Abstract
The profound metabolic reprogramming that occurs in cancer cells has been investigated primarily in two-dimensional cell cultures, which fail to recapitulate spatial aspects of cell-to-cell interactions as well as tissue gradients present in three-dimensional (3D) tumours. Here, we describe an engineered model to assemble 3D tumours by rolling a scaffold-tumour composite strip. By unrolling the strip, the model can be rapidly disassembled for snap-shot analysis, allowing spatial mapping of cell metabolism in concert with cell phenotype. We also show that the establishment of oxygen gradients within samples are shaped by oxygen-dependent signalling pathways, as well as the consequential variations in cell growth, response to hypoxic gradients extending from normoxia to severe hypoxia, and therapy responsiveness, are consistent with tumours in vivo. Moreover, by using liquid chromatography tandem mass spectrometry, we mapped cellular metabolism and identified spatially defined metabolic signatures of cancer cells to reveal both known and novel metabolic responses to hypoxia.
Keywords: Tumour heterogeneity, microenvironment, metabolomics, hypoxia inducible factor, hypoxia, cancer, tissue mimetic platform, tissue engineering
In order to sustain the metabolic needs associated with increased growth, cancer cells undergo profound metabolic changes, which are regulated both genetically and by cues from the local microenvironment1. The tumour microenvironment is heterogeneous and dynamic. A balance of molecular transport and cellular consumption/secretion establish cell-dependent spatial and temporal gradients of small molecules, metabolites, and other proteins. Oxygen is a key small molecule in the tumour microenvironment and regions of low oxygen (hypoxia) are a feature of most solid tumours. Regions of hypoxia often develop due to insufficient or dysfunctional vasculature2,3 and are associated with greater therapy resistance and acquisition of phenotypes that drive poor clinical prognosis4. In order to survive in these hypoxic regions, cancer cells adapt their metabolic behaviour, including their use of oxygen, to ensure continued survival and growth despite limited oxygen availability. Consequently these spatial variations in cellular oxygen uptake rate lead to complex dynamic gradients in oxygen and other metabolite levels within the tumour, which are dependent on both oxygen supply and cellular consumption. Response to hypoxia is regulated by several signalling pathways, including the hypoxia inducible factors (HIFs), which increase glucose uptake, glycolysis and lactate production, and concomitantly decrease mitochondrial oxygen consumption5. This reduction in oxygen consumption is hypothesized to allow increased delivery of oxygen to cells that are located further away from blood vessels, and thus prevent more severe hypoxia that would otherwise be toxic. More severe hypoxia causes defects in protein folding in the ER, resulting in activation of the unfolded protein response (UPR)6,7 to promote cell survival8.
While spatial dynamics in oxygen availability are expected to have a major influence on tumour cell metabolism1,9, assessing the role of hypoxia on metabolic response within a solid tumour presents an experimental challenge: metabolite turnover is rapid compared to changes in mRNA and proteins and consequently, profiling intracellular metabolites requires rapid quenching of metabolic activity prior to analysis. Studies on metabolic adaptations in tumour cells are therefore currently limited to homogeneous 2D or suspension cultures that facilitate rapid collection of metabolite samples for analysis. Alternative experimental models that better recapitulate the heterogeneous and dynamic cell-defined 3D microenvironment of a tumour such as in vitro spheroid cultures10, tissue engineered tumours11,12, and modular tissues13,14 are available, but none of these enables the rapid collection of cells from defined tumour locations to spatially correlate different tumour microenvironments to real-time snap-shot molecular or metabolic signatures.
Here we describe an engineered-tumour that is assembled by rolling a single component biocomposite sheet and that can be rapidly unrolled and disassembled to allow isolation of cells from specific tumour locations. Cell defined oxygen and metabolite gradients are established within our tumour model and lead to spatially differential cellular responses that accurately mimic those observed in tumours in vivo. Using our system we perform spatial mapping of cellular metabolism in 3D and identify spatial metabolic signatures that contain both HIF-dependent and HIF-independent components. We find that in the absence of HIF, oxygen gradients are significantly perturbed and cell metabolism is deregulated, highlighting the complex impact of metabolic adaptation to low oxygen and the resulting changes in global metabolite gradients within the tumour microenvironment.
Rollable biocomposite design
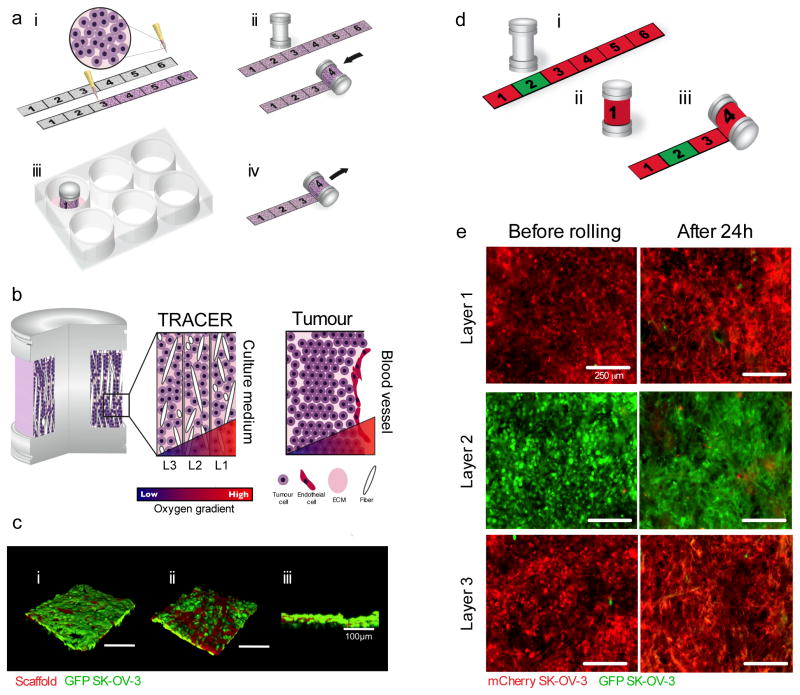
We assemble our engineered Tumour Roll for Analysis of Cellular Environment and Response (TRACER) in three steps (Figure 1a) i) generating a dense tumour biocomposite by infiltrating a suspension of tumour cells (1×108 cells/ml) in type I collagen into a thin (34.19 μm±4.38 μm, Supplementary Figure 1) porous cellulose scaffold strip where the cells are not nutrient limited (remain viable (Supplementary Figure 2)), and culturing for 24h to allow remodelling and compaction by the encapsulated cells (Figure 1Ai, Supplementary Figure 3), (ii) rolling the biocomposite on an oxygen impermeable 6 mm diameter metallic core to generate a layered configuration (Figure 1aiii) that mimics the length scales (100–200 μm) over which oxygen gradients occur in vivo15 and iii) submerging the TRACER in culture medium (Figure 1aiii). In the TRACER, oxygen and nutrients from the medium are only available to cells by diffusion from the culture medium surrounding the TRACER and therefore cellular consumption of oxygen and nutrients was predicted to generate gradients with progressively lower levels in each layer, mimicking the gradients seen in tumours at progressively further distances from a blood vessel (Figure 1b). Rapid disassembly (<1s) of the TRACER after the desired culture time is achieved by unrolling the biocomposite back into the thin strip (Fig 1aiv) and snap-freezing or fixing cells for snapshot analysis. Alternatively, live cells can be retrieved from different regions of the thin biocomposite strip for secondary assays involving live cells. A 3.2mg/mL collagen gel was selected as the encapsulation matrix because it is abundant in tumours and cultures at this concentration have been shown previously to produce cell behaviours that recapitulate in vivo tumour phenotypes16. Cellulose was selected as the scaffold material on the basis that it does not affect cell survival (Supplementary Figure 2) and that its interconnected porous structure produces mechanical interlocking between the cell matrix phase and the scaffold fibres critical for maintaining the integrity of the composite (Figure 1c and Supplementary Figures 1 and 4). Importantly, this ensures the unrolling procedure does not cause significant damage of the layers (Figure 1d and e, Supplementary Figure 5), even after multiple days of rolled culture (Supplementary Figure 5), allowing accurate isolation of cells from specific locations within the engineered tumours. Furthermore, cell morphology in the biocomposite was indistinguishable from cells in collagen-only gels: cells remained encapsulated and formed junctions with neighbouring cells within the collagen phase and did not grow along the scaffold fibres (Supplementary Figures 1, 6 and 7) suggesting the scaffold fibres have minimal negative impact on cell organization and primarily provide structural integrity to the layer to enable unrolling. The 3D spatial location of the cells in the TRACER is then easily mapped to their position along the unrolled strip to facilitate collection of populations of cells from different layers and hence locations within the 3D tumour. Importantly, at the cell densities and over the culture time frames used here, no significant migration occurred between adjacent layers over 72h (Supplementary Figure 8). The TRACER therefore provides an excellent platform to investigate variations in cancer cell behaviour in three dimensions and in a quasi-physiological environment.
