ABSTRACT
A neoadjuvant clinical trial was previously conducted in patients with resectable colorectal cancer liver metastases (CRLM). At a median follow up of 28 months, 20/33 patients were dead of disease, 8 were alive with disease and 5 were alive with no evidence of disease. To shed further insight into biological features accounting for different outcomes, the expression of CXCR4–CXCL12–CXCR7, TLR2–TLR4, and the programmed death receptor-1 (PD-1)/programmed death-1 ligand (PD-L1) was evaluated in excised liver metastases. Expression profiles were assessed through qPCR in metastatic and unaffected liver tissue of 33 CRLM neoadjuvant-treated patients. CXCR4 and CXCR7, TLR2/TLR4, and PD-1/PD-L1 mRNA were significantly overexpressed in metastatic compared to unaffected liver tissues. CXCR4 protein was negative/low in 10/31, and high in 21/31, CXCR7 was negative/low in 16/31 and high in 15/31, CXCL12 was negative/low in 14/31 and high in 17/31 CRLM. PD-1 was negative in 19/30 and positive in 11/30, PD-L1 was negative/low in 24/30 and high in 6/30 CRLM. Stromal PD-L1 expression, affected the progression-free survival (PFS) in the CRLM population. Patients overexpressing CXCR4 experienced a worse PFS and cancer specific survival (CSS) (p = 0.001 and p = 0.0008); in these patients, KRAS mutation identified a subgroup with a significantly worse CSS (p < 0.01). Thus, CXCR4 and PD-L1 expression discriminate patients with the worse PFS within the CRLM evaluated patients. Within the CXCR4 high expressing patients carrying Mut-KRAS in CRLM identifies the worst prognostic group. Thus, CXCR4 targeting plus anti-PD-1 therapy should be explored to improve the prognosis of Mut-KRAS-high CXCR4-CRLMs.
KEYWORDS: CXCR4–CXCL12–CXCR7 axis, colorectal cancer liver metastases (CRLM), KRAS mutation, PD-1/PD-L1, toll-like receptors (TLRs)
Introduction
Despite significant improvements in management of colorectal cancer (CRC), the 5-y survival rate for patients with metastatic CRC remains poor.1 About 65% of CRC patients develop distant metastasis with the liver being the most commonly involved site.2,3 Surgical resection is the only available treatment in patients with colorectal cancer liver metastases (CRLM)4 that improves 5-y survival rates ranging from 27% to 58%.5 Resectability criteria have been expanded to include all hepatic lesions that can be removed with a negative margin leaving behind an appropriate liver volume or liver functional reserve.6 In patients with unresectable CRLM, standard chemotherapy regimens which combine 5-fluorouracil with oxaliplatin or irinotecan (i.e., FOLFOX or FOLFIRI, respectively) facilitate secondary resection.7 New antiangiogenic targeted therapies, such as bevacizumab, aflibercept, and regorafenib, in combination with neoadjuvant and conversion chemotherapy may improve response rates and increase the proportion of patients eligible for surgical resection.4,8,9 As non-surgical alternative, radiofrequency ablation was reported. Radiofrequency ablation was evaluated in patients with Perioperative FOLFOX in the EPOC/CLOCC trial showing a higher local recurrence rate for RFA procedures, when lesion size exceeded 3 cm.10 Preoperative treatment of resectable liver metastases from CRC was recently conducted in a prospective phase II study with the intent to assess the feasibility and activity of bevacizumab plus FOLFIRI.11 At a median follow up of 28 months (mo), 20/33 patients (60.6%) were dead of disease (DOD), 8/33 patients (24.2%) were alive with disease (AWD), and 5/33 patients (15.2%) were alive with no evidence of disease (NED). To shed further insight into biological features accounting for that, a study evaluating the expression of CXCR4–CXCL12–CXCR7 pathway, TLR2–TLR4, and the programmed death receptor-1 (PD-1)/programmed death-1 ligand (PD-L1) was conducted. The chemokine receptor CXCR4 was previously described in primary CRC12 and in liver metastasis13-15 whereas CXCR7 was described in secondary lesions of CRC in sites other than liver.16 In locally advanced rectal cancer patients, high CXCR4 correlated with a shorter relapse-free and cancer specific survival and high CXCR4/N+ identified the worst prognostic category.17 Exosomes derived from HT29 human colon cancer cells, increased CXCL12 expression in the metastatic microenvironment, attracting cancer cells and other stromal cells expressing CXCR4.18 Interaction between CXCR4 and innate immunity were previously reported. LPS-induced inflammation activates TLR4 that in turn induces CXCR4 and/or CXCR7 expression in tumor cells, enhancing the response to CXCL12 to promote invasion and cell dissemination.19 LPS exposure induced CXCR7 expression in human colon cancer cell lines SW480 and Colo 205 expressing TLR4/myeloid differential protein (MD-2).19 Moreover, N15P polypeptide, a new CXCR4 antagonist derived from the Kaposi sarcoma secreted cytokine vMIP-II reversed the LPS-induced inflammation in human PBMC.20,21
Targeting PD-1 T-cell coreceptor and its ligand B7-H1/PD-L1 induce durable tumor responses22 nevertheless only a minority of patients respond, and it has been unclear which patients and which tumors are the best candidates for this therapy.23,24 In colorectal cancer, high infiltrate of CD8+ CTL and T helper 1 (Th1)-type cells constitutes a favorable prognostic factor.25,26 Llosa et al. demonstrate that tumors with a high Th1/CTL infiltrate had defects in mismatch repair (MMR), resulting in microsatellite instability (MSI).27 The high neoantigen burden might be one explanation for the high level of tumor-infiltrating lymphocytes (TILs) in MSI tumors. Tumors with MSI had significant upregulation of PD-1 and PD-L1.28 Recently, Galon et al. demonstrated the presence of functional mutation-specific cytotoxic T cells and the superiority of Immunoscore over microsatellite instability in predicting survival in microsatellite-instable tumors. MSI, and a subgroup of microsatellite stability (MSS) tumors with a prominent expression of Th1, cytotoxic genes, cytokines, and chemokines, have a better prognosis than MSS.29 With the intent to identify prognostic/predictive pattern for CRLM neoadjuvant-treated patients, the expression of CXCR4–CXCL12–CXCR7 as well as for TLR2 and TLR4 and immune checkpoint PD-1 /PD-L1 was evaluated.
