Abstract
Background:
CD52 is a small glycoprotein with a GPI anchor at its C-terminus. CD52 is expressed by Normal and malignant T and B lymphocytes and monocytes. There are detectable amounts of soluble CD52 in plasma of patients with CLL and could be used as a tumor marker. Although the biological function of CD52 is unknown but it seems that CD52 may be involved in migration and activation of T-cells .The aim of this study was to clone and express human CD52 gene in CHO cell line and studying its function in more details
Methods:
Based on GenBank databases two specific primers were designed for amplification of cd52 gene. Total RNA was extracted from Raji cell line and cDNA synthesized. Amplified fragment was cloned in pBudCE4.1 vector. The new construct was transfected to CHO-K1 cell line using electroporation method. Expression of recombinant CD52 protein was evaluated by Real time PCR and flow cytometry methods.
Results:
Amplification of CD52 gene using specific primers on Raji cDNA showed a 209 bp band. New construct was confirmed by PCR and restriction pattern and sequence analysis. The new construct was designated as pBudKT1. RT-PCR analysis detected cd52 mRNAs in transfected cells and Flow cytometry Results showed that 78.4 % of cells represented CD52 in their surfaces.
Conclusion:
In conclusion, we established a human CD52 positive cell line, CHO-CD52, and the protein was expressed on the membrane. Cloning of the CD52 gene could be the first step for the production of therapeutic monoclonal antibodies and detection systems for soluble CD52 in biological fluids
Key Words: CD52, Recombinant DNA, Therapeutic and diagnostic proteins
Introduction
CD52 is a small glycoprotein consisting of 12 aa, with a GPI anchor at its C-terminus. The gene is located on chromosome 1 and has 2 alleles with identical phenotypes (1-3). It was first characterized as a human leukocyte differentiation antigen(CDw52) (3). CD52 is expressed by Normal and malignant T and B lymphocytes, monocytes, eosinophils, epithelial cells of the epididymis and seminal vesicles express CD52 as surface antigen(1, 4, 5). There are detectable amounts of soluble CD52 (sCD52), released by phospholipase C, in plasma of patients with CLL and so could be used as a tumor marker. Plasma levels of sCD52 reflect the clinical features of CLL disease(6). Although the biological function of CD52 is unknown, but it seems that CD52 may be involved in migration and activation of T-cells (4,7). CD52 is the molecular target of Alemtuzumab (CAMPATH-1) antibody. Alemtuzumab is a humanized monoclonal antibody and has FDA approved for therapeutic use of various hematologic neoplasms, particularly in chronic lymphocytic leukemia (CLL) (5,6).
In the present study, we cloned the cd52 gene from B-cell lymphoma line (Raji) and CHO cell line was used as host to express the gene. In future studies recombinant protein will be evaluated to determine more details about its function and also to develop ELISA kit for detection of its soluble form in patients’ plasma.
Materials and Methods
Microorganisms, Cell lines and growth conditions
Escherichia coli DH5α (CinnaGen, Iran) as a host and pBudCE4.1 (Invitrogen, USA) as an expression vector were used. E. coli was cultured in LB medium at appropriate temperature (37 °C) with shaking (150 rpm). Lymphoma cell lines were grown in T-25 tissue-culture flasks in RPMI medium (Bio-Idea, Iran) supplemented with 10% Fetal Bovine Serum (FBS) (Gibco, Germany) and incubated at 37 °C in a humidified incubator with 5% CO2. CHO-K1 cells were maintained in RPMI 1640 supplemented with 10% (v/v) fetal bovine serum. Adherent cells were obtained with 1% trypsin-EDTA solution (Bio-IDEA, Iran). Based on MTT assay the selection medium was made by adding G418 sulfate (500 µg/ml).
