ABSTRACT
We recently completed a phase I/IIa trial of RNActive® CV9201, a novel mRNA-based therapeutic vaccine targeting five tumor-associated antigens in non-small cell lung cancer (NSCLC) patients. The aim of the study presented here was to comprehensively analyze changes in peripheral blood during the vaccination period and to generate hypotheses facilitating the identification of potential biomarkers correlating with differential clinical outcomes post RNActive® immunotherapy. We performed whole-genome expression profiling in a subgroup of 22 stage IV NSCLC patients before and after initiation of treatment with CV9201. Utilizing an analytic approach based on blood transcriptional modules (BTMs), a previously described, sensitive tool for blood transcriptome data analysis, patients segregated into two major clusters based on transcriptional changes post RNActive® treatment. The first group of patients was characterized by the upregulation of an expression signature associated with myeloid cells and inflammation, whereas the other group exhibited an expression signature associated with T and NK cells. Patients with an enrichment of T and NK cell modules after treatment compared to baseline exhibited significantly longer progression-free and overall survival compared to patients with an upregulation of myeloid cell and inflammatory modules. Notably, these gene expression signatures were mutually exclusive and inversely correlated. Furthermore, our findings correlated with phenotypic data derived by flow cytometry as well as the neutrophil-to-lymphocyte ratio. Our study thus demonstrates non-overlapping, distinct transcriptional profiles correlating with survival warranting further validation for the development of biomarker candidates for mRNA-based immunotherapy.
Keywords: Biomarker candidates, cancer immunotherapy, mRNA-based vaccines, NSCLC, systems immunology, transcriptomics
Introduction
Lung cancer is the leading cause of cancer-related deaths world wide and 85–90% of those malignancies are classified as non-small cell lung cancer (NSCLC).1 For decades, clinical outcomes of patients with advanced NSCLC were poor with 5-y survival rates of less than 5%.2,3 However, immunotherapeutic approaches, in particular checkpoint inhibitors, have recently dramatically impacted the field of cancer therapy.4 The aim of immunotherapy is to induce and activate host immune responses against the tumor.5 Antibodies blocking the interaction of inhibitory T cell receptors CTLA-4 or PD-1 with their ligands were demonstrated to improve survival rates in patients with metastasized melanoma and NSCLC.6 In addition, encouraging long-lasting responses have been observed in a variety of solid tumors and hematologic malignancies after treatment with antibodies blocking PD-1 or PD-L1.7
Whereas these clinical results are highly encouraging, a substantial portion of patients fail to benefit from checkpoint inhibition treatment. Furthermore, immune-mediated adverse effects occur even in patients who do not respond to treatment.8 Combined blockade of PD-1 and CTLA-4 has resulted in improved response rate and progression-free survival in melanoma patients compared to CTLA-4 blockade alone and in patients with PD-L1 negative tumors also compared to PD-1 blockade. However, severe immune-mediated side effects were more frequent with the combined treatment.9 A likely cause for immune-mediated adverse effects is non-specific T cell activation and inhibition of regulatory CD4+ T cells.10 Also, checkpoint inhibition requires that tumor-specific T cells are present prior to the treatment in order to result in antitumor activity.4 Central immunological tolerance mechanisms delete many T cells with high avidities to tumor antigens, resulting in a depleted tumor-specific T cell repertoire too limited to robustly control the tumor.5 Nonetheless, spontaneous T cell responses against tumor antigens frequently develop in most cancer patients11 and the depleted T cell repertoire can still be effectively harnessed for clinical benefit. In this context, active therapeutic vaccination, which aims at priming novel tumor-specific immune responses in addition to boosting pre-existing ones, could play a critical role to alleviate this problem.12 Due to its specific mechanism of action, active immunotherapy could enhance the antitumor effects of checkpoint inhibition without the substantial increase in immune-mediated adverse effects seen with combined checkpoint blockade.