Figure 1. Schematic of TRACER design concept.
a- Concept of the TRACER: (i) cells are infiltrated into scaffold to form a biocomposite strip, (ii) the biocomposite strip is rolled on to an oxygen impermeable core, (iii) the TRACER is cultured for desired time, (iv) the TRACER is unrolled for analysis and location of the cells in 3D determined by their position along the strip. b - Geometry comparison of TRACER and tumours. Oxygen and nutrient consumption through the layers of the TRACER is predicted to generate gradients mimicking those observed in tumours in vivo at progressively further distances from a blood vessel. c - Confocal 3D reconstruction of GFP-SK-OV-3 cells in a biocomposite layer before rolling imaged from both scaffold sides (i-ii) and associated side profile (iii). Fibers, visualized through auto-fluorescence, shown in red. The side of the scaffold in contact with the mold surface (ci) during fabrication exhibits a smoother profile than the side of the scaffold not in contact with the mold (cii) Scale bars are 100μm. d -Biocomposite strips containing GFP and mCherry labelled cells were generated (i) and cultured in a rolled configuration (ii) for 24h and then unrolled for analysis (iii). e- The number of green cells transferred to neighbouring red layers during a 24h culturing period and after unrolling was assessed using fluorescence microscopy. Minimal cell transfer was observed. Both sides of the strip were similar. Scale bars are 250μm.
Cellular behaviour is consistent with tumours in vivo
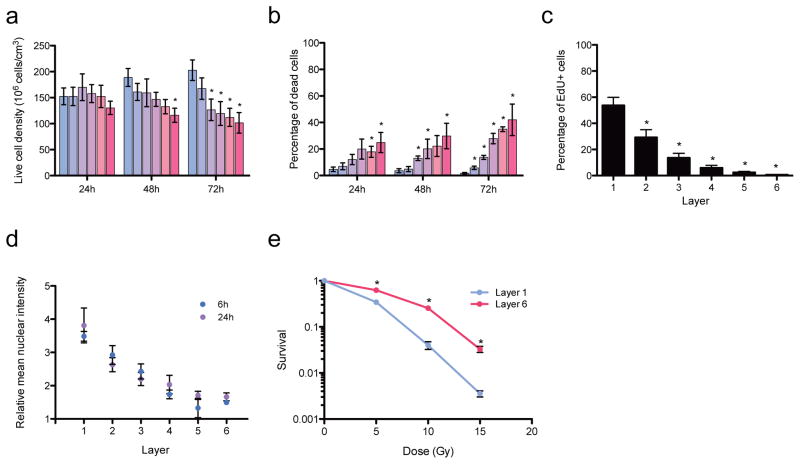
We generated 6-layer thick (~200 microns total thickness) TRACERs using the human ovarian cancer cell line SK-OV-3. Cells in the TRACER system preserved known spatial features observed in in vivo tumours. For instance, after 72h the number of viable cells was lower in the inner hypoxic and nutrient limited layers, similar to observations in tumours in vivo (Figure 2a,)3,11, and significantly increased cell death was observed in the deeper layers (4–6) as time progressed (Figure 2b, Supplementary Figure 9). Cell proliferation was also significantly reduced in the inner layers (Figure 2c), likely in response to the limited oxygen or nutrient availability, as is observed in tumours in vivo 3,11. Importantly, variations in cellular response to therapy in different regions of the TRACER mimicked those of in vivo tumours. Treatment of the TRACER with doxorubicin showed decreasing drug concentration levels from layers 1 to 6 as expected, due to known penetration limitations of this drug within tumours17 (Figure 2d). In these experiments, doxorubicin concentration begins to plateau between layers 3 and 4 (~100–150 microns deep) similar to doxorubicin penetration length scales observed in mouse xenografts18. Exposure of the TRACER to radiotherapy mimicked therapy resistance in the deeper layers, characteristic of hypoxic tumours in vivo19 with significantly less cell death occurring in response to the radiation dose in deeper layers as assessed by increased clonogenic potential (Figure 2e, Supplementary Figure 10). Together these data demonstrate that cells in the TRACER system display relevant behaviours consistent with in vivo tumours.
Figure 2. Cellular behaviour within the TRACER.
a - Number of live cells in each TRACER layer after 1, 2 or 3 days. At day 2, L6 is significantly different from L1 (p=0.011). At day 3, layers 3, 4, 5, and 6 are significantly different from L1 (p<0.02). Error bars are SEM with n = 3 TRACERs. b -percentage of dead cells in each TRACER layer after 1, 2 or 3 days. At day 1, layers 5 and 6 are significantly different than L1 (p<0.01). At day 2, Layers 3, 4 and 6 are significantly different than L1 (p<0.026). At day 3, all layers are significantly different than L1 (p<0.025). c- Percentage of proliferating cells in each TRACER layer over 3 days. All layers are statistically different from layer 1 (p<0.05). Graphs a and b were produced from quantitative analysis of confocal images stained for nuclei (DRAQ5), dead cells (ethidium homodimer-1) (Supplementary Figure 9) while graph c was produced counting EdU labelled cells using flow cytometry. Error bars for graphs ac are SEM with n = 3 TRACERs. d - Relative nuclear doxorubicin binding in each TRACER layer after a 30 min doxorubicin treatment following 6 or 24h of assembled culture. Data normalized to non-treated TRACERs. Error bars are SEM with n = 3 TRACERs at 6h and n = 4 TRACERs at 24h. e – Clonogenic survival of cells from layer 1 and layer 6 of TRACERs dosed with 5, 10 or 15Gy X-ray radiation. Data normalized to L1 or L6 of a non-treated TRACER. Single layer control data from cultures in set hypoxia levels in Supplementary Figure 10. Error bars are SEM with n = 9 measurements from 2 TRACERs. * indicates significant with p≤0.05.