RNA extraction and cDNA synthesis
2×106 cells were harvested and washed two times by PBS. Total RNA was isolated by High Pure RNA
Cloning in eukaryotic expression vector
Based on GenBank databases two specific primers (CD52f and CD52r) were designed. For subsequent cloning, PstI and XbaI restriction sites were added at the 5’-end of primers respectively (Table 1). A nucleotide (G) was inserted after PstI site and Kozak sequence (ACC) was added to the forward primer before ATG codon. 1 µl of the cDNA was used in PCR, using Pfu DNA polymerase (Thermo Fisher Scientific, USA). PCR products were separated by agarose gel electrophoresis and products were purified by high pure PCR product purification kit (Roche, Germany). Purified fragments were ligated into pBudCE4.1 at PstI / XbaI restriction sites. Transformed colonies were selected on Zeocin agar plates (30 µg/µl) and verified by colony PCR and restriction enzyme patterns. Recombinant plasmids were confirmed by sequencing using universal primers, T7f and pBudCE4.1r. (Bioneer, Korea). DNA manipulation and transformation method were based on the procedures described by Sambrook and Russell (8).
Table 1.
List of primers (restriction sites were showed in bold)
| Primers | Sequence 5′to 3′ | Orientation | 5′ cloning site |
|---|---|---|---|
| A) Primers used for amplification of cd52 gene | |||
| CD52 F | 5′-AACTGCAGGGCCACCATGAAGCGCTTCCTC-3 | Sense | PstI |
| CD52 R | 5′-CCTCTAGATCAACTGAAGCAGAAGAG-3′ | Anti-sense | XbaI |
| B) Primers used for Real time PCR | |||
| RT CD52 F | 5′- TTCCTCTTCCTCCTACTCACC-3′ | Sense | - |
| RT CD52 R | 5′-ATGCCTCCGCTTATGTTGCT-3′ | Anti-sense | - |
| CHO b2m F | 5′-TGTTGAAGAATGGAAAGAAGATGGA-3′ | Sense | - |
| CHO b2m R | 5′-CGGTCGCAGTGGGTGTAAA-3′ | Anti-sense | - |
| C) Universal primers | |||
| T7f | 5′-GTA AAA CGA CGG CCA GT | Sense | - |
| pBudCE4.1r | 5′-CAG GAA ACA GCT ATG AC | Anti-sense | - |
Sequence analysis
DNA fragments were sequenced by a Commercial Service (Bioneer). Alignment of nucleotide sequence was done by BLASTN Network Service (NCBI)
MTT Assay
To determine the appropriate concentration of Zeocin, MTT assay was done (9). Approximately 3×105-5×105 of passage 3-5 CHO-K1 cell line were cultured in 96-well flat bottom plates with 10% RPMI culture medium in triplicates. After 24 h, the culture media were completely removed and different concentration of Zeocin (200, 300, 400, 500, 600, 800, 1000 mg/ml) in complete culture medium was added, pure medium was considered as negative control. Media was replenished every 3-4 days. After 14 days the media was completely removed and 200 µl of 0.1 % MTT solution was added to each well. Cells were incubated for 4h at 37 °C in a 5% humidified CO2 incubator. After incubation, 200 µl of MTT solvent (DMSO) were added to each complete dried well. The absorbance of the plate was read at 495 nm.
Expression of human CD52 in CHO-K1
CHO-K1 cells were passaged 1 to 2 days before the electroporation, maintaining the cell density of about 75–85% confluency on the day of the experiment. Cells should be on passage 3-7. Before electroporation, the cells were harvested, washed with PBS, counted, and resuspended in PRMI 1640 at a density of 2-4 x 106 viable cells /ml. 0.8 ml cell suspension was gently mixed with 20 µg sterile Plasmid DNA. The cells were electroporated in 0.4 cm cuvettes (Bio-Rad, USA) with the Gene Pulser II Xcell electroporation system (Bio-Rad, USA) with 300 V and 950 µF using a single exponentially decaying pulse. Three replicates were performed for each condition. The electroporated cell suspension was then immediately transferred into 2 ml medium for recovery.