Results
Patients features
Neoadjuvant-treated CRLMs patients from Phase 2 Clinical trial11 were evaluated (Table S1). The objective responses to neoadjuvant therapy were: partial response (PR) in 28/33 patients (85 %) and stable disease (SD) in 4 patients (12%). The overall response rate (ORR) was 97%, with one patient with progressive disease (PD) (3%). The neoadjuvant response did not affect cancer specific survival (CSS) (Fig. S1). At a median follow up of 28 mo, 20/33 (60.4%) patients were DOD, 8/33 (24.2%) AWD, and 5/33 (15.2%) were NED; median progression-free survival (PFS) was 18 mo (range 5–66) and CSS 28 mo (range 10–66).
CXCR4–CXCR7, TLR2–TLR4, and PD-1/PD-L1 upregulation in neoadjuvant-treated CRLM
CXCR4, CXCL12, CXCR7, TLR2, TLR4, PD-1, and PD-L1 mRNA levels were evaluated in 33 CRLM homogeneously treated patients11 by Real-time PCR analysis. The analysis was conducted on the metastatic tissue (T) and on the surrounding unaffected tissue (H), and then, the T/H ratio was calculated. CXCR4 and CXCR7 significantly increased in metastatic tissue compared to unaffected liver tissue in CRLMs (p = 0.0013 and p = 0.0080, respectively), whereas CXCL12 mRNA was not significantly different between T and H (Fig. 1A). TLR-2 and TLR-4 mRNA significantly increased in metastatic compared to unaffected liver tissue (p = 0.0032 and p = 0.0044, respectively; Fig. 1B). Similarly, PD-1 and PD-L1 mRNA dramatically increased in metastatic compared to unaffected liver tissue (p = 0.0004 and p = 0.0100, respectively; Fig. 1C). Correlation analyses displayed that CXCR4 mRNA correlated with CXCR7 and CXCL12. TLR-2 mRNA correlated with TLR-4, PD-L1, and CXCR7 (Table S2). Although KRAS mutations are referred as poor prognostic markers in primary CRCs and predictive anti-EGFR resistance, the prognostic value of KRAS remains controversial in CRLMs patients.30 Correlations between mRNA expression and PFS/CSS for CXCR4, CXCL12, CXCR7, TLR2, TLR4, PD-1, and PD-L1 are summarized in Fig. S2.
Figure 1.
CXCR4–CXCR7, TLR2–TLR4, and PD-1/PD-L1 upregulation in neoadjuvant-treated CRLM patients. mRNA expression was measured using real-time PCR in paired liver tissues (H) and colorectal liver metastasis (T), respectively. (A) CXCR4, CXCR7 and CXCL12 mRNA expression; (B) TLR 2 and 4 mRNA expression. (C) PD-1 and PD-L1 mRNA expression. The y-axis values represent the ratio between the values of each gene in CRLM/unaffected liver normalized to GUSB RNA expression. Each assay was performed in triplicate. CRLM: colorectal liver metastasis; PCR: polymerase chain reaction. Data are presented as box and whisker plots: boxes extend from the 25th to 75th percentile, with a black line at the population median. Statistical significance was determined by Mann–Whitney U nonparametric test, p < 0.05.
CXCR4 and CXCR7 protein correlate with outcome in neoadjuvant-treated CRLMs patients
CXCR4–CXCL12–CXCR7 axis was evaluated through IHC. CXCR4 was negative/low in 10/31 patients (32.3%), and high in 21/31 (67.7%). In Fig. 2A–C, an example of negative/low-and high CXCR4 staining is reported; CXCR7 was negative/low in 16/31 (51.6%) and high in 15/31 (48.4%). In Fig. 2D–F an example of negative/low and high CXCR7 is reported. CXCL12 was negative/low in 14/31 patients (45.2%) and high in 17/31 (54.8%). In Fig. 2G–I, an example of low and high CXCL12 is reported (Fig. 2). Kaplan–Meier curves revealed that patients overexpressing CXCR4 displayed a worse PFS and CSS (p = 0.001 and p = 0.0008, respectively) (Fig. 3A and B). Within the CXCR4 high expressing population, the evaluation of PD-L1 T/H RNA ratio discriminate patients with worse PFS, where higher PD-L1 T/H ratio stands for lower stromal PD-L1 detected in the peritumoral tissue (p = 0.0079) (Fig. 3C). Patients overexpressing CXCR7 displayed a marginal significance with worse PFS (p = 0.050; Fig. S3). The evaluation of CXCL12 did not determine a significant separation of patients' population in term of PFS or CSS (Fig. S3).
Figure 2.
CXCR4, CXCR7, and CXCL12 expression in CRLMs. Representative microphotograph (200× magnification) for CXCR4, CXCR7 and CXCL12 IHC staining in CRLM as (A, B) Diffuse low and moderate and (C) diffuse high cytoplasmic and/or nuclear staining for CXCR4: Arrow shows area with staining in stromal cells of microenvironment. (D, E) Negative/low, and (F) high CXCR7 cytoplasmic staining. Arrow shows staining in stromal cells of microenvironment. (G) Negative low and (H, I) high membrane and cytoplasmic CXCL12 staining.
Figure 3.
CXCR4 protein level affects CRLMs survival. (A, B) PFS and CSS Kaplan–Meier curves according to CXCR4 expression in CRLM patients (PFS: negative- low CXCR4, N = 10, median survival 46 mo vs high CXCR4, N = 21, median survival 14 mo, p = 0.001) (CSS: negative-low CXCR4, N = 10, median survival 46 mo vs high CXCR4, N = 21, median survival 28 mo, p = 0.0008). (C) PFS Kaplan–Meier curves according to PD-L1 mRNA tumoral expression (T)/PD-L1 mRNA healthy tissue (H) (PFS low <1; N = 7, median survival 10 mo vs high >1; N = 13, median survival 18 mo, p = 0.0079).
To further characterize the subgroup of patients with worse prognosis, KRAS mutational status was considered in the CXCR4-overexpressing patients. Within 21 CXCR4-overexpressing patients, the KRAS status was available for 18. In this subgroup, KRAS status become very significant (p = 0.0061) for CSS (Fig. 4A). Conversely, Mut-KRAS patients considered in the entire population (10 Mut-KRAS/25 KRAS status available) expressed high level of CXCR4 (Fig. 4B), KRAS mutations alone did not affect survival (Fig. S4A). Of note CXCR7 showed a trend correlation with KRAS (Fig. S4B).
Figure 4.
