Abstract
Recent advances in genome editing with programmable nucleases have opened up new avenues for multiple applications, from basic research to clinical therapy. The ease of use of the technology—and particularly clustered regularly interspaced short palindromic repeats (CRISPR)—will allow us to improve our understanding of genomic variation in disease processes via cellular and animal models. Here, we highlight the progress made in correcting gene mutations in monogenic hereditary disorders and discuss various CRISPR-associated applications, such as cancer research, synthetic biology, and gene therapy using induced pluripotent stem cells. The challenges, ethical issues, and future prospects of CRISPR-based systems for human research are also discussed.
Keywords: Clustered regularly interspaced short palindromic repeats-Cas9, Clustered regularly interspaced short palindromic repeats, Gene editing, Induced pluripotent stem cells, Genetic therapy
INTRODUCTION
Socioeconomic burden of rare genetic diseases is increasing. There have been numerous attempts to treat genetic diseases with various methods. However, they were not overly successful till now. Recently, the technology of clustered regularly interspaced short palindromic repeats (CRISPR) emerges as a promising tool to correct genetic abnormalities. This technique is being heralded for precision and accuracy in genetic editing. In this review, we recapitulate the history and recent progress made in the area of CRISPR technology. In the first part of the review, we summarize the history and action mechanism of CRISPR. In the second part of the review, we deliberate upon assorted clinical applications of CRISPR, from the standpoint of recent feasibility and future possibilities. In the third part, we discuss about future perspective of CRISPR technology. Ideal combination of CRISPR technology and induced pluripotent stem cell (iPSC) may bring new CRISPR-based clinical applications into clinics in near future.
PART 1. HISTORY AND MECHANISMS OF ACTION
In light of the heterogeneity of disease manifestations among patients, the field of precision medicine has attracted a great deal of interest, especially following a new initiative launched in 2015. The Precision Medicine Initiative (PMI), which involves investment in medical research in the United States on a national scale, envisions treatment and prevention of diseases on an individual basis, according to differences in genes and environmental and lifestyle factors [1,2]. The short-term goals of the PMI include combating cancers, while the accrual of knowledge pertaining to health and diseases represents the long-term focus. The aggregation of personalized information pertaining to genomics, proteomics, and phenotypical parameters is expected not only to provide a better understanding of health and disease, but also to change our approach to risk assessment, diagnostic tests, and therapeutic interventions [3]. However, despite major progress in gene sequencing and profiling, based on advances in technologies such as high-throughput sequencing, precise gene editing remains challenging; this difficulty has hindered the translation of information into clinical applications. As a consequence, the demand for targeted, straightforward, and affordable genetic engineering tools continues to grow.
Over the past several decades, scientists have revolutionized genetic engineering techniques to allow modulation of their function. Since Watson and Crick elucidated the structure of the DNA double helix, many researchers have focused on changing the genome according to particular scientific needs. To achieve this, a platform that can identify the target sequence of interest, specifically cleave that region of the DNA, and alter the sequence at the cleavage site is required. Endogenous site-specific DNA-protein complexes and natural DNA repair pathways from multiple species have been exploited to create various gene engineering toolkits. Among these, the most rapidly evolving technology involves CRISPR and CRISPR-associated nuclease 9 (Cas9) (CRISPR-Cas9), which was selected as Science’s Breakthrough of the Year in 2015.
CRISPR-Cas9 in the bacterial adaptive immune system
CRISPR is derived from the prokaryotic adaptive immune system (Fig. 1) [4-36]. The distinctive clustered repeats were originally recognized in Escherichia coli by Ishino et al. [5] in 1987, and were later found to include unique barcode-like sequences of viral or plasmid origin, termed spacers (Fig. 2) [6-8]. In 2007, the hypothesized role of the repeats in adaptive defense was confirmed by experimental demonstration of spacer integration into the bacterial genome following phage challenge, as well as alteration of sensitivity to subsequent phage infection dependent upon the spacer content [4]. Subsequent studies revealed that CRISPR works in sync with the Cas gene, in the vicinity of the CRISPR locus, to cleave DNA or RNA sequences [9,10] targeted by a small guide RNA [11]. Based on these findings, multiple studies sought to identify the components of the CRISPR-Cas system and apply this knowledge to sequence-specific gene engineering.
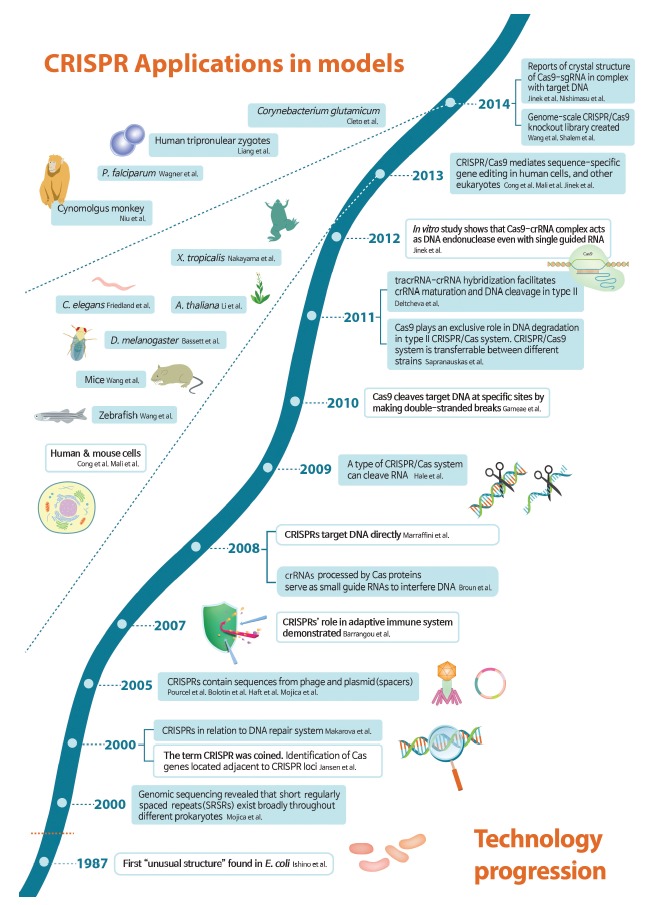
Figure 1.
Timeline of technological progression of clustered regularly interspaced short palindromic repeats (CRISPR) and its application in model organisms. Key developments are shown and major breakthroughs are highlighted in white boxes. While the CRISPR story starts in 1987, the name was coined in 2000, and CRISPR’s role in adaptive immune system was demonstrated in 2007. A key insight in 2012 that CRISPR-associated nuclease 9 (Cas9) is an RNA-guided DNA endonuclease led to an explosion of papers related to CRISPR gene-editing technology. From 2013, CRISPR was successfully applied in modification of genes in humans and other various organisms [4-36]. sgRNA, single guide RNA; P. falciparum, Plasmodium falciparum; X. tropicalis, Xenopus tropicalis; C. elegans, Caenorhabditis elegans; A. thaliana, Arabidopsis thaliana; D. melanogaster, Drosophila melanogaster; tracrRNA, trans-acting CRISPR RNA; crRNA, CRISPR RNA; E. coli, Escherichia coli.
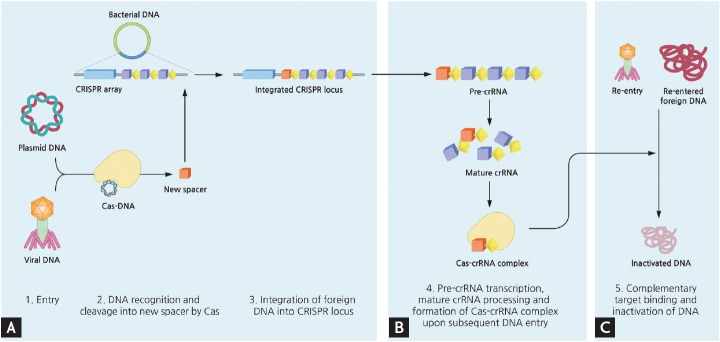
Figure 2.
Simplified mechanism of microbial adaptive immune system using clustered regularly interspaced short palindromic repeats (CRISPR). Upon entry of foreign DNA into bacteria, CRISPR-associated (Cas) enzymes acquire new spacers from the exogenous sequence and integrate this spacer unit into the leader end of CRISPR locus within bacterial genome. The transcript of CRISPR array is further processed, and when another corresponding invasion occurs this mature CRISPR RNA (crRNA) act as a guide by Cas complex to degrade matching DNA. The detailed mechanisms of each type of CRISPR systems vary slightly. (A) Acquisition. (B) crRNA biogenesis. (C) Interference.
Mechanism underlying CRISPR-Cas9 gene editing
There are six putative CRISPR systems; the three main types (types I to III) were discovered first, with three additional types (types IV to VI) being identified more recently [37,38]. During the processes of immunity, adaptation, expression, and interference, each type acts according to distinct mechanisms to ensure DNA recognition and cleavage [39].
Type I uses a large complex of Cas proteins, encoded by the Cas3 gene, which show separate helicase and DNase activities. Similarly, type III uses repeat-associated mysterious proteins, which constitute a large superfamily of Cas proteins. Types I, III, and IV are categorized as class 1 based on their multi-subunit effector complexes. By contrast, class 2 systems (comprising types II, V, and VI) each have a single-subunit effector. Type II uses only a single protein (Cas9) for its nuclease activity; the same is true for types V and VI, but with Cas9-like proteins. Owing to their simplicity, class 2 systems have been adopted for genome engineering [40-42], and only the bacterial type II CRISPR-Cas9 system has been utilized for RNA-guided engineering nucleases [43,44].