Real-time PCR
Following 48 h of transfection, expression of CD52 molecule was evaluated by Real-time PCR. The cells were collected and RNA extraction and cDNA synthesis was performed as mentioned before. Specific primers for cd52 (NM_001803.2) and β2 microglobulin (b2m) (NM_001246674.2) genes were designed using Allel ID 7.0 (Table1). Quantitative Real Time PCR was performed in duplicate for each sample in addition to a negative control and 4 standard samples using Power SYBR Green PCR Master Mix (2X) (ABI, UK) on ABI Step-One Real-Time thermal cycler (USA). The PCR mixture was containing 10 µl of 2x SYBR Green Master Mix, 4 pmol of each primer and 1µl of cDNA in final volume of 20 µl. PCR amplification was carried out with initial denaturation of 94 °C for 10 min, followed by 40 cycles of 94 °C for 15 s, 60 °C for 1 min. The specificity of the amplifications was determined, based on melting curves resulted by heating the amplicons from 60 to 90 °C. cd52 expression level compared to b2m housekeeping gene. The relative expression of the cd52 gene was calculated based on the 2-ΔCt method.
Flow cytometry
Following 48h of transfection, expression of CD52 molecule was assessed by flow cytometry method. The cells were collected by treatment with 1% trypsin–EDTA (Bio-IDEA, Iran) and washed twice with PBS, then stained with fluorescein isothiocyanate (FITC)-conjugated mouse anti-human CD52 antibody (Biolegend, USA). For negative control, untransfected CHO-K1 cells were stained with the same antibody. As Isotype control the cells were stained with FITC labeled mouse IgG with other specificity. The cells were incubated for 1 h at 4 °C, and then washed twice with PBS. Approximately 20,000 events were collected (BD FACSCalibur flow cytometer, USA). Data analysis was performed using FlowJo analysis software (version 7, 6, 2).
Stable transfection and characterization
For generation of stable cell lines, following characterization of the CHO-K1 cells by RT PCR and flow cytometry, transfected cells were cultured with selection-medium (Zeocin concentration 600 µg/ml) from the second day of transfection for approximately 2 months, then the CHO-CD52 cells were maintained in the culture medium containing 250 µg/ml Zeocin.
Results
In order to clone the cd52 gene two specific primers (CD52f and CD52r) were designed based on GenBank databases. CD52 is expressed on most B and T cell malignancies(10), so some B and T cell lines including Raji, MLA-144 and Jurkat cell lines were selected to isolate the cd52 gene. PCR with specific primers showed a 209 bp band and according to the result Raji cell line was selected (Data not shown). Amplified fragment was confirmed by restriction pattern using XcmI enzyme (PCR Based RFLP, PBR) (Fig. 1). pfu amplified DNA was cloned into the PstI/XbaI sites of pBudCE4.1 eukaryotic expression vector and confirmed by PCR and restriction patterns. The open reading frame was confirmed by nucleotide sequence analysis and revealed 100% homology with databases in GenBank without frame shift (data not shown). New construct was designated pBudKT1. The sequence was deposited in GenBank as accession number KP176645.1.
Fig. 1.
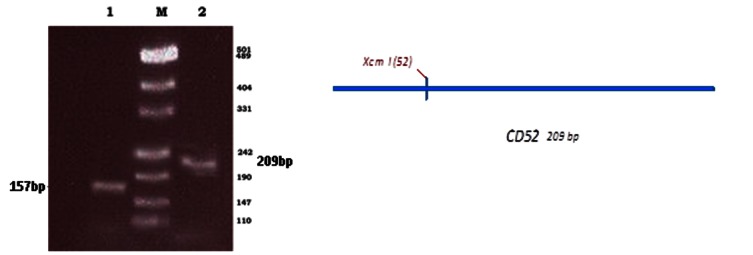
Restriction pattern analysis of cd52 DNA. Line 1: digestion of PCR product using XcmI enzyme, line 2: PCR product, M: DNA size marker.Schematic representation of expected restriction pattern of cd52 sequence. XcmI restriction site is indicated on the map
In order to express CD52, the new construct was transfected into CHO-K1 cells by electroporation method. 48 hour after transfection, CD52 expression was evaluated by Real Time PCR and Flow cytometry methods. RT-PCR analysis detected cd52 mRNAs in transfected cells. Cycle threshold (CT) for transfected cells was 14.43 while the expression level of cd52 gene in untransfected CHO-K1 was undetectable. Based on 2-ΔCt method, expression of CD52 was 3.7 fold in comparison to b2m gene expression.