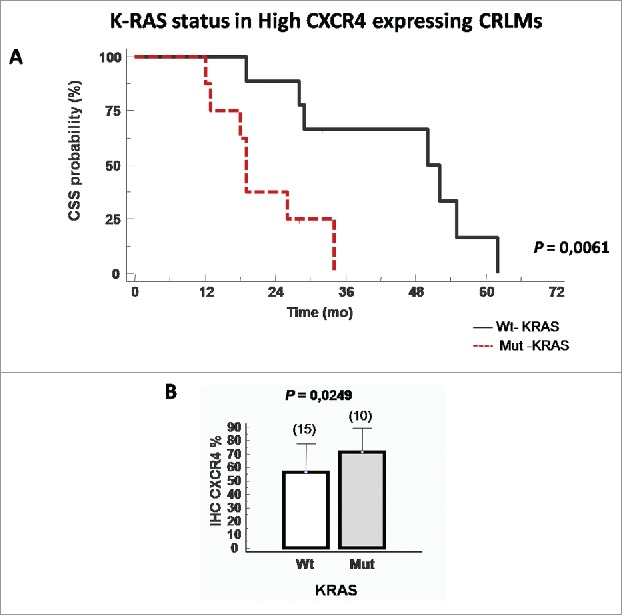
CXCR4 protein correlates with K-RAS mutational status. (A) Cancer specific survival (CSS) Kaplan–Meier curves described the patients groups at high and low risk of death according to KRAS mutational status. (CSS wt-KRAS; N = 10, median survival 51 mo vs mut-KRAS; N = 8, median survival 19 mo, p = 0.0061). (B) Correlation between CXCR4 value and KRAS mutation.
PD-L1 but not PD-1 protein expression affects CRLM progression free survival?
The protein level of PD-1 and PD-L1 was then evaluated in CRLM samples. PD-1 was focally expressed in tumor immune infiltrating cells. PD-1 staining was negative in 19/30 (63.3%) and positive in 11/30 (36.6%) (Fig. 5A and B). In Fig. 5C and D, the PD-L1 staining is shown. PD-L1 was moderate-highly expressed in stromal cells and hardly detectable in neoplastic cells. PD-L1 staining was conducted with two antibodies 5H1 and SP142. The results were comparable and the staining localized on membrane with the presence or not of cytoplasmic.31 Nevertheless, the staining obtained with Sp142 clone (Fig. 5C and D) more selectively identified PD-L1 membrane localization. PD-L1 was categorized as negative/low in 24/30 (80.0%) and high in 6/30 (20.0%). PD-1 and PD-L1 protein expression did not affect prognosis (Fig. S3C and D). PD-L1 stromal expression was also evaluated (macrophages, fibroblasts) trough IHC on the entire cohort (Fig. S5A–D). PD-L1 expression was low in 18/32 and high in the stroma of 14/32 patients. The implication of stromal PD-L1 expression on prognosis was evaluated and PFS curves significantly separated according to stromal PD-L1 expression, (p = 0.005; HR 0.3770; 95% confidence interval (CI) 0.1025–0.6665). CSS curves did not significantly separate according to stromal PD-L1 expression (Fig. S5E and F).
Figure 5.
PD-L1 and PD-1 protein expression. Representative microphotograph for PD-1 and PD-L1 IHC staining in CRLM (200× magnification). (A) Negative and (B) focal PD-1 immunoreactivity on microenvironment immune cells at tumor periphery (inset detail: 400× magnification). (C) Negative or (D) diffuse PD-L1 membrane with or without cytoplasmic staining on tumor and stromal cells (inset : 400× magnification).
In multivariable Cox model analyses, gender and the size of liver metastasis were independently correlated to CSS. PDL1 T/H and CXCR4 were independently correlated to PFS (Table 1).
Table 1.
Univariate and multivariate Cox analyses on survival models in patients with resecable CRLM neoadjuvant bevacizumab plus FOLFIRI.
| PFS |
CSS |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Univariate analysis | Multivariate analysis | Univariate analysis | Multivariate analysis | |||||||
| Variable | N | HR (95% CI) | p value | HR (95% CI) | p value | N | HR (95% CI) | p value | HR (95% CI) | p value |
| Age (years; ≤ median/ > median) | 19/14 | 1.476 (0.686–3.273) | 0.309 | 19/14 | 1.057 (0.417–2.677) | 0.903 | ||||
| Gender (male/female) | 20/13 | 2.821 (1.345–6.541) | 0.007 | 20/13 | 4.038 (1.503–9.870) | 0.005 | 12.404 (1.468–104.790) | 0.021 | ||
| KRAS mutational status (Wt/Mut) | 16/10 | 0.6216 (0.220–1.560) | 0.285 | 16/10 | 0.401 (0.090–1.027) | 0.055 | ||||
| Liver metastasis (metachronus/synchronus) | 11/22 | 0.611 (0.275–1.350) | 0.223 | 11/22 | 0.418 (0.175–1.167) | 0.101 | ||||
| Number of metastatic lesions (≤ 3/ > 3) | 20/13 | 0.550 (0.201–1.169) | 0.107 | 20/13 | 0.543 (0.189–1.338) | 0.168 | ||||
| Size of metastatic lesions (cm) (≤ 3/ > 3) | 14/19 | 0.413 (0.169–0.888) | 0.025 | 14/19 | 0.079 (0.062–0.480) | 0.001 | ||||
| Metastases surgical outcome (R0/R1–R2) | 28/5 | 0.569 (0.098–2.226) | 0.341 | 28/5 | 0.305 (0.028–0.654) | 0.013 | 3.507 (0.930–13.217) | 0.065 | ||
| CXCR4 expression (negative-low/high) | 10/21 | 0.232 (0.091–0.529) | 0.001 | 3.405 (1.701–17.338) | 0.004 | 10/21 | 0.079 (0.062–0.480) | 0.0008 | ||
| CXCR7 expression (negative-low/high) | 16/14 | 0.479 (0.179–1.002) | 0.050 | 16/14 | 0.444 (0.162–1.144) | 0.092 | ||||
| PD-L1 mRNA T/H ratio (low < 1/high > 1) | 10/18 | 2.024 (0.882–7.309) | 0.084 | 0.315 (0.142–0.869) | 0.026 | 10/18 | 1.963 (0.728–6.816) | 0.160 | ||
Discussion
To explore on the biological heterogeneity of colorectal cancer, the expression of CXCR4–CXCL12–CXCR7, TLR2–TLR4, and PD-1/PD-L1 was evaluated in patients enrolled in a previous institutional trial.11 Preoperative treatment followed by surgery was conducted in 33 patients carrying resectable CRLM. At a median follow up of 28 mo, 20 were dead of disease, 8 were alive with disease, and 5 patients were alive in absence of disease. CXCR4–CXCR7, TLR2–TLR4, and PD-1/PD-L1 were significantly overexpressed in liver metastasis compared to neighboring unaffected liver tissues. As a study limitation, since pre-operative therapy was proposed to all resectable CRLMs carrying patients, expression of CXCR4–CXCR7, TLR2–TLR4, and PD-1/PD-L1 in CRLMs from patient not subjected to neoadjuvant therapy was unavailable. At the protein level, high CXCR4 identified a poor prognostic subgroup; moreover, within this group, the patients with mutant KRAS showed the worst prognosis. KRAS mutations are considered driver mutations and a poor prognostic factor in primary CRC32,33 with no significant associations between KRAS mutation and metastatic progression, proliferative index, or survival.34 In accordance with the reported finding, Alamo reported that colorectal cancer carrying KRAS G12V revealed higher aggressiveness and overexpressed CXCR4.35 KRAS was reported to be informative for prognosis only among patients who received preoperative chemotherapy and not for patients undergone upfront surgery.36 Thus this is the first report of a significant correlation between KRAS mutant status, CXCR4 and prognosis in CRLM.