As noted above, type II CRISPR-Cas9 systems use a single endonuclease, Cas9. This enzyme acts in concert with two guide RNAs: CRISPR RNA (crRNA) and trans-acting CRISPR RNA (tracrRNA) (Fig. 3) [45]. To simplify the system and improve its utility, scientists employed a linker loop to engineer a dual tracrRNA:crRNA (called single guide RNA [sgRNA]), which participates in sequence-specific DNA cleavage with Cas9 [12]. Short 2 to 5 bp conserved sequences known as proto-spacer adjacent motifs, located on the side opposite to that of the RNA-DNA hybridization, are required for Cas9-DNA recognition [6]. Once recognition occurs, double-stranded DNA cleavage is performed by two Cas9 domains: the HNH domain, which cleaves the strand complementary to the crRNA-guide sequence, and the RuvC-like domain, which cleaves the noncomplementary strand [12]. Via this mechanism, programmed nucleases with customized sgRNA can cleave genomic DNA at specific loci, enabling precise genome editing.
Figure 3.
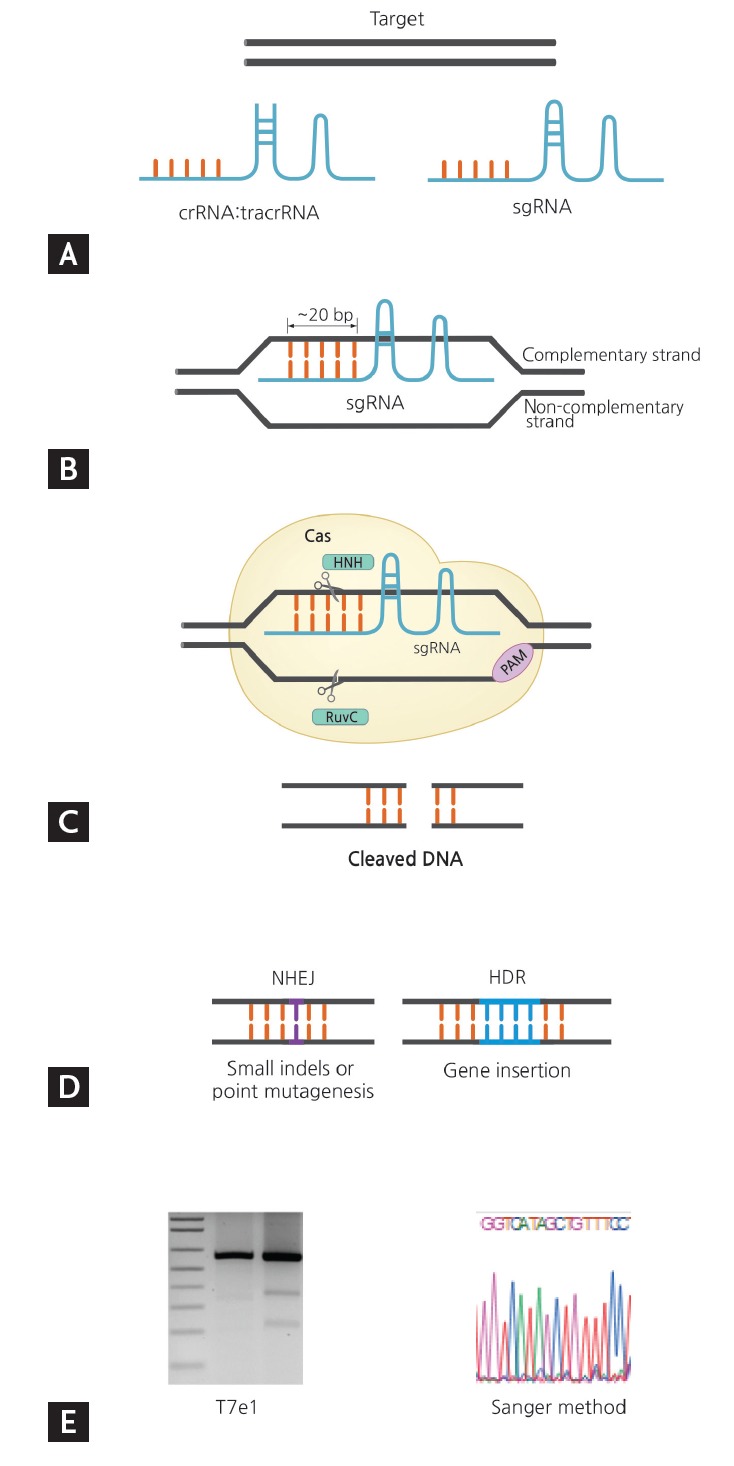
Overview of clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR-associated nuclease 9 (Cas9) gene editing from target selection and guide design to validation. (A) Select gene of interest and design guide RNA. (B) Base pairing of sgRNA: genomic DNA. (C) Detection of PAM by Cas and cleavage of gene of interest by Cas domains HNH and RuvC. (D) Formation of nuclease-induced double strand breaks (DSB). (E) Validation of gene editing. sgRNA, single guide RNA; crRNA, CRISPR RNA; tracrRNA, trans-acting CRISPR RNA; PAM, proto-spacer adjacent motif; NHEJ, nonhomologous end joining; HDR, homology-directed repair.
Cas9 protein can be easily re-targeted to new DNA sequences by changing a small portion of the sequence of the accompanying RNA guide, which base-pairs directly with target DNA [46]. Another potential advantage of Cas9 is its ability to introduce several double-strand breaks (DSBs) within the same genome (also referred to as multiplexing) via the expression of multiple guide RNAs [13,14].
Earlier approaches to gene editing
Prior to the advent of CRISPR technology, biologists used several generations of tools, all of which employed site-specific DNA DSBs for genome editing. To date, four types of DNA-binding nuclease have been developed: meganucleases, zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and the most recently identified nuclease, Cas9.
Each of the previous tools had unique limitations. Meganucleases are like restriction enzymes but are programmed to target DNA sequences 14 to 40 bp in length. Owing to several shortcomings, including their lack of specificity in DNA recognition and the need to fuse the recognition and cleavage domains, meganucleases were used only briefly [47]. ZFNs and TALENs function according to similar principles, but differ in that their binding domains consist of three- and one-nucleotide recognition modules, respectively. These enzymes have separate DNA-binding domains and nonspecific cleavage domains, namely, FokI endonucleases, making them more efficient than meganucleases [48,49].
However, the construction of ZFNs remains a challenge due to the need to account for context-dependent binding preferences [50], notwithstanding previous efforts to circumvent this shortcoming. TALENs, despite having the advantage of one-to-one binding between the Transcription activator-like effector (TALE) unit and each base pair [51], require painstaking molecular biology cloning methods to synthesize highly conserved and repetitive TALE structures [52]. Consequently, the comparatively facile protein engineering of CRISPR makes this approach much more affordable and practical compared with precursor technologies (Table 1).
Table 1.
Comparison of different programmable nucleases
| Variable | ZFN | TALEN | CRISPR |
|---|---|---|---|
| DNA-binding moiety | Protein | Protein | RNA |
| Target site size, bp | 18–36 | 30–40 | 22 |
| Nuclease | FokI | FokI | Cas |
| Cytotoxicity | Variable to high | Low | Low |
| Design availability | More complex | Complex | Simple |
| Ease of multiplexing | Low | Low | High |
ZFN, zinc f inger nuclease; TALEN, transcription activator-like effector nuclease; CRISPR, clustered regularly interspaced short palindromic repeats.
PART 2. APPLICATIONS OF CRISPR
Basic application of the CRISPR-Cas9 system
When DSBs are introduced, the lesion may be corrected by one of two major repair pathways: homology-directed repair (HDR) or nonhomologous end joining (NHEJ). HDR allows the exchange of genetic information between DNA molecules with similar sequences, whereas NHEJ forms short insertions or deletions (indels) in the target sequence. NHEJ does not require a repair template, but the resultant indels can cause frameshift mutations that lead to the production of nonfunctional, incomplete proteins, or to micro-RNA degradation by nonsense-mediated decay. On the other hand, the HDR machinery can repair DNA using exogenous single- or double-stranded DNA templates with sequence similarity to the DSB site. Thus, exploitation of HDR has allowed researchers to insert new genetic information at a target site, or to perform direct replacement of a mutated gene [45,46].
Although this approach was revolutionary, the natural process of HDR is inefficient because it requires selection and screening to identify the one-in-a-million cell in which homologous recombination has exchanged the wild-type gene for the desired modified version. However, CRISPR-Cas9 technology allows the inducible formation of DSBs, so that scientists can modify gene expression at the repair sitex; thus, opening a new avenue in genome editing [42].
Cell-based and in vivo animal studies
The applications of CRISPR-Cas9 have expanded into fields such as agricultural products, livestock, disease modeling, and therapeutics. In this section, we focus on the therapeutic aspects of gene-based diseases, especially monogenic disorders (Fig. 4).
Figure 4.
