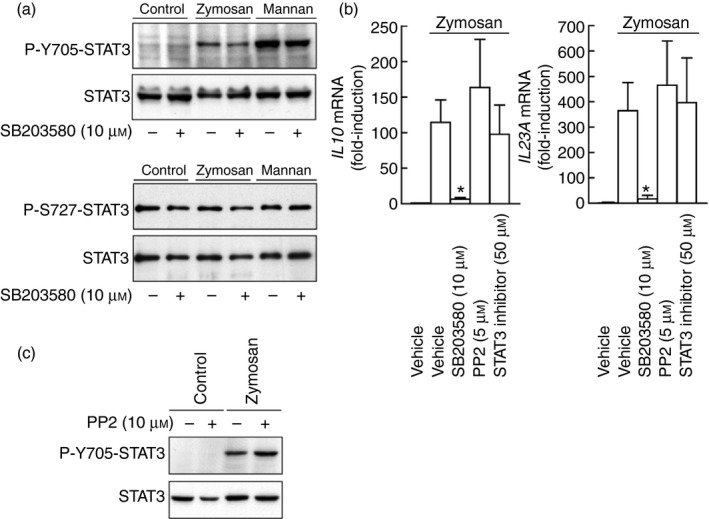
Effect of kinase and signal transducer and activator of transcription 3 (STAT3) inhibitors. (a) Effect of p38 mitogen‐activated protein kinase (MAPK) inhibition on P‐Y705‐STAT3 and P‐Ser727‐STAT3 phosphorylation. (b) Effect of the p38 MAPK inhibitor SB203580, the Src kinase inhibitor PP2, and the STAT3 inhibitor on IL10 and IL23A induction. (c) Effect of Src kinase inhibitor PP2 on P‐Y705‐STAT3 phosphorylation. Results show mean ± SEM of three independent experiments. *P < 0·05.
IRF5 displays different effects in the induction of IL‐10 elicited by LPS and zymosan
Because the effect of IRF3 may be countered by IRF5, which uncouples the binding of RNA polymerase II to the Il10 promoter, the effect of IRF5 deletion was addressed.44 The initial hypothesis was that the effect of IRF5 might prevail in the absence of IRF3. Experiments disclosed that Irf5 −/− mice showed a significant reduction of Ifnb1 and Il12a mRNA in response to zymosan compared with WT mice, whereas no significant changes were observed in Il10 and Il23a mRNA (Fig. 9a). The assay of IL‐10 protein did not show differences in response to zymosan, mannan and Candida conidia between Irf5 −/− and WT mice (Fig. 9b), whereas the production of IL‐10 and the expression of Il10 and Ifnb1 mRNA in response to LPS was reduced in the Irf5 −/− animals (Fig. 9c). Of note, GM‐CSF macrophages from Irf5 −/− mice showed a reduced expression of Ifnb1 and Il10 mRNA in response to zymosan (Fig. 9d). No evidence of type I IFN cytokine signature was observed, as judged from the absence of Cxcl9 and Cxcl10 induction in response to zymosan (data not shown). The assay of transcription factor binding to the IL10 promoter showed a significant binding of P‐Y705‐STAT3 to the most proximal IFN‐γ‐activated (GAS)‐site (Fig. 10a). This was not observed in response to β‐glucan and agrees with the limited extent of Y705‐STAT3 phosphorylation elicited by this stimulus. To confirm the specificity of the results, a region in the proximal promoter of IL23A lacking GAAA core sequences showed no binding when the assay was carried out with the set of primers indicated in the Supplementary material (Table S1). Unexpectedly, the effect of mannan was less prominent than that elicited by zymosan, although it was a robust activator of Y705‐STAT3 phosphorylation. A likely explanation could be that mannan also activates STAT1 and this might compete for binding at the same site. Binding of IRF3 and IRF5 to the IRF site increased significantly in response to both mannan and zymosan (Fig. 10b, c), the response to mannan being more robust. The binding of IRF1 was addressed because it is involved in the autocrine loop started by TNF‐α that activates type I IFN‐response genes. However, binding of IRF1 occurred to a low extent (Fig. 10d) and blockade of TNF‐α with the soluble TNF receptor etanercept showed some degree of inhibition of Y705‐STAT3 phosphorylation (Fig. 10e). Taken together, these data indicate that induction of Ifnb1 and Il12a by zymosan in BMDC depends on IRF5, whereas the induction of Il10 and Il23a mRNA is not affected. Conversely, the production of IL‐10 protein by LPS was significantly reduced in Irf5 −/− mice. IRF5 seems also involved in the induction of Ifnb1 and Il10 mRNA in GM‐CSF macrophages. Given that IRF5 can bind to the IL10 promoter, it is possible that their functions might be exerted through either a direct effect at the promoter level or indirectly through Ifnb1 induction.