Oxygen gradient dynamics reveal HIF-dependent adaptations
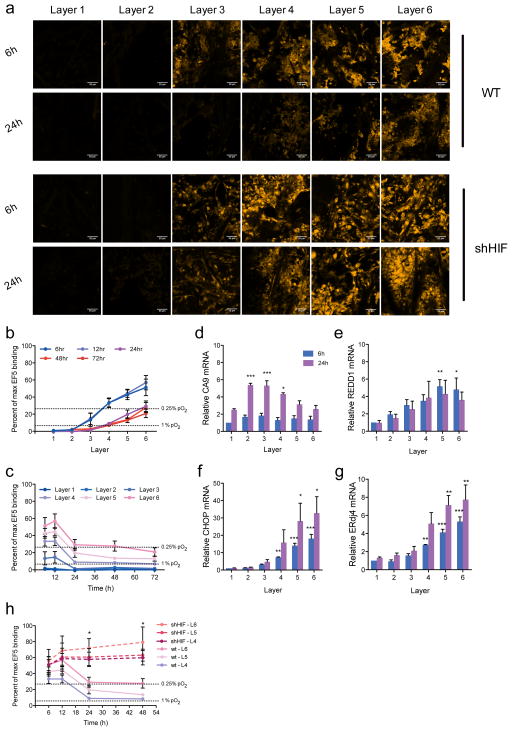
We evaluated the presence of oxygen gradients within the TRACER by quantifying levels of EF5 binding, which forms adducts in viable cells following enzymatic reduction in an oxygen dependent manner20,21. Hypoxic regions were not observed within single biocomposite layers in non-rolled constructs despite the high cell density because of the thin dimensions of the scaffold (34.19um ±4.38 μm, Supplementary Figure 1 and 3) and cells within unrolled strips remained viable and proliferative over at least 3 days (Supplementary Figure 2). A cellular-consumption-generated oxygen gradient developed rapidly however, when strips were rolled into TRACERs. Gradients in EF5 binding (a measure of oxygen concentration) were observed from the outer to the inner layers of the TRACER by 6h (Figure 3a–b, Supplementary Figure 11). As expected, since oxygen gradient generation occurs due to cellular consumption, gradients were dependent on cell type (Figure 3b versus Supplementary Figure 11) cell density (Supplementary Figure 12) and were homogeneous between the edge and the centre regions of each TRACER layer (Supplementary Figure 13). EF5 binding indicated that the deep layers (layers 4–6) were severely hypoxic (O2 <0.1%) at 6 and 12h. Interestingly our system revealed a progressive decrease in the level of hypoxia in these deep layers over time, with evidence of increased oxygenation in all layers between 12 and 24h, after which oxygen levels remained stable between 24 and 48h (Figure 3b – c). This is consistent with the known adaptive hypoxia response mediated by HIF, which decreases oxygen consumption as described above5.
Figure 3. Oxygen gradients within the TRACER over time.
a- EF5 (yellow) staining in layers 1–6 of the TRACER after 6h, and 24h in WT and shHIF cells. All scale bars 100 microns. Gradients in EF5 staining (and hence oxygen levels) are clearly visible at 6 and 24h. b – Percentage of maximum EF5 binding (inversely correlated with oxygen levels) in WT cells quantified from nuclear EF5 fluorescence in each layer between 6 and 72h in culture. c - Percentage of maximum EF5 binding in WT cells as a function of time. Error bars in b-c are SEM (n=3). Dashed lines indicate conversions to hypoxia levels based on previously reported correlations30. d–g -Quantitative reverse transcriptase–PCR analysis of HIF target genes CA9 and REDD1 and the UPR response genes CHOP and ErDj4 relative to housekeeping gene RPL13a in each layer (L1 (outer) to L6 (inner)) after 6h (blue bar) and 24h (magenta bars). Data normalized to 6h, layer 1. Error bars are standard deviation (n=3 TRACERs). P-values obtained with one-way ANOVA, Dunnett post-test. *P<0.05, **P<0.01,***P<0.001. Single layer controls provided in Supplementary Figure 14. h - Percentage of maximum EF5 binding as a function of time for TRACERs containing WT (solid lines) or shHIF (dashed lines) cells. Error bars are SEM (n=3).
To explore the hypoxia response in our system, we investigated the expression of well-established HIF and UPR target genes in the different layers and at different time points. Cells in the TRACER demonstrated a robust HIF response in a time and layer-specific fashion: the HIF target gene CA9 was induced in mild hypoxia (middle layers 2–4) after 24h of rolled culture, whilst the HIF target gene REDD1 and the unfolded protein response (UPR) target genes (CHOP, ERDJ4) were induced by more severe hypoxia (layers 4–6) (Figure 3d–g, and Supplementary Figure 14) after both 6 (blue bars) and 24h (magenta bars). Immunostaining for HIF1α also revealed increased nuclear intensities in the inner hypoxic layers (Supplementary Figure 15). We tested whether the adaptation and subsequent changes in oxygen levels observed at 12h and beyond in the TRACER were HIF dependent by generating TRACERs from isogenic cells stably expressing a short-hairpin RNA (shRNA) against HIF1α (shHIF) (Supplementary Figure 16) and quantified oxygen gradient dynamics. As seen in figure 3h, in the absence of HIF no cellular adaptation to hypoxia occurred at 12h and significantly higher levels of hypoxia were present in the deeper layers (L4-6) after 12h (Figure 3a). Therefore, TRACER does not only allow the establishment of cellular-consumption-defined oxygen gradients from normoxia to severe hypoxia as observed in tumours, but also provides the capability to easily monitor the dynamic response to these gradients over time.