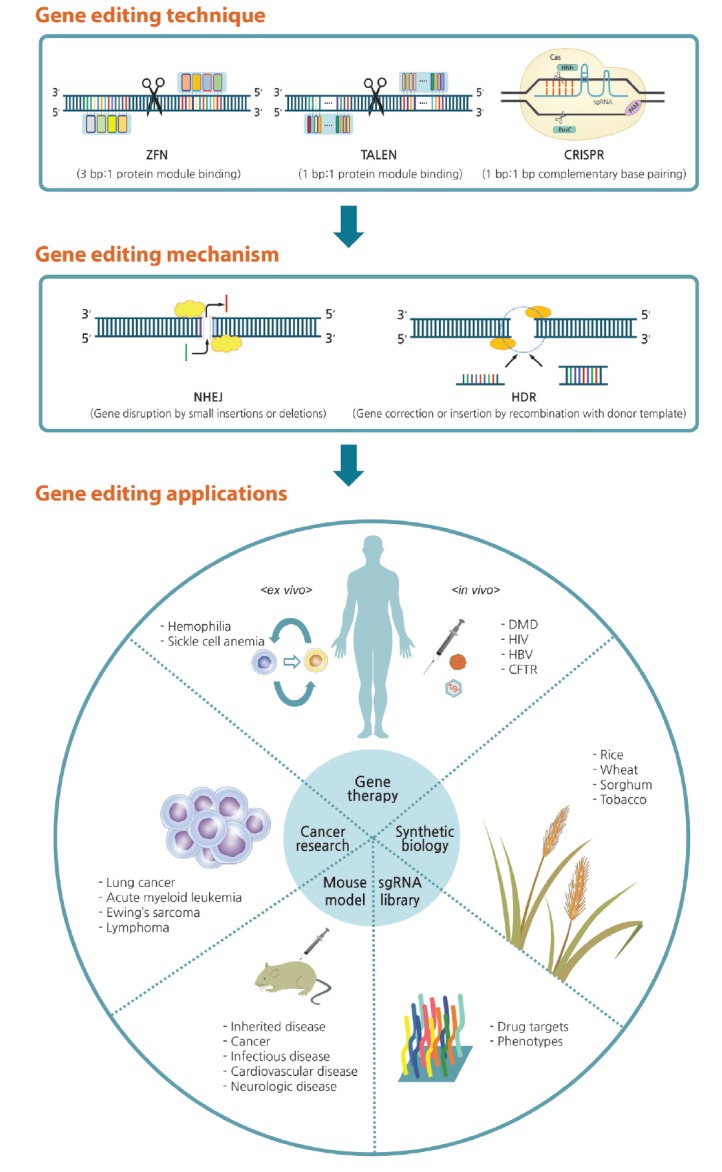
Overview of gene editing and its applications. Genetic defects can be corrected via gene editing with zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and the clustered regularly interspaced short palindromic repeats (CRISPR) system. When double-strand breaks occur, the lesion can be corrected by either nonhomologous end joining (NHEJ) or homology-directed repair (HDR) pathways. Arising from this technique, gene editing can be applied in various fields of research and biotechnology. sgRNA, single guide RNA; PAM, proto-spacer adjacent motif; DMD, Duchenne muscular dystrophy; HIV, human immunodeficiency virus; HBV, hepatitis B virus; CFTR, cystic fibrosis transmembrane conductance regulator.
In gene therapy, genes in diseased cells and tissues can be corrected by two approaches: ex vivo and in vivo editing [46]. In ex vivo therapy, the target cell population is removed from the body, modified using a programmable nuclease, and then transplanted back into the original host; thereby, preventing complications due to immunological rejection. By contrast, in vivo editing therapy involves direct transfer of genome-editing reagents, such as a programmable nuclease and donor templates, into the human body [53]. Each approach has advantages and disadvantages, and they are implemented differently to treat particular disorders. There has been examples of gene-editing techniques applied in disease cell lines (Table 2) [54-77] and in disease mouse models (Table 3) [60,63-66,78-88]. Furthermore, scientists have reported series of therapeutic applications with genome editing using stem cell (Table 4) [89-111].
Table 2.
Examples of gene-editing techniques applied in cell lines
| Cell lines | Disease | Target gene | Strategy | Delivery | Nuclease | Reference |
|---|---|---|---|---|---|---|
| HBE, CFTE | Cystic fibrosis | CFTR | HDR-mediated cDNA knock-in | Plasmid | ZFN | Lee et al. (2012) [71] |
| K-562, Hep3B | Hemophilia B | hF9 | HDR-mediated addition of corrective cDNA | AAV | ZFN | Li et al. (2011) [72] |
| K-562 | Sickle-cell anemia | HBB | HDR-mediated cDNA knock-in | Plasmid | TALEN | Voit et al. (2014) [58] |
| K- 562, hCD4+ T cells, HEK-293, lymphoblastoid cells, Jurkat cells, hESCs | SCID | IL2Rγ | HDR-mediated cDNA knock-in | IDLV, mRNA | ZFN/TALEN | Lombardo et al. (2007) [73] |
| Urnov et al. (2005) [74] | ||||||
| Matsubara et al. (2014) [75] | ||||||
| Genovese et al. (2014) [76] | ||||||
| Immortalized patient myoblasts | DMD | DMD | Excision of exons 51 or 45–55, restoring the reading frame | Plasmid | ZFN/CRISPR | Ousterout et al. (2015) [56] |
| Ousterout et al. (2015) [57] | ||||||
| Addition of the microdystrophin gene | Plasmid | ZFN | Benabdallah et al. (2013) [55] | |||
| NHEJ restoration of reading frame | Plasmid | TALEN | Ousterout et al. (2013) [54] | |||
| Patient fibroblasts | Epidermolysis bullosa | COL7A1 | HDR using a ssODN | Plasmid | TALEN | Osborn et al. (2013) [77] |
| SiHa, C33-A, Caski | HPV | E6, E7 | NHEJ-mediated disruption of promoter, E6, and E7 gene | Plasmid | CRISPR | Zhen et al. (2014) [67] |
| Kennedy et al. (2014) [68] | ||||||
| Hu et al. (2014) [70] | ||||||
| Yu et al. (2015) [69] | ||||||
| Huh7, HepG2, HepAD38, HepaRG | HBV | Multiple | NHEJ-mediated disruption of multiple genes | Plasmid | CRISPR | Lin et al. (2014) [60] |
| Seeger et al. (2014) [62] | ||||||
| Zhen et al. (2015) [64] | ||||||
| Dong et al. (2015) [65] | ||||||
| Liu et al. (2015) [66] | ||||||
| Kennedy et al. (2015) [61] | ||||||
| Ramanan et al. (2015) [63] | ||||||
| CHME5, HeLa. TZM-b1, U1 | HIV | LTR U3 region | NHEJ-mediated disruption of viral genes | Plasmid | CRISPR | Hu et al. (2014) [59] |
HDR, homology-directed repair; ZFN, zinc finger nuclease; hF9, human F9; AAV, adeno-associated virus; TALEN, transcription activator-like effector nuclease; SCID, severe combined immunodeficiency; IDLV, integration-deficient lentiviral vector; DMD, Duchenne muscular dystrophy; CRISPR, clustered regularly interspaced short palindromic repeats; NHEJ, nonhomologous end joining; ssODN, single-stranded oligonucleotide; HPV, human papilloma virus; HBV, hepatitis B virus; HIV, human immunodeficiency virus; LTR U3, long terminal repeat U3.
Table 3.
Examples of therapeutic applications of genome editing in mouse model
| Disease | Target gene | Strategy | Delivery | Model | Nuclease | Reference |
|---|---|---|---|---|---|---|
| Hemophilia B | hF9 | HDR-mediated addition of corrective cDNA | AAV | Humanized neonatal, adult mice | ZFN | Li et al. (2011) [82] |
| Anguela et al. (2013) [83] | ||||||
| Hemophilia A, B | mAlb | HDR-mediated insertion of F8 and F9 cDNA, respectively | AAV | Humanized adult mice | Sharma et al. (2015) [84] | |
| Hereditary tyrosinemia I | Fah | HDR of point mutation | Hydrodynamic injection | Adult mouse model | CRISPR | Yin et al. (2014) [85] |
| Cataract | Crygc | HDR-mediated correction | Plasmid | Zygote, mouse SSC | CRISPR | Wu et al. (2015) [81] |
| DMD | Exon 23 of dmd gene | HDR using a ssODN | Cas9, sgRNA | Zygote | CRISPR | Long et al. (2014) [86] |
| NHEJ-mediated disruption of exon 23 | AAV | Adult or neonatal | CRISPR | Xu et al. (2016) [87], | ||
| Nelson et al. (2016) [78] | ||||||
| Tabebordbar et al. (2016) [79] | ||||||
| Long et al. (2016) [80] | ||||||
| NHEJ-mediated disruption of exon 23 | Plasmid | Adult | CRISPR | Xu et al. (2016) [87] | ||
| HBV | Multiple | NHEJ-mediated disruption of multiple genes | Hydrodynamic injection, Plasmid | Adult | CRISPR | Lin et al. (2014) [60] |
| Zhen et al. (2015) [64] | ||||||
| Dong et al. (2015) [65] | ||||||
| Liu et al. (2015) [66] | ||||||
| Ramanan et al. (2015) [63] | ||||||
| Cardiovascular disease | Pcsk9 | NHEJ-mediated disruption of PCSK9 | Cas9, sgRNA | Adult | CRISPR | Ding et al. (2014) [88] |
hF9, human F9; HDR, homology-directed repair; AAV, adeno-associated virus; ZFN, zinc finger nuclease; CRISPR, clustered regularly interspaced short palindromic repeats; SSC, spermatogonial stem cell; DMD, Duchenne muscular dystrophy; ssODN, single-stranded oligonucleotide; Cas9, CRISPR associated protein 9; sgRNA, single guide RNA; NHEJ, nonhomologous end joining; HBV, hepatitis B virus.