Figure 9.
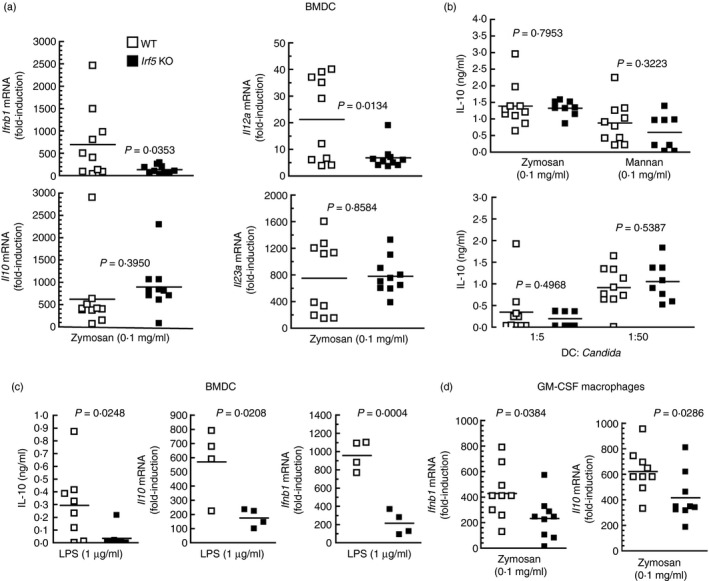
Effect of Irf5 deletion on interleukin‐10 (IL‐10) production by mouse bone‐marrow‐derived dendritic cells (BMDC) and granulocyte–macrophage colony‐stimulating factor (GM‐CSF) macrophages. (a–c) Bone marrow cells were cultured in medium supplemented with murine GM‐CSF. On day 5, the floating cells (BMDC) were collected and plated before stimulation with the indicated stimuli. (d) Adherent cells (GM‐CSF macrophages) were processed separately and stimulated with 0·1 mg/ml zymosan under the same conditions as BMDC. The stimulation was maintained for 4 hr for collection of RNA or 18 hr for the assay of IL‐10 protein.
Figure 10.
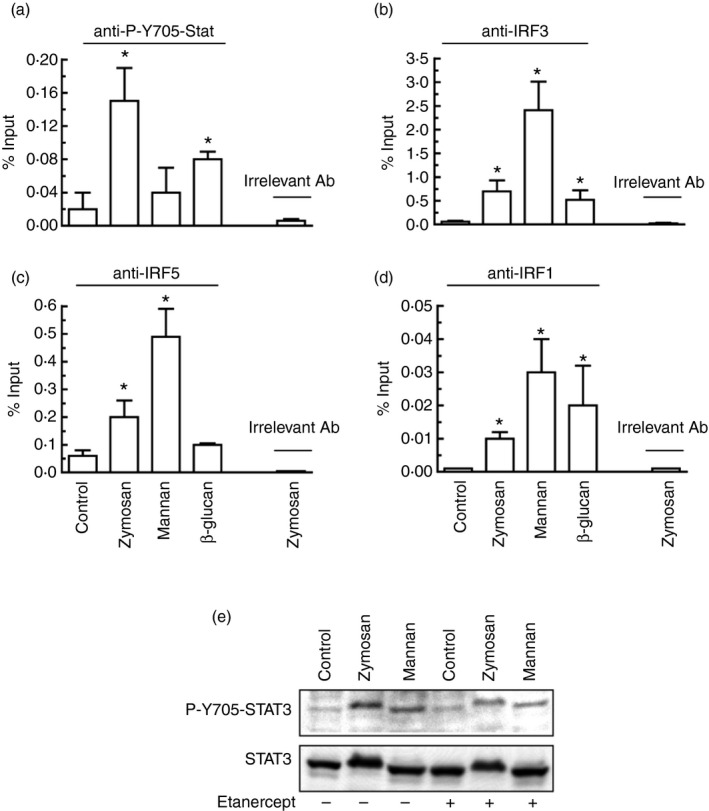
Binding of P‐Y705‐STAT3 and interferon regulatory factors 1/3/5 (IRF1/3/5) to the proximal GAS site of the human IL10 promoter. (a–d) Binding of P‐Y705‐STAT3 was studied at 6 hr after addition of the stimuli. Binding of the different IRF was studied at 4 hr after addition of the stimuli. The selection of times was carried out according to the time–course of Y705‐STAT3 phosphorylation and with reports on optimal binding of IRF. The specificity of the response was confirmed by the absence of antibody binding to a region in the proximal promoter of IL23A lacking GAAA core sequences and the results with irrelevant antibody. (e) Effect on Y705‐STAT3 phosphorylation of the blockade of tumour necrosis factor‐α (TNF‐α) action with 1 μg/ml etanercept. Results represent mean ± SEM of three to five independent experiments. *P < 0·05 compared with controls.