Although the expression of TLR2–TLR4 was not significantly different among the evaluated patients an interesting link between CXCR4 and TLR4 recently emerged. Searching through the publicly available TCGA,37,38 it was discovered that CXCR4 expression on colorectal cancers significantly correlated with TLR4-accessory molecule RP105 (CD180) with Spearman's rho = 0.6173 (p < 0.001) and Bruton's tyrosine kinase (BTK) gene Spearman's rho = 0.6172 (p < 0.001) (Fig. S6). The CXCR4 ligand, CXCL12, induces BTK phosphorylation and downstream MAPK signaling in primary acute myeloid leukemia (AML); the BTK inhibitor drug, Ibrutinib,39 interrupts the interaction between the tumor cell and the microenvironment that protects leukemic cells,40 and CXCR4 mutations confer resistance to Ibrutinib.41 Thus, CXCR4 targeting may regulate TLR4 and BTK pathways both with biological implications in mCRC. However, there was no significant correlation between CXCR4 and TLR4 expression in CRLM patients, and light but significant correlation was observed between CXCR7 and TLR2. TLR2 and CXCR7 are novel target genes repressed by Hypermethylated in cancer 1 (HIC1). HIC1 is a prototypic tumor suppressor gene frequently inactivated by DNA methylation in solid tumors. Recent data indicated that HIC1 depletion in the intestinal epithelium resulted in increased TLR2 that promoted proliferation of colonic tumors induced by chemical carcinogenesis.42 Moreover, genome-wide expression profiling analyses to identify new HIC1 target genes, using HIC1-deficient U2OS human osteosarcoma cells, identified CXCR7 as putative direct target genes.43 PD-L1 was mainly expressed in stromal cells of CRLM and showed diffuse faint immunoreactivity in neoplastic cell as previously reported (http://www.proteinatlas.org/); when the PD-L1 mRNA was evaluated in the subgroup of CXCR4 high expressing tumors, the higher PD-L1 expression in healthy tissues decreased the tumor/healthy ratio in patients with a worse disease free survival suggesting that stromal PD-L1 expression may represent a more valuable indicator of anti-PD-1 resistance compared to tumoral PD-L1 expression. Distinct patterns of PDL1 expression may be observed not only in different tumor types, but also in individual cases within the same tumor type.31 The mixture of cytoplasmic and membranous PD-L1 staining made its interpretation difficult and unclear.31,44-46 More clearly, PD-L1 expression results on infiltrating immune cells immediately adjacent to tumor cells.31 Since evidence demonstrated that CXCR4 antagonism may favor the efficacy of anti-PD-1/PD-L1 therapy that modulates T cell access,47 targeting CXCR4 could affect intratumoral PD-1/PD-L1 levels.
Recent data on the role of T cell infiltration in liver metastases showed a strong association between the local immune cell profile (based on granzyme B and CD8+, CD3+ T cells and FOXP3+ lymphocytes) and chemotherapy outcome in CRLM. TIL densities at the invasive margin of liver metastases allowed the prediction of response to chemotherapy with a sensitivity of 79% and specificity of 100%.48 Albeit limited by the sample size, the present study demonstrated that high CXCR4 and Mut-KRAS identifies the worst prognostic group within a homogeneous cohort of mCRC with resectable liver metastases undergone to common neoadjuvant therapy. We speculate that the new class of CXCR4 antagonists recently developed49 may be highly effective in Mut-KRAS CRLMs targeting CXCR4-positive cancer cells and improving T cell access by reducing tolerogenic effect and improving immune response in CRLM patients.
Material and methods
Patient and treatment
From October 2007 to December 2009, 33 patients from a phase 2 trial,11 aged 18–75 y, PS 0–1, with resectable CRLMs were treated with FOLFIRI plus bevacizumab preoperative treatment. FOLFIRI was administered intravenously (i.v.) every 14 d with irinotecan 180 mg/m2 i.v. infusion on day 1, leucovorin (200 mg/m2) by i.v. infusion on day 1, 5-fluorouracil (400 mg/m2) by i.v. bolus on day 1, 5-fluorouracil (2,400 mg/m2) 46-h continuous infusion; bevacizumab was administered at 5 mg/kg by i.v. infusion over 90 min at the first cycle, and then, if tolerated, over 60 min. The treatment was administered every 14 d, for 7 cycles; bevacizumab was stopped at cycle 6 to prolong the bevacizumab-free interval and to reduce the risk of surgical bleeding. After restaging, patients whose liver metastases were still confirmed as potentially resectable underwent surgery. Following surgery, four further cycles of FOLFIRI plus bevacizumab were planned, with the same dose and schedule planned. Liver resection was defined as radical (R0) according to both macroscopic description of surgery and histological evaluation. Radical surgery was defined as a margin of at least 1 mm. Thus, R0 corresponds to resection for cure or complete remission, R1 corresponds to microscopic residual tumor (≤1 mm), and R2 corresponds to macroscopic residual tumor.