Table 4.
Examples of therapeutic applications of genome editing using stem cell
| Disease | Target gene | Strategy | Delivery | Study model | Nuclease | Reference |
|---|---|---|---|---|---|---|
| Cystic fibrosis | CFTR | HDR-mediated correction of CFTR deltaF508 mutation | Plasmid | Patient iPSCs | ZFN/TALEN/CRISPR | Crane et al. (2015) [101] |
| Sargent et al. (2014) [102] | ||||||
| Firth et al. (2015) [103] | ||||||
| HDR-mediated cDNA knock-in | Plasmid | Intestinal organoid | CRISPR | Schwank et al. (2013) [104] | ||
| Small/short DNA fragments-mediated correction of CFTR deltaF508 mutation | Plasmid | Patient iPSCs | TALEN | Suzuki et al. (2016) [105] | ||
| Hemophilia A | hF8 | NHEJ-mediated correction of inversion | Plasmid, Cas9, gRNA | Patient iPSCs | TALEN/CRISPR | Park et al. (2014) [90] |
| Park et al. (2015) [91] | ||||||
| Sickle-cell anemia | HBB | HDR-mediated correction | Plasmid | Patient iPSCs | ZFN/TALEN/CRISPR | Sebastiano et al. (2011) [93] |
| Sun et al. (2014) [94] | ||||||
| Huang et al. (2015) [95] | ||||||
| mRNA | Patient CD34+ HSCs | ZFN | Hoban et al. (2015) [92] | |||
| NHEJ-mediated deletion of enhancer BCL11A | mRNA, lentivirus | Adult CD34+ HSCs, immortalized human CD34+ HSPCs | ZFN/TALEN/CRISPR | Vierstra et al. (2015) [106] | ||
| Canver et al. (2015) [107] | ||||||
| β-Thalassemia | HDR-mediated correction | Plasmid | Patient iPSCs | ZFN/TALEN/CRISPR | Ma et al. (2015) [97] | |
| Xie et al. (2014) [96] | ||||||
| Sun et al. (2014) [94] | ||||||
| SCID | Prkdc | HDR of point mutation | Plasmid | - | ZFN | Rahman et al. (2015) [108] |
| IL2Rγ | HDR-mediated correction | Plasmid | Patient iPSCs | TALEN | Menon et al. (2015) [109] | |
| DMD | Exon 45 of dmd gene | Disruption of the splicing acceptor to skip exon 45/NHEJ restoration of reading frame/HDR-mediated exon 44 cDNA knock-in | Plasmid | Patient iPSCs | TALEN/CRISPR | Li et al. (2015) [89] |
| Epidermolysis bullosa | COL7A1 | Repairing the COL7A1 locus by HR | AAV | Patient iPSCs | AAV-DJ | Sebastiano et al. (2014) [100] |
| HIV | CCR5 or CXCR4 | NHEJ-mediated disruption of CCR5 | AAV | Normal iPSCs, human CD34+ HSPCs | ZFN/TALEN/CRISPR | Holt et al. (2010) [110] |
| Wilen et al. (2011) [99] | ||||||
| Li et al. (2013) [111] | ||||||
| Ye et al. (2014) [98] |
HDR, homology-directed repair; iPSC, induced pluripotent stem cell; AAV, adeno-associated virus; ZFN, zinc finger nuclease; TALEN, transcription activator-like effector nuclease; CRISPR, clustered regularly interspaced short palindromic repeats; NHEJ, nonhomologous end joining; Cas9, CRISPR associated protein 9; HSC, hematopoietic stem cell; SCID, severe combined immunodeficiency; DMD, Duchenne muscular dystrophy; HIV, human immunodeficiency virus; CCR5, cinnamoyl-CoA reductase 5; CXCR4, C-X-C chemokine receptor type 4; HSPC, hematopoietic stem and progenitor cell.
Inactivation or correction of deleterious mutations
Duchenne muscular dystrophy
Duchenne muscular dystrophy (DMD) is the most prevalent fatal genetic disease passed on through the X chromosome. The gene dystrophin consists of 79 exons, and several types of mutation in exon sequences lead to DMD. Currently, there is no effective treatment for DMD, but genome editing has the potential to restore expression of a modified dystrophin gene [53].
Efforts aimed at correction of the dystrophin gene in immortalized patient myoblasts with ZFNs and TALENs were initiated in 2013 [54,55]. Because 13% of DMD patients have a mutation in exon 51, the introduction of indels into, or complete excision of, exon 51 can restore dystrophin expression [56]. In one study, permanent removal of exons 45 to 55 by multiplexed Cas9 was therapeutically applicable in 62% of patients [57].
Mouse models can provide data that is relevant to in vivo human therapy. For example, the Mdx model mouse harbors a mutation in exon 23 of the dystrophin gene. Local and systemic delivery of gene correction to Mdx mice using adeno-associated virus (AAV) vector and the CRISPR-Cas9 system resulted in 2% to 100% correction, and the therapeutic benefits were predicted to be 15% to 20% [78-81].
Genome editing has also been effective in DMD gene therapy in patients lacking exon 44; in this case, the correction was performed ex vivo in induced pluripotent stem cells (iPSCs). Three correction strategies were tested: skipping of exon 45, introduction of small indels resulting in a frameshift in exon 45, and knock-in of the missing exon 44 to restore the full protein coding region; the last of these strategies was the most effective. The corrected iPSCs successfully differentiated into muscles and expressed functional protein [89].
Insertion of corrective or protective mutation
Hemophilia
Hemophilia is caused by different genetic mutations— in coagulation factor VIII (F8) for hemophilia A, and in coagulation factor XI (F9) for hemophilia B. Gene therapy is an option for treating hemophilia because correction of the defective gene results in permanent expression of functional protein, and even 1% wild-type expression of coagulation factor VIII or XI is sufficient to confer a therapeutic effect [112,113].
The first successful in vivo gene targeting of hemophilia was achieved in a hemophilia B neonate mouse [82]. Using a ZFN pair to target the defective human F9 (hF9) gene, and AAV as the delivery vector, donor cDNA was inserted into the mouse genome. A similar approach in adult hF9 mice resulted in stable production of human factor IX [83].
Hemophilia A, which is more prevalent than hemophilia B, involves a more complex type of mutation, making it harder to edit the gene. However, chromosomal inversion at the F8 gene is a common cause of hemophilia, and TALENs were previously shown to be able to correct this rearrangement [90]. Thus, the CRISPR-Cas9 system was used to target each side of the ~600 kb inversion and correct the mutation in iPSCs derived from hemophilia A patients [91].
Sickle-cell anemia and β-thalassemia
Sickle-cell anemia and β-thalassemia are both caused by mutations in the HBB gene, resulting in an inappropriate level of the β-globin chain of hemoglobin. Editing the β-globin locus by targeted nucleases represents a new strategy for permanently curing hemoglobinopathies. TALENs programmed to target the β-globin locus were used for HDR-mediated full-length cDNA knockin in K562 cells [58], and ZFNs were used to correct a sickle-cell anemia-associated point mutation in CD34+ hematopoietic stem progenitor cells [92]. These hemoglobinopathies are particularly advantageous for gene therapy because extracted patient iPSCs can be differentiated into hematopoietic stem cells, which can in turn be inserted back into patients by autologous transplantation. This strategy has already been implemented with all ZFNs, TALENs, and CRISPR-Cas9 in both sickle-cell anemia and β-thalassemia patients [93-97].
Disruption of viral DNA
Human immunodeficiency virus
The most advanced gene-editing strategy is the ex vivo modification of T cells to knock out the cinnamoyl-CoA reductase 5 (CCR5) gene, resulting in resistance to human immunodeficiency virus (HIV) infection. This is one of the few cases in which a treatment that exploits the gene-editing machinery has been used in clinical trials (Table 5). This idea was clinically validated when an HIV-infected patient received a stem cell transplant from a donor with a homozygous deletion in the CCR5 allele, resulting in undetectable levels of HIV and restoration of normal CD4+ T cell counts [114].
Table 5.
Ongoing and completed clinical trials adopting gene-editing technology
| Nuclease | Disease | Status | Phase | Title |
|---|---|---|---|---|
| ZFN | HIV | Completed | I | Autologous T cells genetically modified at the CCR5 gene by ZFN SB-728 for HIV |
| Dose escalation study of autologous T cells genetically modified at the CCR5 gene by ZFN in HIV-infected patients | ||||
| I/II | Study of autologous T cells genetically modified at the CCR5 gene by ZFN in HIV-infected subjects | |||
| Active | I/II | Repeat doses of SB-728mR-T after cyclophosphamide conditioning in HIV-infected subjects on HAART | ||
| Recruiting | I | Safety study of ZFN CCR5-modified hematopoietic stem/ progenitor cells in HIV-1 infected patients | ||
| I/II | Dose escalation study of cyclophosphamide in HIV-infected subjects on HAART receiving SB-722-T | |||
| Hemophilia B | Not yet recruiting | I | Ascending dose study of genome editing using the ZFP therapeutic SB-FIX in subjects with severe hemophilia B |
ZFN, zinc finger nuclease; HIV, human immunodeficiency virus; CCR5, cinnamoyl-CoA reductase 5.