Discussion
Our data underscore the distinct signalling routes involved in the expression of IL‐10 triggered by bacterial and fungal patterns, disclose some mechanisms whereby ITAM‐associated receptors interfere with the function of the IFN‐α/β receptor, and provide some clues about the mechanism of activation of STAT3 by the secondary mediators induced by fungal patterns. The genetic approach using Irf3 −/−, Irf5 −/− and Ifnar1 −/− mice proved that the IFN‐β‐dependent autocrine cycle observed in the LPS/TLR4 route is not involved in the production of IL‐10 elicited by both Candida and zymosan. Given that zymosan is a strong inducer of Ifnb1 expression, there is not a clear reason to explain the differences between both models. The induction of Ifnb1 expression by zymosan has been associated with the presence of yeast nucleic acid and engagement of TLR7 and TLR9.45 However, zymosan did not induce the expression of CXCL9 and CXCL10 mRNA and blunted the induction of those cytokines by LPS.31 These results can be explained by the induction of SOCS1 and SOCS3, and the robust activation of STAT3, which suppresses STAT1 function by allowing the formation of STAT1/STAT3 heterodimers.46 These results agree with reports on the transcriptome of macrophages stimulated with Candida, where Il10, Il23a and Socs3 mRNA were expressed to a large extent in the absence of an IFN‐cytokine signature.43 Together, our data do not support the involvement of STAT3 in the induction of IL10 expression by fungal patterns, and the low production of IL‐10 elicited by mannose‐depleted zymosan agrees with the involvement of receptors that recognize mannose‐based patterns.7, 13, 47 Because STAT3 activation did not occur immediately after stimulation, the identification of secondary mediators other than type I IFN was necessary. Interleukin‐10 could be a possible effector, but a central role was ruled out in view of the high concentration of the cytokine required to induce P‐Y705‐STAT3 and the limited effect of a blocking antibody. A similar reasoning was assigned to IL‐6. The involvement of PGE2 and PAF was also considered. Although these mediators may be necessary for the activation of STAT3, they are not sufficient in view of our inability to show Y705‐STAT3 phosphorylation in response to these agonists. A possible explanation might be that they are produced at different times after zymosan challenge and this makes it difficult to reproduce the conditions for Y705‐STAT3 phosphorylation to occur. Although GM‐CSF is a component of the cytokine‐signature of zymosan, the effect of both exogenous and endogenous GM‐CSF was discarded using distinct approaches. Consistent with the notion that JAK inhibition blunts the IFN‐β feedback loop and enhances the production of pro‐inflammatory cytokines,47 JAK inhibitors enhanced IL23A mRNA expression, although they did not influence IL10 induction in response to zymosan. The involvement of Src kinases was ruled out because of the limited effect of the Src inhibitor PP2. Taking into account the most recent notions highlighting the central role of the energetic metabolism in the innate immune response, the chance that STAT3 phosphorylation might be the result of pyruvate kinase M2 activity is a challenging hypothesis.48 Likewise, recent research has disclosed the role of mTORC1 complex as a central element of the late phase of the TNF‐α response by regulating downstream genes that inhibit inflammatory signalling and nuclear factor‐κB activation. This route has a rigorous sensitivity to the metabolic state and oxygen levels and depends on IL‐10 induction and a delayed activation of STAT3.49
Strong activation of ITAM‐containing receptors by high‐avidity cross‐linking has been found to suppress IFNAR signalling via protein kinase C‐mediated recruitment of SHP2 and the induction of SOCS3 and repressors of gene transcription such as HES1 and STAT3.30 On this basis, Y705‐STAT3 phosphorylation might depend on a network of secondary mediators, rather than on a direct activation of JAK. For instance, the phosphorylation of Y705‐STAT3 and the production of IL‐10 by FcγR and integrins have been related to the activation of MSK, which are downstream from p38 MAPK.7, 50 However, our experiments showed that whereas p38 MAPK inhibition was associated with a strong down‐regulation of IL10 mRNA, only a modest inhibition of Y705‐STAT3 phosphorylation was observed.