Metabolomic signature mapping in 3D
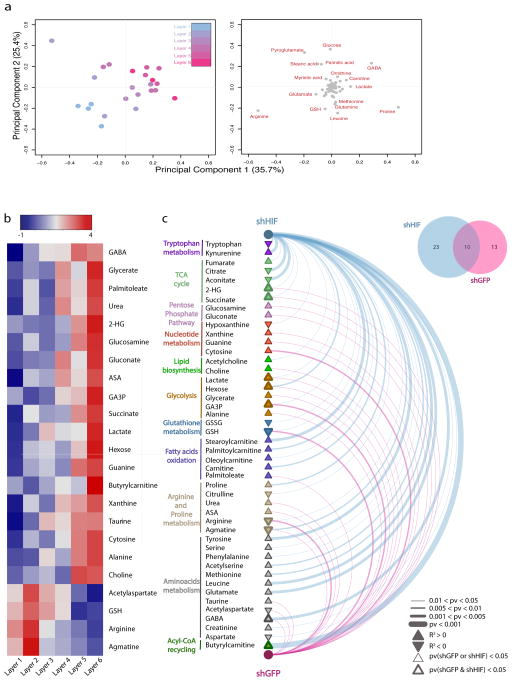
We exploited the ability to rapidly disassemble the TRACER to better understand spatial variations in cellular metabolism in the presence of graded oxygen profiles. Specifically we performed a layer-specific (LS) Liquid Chromatography –Mass Spectrometry (LCMS)-based metabolomic analysis of 88 metabolites in each layer. Metabolite intensities were subjected to unsupervised hierarchical clustering, which revealed a clear separation of samples across the TRACER layers (Supplementary Figure 17). Furthermore, an unsupervised principal component analysis (PCA) showed that samples from specific TRACER layers sequentially distributed across the PCA score plot in the expected order (Figure 4a). Together these results indicate that the different environmental conditions experienced by cells in each TRACER layer induce distinct, layer-specific metabolic signatures.
Figure 4. Layer-specific metabolomic analysis and correlation with hypoxia in the TRACER.
a - Principal component analysis (PCA) of the metabolomic signature of cells in the indicated TRACER layers. Score plot (left) and loading plot (right) of principal components 1 and 2 are indicated. Samples in the score plot are color-coded according to the layer of origin. Metabolites that covariate with samples clustering are highlighted in red in the loading plot. b -Heatmap representation of metabolite intensities per layer for metabolites that show a significant correlation (Pearson pv < 0.05) with hypoxia levels. Hypoxia levels reported in Figure 3 were linearly transformed by applying a probit transformation (see methods) and Pearson correlation analysis was performed to test for non-zero correlations between the transformed hypoxia levels and metabolite intensities in each layer. c - Arc plot of metabolites showing significant correlation with hypoxia across the TRACER containing shGFP cells (violet) and shHIF cells (blue). Line thickness is indicative of p-value significance. Metabolite nodes are represented as filled triangles, where pathway are colour coded and the border thickness indicates whether each metabolite was found significant in shGFP or shHIF alone (thin border) or both in shGFP and shHIF (thick border). The Venn diagram in the top right corner indicates the overlap proportion of significant metabolites in shGFP and shHIF. Measurements are generated from n = 4 TRACERs.
We identified metabolites whose intensity across the TRACER layers correlated significantly with levels of hypoxia quantified using EF5 binding by calculating the linear regression coefficient of normalised metabolite intensity versus level of EF5 binding (a surrogate of hypoxia) for each metabolite. Of the 88 metabolites, 23 showed a statistically significant correlation (Pearson p-value ≤ 0.05) with EF5 levels in shGFP cells (Figure 4b) including metabolites associated with glucose metabolism (lactate, hexose, glycerate, GA3P, and alanine) and mitochondrial function (succinate) consistent with a glycolytic switch under hypoxia22. We also observed changes in several urea cycle metabolites, including a decrease in arginine and an accumulation of argininosuccinate (ASA) in the hypoxic layers, consistent with known hypoxic inhibition of the urea cycle enzyme Argininosuccinate Lyase23. Furthermore, 2-hydroxyglutarate, a metabolite recognized for its oncogenic properties24, accumulated in the innermost layers of the TRACER, in line with recent reports25. Finally, decreased levels of reduced glutathione (GSH) were observed in inner layers, consistent with increased oxidative stress under hypoxia26. Together, these results suggest that the TRACER appears to faithfully recapitulate known metabolic responses of cancer cells to hypoxic microenvironments.
HIF-dependent and HIF-independent metabolic signatures
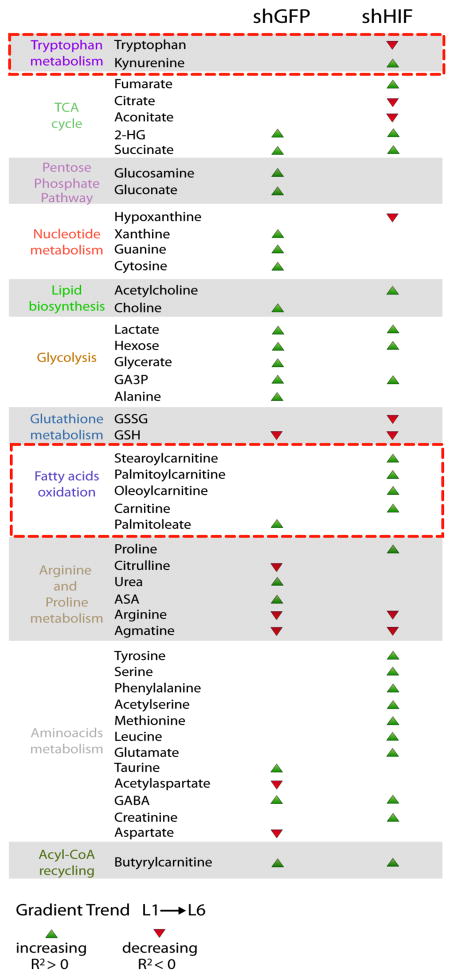
Since HIF has a key role in the metabolic adaptation to hypoxia and we have shown that its ablation disrupts the oxygen gradients in the TRACER (Figure 3h), we performed LS-metabolomic analyses on TRACERs derived from both control (shGFP) and knockdown (shHIF) cells and assessed metabolic adaptation to varying oxygen levels by performing correlation analysis accounting for the different distributions of oxygen in the shGFP and shHIF TRACERs. This analysis allowed us to disentangle the effects of hypoxia per se and HIF activation on cellular metabolism in a 3D environment containing cell-generated metabolite gradients. Metabolites were differentiated based on those that form spatial gradients in a HIF-dependent manner (occurring only in shGFP OR shHIF TRACERs), versus those that form spatial gradients that are HIF-independent (occurring in both shGFP AND shHIF) (Figure 4c, Supplementary Figure 18 and Table 1). We also quantified fold changes in metabolite levels in layer 1 between shGFP and shHIF cells to account for potential effects of HIF in oxygenated conditions (Supplementary Table 1). Among the HIF-independent metabolites that followed oxygen gradients we identified the glycolytic metabolites lactate, GA3P, and hexose; and the TCA cycle metabolite succinate. On the other hand, several metabolites showed significant trends only in the absence of HIF, which may indicate a role for HIF in their metabolism. Among these, the TCA cycle metabolites aconitate and citrate significantly decreased in the hypoxic inner layers only in shHIF cells. Furthermore, several long chain fatty acyl-carnitines, including stearoylcarnitine and palmitoylcarnitine, showed marked increase in deeper layers of shHIF cells. We also observed a significant depletion of tryptophan and concomitant accumulation of kynurenine in hypoxic layers of shHIF cells and depletion of hypoxanthine and accumulation of xanthine in the hypoxic layers of shHIF cells. Together these data demonstrate that the TRACER can be used to dissect the spatial environmental metabolic reprogramming that occurs during cellular adaptations to hypoxia gradients in 3D.