One study demonstrated the safety of infusion of ZFN-modified autologous CD4+ T cells bearing a deletion of the CCR5 gene into HIV-positive human patients [115]. Building on the results obtained with ZFNs, multiple efforts were made using similar gene-editing strategies to knock out CCR5 using TALENs or CRISPR-Cas9. Wild-type iPSCs were seamlessly modified by NHEJ-mediated deletion of the CCR5 gene, at an average rate of 14% with TALENs and 33% with CRISPR [98]. To increase resistance to HIV infection, other genes were targeted in addition to CCR5, including C-X-C chemokine receptor type 4 (CXCR4), which encodes a coreceptor, and PC4 and SFRS1 interacting protein 1 (PSIP1), which encodes the lens epithelium-derived growth factor (LEDGF)/p75 protein required for HIV integration [99,116,117].
To reduce adverse off-target effects, several studies have attempted to eliminate the integrated HIV-1 genome. In one study, for example, the HIV-1 long terminal repeat (LTR) U3 region was efficiently targeted by the CRISPR-Cas9 system, resulting in inactivation of viral gene expression and replication in CHME5, HeLa, TZM-b1, and U1 cells [59].
Beyond HIV, programmable gene-editing nucleases have also been applied to other viral pathogens. For example, in Huh7, HepG2, HepAD38, and HepaRG cells transfected with a hepatitis B virus (HBV) expression vector, targeted editing of multiple genes was used to reduce the production of HBV core and surface proteins. HBV-expressing templates were disrupted by CRISPR-Cas9 both in vitro and in vivo [60-66]. Likewise, the E6 and E7 genes of human papillomavirus were also targeted by CRISPR-Cas9 [67-70].
There are increasing numbers of ongoing and completed clinical trials adopting gene-editing technology (Table 5).
PART 3. FUTURE PERSPECTIVES
Cancer research
All cancers harbor multiple mutations that cause cells to grow progressively and express malignant phenotypes. These mutations can be categorized into four types: oncogenes, tumor suppressors, epigenetic factors and control loci, and chemoresistance genes. The CRISPR-Cas9 system represents a powerful, highly specific and adaptable tool for correcting such mutations and treating the cancers that contain them [118]. While oncogenic changes occur in many cancers and play important roles in malignant cell proliferation, oncogenes such as the receptor tyrosine kinase Erb2 can be targeted directly by CRISPR-Cas9 [119].
From another perspective, it is possible to utilize CRISPR-Cas9 to introduce cancer-causing mutations in human cell lines and animal models. In this context, the following cell lines have been constructed to date: lung cancer [120], acute myeloid leukemia [121], liver cancer [122], and pancreatic cancer [123].
Animal models
CRISPR-Cas9 technology can be applied to animal models for the study of both cancers and other inherited diseases. Heritable gene modification can be achieved by injecting CRISPR-Cas9, targeting one or multiple alleles, directly into fertilized zygotes [42]. Among transgenic animal models, mice are the most widely used in experiments because of the relatively short time required to generate mutants; however, non-human primate models have been created successfully by multiplex gene targeting, potentially generating superior systems for the study of complex human diseases, for example, neurodegenerative disorders [124]. Nonetheless, mouse models remain the most cost-effective. Moreover, mice are amenable to large-scale in vivo mutagenesis studies, especially when highly specific targeted editing can avoid the confounding effects of off-target mutagenesis [125].
Synthetic biology
The applications of the CRISPR-Cas9 system to synthetic biology include all concepts related to synthetic gene circuits in living cells. Because synthetic gene circuits consist of sensors, processors, and actuators, synthetic biology has the potential not only to advance basic research, but also to enable practical applications in medicine, biofuel production, and synthesis of commodity chemicals [126]. The most practical applications of the CRISPR-Cas9 system have been in plants, especially crops such as rice, wheat, sorghum, and tobacco. For example, CRISPR-Cas9 was used to target and knock out the mildew-resistance locus (MLO) genes, which encode proteins that repress the defense against powdery mildew disease in hexaploid bread wheat [127].
sgRNA library
CRISPR-Cas9 can also be applied to the systematic analysis of gene functions in human cells. A lentiviral sgRNA library was developed against genes identified by functional screening and high-throughput sequencing analysis. This powerful loss-of-function library screening is expected to facilitate discovery of genes that participate in various biological processes, including drug targeting, toxicity, and expression of certain phenotypes [128].
Induced pluripotent stem cells
iPSCs, which are very similar to embryonic stem cells, are pluripotent cells with a high self-renewal rate that can differentiate into almost all cell types; however, their utilization is associated with significantly less ethical controversy than that of their embryonic counterparts. Recent advances in stem cell technology are likely to provide great benefits to the clinical use of iPSCs in clinical applications [129].
As mentioned above, iPSCs have a major advantage for personalized medicine because they can be derived from the patients themselves, and can therefore avoid immune rejection when transplanted. Ex vivo therapy includes correction of patient-derived iPSCs through gene editing, as well as differentiation into nonrenewable cell types such as neurons and cardiomyocytes (Fig. 5) [100].
Figure 5.
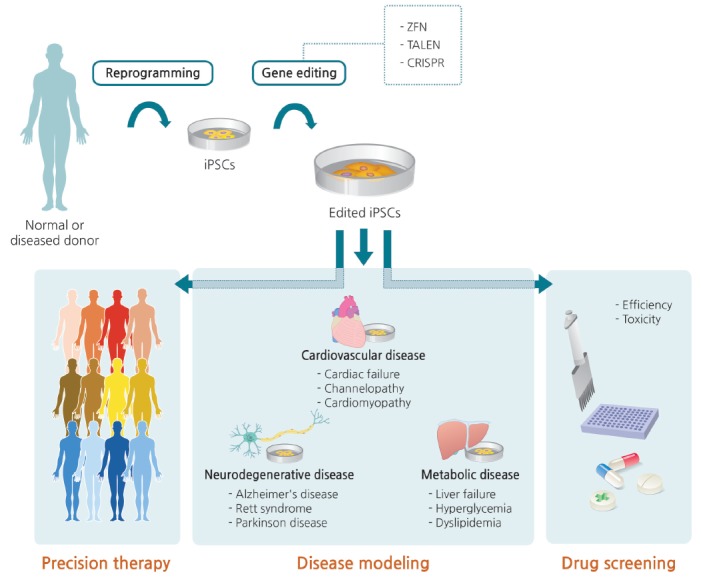
Generation of edited induced pluripotent stem cells (iPSCs) and clinical applications thereof. Somatic cells isolated from a normal person or patient are reprogrammed into iPSCs. Normal sequence can be disrupted or genetic defects can be corrected via gene editing. iPSCs with edited modifications are differentiated into various target cells for disease modeling, which can provide a useful channel for precision therapy and drug screening. ZFN, zinc finger nuclease; TALEN, transcription activator-like effector nuclease; CRISPR, clustered regularly interspaced short palindromic repeats.
In addition to this type of precision therapy, human iPSC lines with genotypes characteristic of specific diseases could be used to understand pathogenic mechanisms. Disease modeling and drug efficiency/toxicity testing with iPSCs not only increase the accuracy of disease simulation, but are also less expensive than generating animal models. However, care must be taken when interpreting the results of phenotypic comparisons between patient iPSC-derived cells and healthy control cells. Specifically, the results are vulnerable to confounding variables that might influence the phenotypes of interest, including epigenetic status and unmatched age, gender, and ethnicity. In this respect, gene editing is the only way to distinguish changes that are specifically relevant to a given disease [130].
The CRISPR-Cas9 system enables simultaneous knockout of multiple genes, as well as knock-in of specific alleles in iPSCs, distinguishing it from earlier gene-editing technologies such as ZFNs and TALENs. An isogenic human iPSC cell line precisely corrected by the CRISPR-Cas9 system was recently constructed, despite the handling difficulties associated with gene editing of human stem cells [131]. In the future, the use of CRISPR-Cas9 with iPSCs will lead to novel combinations of gene and cell therapies [132].
Areas of technical improvement: DSB repair, nucleases, delivery
Prior to the clinical application of CRISPR-Cas9 in human patients, the safety and efficacy of the system must be validated. The specificity and efficiency of genome-editing tools can be improved by targeting DSB repair pathways, modifying nucleases, and changing the mode of delivery. We briefly discuss each topic below.
DSB repair
As noted previously, DNA editing rates are currently determined by the two major endogenous DSB repair pathways. HDR is more suitable for gene correction or gene insertion than NHEJ, which creates indels that generally induce a loss of function. NHEJ is more efficient because it is active throughout the cell cycle and does not require a repair template; by contrast, the rate of HDR is inherently low. In addition, some HDR components are expressed only during the S/G2 phase, limiting the use of HDR-based editing approaches to dividing cells and preventing their use in post-mitotic cells, such as neurons and cardiac myocytes. For this reason, controlling the efficiency of HDR has become a major focus of efforts aimed at increasing the effectiveness of gene correction [46].
One strategy for improving the efficiency of HDR is the suppression of NHEJ during DSB repair. Suppression of a key enzyme in the NHEJ pathway increases the efficiency of HDR-mediated genome editing up to 19-fold [133,134]. Another strategy involves the induction of HDR-like corrections in post-mitotic cells via a novel non-HDR-based pathway: microhomology-mediated end joining [135,136]. Using microhomologous sequences (5 to 25 bp), the so-called PITCH (Precise Integration into Target Chromosome) system can produce precise gene knock-ins. Meanwhile, the lower success rate of CRISPR-Cas9 relative to TALEN could be overcome by the generation of sticky, instead of blunt, ends [137].