Binding of IRF3 to the seven GAS‐sites detected in the −2000/+2000 nucleotides of the transcription start site in the Il10 promoter has been associated with Il10 transcription.51 Given the down‐regulatory effect of IRF5 on Il10 expression ascertained in some systems,44 we posited that IRF3 and IRF5 may compete for binding to the GAS‐sites and exert opposing effects on transcription. On this basis, IRF3 deletion might leave unrestrained the effect of IRF5 and enhance the expression of phenotypic markers of M1 macrophages.52 Taken collectively, experiments showed that the function of IRF5 is complex and may be cell‐ and stimulus‐dependent. A recent report showed that IRF5 is necessary for IL‐10 production in response to TLR7 and TLR9 stimulation in mouse GM‐CSF macrophages and DC, whereas the response to TLR2 and TLR4 ligands did not reach statistical significance, although it was lower in Irf5 −/− mice.53 In summary, we observed a robust activation of STAT3 and production of IL‐10 in human and mouse DC stimulated with zymosan that depends on its mannose‐based component. The effect on STAT3 phosphorylation is explained by an interplay of secondary mediators, which includes lipids, TNF‐α and p38 MAPK‐dependent reactions, but is independent on a feed‐forward mechanism involving IRF3 and type I IFN signalling. These results highlight the different transcriptional programmes involved in IL‐10 production by bacterial and fungal pathogen‐associated molecular patterns and disclose a mechanism whereby activation of ITAM‐containing receptors by fungal pathogen‐associated molecular patterns may suppress IFNAR signalling.
Disclosures
The authors declare that they have no conflict of interest.
Supporting information
Table S1. Primers used for chromatin immunoprecipitation and real time RT‐PCR in human and murine dendritic cells.
Acknowledgements
Dr T. Taniguchi and Dr G. Cheng are thanked for the Irf3 −/− mice. Dr C. Ardavin and Dr U. Kalinke are thanked for providing the Ifnar1 −/− mice. Staff from Centro de Hemoterapia de Castilla y León are thanked for help with blood cell purification. We thank the Wellcome Trust Sanger Institute Mouse Genetics Project (Sanger MGP) and its funders for providing the mutant mouse line (Allele: Irf5). Funding and associated primary phenotypic information may be found at www.sanger.ac.uk/mouseportal. This work was supported by grants from Plan Nacional de Salud y Farmacia (grants SAF2013‐44521‐R to MSC, and SAF2011‐29244 and SAF2014‐56819‐R to AC), Consejería de Sanidad de Castilla y León, Comunidad de Madrid I+D (grant S2010/BMD‐2350 to AC), Fundación Ramón Areces, and Red Temática de Investigación Cardiovascular.
Contributor Information
Mariano Sánchez Crespo, Email: mscres@ibgm.uva.es.
Nieves Fernández, Email: nieves@ibgm.uva.es.