Real-time PCR
RNA was extracted from metastatic and paired adjacent non-tumor liver tissues from fresh samples acquired upon surgery. A safety margin ≤1 cm is generally accepted with regard to the safety surgical margin in CRLMs. The distance 1 cm was the minimal distance between tumor metastasis and normal-appearing liver tissue sampled. Total RNA was isolated using the RNeasy Mini kit (Qiagen). Real time-PCR was carried out using about 10 ng of reverse transcribed cDNA in 25 μL final of SYBR Green reaction mixture. An ABI Prism 7000 (Applied Biosystems) robocycler was used for the amplification. Relative gene expression was calculated in the following formula using GUSB gene as endogenous control: 2−ΔΔCt. The primer pairs are subjected to the specificity checking process through Primer-BLAST publicly available tool at http://www.ncbi.nlm.nih.gov/tools/primer-blast.50
Primers used in this study were as follows:
GUSB: forward: 5′-AGCCAGTTCCTCATCAATGG-3′, reverse: 5′-GGTAGTGGCTGGTACGGAAA-3′;
CXCR4: forward: 5′-TGAGAAGCATGACGGACAAG-3′; reverse: 5′-AGGGAAGCGTGATGACAAAG-3′.
CXCR7: forward: 5′-GATTGCCCGCCTCAGAAC-3′, reverse: 5′-GCAGGACGCTTTTGTTGG-3′.
CXCL12: forward: 5′-TGTGGCACTCAGATACCGACT-3′, reverse: 5′-CCCACAGAGCCAATCACT-3′.
TLR2: forward: 5′-CCTGGCCCTCTCTACAAACTT-3′, reverse: 5′-ACTGTGTATTCGTGTGCTGGATA-3′.
TLR4: forward: 5′-TGAGCAGTCGTGCTGGTATC-3′, reverse: 5′-CAGGGCTTTTCTGAGTCGTC-3′.
PD-1: forward: 5′-GCCACCATTGTCTTTCCTAG-3′, reverse: 5′- AAGAGCAGTGTCCATCCTCAG -3′.
PD-L1: forward: 5′-GCCTCCAAGCAAATCATCCA-3′, reverse: 5′- CATTGAGTGGAGGCAAAGGG -3′.
Each sample was tested in triplicate for analysis of relative gene expression.
Immunohistochemistry
CXCR4, CXCR7, CXCL12, PD-L1, PD-1 were evaluated through immunohistochemistry (IHC) on formalin-fixed and paraffin-embedded (FFPE) tissue blocks derived from surgically collected liver samples. For CXCR4 (mab172, [44716], dilution 1:1000 R&D Systems), CXCL12 (1:50 dilution, MAB 350 mouse monoclonal anti-Hu-CXCL12 [79018] R&D Systems), and CXCR7 (1:50 dilution, MAB42273 mouse monoclonal anti-Hu CXCR7 [11G8] R&D Systems), heat induced epitope retrieval was performed. The staining was categorized in semiquantitative classes based on the rate of stained cancer cells: CXCR4, CXCR7, and CXCL12 were graded as negative/low (0–50%) and high (>50%).17 For B7-H1/PD-L1, both non-commercial and a commercially available antibodies, mouse monoclonal antibody made in Dr. Lieping Chen's lab, clone 5H1, 1:500 dilution51,52 and rabbit monoclonal antibody, clone SP142, 1:200 dilution,44 Spring Bioscience) were used with prior pressure cooker to 110°C × 10 min Antigen Retrieval (Ventana Medical Systems, Cell Conditioning Solution CC1, pH 9.0).44 PD-1 was detected with mouse monoclonal antibody PD-1) (Ventana Medical Systems, NAT105). The extent of staining was categorized in semiquantitative classes based on the rate at which tumor cells exhibit complete and/or partial circumferential linear plasma membrane with or without cytoplasmic at any intensity for PD-L1 as negative/low (0% to <5%) and positive (>5%).31,46,52 Positive PD-L1 and PD-1 stromal cells were counted in 5 HPF (0.028 mm/HPF; Olympus BX41) on three distinct tumor stroma areas.53 Stromal PD-L1 extent of staining was categorized in semiquantitative classes based on the rate cells as negative (0% to <15%) and positive (>15%). PD-1 was categorized in semiquantitative classes as immune cell/HPF scored negative or positive. Semiquantitative classes were chosen with a double-blind method by our two pathologists (FT and GB) after consensus discussion and careful revision of all slides.
Expression data from TCGA
Public TCGA data repositories (https://tcga-data.nci.nih.gov) that provides visualization, analysis, and download of large-scale cancer genomics data sets38 were used to obtain colorectal cancer sample data. Expression of mRNA for the selected markers and the associated patient clinical data were analyzed. mRNA expression levels of 20,532 genes (RNA Seq V2) for 433/623 patients were obtained. This was downloaded from The Cancer Genome Atlas Network (TCGA) data portal and cBioPortal (http://www.cbioportal.org)37,54 and used to correlate mRNA expression.
Statistical analysis
Results for continuous variables are presented as means ± standard deviation unless stated otherwise, categorical variables as absolute numbers and percentages; significance was determined using the Mann–Whitney test. Statistical analysis was performed using the MedCalc Statistical Software version 12.3.0 (Microsoft, Inc., Belgium). The Spearman correlation test was used to evaluate the association between markers expressions. PFS was defined as the time from initial treatment to the documented local or distant recurrence (whichever occurred first) or last follow-up. CSS was defined as the time from initial treatment until death for cancer-related cause or to last follow-up. Survival curves were estimated with their 95% CI by the product-limit method of Kaplan–Meier and compared by the log-rank test. Hazard ratios (HRs) with their 95% CI were derived from Cox regression analysis. Univariate analysis was done with the log-rank test. Cox proportional hazards regression was used to analyze the effect of several risk factors on RFS and CSS. Risk factors (covariates) were considered dichotomous. Based on univariate Cox significance level of 0.1, clinical variables and biomarkers were incorporated into the Cox models for multivariate analysis; a backward selection was then applied to construct the final multivariate model. All statistical tests were two tailed and p values less of than 0.05 were considered significant.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by grants from the Associazione Italiana per la Ricerca sul Cancro (AIRC) AIRC-IG 13192 and Ricerca corrente 2014: “Ruolo dell'asse CXCR4-CXCL12-CXCR7 e dell'EMT nel carcinoma rettale: implicazioni per la prognosi ed il trattamento.”
Ethical statement
Tissues samples from patients were obtained with their informed consent in accordance with the Helsinki Declaration as revised in 2000. The study was approved by Istituto Nazionale Tumori, “Fondazione G. Pascale” Ethics committee (CE 689-24/10/2007).