Nucleases
From a clinical standpoint, highly specific gene engineering technology is essential because specificity is correlated with safety. Unexpected off-target mutations may cause cells to become carcinogenic or functionally impotent. Because genetic modifications are permanent, multiple ongoing research efforts are devoted to the reduction of off-target effects.
One strategy for achieving this goal is to improve the targeting specificity of Cas9. Careful design of sgRNA, including avoidance of poly-G/poly-C-rich targets, as well as tight control of the amount and duration of Cas9 and sgRNA expression, are both important for high specificity [138]. The use of modified Cas9 with two separate sgRNAs that each generate a single-strand nick on opposing DNA strands can potentially reduce off-target activity by 50- to 1,500-fold in cell lines [139,140]. Additionally, truncation of the guide RNA, with a target-complementary region shorter than 20 nucleotides in length, can decrease off-target activity by 5,000-fold or more [141]. Moreover, a fusion protein of catalytically inactive Cas9 and FokI nuclease (fCas9) can recognize the target DNA site with 140-fold greater specificity than the wild-type protein in human cells [142,143].
Delivery
The delivery of gene-editing tools to target cells is another major challenge with respect to efficacy and specificity. Both viral and nonviral delivery methods are currently being evaluated for the introduction of Cas9 into target cells ex vivo or in vivo. Depending on the mode of delivery and the duration of nuclease expression, both off-target activities and immune reactions are possible.
Viral vectors, such as AAV or lentivirus, represent the most common delivery systems, and these vectors have recently been approved for clinical use. In particular, AAV, which was recently clinically approved, is an attractive candidate for in vivo use because of its low degree of immune stimulation, well-characterized serotypes, and ability to target diverse tissues such as eye, brain liver, and muscle [144]. However, one challenge of using AAV is its relatively small packaging capacity (4.7 kb). Consequently, efforts are underway to deliver the Cas9 and sgRNA coding sequences using two separate AAV vectors. Alternatively, these size constraints can be sidestepped by creating a shorter Cas9 ortholog. In addition to size incompatibility, viral vectors have the drawback of possible constitutive nuclease expression, resulting in cell toxicity and genomic instability [145].
A variety of nonviral methods exist for both in vivo and ex vivo delivery of CRISPR-Cas9 in the form of mRNA or proteins [146]; these include electroporation, hydrodynamic delivery, and the use of liposomes [147]. Because mRNA and proteins introduced by these methods are present in cells only transiently, nonviral delivery systems are expected to decrease the frequency of off-target effects and cell toxicity relative to viral systems.
Ethics
As promising as CRISPR sounds, a variety of concerns have been expressed about this technique. In early 2015, a Chinese research group used CRISPR-Cas9 to perform editing on nonviable human trinuclear zygotes, stimulating a vigorous discussion of the ethical implications. The International Summit on Human Gene Editing in Washington was convened in late 2015, providing a forum in which scientists could achieve a consensus on human germline engineering.
The CRISPR-Cas9 toolbox has many advantages and it can be used to correct many defects that occur systemically or from birth, including cystic fibrosis and Huntington’s disease. However, it remains unclear how we should set boundaries regarding which human traits are appropriate for editing. In addition, few would argue that a number of safety and efficacy concerns about CRISPR-Cas9 remain to be resolved. Especially in light of the possibility that undesirable parties could use this technology for eugenics, it would be irresponsible to allow modification of human embryos. In this respect, we must confront the need for robust regulation, even as arguments rage between the advocates of caution and progress.
CONCLUSIONS
Recent advances in genome editing with CRISPR is very rapid from basic research to clinical therapy. Development of CRISPR may widen the opportunity of iPSC application in real clinic. To deeply understand cutting-edge CRISPR technology can promote gene-editing applications in fields of cancer research, synthetic biology, and gene therapy using induced pluripotent stem cells.
Acknowledgments
This work was supported by a grant from the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Science, ICT and Future Planning (2013R1A1A1076125).
Footnotes
No potential conflict of interest relevant to this article was reported.
REFERENCES
- 1.Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;372:793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lander ES. Cutting the Gordian helix: regulating genomic testing in the era of precision medicine. N Engl J Med. 2015;372:1185–1186. doi: 10.1056/NEJMp1501964. [DOI] [PubMed] [Google Scholar]
- 3.Jameson JL, Longo DL. Precision medicine: personalized, problematic, and promising. N Engl J Med. 2015;372:2229–2234. doi: 10.1056/NEJMsb1503104. [DOI] [PubMed] [Google Scholar]
- 4.Barrangou R, Fremaux C, Deveau H, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 5.Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol. 1987;169:5429–5433. doi: 10.1128/jb.169.12.5429-5433.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151(Pt 8):2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 7.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 8.Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151(Pt 3):653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- 9.Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hale CR, Zhao P, Olson S, et al. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brouns SJ, Jore MM, Lundgren M, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mojica FJ, Diez-Villasenor C, Soria E, Juez G. Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria. Mol Microbiol. 2000;36:244–246. doi: 10.1046/j.1365-2958.2000.01838.x. [DOI] [PubMed] [Google Scholar]
- 16.Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43:1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- 17.Makarova KS, Aravind L, Grishin NV, Rogozin IB, Koonin EV. A DNA repair system specific for thermophilic Archaea and bacteria predicted by genomic context analysis. Nucleic Acids Res. 2002;30:482–496. doi: 10.1093/nar/30.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garneau JE, Dupuis ME, Villion M, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 20.Deltcheva E, Chylinski K, Sharma CM, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sapranauskas R, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39:9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNAprogrammed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hwang WY, Fu Y, Reyon D, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang H, Yang H, Shivalila CS, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bassett AR, Tibbit C, Ponting CP, Liu JL. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep. 2013;4:220–228. doi: 10.1016/j.celrep.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friedland AE, Tzur YB, Esvelt KM, Colaiacovo MP, Church GM, Calarco JA. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods. 2013;10:741–743. doi: 10.1038/nmeth.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li JF, Norville JE, Aach J, et al. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol. 2013;31:688–691. doi: 10.1038/nbt.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakayama T, Fish MB, Fisher M, Oomen-Hajagos J, Thomsen GH, Grainger RM. Simple and efficient CRISPR/Cas9-mediated targeted mutagenesis in Xenopus tropicalis. Genesis. 2013;51:835–843. doi: 10.1002/dvg.22720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jinek M, Jiang F, Taylor DW, et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343:1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishimasu H, Ran FA, Hsu PD, et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shalem O, Sanjana NE, Hartenian E, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niu Y, Shen B, Cui Y, et al. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014;156:836–843. doi: 10.1016/j.cell.2014.01.027. [DOI] [PubMed] [Google Scholar]
- 34.Wagner JC, Platt RJ, Goldfless SJ, Zhang F, Niles JC. Efficient CRISPR-Cas9-mediated genome editing in Plasmodium falciparum. Nat Methods. 2014;11:915–918. doi: 10.1038/nmeth.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang P, Xu Y, Zhang X, et al. CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein Cell. 2015;6:363–372. doi: 10.1007/s13238-015-0153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cleto S, Jensen JV, Wendisch VF, Lu TK. Corynebacterium glutamicum metabolic engineering with CRISPR interference (CRISPRi) ACS Synth Biol. 2016;5:375–385. doi: 10.1021/acssynbio.5b00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wright AV, Nunez JK, Doudna JA. Biology and applications of CRISPR systems: harnessing nature’s toolbox for genome engineering. Cell. 2016;164:29–44. doi: 10.1016/j.cell.2015.12.035. [DOI] [PubMed] [Google Scholar]
- 38.Shmakov S, Abudayyeh OO, Makarova KS, et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol Cell. 2015;60:385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doudna JA, Charpentier E. Genome editing: the new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 40.Makarova KS, Haft DH, Barrangou R, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Makarova KS, Wolf YI, Alkhnbashi OS, et al. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lander ES. The heroes of CRISPR. Cell. 2016;164:18–28. doi: 10.1016/j.cell.2015.12.041. [DOI] [PubMed] [Google Scholar]
- 43.Charpentier E, Richter H, van der Oost J, White MF. Biogenesis pathways of RNA guides in archaeal and bacterial CRISPR-Cas adaptive immunity. FEMS Microbiol Rev. 2015;39:428–441. doi: 10.1093/femsre/fuv023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim H, Kim JS. A guide to genome engineering with programmable nucleases. Nat Rev Genet. 2014;15:321–334. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- 45.Charpentier E, Doudna JA. Biotechnology: rewriting a genome. Nature. 2013;495:50–51. doi: 10.1038/495050a. [DOI] [PubMed] [Google Scholar]