Table 1. Metabolites with significant correlations with level of EF5 binding for shGFP and shHIF cells.
Green arrows indicate a positive correlation with EF5 binding (i.e. a gradient of increasing metabolite concentration from normoxia to hypoxia) and red arrows indicate a significant negative correlation with EF5 binding (ie a gradient of decreasing metabolite concentration from normoxia to hypoxia). When no arrow is present that metabolite did not show a significant correlation with EF5 binding in those samples. The red boxes highlight potentially novel pathways regulated by HIF identified from our analysis.
Outlook
Understanding links between metabolism and cancer biology are limited by the lack of appropriate tools to recapitulate tumour microenvironment in vitro, while simultaneously enabling acquisition of spatial microenvironmental and cellular response data. Here, we have developed a unique engineered tumour model that preserves complex cell-to-cell interactions and microenvironmental variation, which cannot be achieved in experimental systems where microenvironmental gradients are studied in separated conditions, and that enables rapid isolation of cells from different locations within the 3D microenvironment on-demand for downstream functional analyses. While our approach here does not account for cell migration through layers (and hence microenvironments) during culture, this feature could be introduced if needed through cell labelling procedures. However, at the cell densities used here no significant migration occurred between adjacent layers over 72h. For applications where cell motility or invasion is substantial and not desired, barrier layers to prevent cell migration can also be incorporated. Furthermore, for the use of TRACER to study tumour cell migration and invasion likely requires longer culture periods and potentially in inclusion of supporting tumour cell types, such as cancer associated fibroblasts, to induce invasive phenotypes in the tumour cells.
Key features of our design include i) a one component piece construction strategy that enables easy and rapid tumour disassembly and easy registration between the thin strip and thick 3D construct configurations, ii) the use of a fibrous scaffold material that interlocks the fragile gel-cell phase: this provides structural support, which unlike other materials we tested (Supplementary Figure 4) ensures robust separation of the construct layers and definition of a dominant fracture plane during disassembly and hence no transfer of the cellular phase between layers, and iii) sub-100 micron scaffold thicknesses to ensure single layers are not nutrient limited, even at the high cell densities used here that mimic dense solid tumours. It is therefore possible to mature the unrolled strip (for example to allow junction formation) for several days before beginning rolled culture if desired.
Our design enables the establishment of heterogeneous, cell-derived (by consumption and secretion) microenvironments and provides the opportunity to tune these microenvironments: combinations of cell types, ECM composition and concentrations (hence mechanical properties), and potentially exogenous growth factors can be incorporated and patterned within the scaffold strips to generate controlled heterogeneity in the assembled 3D structure designed to mimic specific aspects of tumour organization in vivo. Furthermore, our design provides flexibility in the amount of cellular material that can be retrieved for analysis from each layer: by simply changing the geometry of the bioreactor core the area (and hence number of cells, Supplementary Table 2) associated with any specific layer can be matched to the requirements of a particular end-point assay. By enabling both an accurate design of the structure and microenvironment of a solid tumour in vitro, in combination with the capability to rapidly isolate cell populations from specific microenvironments, the TRACER provides an invaluable and accessible platform for simultaneous snap-shot characterization of cellular behaviour and local microenvironment.
Here we showcase the potential of the TRACER by characterising spatial metabolic adaptations of tumour cells to hypoxia in a quasi-physiological context. Our metabolomic analysis revealed that cells in TRACER maintain distinct metabolic signatures (Figure 4) and that the metabolic signature of the innermost layers recapitulates key features of hypoxic cells, such as increased glycolysis, deregulation of TCA cycle, and decreased fatty acid oxidation (Figure 4b). Indeed, besides the accumulation of lactate, a hallmark of aerobic glycolysis, we have evidence of a complex regulation of mitochondrial metabolism by oxygen. For instance, the TCA cycle metabolite succinate accumulated in oxygen-limiting conditions, in line with our recent finding of accumulation of succinate during ischaemia27. Furthermore, we were also able to determine the global impact of HIF, a master hypoxia regulator on these spatial metabolic adaptations to hypoxia (Figure 4c). Of note, the metabolic changes in glycolysis and mitochondrial metabolism appear to be by and large HIF-independent, suggesting that the underpinning metabolic pathways might be under biochemical, rather than genetic, control. Our analysis enabled the identification of potentially novel pathways regulated by HIF. For instance, the depletion of tryptophan coupled to accumulation of kynurenine in hypoxic layers within TRACERs containing shHIF cells may suggest a HIF-dependent regulation of the tryptophan catabolizing dioxygenase Indoleamine Dioxygenase IDO1. Although a decrease of IDO under hypoxia has already been demonstrated28, our results suggest that HIF impacts the level or activity of this enzyme, which could be important for creating immunosuppressive environments 29. Intriguingly, we also observed the depletion of hypoxanthine and an accumulation of xanthine in the hypoxic layers of shHIF cells, consistent with an increased activity of xanthine oxidase (XO) in the absence of HIF. These results suggest that HIF could potentially control oxygen availability not only via keeping in check mitochondrial function but also by regulating the activity of oxygen dependent enzymes, such as XO and IDO. Our data therefore highlight both known metabolic hypoxia-reprogramming responses and identify novel signatures for future investigations.
Although our focus was on hypoxia, our dataset could also be correlated with other small molecule gradients such as lactate and glucose to assess metabolic adaptation of tumour cells to other key molecules that impact cell metabolism in the context of cancer. Beyond studies of metabolic reprogramming, we envision the TRACER will facilitate novel types of mechanistic studies in a wide range of applications such as drug screenings to understand cancer cell liability to new anti-hypoxic drugs and identification of effective combination therapies.
Materials and methods
Cell culture
Experiments were conducting using the ovarian adenocarcinoma cell line SK-OV-3 (ATCC, Manassa, USA) and the pancreatic cell line KP4 (JCRB Cellbank). SK-OV-3 cells were maintained in McCoys 5A medium (Life Technologies, Burlington, Canada) (SK-OV-3) or DMEM (Sigma Aldrich, St. Louis, USA) (KP4) containing 10% fetal bovine serum (PAA Laboratories, Toronto, Canada), and 1μg/ml penicillin and streptomycin (Sigma Aldrich, St. Louis, USA). SK-OV-3 stable knockdown cells were made by infecting with lentivirus pLKO.1 shGFP (TRCN0000072179) and shHIF1a (TRCN0000003810) in the presence of 8μg/ml polybrene and after 24h selected with 4μg/ml of puromycin for two days.