Among vaccination platforms currently under evaluation in clinical trials, mRNA-based vaccines represent highly attractive candidates.13-15 RNActive® is a novel mRNA-based vaccination technology with self-adjuvanting activity, engineered with chemically unmodified, natural nucleotides. The RNActive® cancer immunotherapies employed in our studies consist of two components: free and protamine-complexed, sequence-optimized, full-length mRNA to support both high antigen expression and immune stimulation, predominantly mediated by toll-like receptor 7 (TLR-7).16-18 In pre-clinical experiments, intradermal injection of the two-component mRNA vaccines encoding tumor-associated antigens induced boostable and balanced effector and memory immune responses including antigen-specific CD4+ T helper cells, cytotoxic CD8+ T cells, memory T cells, and antibody-producing B cells. These responses were associated with strong antitumor effects and complete protection against antigen-positive tumor cells.19-24 Further pre-clinical studies provided evidence for synergistic effects of the two-component mRNA vaccine combined with radio therapy as well as checkpoint-inhibiting anti-CTLA-4 antibodies.18,25
Moreover, we have successfully completed a first-in-man phase I/IIa trial in patients with advanced castration-resistant prostate cancer. This study showed that repeated immunizations with two-component mRNA vaccines, encoding prostate cancer-associated antigens, induced vaccine-specific immune responses and were well tolerated showing a favorable safety profile.26 We have also successfully completed a phase I/IIa trial, conducted in patients with advanced NSCLC. In this trial, patients were treated with RNActive® CV9201 vaccine, which targets five known NSCLC-associated cancer antigens: cancer/testis antigen 1B (New York esophageal squamous cell carcinoma, NY-ESO1), melanoma antigen family C1 (MAGE-C1), MAGE-C2, baculoviral inhibitor of apoptosis repeat-containing 5 (survivin), and trophoblast glycoprotein (5T4).
These pioneering studies represent some of the first human trials employing mRNA-based vaccine technology to treat highly aggressive cancer, such as NSCLC. To investigate potential mechanisms of action and to identify immune correlates of protection, we sought to comprehensively monitor gene expression changes post vaccination. We therefore performed whole-genome transcriptome analyses of peripheral blood mononuclear cell (PBMC) samples derived before and after initiation of mRNA vaccination in NSCLC patients. Gene expression data can elucidate the underlying biology of vaccine-associated changes in blood, including alterations of cell subset composition and biological processes including signaling pathways, proliferation, apoptosis, cytotoxicity and others. Furthermore, transcriptome analyses are frequently performed in vaccine studies to identify correlates of immune protection, which can be further assessed as potential biomarker candidates.27-31 Samples for gene expression profiling described in this manuscript were available from 22 of the 32 stage IV NSCLC patients enrolled in the phase IIa part of the clinical trial. These patients had been treated with CV9201 at a dose of 320 µg of each individual mRNA component encoding one of the five antigens and at a total dose of 1,600 µg per application. Since the primary aim of this first-in man dose escalation clinical trial was to assess safety and immunogenicity of CV9201, no placebo arm was included in the study. The aim of this exploratory gene expression profiling analysis was to generate hypotheses regarding biological changes in peripheral blood after repeated mRNA vaccinations compared to baseline.
Utilizing blood transcriptional modules (BTMs) previously established by Li et al.32, we show an enrichment of distinct modules in the post vaccine samples. In comparison with baseline, we found an upregulation of transcriptional modules consistent with myeloid cells and inflammation in one group of patients and an upregulation of T and NK cell modules in another group of patients. Notably, these transcriptional changes were mutually exclusive and associated with different clinical outcomes. The transcriptional signatures identified in our study require further evaluation in controlled studies to explore their potential as biomarker candidates associated with mRNA-based immunotherapy. Moreover, these data will help inform future immunotherapeutic approaches in cancer trials.
Results
Enrichment of transcriptional modules consistent with adaptive immunity post vaccination
Demographic and clinical data of the 22 stage IV NSCLC patients investigated in this gene expression study is provided in Table 1 and Table S1. Patients received intradermal vaccinations at weeks 1, 2, 3, 5 and 7 with a total dose of 1,600 μg mRNA per vaccination (320 µg per antigen). Blood sampling by venipuncture for PBMC analysis was performed at day 0 (baseline), week 5 (2 weeks post third vaccination) and week 9 (2 weeks post 5th vaccination). Within the first 17 mo after initiation of mRNA treatment, 15 deaths had been reported in our study subgroup (Fig. S1A). Notably, the next death occurred 15 mo later and six other patients were still alive at their final follow-up visit. Due to this gap of 15 mo, we defined patients with a survival exceeding 30 mo as long-term survivors, whereas the remaining patients were classified as short-term survivors. The median survival of the long-term survivors at their last visit was 35 mo, whereas the median survival of the short-term survivors was 7.4 mo. We first sought to identify gene expression differences between short-term and long-term survivors.