Expression of recombinant CD52 in CHO-CD52 cells was evaluated by flow cytometry analysis. After staining cells with FITC conjugated anti-CD52 antibody 20,000 cells were read in FACSCalibur. Results showed that 78.4 % of cells represented CD52 in their surfaces (Fig. 2).
Fig. 2.

Flow cytometry results of heterologous expression of CD52 in CHO-K1 cell line. A) Untransfected CHO-K1 cell line B) Transfect CHO-K1 with pBudKT1 construct. Gray filled line showed cells stained with specific anti-CD52 antibody, dotted line shoed Isotype control and solid line showed unstained cells.
Discussion
CD52 is a small membrane glycoprotein which is expressed on the surface of lymphocytes, monocytes, macrophages and some lymphomas and leukemias (10, 11). Also a subpopulation of Treg cells have been identified which express CD52 on their surfaces. These cells halt the immune response by soluble factors such as CD52, independent of cell–cell contact (12). Despite low diagnostic value, CD52 is a good target for treatment of hematopoietic and nonhematopoietic disease (10). Anti-CD52 mAbs, Campath-1H in particular, mediate depletion of lymphocytes in vivo by ADCC, CDC (10, 11, 13). They have been developed as Therapeutic agents in treatment of lymphoid malignancy and autoimmune disease (12, 14). This molecule has been expressed in 1996 by Xia and colleagues in JURKAT and CHO cell lines (15).
In this study we cloned cd52 gene in order to investigate undetermined functions of this protein and also to produce ELISA kit. For these purposes two specific primers were designed and Kozak sequence was added in forward primer before ATG codon.
Although PBMC exhibit heterogeneous expression of CD52 (1), B or T cell lines overexpress this molecule on their surfaces (10). So some B and T cell lines were screened to isolate the cd52 gene. Based on PCR results Raji cell line was selected. To avoid the mismatch occurrence, PCR reaction was performed with Pfu polymerase which has proofreading. cd52 gene was ligated into pBudCE4.1 eukaryotic expression vector at PstI/XbaI cloning site. Analysis of cloned gene showed 100% homology with NCBI databases and revealed an open reading frame with 186 bp in length.
CHO-k1 cell line has revealed much efficiency and stability in transfection and recombinant
protein production (16, so we selected this cell line as the host cell. Zeocin antibiotic was used to select genetically engineered cells. Usually for mammalian cells a concentration of 400-800 µg/ml is used for selection and 250 µg/ml for maintenance (17). In order to find the suitable amount of antibiotic concentration MTT assay was done. The results showed that 600 µg/ml is appropriate to select the resistant cells.
Real-time PCR was performed to evaluate the presence of cd52 mRNA in transfected CHO-K1 cell line. Because untransfected sample was undetermined for cd52 expression so we used 2-ΔCt method to calculate the expression levels. Flow cytometry analysis revealed that recombinant CD52 is expressed in transfected cells and confirmed the Real Time PCR results.
After about 2 months of selection with 250 µg/ml Zeocin, we obtained a stable cell line, CHOK1-CD52.
In conclusion, we established a human CD52 positive cell line, CHO-CD52, and the protein was expressed on the membrane. Cloning of the CD52 gene and producing recombinant CD52 protein is attempting to provide first steps for the production of therapeutic monoclonal antibodies as well as facilitating a set of soluble CD52 detection system in biological fluids.
Acknowledgements
This work was supported by a grant from Shiraz University of Medical Sciences (grant number: 92-6698) and in part by Shiraz Institute for Cancer Research (grant number: ICR-100-509). This study was conducted as a requirement for the MSc. student thesis defended by Khadije Tati in Shiraz School of Medicine. The authors would like to thank Mr. Ahmad Hosseini for his help in flow cytometry data analysis.