- 46.Cox DB, Platt RJ, Zhang F. Therapeutic genome editing: prospects and challenges. Nat Med. 2015;21:121–131. doi: 10.1038/nm.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Silva G, Poirot L, Galetto R, et al. Meganucleases and other tools for targeted genome engineering: perspectives and challenges for gene therapy. Curr Gene Ther. 2011;11:11–27. doi: 10.2174/156652311794520111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 49.Christian M, Cermak T, Doyle EL, et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maeder ML, Thibodeau-Beganny S, Osiak A, et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 52.Joung JK, Sander JD. TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol. 2013;14:49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prakash V, Moore M, Yanez-Munoz RJ. Current progress in therapeutic gene editing for monogenic diseases. Mol Ther. 2016;24:465–474. doi: 10.1038/mt.2016.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ousterout DG, Perez-Pinera P, Thakore PI, et al. Reading frame correction by targeted genome editing restores dystrophin expression in cells from Duchenne muscular dystrophy patients. Mol Ther. 2013;21:1718–1726. doi: 10.1038/mt.2013.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Benabdallah BF, Duval A, Rousseau J, et al. Targeted gene addition of microdystrophin in mice skeletal muscle via human myoblast transplantation. Mol Ther Nucleic Acids. 2013;2:e68. doi: 10.1038/mtna.2012.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ousterout DG, Kabadi AM, Thakore PI, et al. Correction of dystrophin expression in cells from Duchenne muscular dystrophy patients through genomic excision of exon 51 by zinc finger nucleases. Mol Ther. 2015;23:523–532. doi: 10.1038/mt.2014.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ousterout DG, Kabadi AM, Thakore PI, Majoros WH, Reddy TE, Gersbach CA. Multiplex CRISPR/Cas9-based genome editing for correction of dystrophin mutations that cause Duchenne muscular dystrophy. Nat Commun. 2015;6:6244. doi: 10.1038/ncomms7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Voit RA, Hendel A, Pruett-Miller SM, Porteus MH. Nuclease-mediated gene editing by homologous recombination of the human globin locus. Nucleic Acids Res. 2014;42:1365–1378. doi: 10.1093/nar/gkt947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hu W, Kaminski R, Yang F, et al. RNA-directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proc Natl Acad Sci U S A. 2014;111:11461–11466. doi: 10.1073/pnas.1405186111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin SR, Yang HC, Kuo YT, et al. The CRISPR/Cas9 system facilitates clearance of the intrahepatic HBV templates in vivo. Mol Ther Nucleic Acids. 2014;3:e186. doi: 10.1038/mtna.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kennedy EM, Bassit LC, Mueller H, et al. Suppression of hepatitis B virus DNA accumulation in chronically infected cells using a bacterial CRISPR/Cas RNA-guided DNA endonuclease. Virology. 2015;476:196–205. doi: 10.1016/j.virol.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Seeger C, Sohn JA. Targeting hepatitis B virus with CRISPR/Cas9. Mol Ther Nucleic Acids. 2014;3:e216. doi: 10.1038/mtna.2014.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ramanan V, Shlomai A, Cox DB, et al. CRISPR/Cas9 cleavage of viral DNA efficiently suppresses hepatitis B virus. Sci Rep. 2015;5:10833. doi: 10.1038/srep10833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhen S, Hua L, Liu YH, et al. Harnessing the clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated Cas9 system to disrupt the hepatitis B virus. Gene Ther. 2015;22:404–412. doi: 10.1038/gt.2015.2. [DOI] [PubMed] [Google Scholar]
- 65.Dong C, Qu L, Wang H, Wei L, Dong Y, Xiong S. Targeting hepatitis B virus cccDNA by CRISPR/Cas9 nuclease efficiently inhibits viral replication. Antiviral Res. 2015;118:110–117. doi: 10.1016/j.antiviral.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 66.Liu X, Hao R, Chen S, Guo D, Chen Y. Inhibition of hepatitis B virus by the CRISPR/Cas9 system via targeting the conserved regions of the viral genome. J Gen Virol. 2015;96:2252–2261. doi: 10.1099/vir.0.000159. [DOI] [PubMed] [Google Scholar]
- 67.Zhen S, Hua L, Takahashi Y, Narita S, Liu YH, Li Y. In vitro and in vivo growth suppression of human papillomavirus 16-positive cervical cancer cells by CRISPR/Cas9. Biochem Biophys Res Commun. 2014;450:1422–1426. doi: 10.1016/j.bbrc.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 68.Kennedy EM, Kornepati AV, Goldstein M, et al. Inactivation of the human papillomavirus E6 or E7 gene in cervical carcinoma cells by using a bacterial CRISPR/Cas RNA-guided endonuclease. J Virol. 2014;88:11965–11972. doi: 10.1128/JVI.01879-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu L, Wang X, Zhu D, et al. Disruption of human papillomavirus 16 E6 gene by clustered regularly interspaced short palindromic repeat/Cas system in human cervical cancer cells. Onco Targets Ther. 2015;8:37–44. doi: 10.2147/OTT.S64092. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 70.Hu Z, Yu L, Zhu D, et al. Disruption of HPV16-E7 by CRISPR/Cas system induces apoptosis and growth inhibition in HPV16 positive human cervical cancer cells. Biomed Res Int. 2014;2014:612823. doi: 10.1155/2014/612823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee CM, Flynn R, Hollywood JA, Scallan MF, Harrison PT. Correction of the deltaF508 mutation in the cystic fibrosis transmembrane conductance regulator gene by zinc-finger nuclease homology-directed repair. Biores Open Access. 2012;1:99–108. doi: 10.1089/biores.2012.0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li XB, Chen J, Deng MJ, Wang F, Du ZW, Zhang JW. Zinc finger protein HZF1 promotes K562 cell proliferation by interacting with and inhibiting INCA1. Mol Med Rep. 2011;4:1131–1137. doi: 10.3892/mmr.2011.564. [DOI] [PubMed] [Google Scholar]
- 73.Lombardo A, Genovese P, Beausejour CM, et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- 74.Urnov FD, Miller JC, Lee YL, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 75.Matsubara Y, Chiba T, Kashimada K, et al. Transcription activator-like effector nuclease-mediated transduction of exogenous gene into IL2RG locus. Sci Rep. 2014;4:5043. doi: 10.1038/srep05043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Genovese P, Schiroli G, Escobar G, et al. Targeted genome editing in human repopulating haematopoietic stem cells. Nature. 2014;510:235–240. doi: 10.1038/nature13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Osborn MJ, Starker CG, McElroy AN, et al. TALEN-based gene correction for epidermolysis bullosa. Mol Ther. 2013;21:1151–1159. doi: 10.1038/mt.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nelson CE, Hakim CH, Ousterout DG, et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351:403–407. doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tabebordbar M, Zhu K, Cheng JK, et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351:407–411. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Long C, Amoasii L, Mireault AA, et al. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016;351:400–403. doi: 10.1126/science.aad5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu Y, Zhou H, Fan X, et al. Correction of a genetic disease by CRISPR-Cas9-mediated gene editing in mouse spermatogonial stem cells. Cell Res. 2015;25:67–79. doi: 10.1038/cr.2014.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li H, Haurigot V, Doyon Y, et al. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature. 2011;475:217–221. doi: 10.1038/nature10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Anguela XM, Sharma R, Doyon Y, et al. Robust ZFN-mediated genome editing in adult hemophilic mice. Blood. 2013;122:3283–3287. doi: 10.1182/blood-2013-04-497354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sharma R, Anguela XM, Doyon Y, et al. In vivo genome editing of the albumin locus as a platform for protein replacement therapy. Blood. 2015;126:1777–1784. doi: 10.1182/blood-2014-12-615492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yin H, Xue W, Chen S, et al. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol. 2014;32:551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Long C, McAnally JR, Shelton JM, Mireault AA, Bassel-Duby R, Olson EN. Prevention of muscular dystrophy in mice by CRISPR/Cas9-mediated editing of germline DNA. Science. 2014;345:1184–1188. doi: 10.1126/science.1254445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xu L, Park KH, Zhao L, et al. CRISPR-mediated genome editing restores dystrophin expression and function in mdx mice. Mol Ther. 2016;24:564–569. doi: 10.1038/mt.2015.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ding Q, Strong A, Patel KM, et al. Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing. Circ Res. 2014;115:488–492. doi: 10.1161/CIRCRESAHA.115.304351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li HL, Fujimoto N, Sasakawa N, et al. Precise correction of the dystrophin gene in duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem Cell Reports. 2015;4:143–154. doi: 10.1016/j.stemcr.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Park CY, Kim J, Kweon J, et al. Targeted inversion and reversion of the blood coagulation factor 8 gene in human iPS cells using TALENs. Proc Natl Acad Sci U S A. 2014;111:9253–9258. doi: 10.1073/pnas.1323941111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Park CY, Kim DH, Son JS, et al. Functional correction of large factor VIII gene chromosomal inversions in hemophilia A patient-derived iPSCs using CRISPR-Cas9. Cell Stem Cell. 2015;17:213–220. doi: 10.1016/j.stem.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 92.Hoban MD, Cost GJ, Mendel MC, et al. Correction of the sickle cell disease mutation in human hematopoietic stem/progenitor cells. Blood. 2015;125:2597–2604. doi: 10.1182/blood-2014-12-615948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sebastiano V, Maeder ML, Angstman JF, et al. In situ genetic correction of the sickle cell anemia mutation in human induced pluripotent stem cells using engineered zinc finger nucleases. Stem Cells. 2011;29:1717–1726. doi: 10.1002/stem.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sun N, Zhao H. Seamless correction of the sickle cell disease mutation of the HBB gene in human induced pluripotent stem cells using TALENs. Biotechnol Bioeng. 2014;111:1048–1053. doi: 10.1002/bit.25018. [DOI] [PubMed] [Google Scholar]