References
- 1. Dillon S, Agrawal S, Banerjee K, Letterio J, Denning TL, Oswald‐Richter K, et al Yeast zymosan, a stimulus for TLR2 and dectin‐1, induces regulatory antigen‐presenting cells and immunological tolerance. J Clin Invest 2006; 116:916–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. LeibundGut‐Landmann S, Gross O, Robinson MJ, Osorio F, Slack EC, Tsoni SV, et al Syk‐ and CARD9‐dependent coupling of innate immunity to the induction of T helper cells that produce interleukin 17. Nat Immunol 2007; 8:630–8. [DOI] [PubMed] [Google Scholar]
- 3. Redford PS, Murray PJ, O′Garra A. The role of IL‐10 in immune regulation during M. tuberculosis infection. Mucosal Immunol 2011; 4:261–70. [DOI] [PubMed] [Google Scholar]
- 4. Alvarez Y, Municio C, Alonso S, Sánchez Crespo M, Fernández N. The induction of IL‐10 by zymosan in dendritic cells depends on CREB activation by the coactivators CREB‐binding protein and TORC2 and autocrine PGE2 . J Immunol 2009; 183:1471–9. [DOI] [PubMed] [Google Scholar]
- 5. Kelly EK, Wang L, Ivashkiv LB. Calcium‐activated pathways and oxidative burst mediate zymosan‐induced signaling and IL‐10 production in human macrophages. J Immunol 2010; 184:5545–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mellett M, Atzei P, Jackson R, O'Neill LA, Moynagh PN. Mal mediates TLR‐induced activation of CREB and expression of IL‐10. J Immunol 2011; 186:4925–35. [DOI] [PubMed] [Google Scholar]
- 7. Elcombe SE, Naqvi S, van den Bosch MWN, MacKenzie KF, Caianfanelli F, Brown GD, et al Dectin‐1 regulates IL‐10 production via a MSK1/2 and CREB dependent pathway and promotes the induction of regulatory macrophage markers. PLoS One 2013; 8:e60086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ouyang W, Rutz S, Crellin NK, Valdez PA, Hymowitz SG. Regulation and functions of the IL‐10 family of cytokines in inflammation and disease. Annu Rev Immunol 2011; 29:71–109. [DOI] [PubMed] [Google Scholar]
- 9. Chang EY, Guo B, Doyle SE, Cheng G. Cutting edge: involvement of type I IFN production and signaling pathway in lipopolysaccharide‐induced IL‐10 production. J Immunol 2007; 178:6705–9. [DOI] [PubMed] [Google Scholar]
- 10. Brown GD, Taylor PR, Reid DM, Willment JA, Williams DL, Martinez‐Pomares L, et al Dectin‐1 is a major β‐glucan receptor on macrophages. J Exp Med 2002; 196:407–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gantner BN, Simmons RM, Canavera SJ, Akira S, Underhill DM. Collaborative induction of inflammatory responses by dectin‐1 and Toll‐like receptor 2. J Exp Med 2003; 197:1107–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brown GD, Herre J, Williams DL, Willment JA, Marshall AS, Gordon SA. Dectin‐1 mediates the biological effects of β‐glucans. J Exp Med 2003; 197:1119–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Robinson MJ, Osorio F, Rosas M, Freitas RP, Schweighoffer E, Gross O, et al Dectin‐2 is a Syk‐coupled pattern recognition receptor crucial for Th17 responses to fungal infection. J Exp Med 2009; 206:2037–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Majer O, Bourgeois C, Zwolanek F, Lassnig C, Kerjaschki D, Mack M, et al Type I interferons promote fatal immunopathology by regulating inflammatory monocytes and neutrophils during Candida infections. PLoS Pathog 2012; 8:e1002811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Casanova JL, Holland SM, Notarangelo LD. Inborn errors of human JAKs and STATs. Immunity 2012; 36:515–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Smeekens SP, Ng A, Kumar V, Johnson MD, Plantinga TS, van Diemen C, et al Functional genomics identifies type I interferon pathway as central for host defense against Candida albicans . Nat Commun 2013; 4:1342. doi: 10.1038/ncomms2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. del Fresno C, Soulat D, Roth S, Blazek K, Udalova I, Sancho D, et al Interferon‐β production via Dectin‐1‐Syk‐IRF5 signaling in dendritic cells is crucial for immunity to C. albicans . Immunity 2013; 38:1176–86. [DOI] [PubMed] [Google Scholar]