Table 1.
Demographic overview of the investigated NSCLC patient cohort. IQR – interquartile range. N.D. – not determined.
| Gender | |
|---|---|
| female | 45.5% (10/22) |
| male | 54.5% (12/22) |
| Age | |
| [years] (IQR) | 64 (55.75 – 73.25) |
| Tumor | |
| Adenocarcinoma | 63.6% (14/22) |
| Squamous cell carcinoma | 18.2% (4/22) |
| Large cell carcinoma | 9.1% (2/22) |
| Mixed | 4.5% (1/22) |
| N.D. | 4.5% (1/22) |
| Metastasis | |
| Lymph node | 63.6% (14/22) |
| Bone | 18.2% (4/22) |
| Overall survival | |
| [days] (IQR) | 345 (194.25–1049.75) |
Traditionally, supervised analysis of whole-genome transcriptional data frequently involves the identification of differentially expressed genes, followed by pathway and annotation enrichment analyses. We initially sought to apply this approach to our expression data. However, after correcting for multiple testing, we were unable to identify differentially expressed genes in a number of comparisons (data not shown). A likely explanation could be that the investigated subset of patients was too heterogeneous with regards to histological characteristics of the tumor, the prior anticancer treatment and concomitant diseases of the patients, resulting in a high transcriptional heterogeneity. To accommodate the high degree of heterogeneity in our samples, we sought to apply a modular approach to our expression data analysis by summarizing coordinately expressed genes into modules. Such approaches provide increased sensitivity over identifying differentially expressed genes on a gene by gene approach by statistical and/or fold change methods.33 We adopted the BTMs developed by Li et al. since these modules were specifically designed to analyze human blood transcriptomes in the context of vaccine-elicited immune responses.32
To investigate the transcriptional differences between short-term and long-term survivors, we performed gene set enrichment analyses (GSEA) utilizing BTMs as gene sets. We found that long-term survivors exhibited an enrichment of transcriptional modules associated with effector lymphocytes, in particular T cells (Figs. S1B–D). This enrichment of T cell modules was observed at both post vaccine time points (week 5 and week 9) and also at baseline (week 0). In addition, NK cell and B cell modules were enriched in long-term survivors at weeks 5 and 9, respectively (Figs. S1C and D).
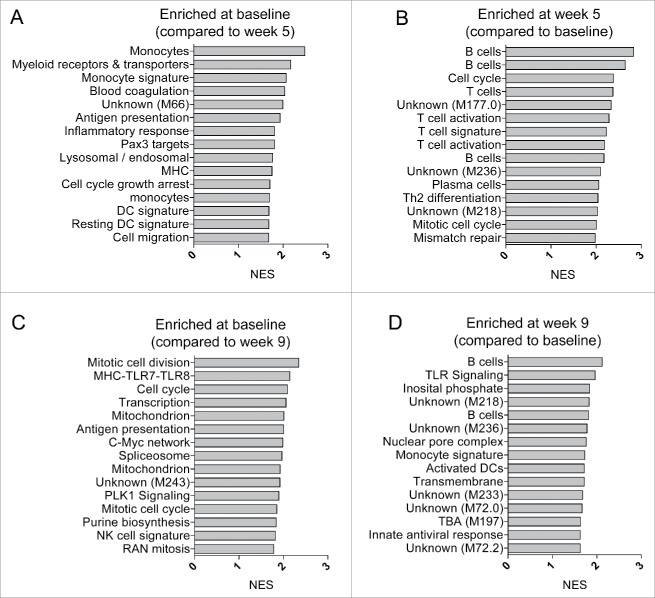
We next sought to determine transcriptional changes post therapeutic vaccination with CV9201 in all 22 subjects. Pre-ranked transcript lists were generated based on paired t-test statistics comparing week 5 samples and week 9 samples with their matching week 0 samples. We performed GSEA on these pre-ranked lists utilizing BTMs as gene sets. Among the top 15 most enriched BTMs at week 5, there was an enrichment of six modules representing T cells, including one module consisting of genes specific for Th2 differentiation and another module with cell cycle genes specific to activated CD4+ T cells (Fig. 1B). We also found four modules reflecting B cells including one module containing genes highly expressed in plasma cells and genes encoding immunoglobulins (Fig. 1B). The enrichment of B cell modules was partially retained in the week 9 samples (Fig. 1B).