TRACER fabrication
Scaffold strips (0.5 cm by 12 cm, thickness 38μm +/− 4μm (SEM, n=3)) were cut from a sheet of scaffold material (Miniminit Products Ltd, Toronto, Canada) the layer numbers and boundaries were laser printed and strips were sterilized by autoclaving. Cells were re-suspended in type 1 collagen (1 × 108 cells/ml) (PurCol 3mg/ml) (Advanced BioMatrix, San Diego, USA) and spotted into a Poly-dimethylsiloxane (PDMS) mold before addition of the scaffold to allow wicking of the cell suspension into the scaffold pores. The collagen-cell mixture was evenly distributed though the scaffold using a custom-built spreading tool. The mold plus infiltrated scaffold were incubated for 45 mins at 37°C to allow gelation of the collagen and then the biocomposite strip was removed and incubated in culture medium for 24h to allow remodelling. To assemble the TRACER, the remodelled biocomposite strip was rolled onto a custom-built aluminum mandrel (a cylinder of height 17mm and diameter 9mm, containing a notch of 5mm width and 2mm depth to house the rolled TRACER). Laser-printed guidelines on the strip facilitated consistent alignment during rolling and a polyethylene clip was used to secure the strip. Rolled TRACERs were mounted and cultured on a custom-built insert within a 6-well plate to ensure constructs remained upright. Culture medium was changed every 8h. To disassemble by unrolling, TRACERs were removed from the culture well with forceps and gently tapped to remove excess medium. TRACERs were then unrolled by first removing the polyethylene clip, gripping the visible end of the strip (edge of layer 1) and pulling the edge upward while the core rested on a sterile surface, causing strip unrolling. The speed of unrolling was not visibly affected by the time in rolled culture.
Quantification of cell density, viability and proliferation
We assessed cell viability in the TRACER using the dead cell stain Ethidium homodimer – 1 from a LIVE/DEAD kit (Molecular Probes, Eugene, USA), proliferation using EdU from the Click-iT EdUAlexa Fluor 488 Imaging Kit (Molecular Probes, Eugene, USA), and labeled all cells using the DRAQ5 nuclear stain (Cell Signaling Technology, Danvers, USA). To quantify the number of total, and dead cells we collected confocal z-stack images (5 micron slices) through the thickness of the layer and performed automated 3D nuclei counting using an Image J macro designed by Vytas Bindokas (University of Chicago, USA). Live cells were determined by subtracting the number of dead cells from total cells. Each layer was imaged in three locations for three biologically independent samples to generate cell count data. We quantified new cells generated in each layer of the TRACER over 3 days by adding EdU to the cell culture medium (10μM) for the entire culture period. EdU diffuses into the TRACER and labels cells as they pass through S phase. To quantify cell proliferation, we extracted cells from the TRACER layers using a digestive enzyme mixture of collagenase, protease, and DNAse. Cells were then Click iT labeled with Alexa Fluro 488 and further labeled with DRAQ5 nuclei stain. The percentage of the population that were EdU positive was determined using a BD LSR II flow cytometry (Becton Dickinson, Ontario, Canada).
Quantification of oxygen gradients
Changes in oxygen levels in each layer of the assembled TRACER were quantified using EF5 staining. Assembled TRACERs were cultured in the presence of 100 μM EF5 for 3.5h, disassembled and immediately fixed in 4% paraformaldehyde (PFA), permeabilized with Triton X, and stained with Cy3 conjugated anti-EF5 antibody (ELK3-51). During the 3.5h culture with EF5, cells in each layer reduce the parent EF5 to generate an unstable product, which will form covalent linkages with cellular components in the absence of oxygen. If there is oxygen present the reduced product is rapidly back oxidized and thus does not form adducts. The level of EF5 staining observed after unrolling is therefore determined by the level of oxygen that was present in that location during the 3.5h period of assembled culture. To estimate levels of hypoxia from EF5 fluorescence levels, we quantified the maximum EF5 binding in our biocomposite by incubating a single layer in anoxia (0.005% oxygen) using a Hypoxystation (Don Whitley Scientific, West Yorkshire, UK) for 20.5h followed by EF5 staining as described above. EF5 intensity was quantified in each layer from six confocal images for three TRACERs per time point.
Confocal imaging and scanning electron microscopy (SEM) analysis
Confocal images were obtained using a Carl Zeiss LSM700. To calibrate the dynamic range for quantifying EF5, the gain was set using the maximum binding positive control and the digital offset was tuned using the negative control (a single layer cultured in 21% oxygen without EF5 then stained with anti-EF5). Similarly, for calibrating the dynamic range for doxorubicin quantification, the gain was set using a single layer cultured in medium containing 25μg/mL doxorubicin for 30 minutes and the digital offset was set using the negative control; a single layer cultured in medium containing no doxorubicin. For HIF1α, the gain was set using the HIF positive control - cells cultured on a glass cover slip and exposed to hypoxia (0.2% pO2) for 4h -to ensure no saturated pixels. For SEM samples were mounted, gold-palladium sputter coated for one minute using a Polaron SC7640 Sputter Coater (Quorum Technologies, UK) and imaged at 15kV using a Hitachi SEM S-3400 (Hitachi High-Technologies Canada Inc., Toronto, Canada). Details of all antibodies used are provided in Supplementary Table 3.
Characterization of hypoxia response
Expression levels of hypoxia response genes in each TRACER layer were characterized using quantitative reverse transcriptase–PCR analysis. RNA was isolated from each layer using TRI Reagent (Sigma-Aldrich, St. Louis, USA) and cDNA was synthesized using qScript cDNA SuperMix as per manufacturer instructions. The quantitative PCR reaction was done using a 1:10 dilution of the cDNA reaction using SYBR® Green-based quantitative PCR. A standard curve for each primer set was generated by diluting cDNA. The relative amount of the target genes was calculated from the Ct and their respective primer standard curves. The relative abundance of the gene of interest was normalized to the housekeeping gene RPL13A in each sample. These normalized values were again normalized to the 6h, Layer 1 sample or the normoxia 21% sample in each plot.
Doxorubicin distribution assessment
Doxorubicin distributions were quantified using confocal analysis of the autofluorescence signal of doxorubicin. TRACERs were cultured with 25μg/mL concentration of Doxorubicin (Princess Margaret Cancer Center Pharmacy, Toronto, Canada) for 30 minutes, disassembled, rinsed once in Phosphate buffered saline (PBS), then washed for 5 minutes in PBS containing DRAQ5 (1:1000) to label nuclei. Samples were then placed on parafilm and frozen on dry ice. Individual layers were sectioned and imaged immediately by confocal. FIJI (National Institute of Health, USA) was used to quantify relative nuclear doxorubicin binding. The mean fluorescence intensity, associated with doxorubicin autofluorescence, within the nuclear region (defined by DRAQ5) was then measured to quantify bound doxorubicin in the nucleus. Mean intensity values for each layer were averaged from six images, for three independent TRACERs.