- 18. Lopez‐Pelaez M, Lamont DJ, Peggie M, Shpiro N, Gray SN, Cohen P. Protein kinase IKKβ‐catalyzed phosphorylation of IRF5 at Ser462 induces its dimerization and nuclear translocation in myeloid cells. Proc Natl Acad Sci USA 2014; 111:17432–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ren J, Chen X, Chen ZJ. IKKβ is an IRF5 kinase that instigates inflammation. Proc Natl Acad Sci USA 2014; 111:17438–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yarilina A, Park‐Min KH, Antoniv T, Hu X, Ivashkiv LB. TNF activates an IRF1‐dependent autocrine loop leading to sustained expression of chemokines and STAT1‐dependent type I interferon‐response genes. Nat Immunol 2008; 9:378–87. [DOI] [PubMed] [Google Scholar]
- 21. Venkatesh D, Ernandez T, Rosetti F, Batal I, Cullere X, Luscinskas FW, et al Endothelial TNF receptor 2 induces IRF1 transcription factor‐dependent interferon‐β autocrine signaling to promote monocyte recruitment. Immunity 2013; 38:1025–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stark GR, Darnell JRE. The JAK‐STAT pathway at twenty. Immunity 2012; 36:503–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Soriano SF, Serrano A, Hernanz‐Falcón P, Martín de Ana A, Monterrubio M, Martínez C, et al Chemokines integrate JAK/STAT and G‐protein pathways during chemotaxis and calcium flux responses. Eur J Immunol 2003; 33:1328–33. [DOI] [PubMed] [Google Scholar]
- 24. Chun KS, Lao HC, Langenbach R. The prostaglandin E2 receptor, EP2, stimulates keratinocyte proliferation in mouse skin by G protein‐dependent and β‐arrestin1‐dependent signaling pathways. J Biol Chem 2010; 285:39672–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lukashova V, Chen Z, Duhé RJ, Rola‐Pleszczynski M, Stanková J. Janus kinase 2 activation by the platelet‐activating factor receptor (PAFR): roles of Tyk2 and PAFR C terminus. J Immunol 2003; 171:3794–800. [DOI] [PubMed] [Google Scholar]
- 26. Chen J, Lan T, Zhang W, Dong L, Kang N, Zhang S, et al Feed‐forward reciprocal activation of PAFR and STAT3 regulates epithelial‐mesenchymal transition in non‐small cell lung cancer. Cancer Res 2015; 75:4198–210. [DOI] [PubMed] [Google Scholar]
- 27. Valera I, Fernandez N, Trinidad AG, Alonso S, Brown GD, Alonso A, et al Costimulation of dectin‐1 and DC‐SIGN triggers the arachidonic acid cascade in human monocyte‐derived dendritic cells. J Immunol 2008; 180:5727–36. [DOI] [PubMed] [Google Scholar]
- 28. Sato M, Suemori H, Hata N, Asagiri M, Ogasawara K, Nakao K, et al Distinct and essential roles of transcription factors IRF‐3 and IRF‐7 in response to viruses for IFN‐α/β gene induction. Immunity 2000; 13:539–48. [DOI] [PubMed] [Google Scholar]
- 29. Barchet W, Cella M, Odermatt B, Asselin‐Paturel C, Colonna M, Kalinke U. Virus‐induced interferon α production by a dendritic cell subset in the absence of feedback signaling in vivo . J Exp Med 2012; 195:507–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang L, Gordon RA, Huynh L, Su X, Park Min KH, Han J, et al Indirect inhibition of Toll‐like receptor and type I interferon responses by ITAM‐coupled receptors and integrins. Immunity 2010; 32:518–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huynh L, Wang L, Shi C, Park‐Min KM, Ivashkiv LB. ITAM‐coupled receptors inhibit IFNAR signaling and alter macrophage responses to TLR4 and Listeria monocytogenes . J Immunol 2012; 188:3447–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Eberle ME, Dalpke AH. Dectin‐1 stimulation induces suppressor of cytokine signaling 1, thereby modulating TLR signaling and T cell responses. J Immunol 2012; 188:5644–54. [DOI] [PubMed] [Google Scholar]
- 33. Alvarez Y, Municio C, Hugo E, Zhu J, Alonso S, Hu X, et al Notch‐ and transducin‐like enhancer of split (TLE)‐dependent histone deacetylation explain interleukin 12 (IL‐12) p70 inhibition by zymosan. J Biol Chem 2011; 286:16583–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Alvarez Y, Rodríguez M, Municio C, Hugo E, Alonso S, Ibarrola N, et al Sirtuin 1 is a key regulator of the IL‐12 p70/IL‐23 balance in human dendritic cells. J Biol Chem 2012; 287:35689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chognard G, Bellemare L, Pelletier AN, Dominguez‐Punaro MC, Beauchamp C, Guyon MJ, et al The dichotomous pattern of IL‐12r and IL‐23R expression elucidates the role of IL‐12 and IL‐23 in inflammation. PLoS One 2014; 9:e89092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Silva CM. Role of STATs as downstream signal transducers in Src family kinase‐mediated tumorigenesis. Oncogene 2004; 23:8017–23. [DOI] [PubMed] [Google Scholar]