Figure 1.
Transcriptional modules consistent with an adaptive immune response profile are enriched at week 5 post initiation of CV9201 treatment. Gene set enrichment analyses contrasting baseline week 0 samples to post vaccine week 5 samples (A), and samples derived from week 5 to 0 (B), week 0 to 9 (C) and week 9 to 0 (D), respectively. Genes were pre-ranked based on t values from paired Student's t-tests. NES – Normalized enrichment scores.
Notably, week 0 in comparison with week 5 samples exhibited an enrichment of BTMs consistent with myeloid cells, dendritic cells, antigen presentation and inflammation (Fig. 1A). In contrast, the baseline week 0 to week 9 revealed a relative enrichment of modules associated with cell cycle and transcriptional activation. The data indicate a transient upregulation of transcriptional modules consistent with adaptive immune responses post therapeutic vaccination and a downregulation of myeloid cell- and inflammation-related modules. Furthermore, these data show that the use of BTMs in combination with GSEA represents a sensitive analytical tool to detect post treatment differences in a subgroup of heterogeneous patients.
Distinct patient clustering based on BTM activity changes at week 5
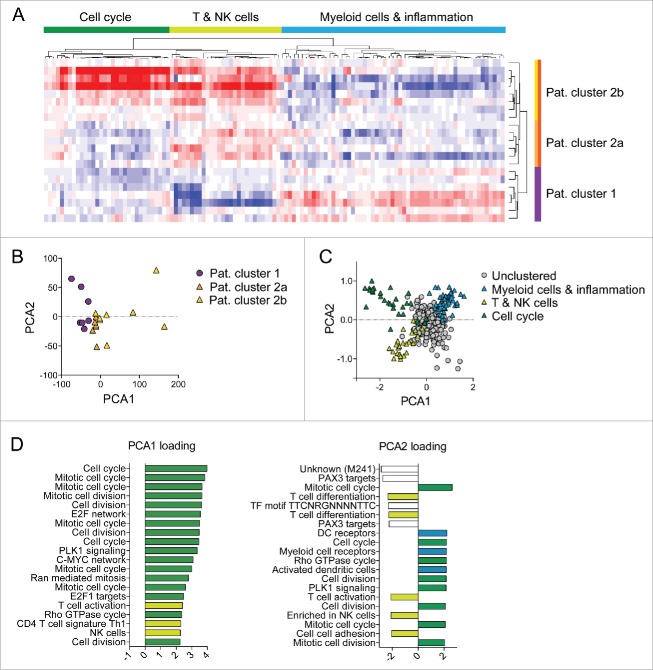
In addition to the supervised approaches, we also applied an unbiased approach to expression data analysis. We first calculated the BTM activity scores for each of the 346 transcriptional modules. These BTM activity scores represent the mean expression values of all genes contained in a single module. We used hierarchical clustering to identify changes in module activity scores between week 0 and 5. Clustering was performed for both subjects and modules. Notably, patients segregated into two distinct groups (Fig. 2A). Importantly, the clustering of patients was predominantly driven by two individual sets of modules. One cluster of modules predominantly contained T and NK cell modules, whereas the other cluster contained modules reflecting myeloid cells, antigen presentation and immune activation. We furthermore noticed that patients upregulating T and NK cell modules at week 5 were further divided into two sub-clusters, based on their expression pattern of cell cycle modules (Fig. 2A). We investigated whether the patient clustering was associated with distinct histological subtypes of NSCLC (Table S1) and were unable to identify such relationships.