References
- 1.Meyerhardt JA, Mayer RJ. Systemic therapy for colorectal cancer. New Engl J Med 2005; 352:476-87; PMID:21527027; http://dx.doi.org/ 10.1056/NEJMra040958 [DOI] [PubMed] [Google Scholar]
- 2.Lordan JT, Karanjia ND, Quiney N, Fawcett WJ, Worthington TR. A 10-year study of outcome following hepatic resection for colorectal liver metastases – The effect of evaluation in a multidisciplinary team setting. Eur J Surg Oncol: J Euro Soc Surg Oncol Brit Assoc Surg Oncol 2009; 35:302-6; PMID:18328668; http://dx.doi.org/ 10.1016/j.ejso.2008.01.028 [DOI] [PubMed] [Google Scholar]
- 3.Pestana C, Reitemeier RJ, Moertel CG, Judd ES, Dockerty MB. The natural history of carcinoma of the colon and rectum. Am J Surg 1964; 108:826-9; PMID:14233766; http://dx.doi.org/ 10.1016/0002-9610(64)90041-8 [DOI] [PubMed] [Google Scholar]
- 4.Garcia-Alfonso P, Ferrer A, Gil S, Duenas R, Perez MT, Molina R, Capdevila J, Safont MJ, Castanon C, Cano JM et al.. Neoadjuvant and conversion treatment of patients with colorectal liver metastasis: the potential role of bevacizumab and other antiangiogenic agents. Target Oncol 2015; PMID:25752908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khan K, Wale A, Brown G, Chau I. Colorectal cancer with liver metastases: neoadjuvant chemotherapy, surgical resection first or palliation alone? World J Gastroenterol 2014; 20:12391-406; PMID:25253940; http://dx.doi.org/ 10.3748/wjg.v20.i35.12391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pawlik TM, Schulick RD, Choti MA. Expanding criteria for resectability of colorectal liver metastases. Oncologist 2008; 13:51-64; PMID:18245012; http://dx.doi.org/ 10.1634/theoncologist.2007-0142 [DOI] [PubMed] [Google Scholar]
- 7.Adam R, Delvart V, Pascal G, Valeanu A, Castaing D, Azoulay D, Giacchetti S, Paule B, Kunstlinger F, Ghemard O et al.. Rescue surgery for unresectable colorectal liver metastases downstaged by chemotherapy: a model to predict long-term survival. Ann Surg 2004; 240:644-57; Discussion 57-8; PMID:15383792; http://dx.doi.org/ 10.1097/01.sla.0000145964.08365.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Artigas V, Marin-Hargreaves G, Marcuello E, Pey A, Gonzalez JA, Rodriguez M, Moral A, Monill JM, Sancho J, Pericay C et al.. Surgical resection of liver metastases from colorectal carcinoma. Experience in Sant Pau Hospital. Cir Esp 2007; 81:339-44; PMID:17553407. [DOI] [PubMed] [Google Scholar]
- 9.Keeley EC, Mehrad B, Strieter RM. Chemokines as mediators of tumor angiogenesis and neovascularization. Exp Cell Res 2011; 317:685-90; PMID:21040721; http://dx.doi.org/ 10.1016/j.yexcr.2010.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanis E, Nordlinger B, Mauer M, Sorbye H, van Coevorden F, Gruenberger T, Schlag PM, Punt CJ, Ledermann J, Ruers TJ. Local recurrence rates after radiofrequency ablation or resection of colorectal liver metastases. Analysis of the European Organisation for Research and Treatment of Cancer #40004 and #40983. Euro J Cancer (Oxford, England: 1990) 2014; 50:912-9; PMID:24411080; http://dx.doi.org/ 10.1016/j.ejca.2013.12.008 [DOI] [PubMed] [Google Scholar]
- 11.Nasti G, Piccirillo MC, Izzo F, Ottaiano A, Albino V, Delrio P, Romano C, Giordano P, Lastoria S, Caraco C et al.. Neoadjuvant FOLFIRI+bevacizumab in patients with resectable liver metastases from colorectal cancer: a phase 2 trial. Brit J Cancer 2013; 108:1566-70; PMID:23558891; http://dx.doi.org/ 10.1038/bjc.2013.140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ottaiano A, Franco R, Aiello Talamanca A, Liguori G, Tatangelo F, Delrio P, Nasti G, Barletta E, Facchini G, Daniele B et al.. Overexpression of both CXC chemokine receptor 4 and vascular endothelial growth factor proteins predicts early distant relapse in stage II-III colorectal cancer patients. Clin Cancer Res 2006; 12:2795-803; PMID:16675573; http://dx.doi.org/ 10.1158/1078-0432.CCR-05-2142 [DOI] [PubMed] [Google Scholar]
- 13.Murakami T, Kawada K, Iwamoto M, Akagami M, Hida K, Nakanishi Y, Kanda K, Kawada M, Seno H, Taketo MM et al.. The role of CXCR3 and CXCR4 in colorectal cancer metastasis. Int J Cancer 2013; 132:276-87; PMID:22689289; http://dx.doi.org/ 10.1002/ijc.27670 [DOI] [PubMed] [Google Scholar]
- 14.Yopp AC, Shia J, Butte JM, Allen PJ, Fong Y, Jarnagin WR, DeMatteo RP, D'Angelica MI. CXCR4 expression predicts patient outcome and recurrence patterns after hepatic resection for colorectal liver metastases. Ann Surg Oncol 2012; 19 Suppl 3:S339-46; PMID:21584832; http://dx.doi.org/ 10.1245/s10434-011-1774-4 [DOI] [PubMed] [Google Scholar]
- 15.Kim J, Mori T, Chen SL, Amersi FF, Martinez SR, Kuo C, Turner RR, Ye X, Bilchik AJ, Morton DL et al.. Chemokine receptor CXCR4 expression in patients with melanoma and colorectal cancer liver metastases and the association with disease outcome. Ann Surg 2006; 244:113-20; PMID:16794396; http://dx.doi.org/ 10.1097/01.sla.0000217690.65909.9c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guillemot E, Karimdjee-Soilihi B, Pradelli E, Benchetrit M, Goguet-Surmenian E, Millet MA, Larbret F, Michiels JF, Birnbaum D, Alemanno P et al.. CXCR7 receptors facilitate the progression of colon carcinoma within lung not within liver. Brit J Cancer 2012; 107:1944-9; PMID:23169289; http://dx.doi.org/; http://dx.doi.org/ 10.1038/bjc.2012.503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D'Alterio C, Avallone A, Tatangelo F, Delrio P, Pecori B, Cella L, Pelella A, D'Armiento FP, Carlomagno C, Bianco F et al.. A prognostic model comprising pT stage, N status, and the chemokine receptors CXCR4 and CXCR7 powerfully predicts outcome in neoadjuvant resistant rectal cancer patients. Int J Cancer 2014; 135:379-90; PMID:24375277; http://dx.doi.org/ 10.1002/ijc.28689 [DOI] [PubMed] [Google Scholar]