Quantification of radiotherapy response
TRACERs were dosed with 0, 5, 10 or 15 Gy of radiation, cells collected from each layer and clonogenic potential was assessed. Acrylic bioreactor cores were used for radiotherapy experiments to reduce attenuation associated with the aluminum core. Acrylic cores generated similar oxygen gradients in the SRT when compared with the aluminum core (Supplementary Figure 19) TRACERs were irradiated in a 225Cx X-RAD (Precision X-Ray, North Branford, USA), which delivers precision-targeted X-Ray radiation. TRACERs were cultured and dosed in custom-made incubation chambers (fashioned from 50mL conical tubes (BD Falcon)) to allow radial arc delivery of dose. TRACERs (within the incubation chamber) were maintained at 37 °C in a water bath prior to dosing using a 4cm × 4cm collimator. Control samples; a single layer cultured in a 20 mL glass scintillation vial containing 3 mL of medium and previously maintained in either 21% oxygen or anoxia using a hypoxic workstations (Don Whitley Scientific, West Yorkshire, UK) prior to vial sealing, were dosed with 5, 10 or 15Gy in a single delivery using the same collimator. After dosing, TRACERs were disassembled, sectioned, and digested in a collagenase (VitaCyte, Indianapolis, USA) solution for 20 minutes followed by the addition of trypsin for 5 minutes. Agitation was achieved through manual pipetting and facilitated the release of cells from the scaffold, and ensured a single cell solution for plating the clonogenic assays. Clonogenic assays were performed by plating cells at appropriate dilutions in 10cm dishes containing 10mL of medium. Colonies were grown for 3 weeks, then fixed/stained with 1% methylene blue and counted. Colonies were considered to contain 50 cells or more. All conditions were normalized to survival using the plating efficiency of the appropriate layer from a TRACER that was cultured for 24h, but not treated with radiation. Controls are normalized to a single layer cultured for 24h in 21% incubator.
LC-MS Metabolomics analysis
TRACERs were cultured as described for 24h, then unrolled and frozen on a parafilm covered metal plate chilled to −80°C on dry ice. This process (unrolling and freezing) took ~3–5 seconds. Sample layers were then sectioned, placed in metabolite extraction solution (50% acetonitile, 30% methanol and 20% water chilled to −80 °C), vortexed briefly, placed on dry ice, and shipped for analysis. We based extraction solution volumes on cell count data, ensuring an extraction volume to cell ratio of 1mL to every 1 million cells. Metabolite profiles were analyzed using LC-MS. For the LC separation, the column used was the sequant Zic-pHilic (150 mm × 2.1 mm i.d. 3.5 μm) with the guard column (20 mm × 2.1 mm i.d. 3.5 μm) from HiChrom, Reading, UK. Mobile phase A: 20 mM ammonium carbonate plus 0.1% ammonia hydroxide in water. Mobile phase B: acetonitrile. The flow rate was kept at 180 μL/minute and gradient was as follows: 0–1 minutes 70% of B, 16 minutes 38% of B, 16.5 minutes 70% of B, 25 minutes 70% of B. The mass spectrometer (Thermo QExactive Orbitrap) was operated in full MS and polarity switching mode. Samples were randomised in order to avoid machine drifts. Spectra were analysed using XCalibur Qual Browser and XCalibur Quan Browser software (Thermo Scientific) by referencing to an internal library of compounds.
HIF1α Staining and Quantification
SK-OV-3 cells were cultured in dishes as a 2D monolayer and in single layer biocomposites and exposed to normoxia (21% pO2) or to hypoxia (0.2% pO2) for 4h prior to fixation in 4% PFA for 10 minutes. TRACERS were cultured for 4h, unrolled and fixed in 4% PFA for 10 minutes. Immunostaning for HIF1α was conducted by first permeabilizing samples for 1h with 2% trition-X followed by washing with PBS, blocking with 5% normal goat serum, and application of rabbit anti-HIF1α (Cat # GTX61608, GeneTex) overnight. Secondary goat anti-rabbit TRITC was applied for 1h. Detection was conducted with a Zeiss LSM 700 by capturing 5um slices and assessing the HIF1α staining intensity in the nuclear regions (identified with the nuclear stain DRAQ5). Images were processed using FIJI.
Cell Morphology and ZO1 Junction Staining
SK-OV-3 cells were cultured in 2D monolayers, in collagen gels, or within a single layer biocomposite for 24, 48, and 72h. Samples were fixed in 4% PFA for 10 minutes, permeablized for 1h with 2% trition-X, and washed with PBS. Actin was detected with Rhodamine phalloidin, while ZO1 was detected using mouse anti-ZO1 followed by secondary labelling with goat anti-mouse 488. Detection was conducted with a Zeiss LSM 700. Images were processed using FIJI.
Layer smoothness and thickness assessment
Single layers or TRACERS containing with SK-OV-3 or KP4 cells were cultured for 0, 1, 2, and 3 days before staining with Calcein AM (Molecular Probes, Eugene, USA) to label live cells. Confocal Z-stacks were collected from each of the samples using a Zeiss LSM 700. Image analysis was conducted using FIJI. To assess layer thickness, Z-stacks were used to generate side profiles, a 50um section of layer was compressed into a single image, and thresholding applied to capture all of the live cell signal. The area of this object was divided by the width of the stack to give an estimate of the stack height or layer thickness. Smoothness was assessed by measuring the length of the top and bottom surfaces of the side profile stack, and normalizing to the control (a single layer, never rolled, cultured for 24h). Three measurements were taken for each of the 6 TRACER layers, per time point.
Statistics
To test for significant differences between two test groups, we first used an F-test to determine if equal variance could be assumed. If equal variance could be assumed, we used a pooled Student’s t-test to identify significant differences between test groups. Otherwise, we used a non-pooled Student’s t-test. ANOVA was used to test for significant differences among multiple test groups. We verified normality using Shapiro-WIlk’s test or QQ plots. Levene’s test for homogeneity was used to test for equal variance among samples. When equal variance could be assumed, the Tukey HSD or a Dunnett post hoc test was used to identify significant differences among multiple test groups. When equal variance could not be assumed, the Games–Howell post hoc test was used to identify significant differences among multiple test groups.
For metabolomics analysis samples were normalised on total metabolite intensity and unsupervised hierarchical clustering was performed using the R package heatmap. plus. Principal component analysis was performed on normalised and pareto-scaled data by using the “explore.data” function of the R package muma. Each metabolite in shGFP and shHIF samples was tested for normality (Shapiro-WIlk’s test) and Welch’s t test or Wilcoxon-Mann Whitney test were applied according to normality. P-values were corrected for multiple testing using Benjamini-Hochberg correction method. To identify metabolites with significant correlations with hypoxia level, hypoxia levels reported in Figure 3 were linearly transformed by applying a probit transformation and Pearson correlation analysis was performed to test for non-zero correlations between the transformed hypoxia levels and metabolite intensities in each layer. Normality was tested with Shapiro Wilk’s test and collinearity was tested by using Ramsey Regression Equation Specification Error Test (RESET). Correlation analysis was performed using the function “cor” of the base R package by applying Pearson’s method. All tests were two-tailed, and P ≤ 0.05 was considered significant. Details of the statistics used for each figure are shown in Supplementary Table 4.