- 37. Yuan ZL, Guan YJ, Chatterjee D, Chin YE. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science 2005; 307:269–73. [DOI] [PubMed] [Google Scholar]
- 38. Rui W, Cherukuri P, Luo J. Activation of Stat3 sequence‐specific DNA binding and transcription by p300/CREB‐binding protein‐mediated acetylation. J Biol Chem 2005; 280:11528–34. [DOI] [PubMed] [Google Scholar]
- 39. Lin A, Wang G, Zhao H, Zhang Y, Han Q, Zhang C, et al TLR4 signaling promotes a COX‐2/PGE2/STAT3 positive feedback loop in hepatocellular carcinoma (HCC) cells. Oncoimmunology 2015; 5:e1074376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sozzani S, Longoni D, Bonecchi R, Luini W, Bersani L, D'Amico G, et al Human monocyte‐derived and CD34+ cell‐derived dendritic cells express functional receptors for platelet activating factor. FEBS Lett 1997; 418:98–100. [DOI] [PubMed] [Google Scholar]
- 41. Sozzani S, Rieppi M, Locati M, Zhou D, Bussolino F, Proost P, et al Synergism between platelet activating factor and C‐C chemokines for arachidonate release in human monocytes. Biochem Biophys Res Commun 1994; 199:761–6. [DOI] [PubMed] [Google Scholar]
- 42. Rodriguez M, Marquez S, Montero O, Alonso S, García Frade J, Sanchez Crespo M, et al Pharmacological inhibition of eicosanoids and platelet‐activating factor signaling impairs zymosan‐induced release of IL‐23 by dendritic cells. Biochem Pharmacol 2016; 102:78–96. [DOI] [PubMed] [Google Scholar]
- 43. Suram S, Silveira LJ, Mahaffey S, Brown GD, Bonventre JV, Williams DL, et al Cytosolic phospholipase A2 α and eicosanoids regulate expression of genes in macrophages involved in host defense and inflammation. PLoS One 2013; 8:e69002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Krausgruber T, Blazek K, Smallie T, Alzabin S, Lockstone H, Sahgal N, et al IRF5 promotes inflammatory macrophage polarization and TH1–TH17 responses. Nat Immunol 2011; 12:231–8. [DOI] [PubMed] [Google Scholar]
- 45. Biondo C, Signorino G, Costa A, Midiri A, Gerace E, Galbo R, et al Recognition of yeast nucleic acids triggers a host‐protective type I interferon response. Eur J Immunol 2011; 41:1969–79. [DOI] [PubMed] [Google Scholar]
- 46. Ho HH, Ivashkiv LB. Role of STAT3 in type I interferon responses: negative regulation of STAT1‐dependent inflammatory gene response. J Biol Chem 2006; 281:14111–8. [DOI] [PubMed] [Google Scholar]
- 47. Pattison MJ, Mackenzie KF, Arthur JS. Inhibition of JAKs in macrophages increases lipopolysaccharide‐induced cytokine production by blocking IL‐10‐mediated feedback. J Immunol 2012; 189:2784–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gao X, Wang H, Yang JJ, Liu X, Liu ZR. Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol Cell 2012; 45:598–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Huynh L, Kusnadi A, Park SH, Murata K, Park‐Min KH, Ivashkiv LB. Opposing regulation of the late phase TNF response by mTORC1‐IL‐10 signaling and hypoxia in human macrophages. Sci Rep 2016; 6:31959. doi:10.1038/srep31959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kim C, Sano Y, Todorova K, Carlson BA, Arpa L, Celada A, et al The kinase p38α serves cell type‐specific inflammatory functions in skin injury and coordinates pro‐ and anti‐inflammatory gene expression. Nat Immunol 2008; 9:1019–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Escors D, Lopes L, Lin R, Hiscott J, Akira S, Davis RJ, et al Targeting dendritic cell signaling to regulate the response to immunization. Blood 2008; 111:3050–61. [DOI] [PubMed] [Google Scholar]
- 52. Paun A, Bankoit R, Joshi T, Pitha PM, Stäger S. Critical role of IRF‐5 in the development of T helper 1 responses to Leishmania donovani infection. PLoS Pathog 2011; 7:e1001246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Watkins AA, Yasuda K, Wilson GE, Aprahamian T, Xie Y, Maganto‐Garcia E, et al IRF5 deficiency ameliorates lupus but promotes atherosclerosis and metabolic dysfunction in a mouse model of lupus‐associated atherosclerosis. J Immunol 2015; 194:1467–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Primers used for chromatin immunoprecipitation and real time RT‐PCR in human and murine dendritic cells.