- 18.Wang X, Ding X, Nan L, Wang Y, Wang J, Yan Z, Zhang W, Sun J, Zhu W, Ni B et al.. Investigation of the roles of exosomes in colorectal cancer liver metastasis. Oncol Rep 2015; 33:2445-53; PMID:25760247; http://dx.doi.org/ 10.3892/or.2015.3843 [DOI] [PubMed] [Google Scholar]
- 19.Xu H, Wu Q, Dang S, Jin M, Xu J, Cheng Y, Pan M, Wu Y, Zhang C, Zhang Y. Alteration of CXCR7 expression mediated by TLR4 promotes tumor cell proliferation and migration in human colorectal carcinoma. PLoS One 2011; 6:e27399; PMID:22180778; http://dx.doi.org/ 10.1371/journal.pone.0027399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mo XM, Sun HX. The anti-inflammatory effect of the CXCR4 antagonist-N15P peptide and its modulation on inflammation-associated mediators in LPS-induced PBMC. Inflammation 2015; 38:1374-83; PMID:25676435; http://dx.doi.org/ 10.1007/s10753-015-0109-1 [DOI] [PubMed] [Google Scholar]
- 21.Fitzgerald KA, Rowe DC, Golenbock DT. Endotoxin recognition and signal transduction by the TLR4/MD2-complex. Microbes Infection/Institut Pasteur 2004; 6:1361-7; PMID:15596121; http://dx.doi.org/ 10.1016/j.micinf.2004.08.015 [DOI] [PubMed] [Google Scholar]
- 22.Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, Drake CG, Camacho LH, Kauh J, Odunsi K et al.. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. New Engl J Med 2012; 366:2455-65; PMID:22658128; http://dx.doi.org/ 10.1056/NEJMoa1200694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB et al.. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. New Eng J Med 2012; 366:2443-54; PMID:22658127; http://dx.doi.org/ 10.1056/NEJMoa1200690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brahmer JR, Drake CG, Wollner I, Powderly JD, Picus J, Sharfman WH, Stankevich E, Pons A, Salay TM, McMiller TL et al.. Phase I study of single-agent anti-programmed death-1 (MDX-1106) in refractory solid tumors: safety, clinical activity, pharmacodynamics, and immunologic correlates. J Clin Oncol: Off J Am Soc Clin Oncol 2010; 28:3167-75; PMID:20516446; http://dx.doi.org/ 10.1200/JCO.2009.26.7609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, Tosolini M, Camus M, Berger A, Wind P et al.. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 2006; 313:1960-4; PMID:17008531; http://dx.doi.org/ 10.1126/science.1129139 [DOI] [PubMed] [Google Scholar]
- 26.Pages F, Galon J, Dieu-Nosjean MC, Tartour E, Sautes-Fridman C, Fridman WH. Immune infiltration in human tumors: a prognostic factor that should not be ignored. Oncogene 2010; 29:1093-102; PMID:19946335; http://dx.doi.org/ 10.1038/onc.2009.416 [DOI] [PubMed] [Google Scholar]
- 27.Llosa NJ, Cruise M, Tam A, Wicks EC, Hechenbleikner EM, Taube JM, Blosser RL, Fan H, Wang H, Luber BS et al.. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Disc 2015; 5:43-51; PMID:25358689; http://dx.doi.org/ 10.1158/2159-8290.CD-14-0863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dudley JC, Lin MT, Le DT, Eshleman JR. Microsatellite instability as a biomarker for PD-1 blockade. Clin Cancer Res: Off J Am Assoc Cancer Res 2016; 22:813-20; PMID:26880610; http://dx.doi.org/ 10.1158/1078-0432.CCR-15-1678 [DOI] [PubMed] [Google Scholar]
- 29.Mlecnik B, Bindea G, Angell HK, Maby P, Angelova M, Tougeron D, Church SE, Lafontaine L, Fischer M, Fredriksen T et al.. Integrative analyses of colorectal cancer show immunoscore is a stronger predictor of patient survival than microsatellite instability. Immunity 2016; 44:698-711; PMID:26982367; http://dx.doi.org/ 10.1016/j.immuni.2016.02.025 [DOI] [PubMed] [Google Scholar]
- 30.Margonis GA, Spolverato G, Kim Y, Karagkounis G, Choti MA, Pawlik TM. Effect of KRAS mutation on long-term outcomes of patients undergoing hepatic resection for colorectal liver metastases. Ann Surg Oncol 2015; 22:4158-65; PMID:26077912; http://dx.doi.org/ 10.1245/s10434-015-4587-z [DOI] [PubMed] [Google Scholar]
- 31.Topalian SL, Taube JM, Anders RA, Pardoll DM. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer 2016; 16:275-87; PMID:27079802; http://dx.doi.org/ 10.1038/nrc.2016.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell 1990; 61:759-67; PMID:2188735; http://dx.doi.org/ 10.1016/0092-8674(90)90186-I [DOI] [PubMed] [Google Scholar]
- 33.Lipsyc M, Yaeger R. Impact of somatic mutations on patterns of metastasis in colorectal cancer. J Gastrointest Oncol 2015; 6:645-9; PMID:26697197; http://dx.doi.org/ 10.3978/j.issn.2078-6891.2015.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spolverato G, Ejaz A, Azad N, Pawlik TM. Surgery for colorectal liver metastases: the evolution of determining prognosis. World J Gastrointest Oncol 2013; 5:207-21; PMID:24363829; http://dx.doi.org/ 10.4251/wjgo.v5.i12.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alamo P, Gallardo A, Di Nicolantonio F, Pavon MA, Casanova I, Trias M, Mangues MA, Lopez-Pousa A, Villaverde A, Vazquez E et al.. Higher metastatic efficiency of KRas G12V than KRas G13D in a colorectal cancer model. FASEB J 2015; 29:464-76; PMID:25359494; http://dx.doi.org/ 10.1096/fj.14-262303 [DOI] [PubMed] [Google Scholar]
- 36.Margonis GA, Kim Y, Sasaki K, Samaha M, Buettner S, Amini N, Pawlik TM. Activating KRAS mutation is prognostic only among patients who receive preoperative chemotherapy before resection of colorectal liver metastases. J Surg Oncol 2016; 114:361-7; PMID:27264476; http://dx.doi.org/ 10.1002/jso.24319 [DOI] [PubMed] [Google Scholar]