Supplementary Material
Acknowledgments
The authors acknowledge Vyntas Bindokas (University of Chicago), Melanie Macasaet-Peralta (Pathology Research Program, UHN), Judy Cathcart (AOMF), James Stewart and Deborah Scollard (STTARR), Battista Calvieri, Steven Doyle, John Soleas, Sahar Javaherian, Camilla Londono, Christine Crossman, Ravi Vellanki, Jianxun Han, Miki Young and Shakir Lakhani (University of Toronto) for technical assistance. This work was funded by a Natural Science and Engineering Council Discovery Accelerator Supplement (RGPIN-314056) to APM, a YSF NSERC fellowship to DR, MRC Cancer Unit Core Funding to CF and EG, Ontario Ministry of Health and Long Term Care (OMOHLTC), the Terry Fox New Frontiers Research Program (PPG09-020005) and the Canadian Institute for Health Research (CIHR grant 201592) grants to BGW, and by a Ontario Graduate Scholarship to DC. The authors have no conflicts of interest to declare.
Footnotes
Author contributions
DR, EG, DC, RM, BW, CF and AM designed experiments, analyzed the data and wrote the manuscript, DR, EG and DC conducted experiments.
References
- 1.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nature reviews. Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 2.Carmeliet P, Jain RK. Principles and mechanisms of vessel normalization for cancer and other angiogenic diseases. Nat Rev Drug Discov. 2011;10:417–427. doi: 10.1038/nrd3455. nrd3455 [pii] [DOI] [PubMed] [Google Scholar]
- 3.Li XF, et al. Visualization of hypoxia in microscopic tumors by immunofluorescent microscopy. Cancer Res. 2007;67:7646–7653. doi: 10.1158/0008-5472.CAN-06-4353. [DOI] [PubMed] [Google Scholar]
- 4.Wouters BG, et al. Modulation of cell death in the tumor microenvironment. Seminars in radiation oncology. 2003;13:31–41. doi: 10.1053/srao.2003.50004. [DOI] [PubMed] [Google Scholar]
- 5.Semenza GL. Regulation of metabolism by hypoxia-inducible factor 1. Cold Spring Harbor symposia on quantitative biology. 2011;76:347–353. doi: 10.1101/sqb.2011.76.010678. [DOI] [PubMed] [Google Scholar]
- 6.Wouters BG, Koritzinsky M. Hypoxia signalling through mTOR and the unfolded protein response in cancer. Nature reviews. Cancer. 2008;8:851–864. doi: 10.1038/nrc2501. [DOI] [PubMed] [Google Scholar]
- 7.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nature reviews. Molecular cell biology. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 8.Bi M, et al. ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. The EMBO journal. 2005;24:3470–3481. doi: 10.1038/sj.emboj.7600777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guillaumond F, et al. Strengthened glycolysis under hypoxia supports tumor symbiosis and hexosamine biosynthesis in pancreatic adenocarcinoma. Proc Natl Acad Sci U S A. 110:3919–3924. doi: 10.1073/pnas.1219555110. 1219555110 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mueller-Klieser W. Multicellular spheroids. A review on cellular aggregates in cancer research. J Cancer Res Clin Oncol. 1987;113:101–122. doi: 10.1007/BF00391431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischbach C, et al. Engineering tumors with 3D scaffolds. Nat Methods. 2007;4:855–860. doi: 10.1038/nmeth1085. nmeth1085 [pii] [DOI] [PubMed] [Google Scholar]
- 12.Infanger DW, Lynch ME, Fischbach C. Engineered Culture Models for Studies of Tumor-Microenvironment Interactions. Annu Rev Biomed Eng. 2013;15:29–53. doi: 10.1146/annurev-bioeng-071811-150028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGuigan AP, Sefton MV. Vascularized organoid engineered by modular assembly enables blood perfusion. Proc Natl Acad Sci U S A. 2006;103:11461–11466. doi: 10.1073/pnas.0602740103. 0602740103 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Derda R, et al. Paper-supported 3D cell culture for tissue-based bioassays. Proc Natl Acad Sci U S A. 2009;106:18457–18462. doi: 10.1073/pnas.0910666106. 0910666106 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carmeliet P, Jain RK. Angiogenesis in cancer and other diseases. Nature. 2000;407:249–257. doi: 10.1038/35025220. [DOI] [PubMed] [Google Scholar]
- 16.Paszek MJ, et al. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 17.Minchinton AI, Tannock IF. Drug penetration in solid tumours. Nature reviews. Cancer. 2006;6:583–592. doi: 10.1038/nrc1893. [DOI] [PubMed] [Google Scholar]
- 18.Primeau AJ, Rendon A, Hedley D, Lilge L, Tannock IF. The distribution of the anticancer drug Doxorubicin in relation to blood vessels in solid tumors. Clin Cancer Res. 2005;11:8782–8788. doi: 10.1158/1078-0432.CCR-05-1664. [DOI] [PubMed] [Google Scholar]
- 19.Horsman MR, Mortensen LS, Petersen JB, Busk M, Overgaard J. Imaging hypoxia to improve radiotherapy outcome. Nature reviews. Clinical oncology. 2012;9:674–687. doi: 10.1038/nrclinonc.2012.171. [DOI] [PubMed] [Google Scholar]
- 20.Waleh NS, et al. Mapping of the vascular endothelial growth factor-producing hypoxic cells in multicellular tumor spheroids using a hypoxia-specific marker. Cancer Res. 1995;55:6222–6226. [PubMed] [Google Scholar]
- 21.Koch CJ. Measurement of absolute oxygen levels in cells and tissues using oxygen sensors and 2-nitroimidazole EF5. Methods Enzymol. 2002;352:3–31. doi: 10.1016/s0076-6879(02)52003-6. [DOI] [PubMed] [Google Scholar]
- 22.Dang CV. Links between metabolism and cancer. Genes & development. 2012;26:877–890. doi: 10.1101/gad.189365.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Latasa MU, et al. Identification of argininosuccinate lyase as a hypoxia-responsive gene in rat hepatocytes. Journal of hepatology. 2000;33:709–715. doi: 10.1016/s0168-8278(00)80300-1. [DOI] [PubMed] [Google Scholar]
- 24.Dang L, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wise DR, et al. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of alpha-ketoglutarate to citrate to support cell growth and viability. Proc Natl Acad Sci U S A. 2011;108:19611–19616. doi: 10.1073/pnas.1117773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tribble DL, Jones DP. Oxygen dependence of oxidative stress. Rate of NADPH supply for maintaining the GSH pool during hypoxia. Biochemical pharmacology. 1990;39:729–736. doi: 10.1016/0006-2952(90)90152-b. [DOI] [PubMed] [Google Scholar]
- 27.Chouchani ET, et al. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature. 2014;515:431–435. doi: 10.1038/nature13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt SK, et al. Regulation of IDO activity by oxygen supply: inhibitory effects on antimicrobial and immunoregulatory functions. PloS one. 2013;8:e63301. doi: 10.1371/journal.pone.0063301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Platten M, Wick W, Van den Eynde BJ. Tryptophan catabolism in cancer: beyond IDO and tryptophan depletion. Cancer Res. 2012;72:5435–5440. doi: 10.1158/0008-5472.CAN-12-0569. [DOI] [PubMed] [Google Scholar]
- 30.Evans SM, Hahn SM, Magarelli DP, Koch CJ. Hypoxic heterogeneity in human tumors: EF5 binding, vasculature, necrosis, and proliferation. Am J Clin Oncol. 2001;24:467–472. doi: 10.1097/00000421-200110000-00011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.