- 95.Huang X, Wang Y, Yan W, et al. Production of gene-corrected adult beta globin protein in human erythrocytes differentiated from patient iPSCs after genome editing of the sickle point mutation. Stem Cells. 2015;33:1470–1479. doi: 10.1002/stem.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xie F, Ye L, Chang JC, et al. Seamless gene correction of beta-thalassemia mutations in patient-specific iPSCs using CRISPR/Cas9 and piggyBac. Genome Res. 2014;24:1526–1533. doi: 10.1101/gr.173427.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ma N, Shan Y, Liao B, et al. Factor-induced reprogramming and zinc finger nuclease-aided gene targeting cause different genome instability in beta-thalassemia induced pluripotent stem cells (iPSCs) J Biol Chem. 2015;290:12079–12089. doi: 10.1074/jbc.M114.624999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ye L, Wang J, Beyer AI, et al. Seamless modification of wild-type induced pluripotent stem cells to the natural CCR5Delta32 mutation confers resistance to HIV infection. Proc Natl Acad Sci U S A. 2014;111:9591–9596. doi: 10.1073/pnas.1407473111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wilen CB, Wang J, Tilton JC, et al. Engineering HIV-resistant human CD4+ T cells with CXCR4-specific zinc-finger nucleases. PLoS Pathog. 2011;7:e1002020. doi: 10.1371/journal.ppat.1002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sebastiano V, Zhen HH, Haddad B, et al. Human COL7A1-corrected induced pluripotent stem cells for the treatment of recessive dystrophic epidermolysis bullosa. Sci Transl Med. 2014;6:264ra163. doi: 10.1126/scitranslmed.3009540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Crane AM, Kramer P, Bui JH, et al. Targeted correction and restored function of the CFTR gene in cystic fibrosis induced pluripotent stem cells. Stem Cell Reports. 2015;4:569–577. doi: 10.1016/j.stemcr.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sargent RG, Suzuki S, Gruenert DC. Nuclease-mediated double-strand break (DSB) enhancement of small fragment homologous recombination (SFHR) gene modification in human-induced pluripotent stem cells (hiPSCs) Methods Mol Biol. 2014;1114:279–290. doi: 10.1007/978-1-62703-761-7_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Firth AL, Menon T, Parker GS, et al. Functional gene correction for cystic fibrosis in lung epithelial cells generated from patient iPSCs. Cell Rep. 2015;12:1385–1390. doi: 10.1016/j.celrep.2015.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schwank G, Koo BK, Sasselli V, et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13:653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 105.Suzuki S, Sargent RG, Illek B, et al. TALENs facilitate single-step seamless SDF correction of F508del CFTR in airway epithelial submucosal gland cell-derived CF-iPSCs. Mol Ther Nucleic Acids. 2016;5:e273. doi: 10.1038/mtna.2015.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Vierstra J, Reik A, Chang KH, et al. Functional footprinting of regulatory DNA. Nat Methods. 2015;12:927–930. doi: 10.1038/nmeth.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Canver MC, Smith EC, Sher F, et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527:192–197. doi: 10.1038/nature15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rahman SH, Kuehle J, Reimann C, et al. Rescue of DNAPK signaling and T-cell differentiation by targeted genome editing in a prkdc deficient iPSC disease model. PLoS Genet. 2015;11:e1005239. doi: 10.1371/journal.pgen.1005239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Menon T, Firth AL, Scripture-Adams DD, et al. Lymphoid regeneration from gene-corrected SCID-X1 subject-derived iPSCs. Cell Stem Cell. 2015;16:367–372. doi: 10.1016/j.stem.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Holt N, Wang J, Kim K, et al. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat Biotechnol. 2010;28:839–847. doi: 10.1038/nbt.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li L, Krymskaya L, Wang J, et al. Genomic editing of the HIV-1 coreceptor CCR5 in adult hematopoietic stem and progenitor cells using zinc finger nucleases. Mol Ther. 2013;21:1259–1269. doi: 10.1038/mt.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nathwani AC, Tuddenham EG, Rangarajan S, et al. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N Engl J Med. 2011;365:2357–2365. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nathwani AC, Reiss UM, Tuddenham EG, et al. Longterm safety and efficacy of factor IX gene therapy in hemophilia B. N Engl J Med. 2014;371:1994–2004. doi: 10.1056/NEJMoa1407309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hutter G, Nowak D, Mossner M, et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med. 2009;360:692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- 115.Tebas P, Stein D, Tang WW, et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med. 2014;370:901–910. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Badia R, Pauls E, Riveira-Munoz E, Clotet B, Este JA, Ballana E. Zinc finger endonuclease targeting PSIP1 inhibits HIV-1 integration. Antimicrob Agents Chemother. 2014;58:4318–4327. doi: 10.1128/AAC.02690-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fadel HJ, Morrison JH, Saenz DT, et al. TALEN knockout of the PSIP1 gene in human cells: analyses of HIV-1 replication and allosteric integrase inhibitor mechanism. J Virol. 2014;88:9704–9717. doi: 10.1128/JVI.01397-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.White MK, Khalili K. CRISPR/Cas9 and cancer targets: future possibilities and present challenges. Oncotarget. 2016;7:12305–12317. doi: 10.18632/oncotarget.7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Brown MT, Cooper JA. Regulation, substrates and functions of src. Biochim Biophys Acta. 1996;1287:121–149. doi: 10.1016/0304-419x(96)00003-0. [DOI] [PubMed] [Google Scholar]
- 120.Choi PS, Meyerson M. Targeted genomic rearrangements using CRISPR/Cas technology. Nat Commun. 2014;5:3728. doi: 10.1038/ncomms4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chen C, Liu Y, Rappaport AR, et al. MLL3 is a haploinsufficient 7q tumor suppressor in acute myeloid leukemia. Cancer Cell. 2014;25:652–665. doi: 10.1016/j.ccr.2014.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Xue W, Chen S, Yin H, et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature. 2014;514:380–384. doi: 10.1038/nature13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chiou SH, Winters IP, Wang J, et al. Pancreatic cancer modeling using retrograde viral vector delivery and in vivo CRISPR/Cas9-mediated somatic genome editing. Genes Dev. 2015;29:1576–1585. doi: 10.1101/gad.264861.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tu Z, Yang W, Yan S, Guo X, Li XJ. CRISPR/Cas9: a powerful genetic engineering tool for establishing large animal models of neurodegenerative diseases. Mol Neurodegener. 2015;10:35. doi: 10.1186/s13024-015-0031-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Jusiak B, Cleto S, Perez-Pinera P, Lu TK. Engineering synthetic gene circuits in living cells with CRISPR technology. Trends Biotechnol. 2016;34:535–547. doi: 10.1016/j.tibtech.2015.12.014. [DOI] [PubMed] [Google Scholar]
- 127.Wang Y, Cheng X, Shan Q, et al. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol. 2014;32:947–951. doi: 10.1038/nbt.2969. [DOI] [PubMed] [Google Scholar]
- 128.Zhou Y, Zhu S, Cai C, et al. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014;509:487–491. doi: 10.1038/nature13166. [DOI] [PubMed] [Google Scholar]
- 129.Diecke S, Jung SM, Lee J, Ju JH. Recent technological updates and clinical applications of induced pluripotent stem cells. Korean J Intern Med. 2014;29:547–557. doi: 10.3904/kjim.2014.29.5.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Musunuru K. Genome editing of human pluripotent stem cells to generate human cellular disease models. Dis Model Mech. 2013;6:896–904. doi: 10.1242/dmm.012054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Grobarczyk B, Franco B, Hanon K, Malgrange B. Generation of isogenic human iPS cell line precisely corrected by genome editing using the CRISPR/Cas9 system. Stem Cell Rev. 2015;11:774–787. doi: 10.1007/s12015-015-9600-1. [DOI] [PubMed] [Google Scholar]
- 132.Orqueda AJ, Gimenez CA, Pereyra-Bonnet F. iPSCs: a minireview from bench to bed, including organoids and the CRISPR system. Stem Cells Int. 2016;2016:5934782. doi: 10.1155/2016/5934782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol. 2015;33:538–542. doi: 10.1038/nbt.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chu VT, Weber T, Wefers B, et al. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol. 2015;33:543–548. doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- 135.Nakade S, Tsubota T, Sakane Y, et al. Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nat Commun. 2014;5:5560. doi: 10.1038/ncomms6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sakuma T, Nakade S, Sakane Y, Suzuki KT, Yamamoto T. MMEJ-assisted gene knock-in using TALENs and CRISPR-Cas9 with the PITCh systems. Nat Protoc. 2016;11:118–133. doi: 10.1038/nprot.2015.140. [DOI] [PubMed] [Google Scholar]
- 137.Ledford H. Bacteria yield new gene cutter. Nature. 2015;526:17. doi: 10.1038/nature.2015.18432. [DOI] [PubMed] [Google Scholar]
- 138.Hsu PD, Scott DA, Weinstein JA, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Mali P, Aach J, Stranges PB, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ran FA, Hsu PD, Lin CY, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol. 2014;32:577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tsai SQ, Wyvekens N, Khayter C, et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32:569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hastie E, Samulski RJ. Adeno-associated virus at 50: a golden anniversary of discovery, research, and gene therapy success: a personal perspective. Hum Gene Ther. 2015;26:257–265. doi: 10.1089/hum.2015.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Moore R, Spinhirne A, Lai MJ, et al. CRISPR-based self-cleaving mechanism for controllable gene delivery in human cells. Nucleic Acids Res. 2015;43:1297–1303. doi: 10.1093/nar/gku1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kormann MS, Hasenpusch G, Aneja MK, et al. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat Biotechnol. 2011;29:154–157. doi: 10.1038/nbt.1733. [DOI] [PubMed] [Google Scholar]
- 147.Zuris JA, Thompson DB, Shu Y, et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat Biotechnol. 2015;33:73–80. doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]