- 37.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E et al.. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012; 2:401-4; PMID:22588877; http://dx.doi.org/ 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E et al.. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013; 6:pl1; PMID:23550210; http://dx.doi.org/ 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zaitseva L, Murray MY, Shafat MS, Lawes MJ, MacEwan DJ, Bowles KM, Rushworth SA. Ibrutinib inhibits SDF1/CXCR4 mediated migration in AML. Oncotarget 2014; 5:9930-8; PMID:25294819; http://dx.doi.org/ 10.18632/oncotarget.2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang BY, Francesco M, De Rooij MF, Magadala P, Steggerda SM, Huang MM, Kuil A, Herman SE, Chang S, Pals ST et al.. Egress of CD19(+)CD5(+) cells into peripheral blood following treatment with the Bruton tyrosine kinase inhibitor ibrutinib in mantle cell lymphoma patients. Blood 2013; 122:2412-24; PMID:23940282; http://dx.doi.org/ 10.1182/blood-2013-02-482125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Treon SP, Tripsas CK, Meid K, Warren D, Varma G, Green R, Argyropoulos KV, Yang G, Cao Y, Xu L et al.. Ibrutinib in previously treated Waldenstrom's macroglobulinemia. New Engl J Med 2015; 372:1430-40; PMID:25853747; http://dx.doi.org/ 10.1056/NEJMoa1501548 [DOI] [PubMed] [Google Scholar]
- 42.Janeckova L, Pospichalova V, Fafilek B, Vojtechova M, Tureckova J, Dobes J, Dubuissez M, Leprince D, Baloghova N, Horazna M et al.. HIC1 tumor suppressor loss potentiates TLR2/NF-kappaB signaling and promotes tissue damage-associated tumorigenesis. Mol Cancer Res 2015; 13:1139-48; PMID:25934696; http://dx.doi.org/ 10.1158/1541-7786.MCR-15-0033 [DOI] [PubMed] [Google Scholar]
- 43.Van Rechem C, Rood BR, Touka M, Pinte S, Jenal M, Guerardel C, Ramsey K, Monte D, Begue A, Tschan MP et al.. Scavenger chemokine (CXC motif) receptor 7 (CXCR7) is a direct target gene of HIC1 (hypermethylated in cancer 1). J Biol Chem 2009; 284:20927-35; PMID:19525223; http://dx.doi.org/ 10.1074/jbc.M109.022350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scognamiglio G, De Chiara A, Di Bonito M, Tatangelo F, Losito NS, Anniciello A, De Cecio R, D'Alterio C, Scala S, Cantile M et al.. Variability in immunohistochemical detection of programmed death ligand 1 (PD-L1) in cancer tissue types. Int J Mol Sci 2016; 17:790; PMID:27213372; http://dx.doi.org/ 10.3390/ijms17050790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen YB, Mu CY, Huang JA. Clinical significance of programmed death-1 ligand-1 expression in patients with non-small cell lung cancer: a 5-year-follow-up study. Tumori 2012; 98:751-5; PMID:23389362; http://dx.doi.org/ 10.1700/1217.13499 [DOI] [PubMed] [Google Scholar]
- 46.Phillips T, Simmons P, Inzunza HD, Cogswell J, Novotny J, Taylor C, Zhang X. Development of an automated PD-L1 immunohistochemistry (IHC) assay for non-small cell lung cancer. Appl Immunohistochem Mol Morphol 2015; 23:541-9; PMID:26317305; http://dx.doi.org/ 10.1097/PAI.0000000000000256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen Y, Ramjiawan RR, Reiberger T, Ng MR, Hato T, Huang Y, Ochiai H, Kitahara S, Unan EC, Reddy TP et al.. CXCR4 inhibition in tumor microenvironment facilitates anti-programmed death receptor-1 immunotherapy in sorafenib-treated hepatocellular carcinoma in mice. Hepatology 2015; 61:1591-602; PMID:25529917; http://dx.doi.org/ 10.1002/hep.27665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Halama N, Michel S, Kloor M, Zoernig I, Benner A, Spille A, Pommerencke T, von Knebel DM, Folprecht G, Luber B et al.. Localization and density of immune cells in the invasive margin of human colorectal cancer liver metastases are prognostic for response to chemotherapy. Cancer Res 2011; 71:5670-7; PMID:21846824; http://dx.doi.org/ 10.1158/0008-5472.CAN-11-0268 [DOI] [PubMed] [Google Scholar]
- 49.Portella L, Vitale R, De Luca S, D'Alterio C, Ierano C, Napolitano M, Riccio A, Polimeno MN, Monfregola L, Barbieri A et al.. Preclinical development of a novel class of CXCR4 antagonist impairing solid tumors growth and metastases. PLoS One 2013; 8:e74548; PMID:24058588; http://dx.doi.org/ 10.1371/journal.pone.0074548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformat 2012; 13:134; PMID:22708584; http://dx.doi.org/ 10.1186/1471-2105-13-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Velcheti V, Schalper KA, Carvajal DE, Anagnostou VK, Syrigos KN, Sznol M, Herbst RS, Gettinger SN, Chen L, Rimm DL. Programmed death ligand-1 expression in non-small cell lung cancer. Lab Investig 2014; 94:107-16; PMID:24217091; http://dx.doi.org/ 10.1038/labinvest.2013.130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB et al.. Safety, activity, and immune correlates of anti–PD-1 antibody in cancer. New Engl J Med 2012; 366:2443-54; PMID:22658127; http://dx.doi.org/ 10.1056/NEJMoa1200690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Llosa NJ, Cruise M, Tam A, Wick EC, Hechenbleikner EM, Taube JM, Blosser L, Fan H, Wang H, Luber B et al.. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov 2015; 5:43-51; PMID:25358689; http://dx.doi.org/ 10.1158/2159-8290.CD-14-0863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Network CGAR. Integrated genomic analyses of ovarian carcinoma. Nature 2011; 474:609-15; PMID:21720365; http://dx.doi.org/ 10.1038/nature10166 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.