References
- 1.Rao SP, Sancho J, Campos-Rivera J, Boutin PM, Severy PB, Weeden T, et al. Human Peripheral Blood Mononuclear Cells Exhibit Heterogeneous CD52 Expression Levels and Show Differential Sensitivity to Alemtuzumab Mediated Cytolysis. PLoS One. 2012;7(6):e39416. doi: 10.1371/journal.pone.0039416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Toh B-H, Kyaw T, Tipping P, Bobik A. Immune regulation by CD52-expressing CD4 T cells. Cellular and Molecular Immunology. 2013;10(5):379–82. doi: 10.1038/cmi.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alina Domaga, a MK. CD52 antigen Ð a review. Med Sci Monit. 2001;7(2):325–31. PubMed PMID: 18226615. [PubMed] [Google Scholar]
- 4.Hu Y, Turner MJ, Shields J, Gale MS, Hutto E, Roberts BL, et al. Investigation of the mechanism of action of alemtuzumab in a human CD52 transgenic mouse model. Immunology. 2009;128(2):260–70. doi: 10.1111/j.1365-2567.2009.03115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoermann G, Blatt K, Greiner G, Putz EM, Berger A, Herrmann H, et al. CD52 is a molecular target in advanced systemic mastocytosis. Faseb J. 2014;28(8):3540–51. doi: 10.1096/fj.14-250894. [DOI] [PubMed] [Google Scholar]
- 6.Albitar M, Do KA, Johnson MM, Giles FJ, Jilani I, O'Brien S, et al. Free circulating soluble CD52 as a tumor marker in chronic lymphocytic leukemia and its implication in therapy with anti-CD52 antibodies. Cancer. 2004;101(5):999–1008. doi: 10.1002/cncr.20477. [DOI] [PubMed] [Google Scholar]
- 7.Toh BH, Kyaw T, Tipping P, Bobik A. Immune regulation by CD52-expressing CD4 T cells. Cell Mol Immunol. 2013;10(5):379–82. doi: 10.1038/cmi.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sambrook J, Fritsch E.F, Manniatis T. Molecular cloning: A laboratory mannual. 2nd. Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 9.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65(1-2):55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 10.Ginaldi L, De Martinis M, Matutes E, Farahat N, Morilla R, Dyer MJ, et al. Levels of expression of CD52 in normal and leukemic B and T cells: correlation with in vivo therapeutic responses to Campath-1H. Leuk Res. 1998;22(2):185–91. doi: 10.1016/s0145-2126(97)00158-6. [DOI] [PubMed] [Google Scholar]
- 11.Bugelski PJ, Martin PL. Concordance of preclinical and clinical pharmacology and toxicology of therapeutic monoclonal antibodies and fusion proteins: cell surface targets. British Journal of Pharmacology. 2012;166(3):823–46. doi: 10.1111/j.1476-5381.2011.01811.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Samten B. CD52 as both a marker and an effector molecule of T cells with regulatory action: Identification of novel regulatory T cells. Cellular and Molecular Immunology. 2013;10(6):456–8. doi: 10.1038/cmi.2013.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Demko S, Summers J, Keegan P, Pazdur R. FDA drug approval summary: alemtuzumab as single-agent treatment for B-cell chronic lymphocytic leukemia. Oncologist. 2008;13(2):167–74. doi: 10.1634/theoncologist.2007-0218. [DOI] [PubMed] [Google Scholar]
- 14.Hale C, Bartholomew M, Taylor V, Stables J, Topley P, Tite J. Recognition of CD52 allelic gene products by CAMPATH-1H antibodies. Immunology. 1996;88(2):183–90. doi: 10.1111/j.1365-2567.1996.tb00003.x. PubMed PMID: 24348139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xia MQ, Tone M, Packman L, Hale G, Waldmann H. Characterization of the CAMPATH-1 (CDw52) antigen: biochemical analysis and cDNA cloning reveal an unusually small peptide backbone. Eur J Immunol. 1991;21(7):1677–84. doi: 10.1002/eji.1830210714. [DOI] [PubMed] [Google Scholar]
- 16.Ning B-T, Tang Y-M. Establishment of the cell line, HeLa-CD14, transfected with the human CD14 gene. Oncology Letters. 2012;3(4):871–4. doi: 10.3892/ol.2012.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Blokland HJM, Hoeksema F, Siep M, Otte AP, Verhees JA. Methods to create a stringent selection system for mammalian cell lines. Cytotechnology. 2011;63(4):371–84. doi: 10.1007/s10616-011-9354-9. [DOI] [PMC free article] [PubMed] [Google Scholar]