Figure 2.
Transcriptional changes post treatment cluster patients into distinct groups. (A) Week 5 to 0 BTM activity score differences were calculated and unsupervised hierarchical clustering was performed. Red indicates up- and blue indicates downregulation of BTM activity scores at week 5 compared to 0. (B, C) Principal component analysis was performed on week 5 to 0 BTM activity scores. Patient (B) and module clustering (C) in PCA space are shown. (D) Weighting coefficients of BTMs for PC1 and PC2 are shown.
We next addressed the question if clustering of patients could be confirmed using principal component analysis (PCA), a widely used method to visualize high-dimension data. Analogously to our unsupervised hierarchical clustering analysis, we used week 5 to week 0 differences of the 346 BTM activity scores as variable values. The first two principal components cumulatively accounted for approximately 51% of variance in our data set. The loading factors of PC1 and PC2 revealed that most of the variance could be attributed to cell cycle modules (Fig. 2D). Thus, patients in PC1 and PC2 space mostly separated based on their differential expression of genes associated with cell cycle and mitosis processes. In addition to these, we observed modules for T and NK cells contributing to PC1 and PC2 as well as modules associated with myeloid and antigen-presenting cells (Fig. 2D). Using the same data set, we also clustered the modules and found distinct clustering of the three sets of BTMs responsible for the clustering of patients (Fig. 2C). In fact, BTMs reflecting monocytes and immune activation on the one hand and BTMs representing T and NK cells on the other hand clustered in opposite corners in the PC1/PC2 projection. We were thus able to confirm the results from the hierarchical clustering using PCA as an independent analysis method.
Non-overlapping transcriptional changes in segregated patients after CV9201 treatment
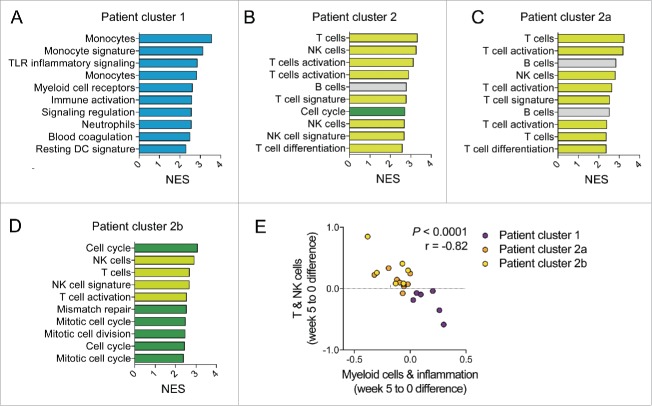
The results from the unsupervised clustering analyses strongly suggested divergent transcriptional changes in distinct subsets of patients. To gain additional insight into the transcriptional changes after vaccination, we separated the patients into two groups according to their clustering pattern. For each of these two groups, we generated ranked transcript lists based on paired t-test statistics comparing week 5 with their matching week 0 samples. GSEA was performed on these lists using BTMs as gene sets. As expected, at week 5, patients belonging to cluster 1 exhibited a strong upregulation of transcriptional modules associated with monocytes, myeloid cells, antigen presentation and immune activation (Fig. 3A). To gain further insight into the single genes enriched within these modules, we extracted the leading edge genes of the top 10 most enriched BTMs. Among the transcripts most frequently enriched in the top modules, we identified genes associated with myeloid cells, including genes encoding for toll-like receptors 2 and 5 (TLR2, TLR5), chemokine receptor type 1 (CCR1), formyl peptide receptors 1 and 2 (FPR1, FPR2), colony stimulating factor 3 receptor (CSF3R) and complement component 5a receptor 1 (C5AR1) (Figs. S2A and B). As expected, all of these genes were upregulated in cluster 1 patients at weeks 5 and 9 compared to baseline (Fig. S2B).
Figure 3.
Transcriptional changes in segregated patients post CV9201 treatment are non-overlapping. Gene set enrichment analyses contrasting post vaccine week 5 to week 0 samples was performed utilizing pre-ranked gene lists based on t values from paired Student's t-tests. Patient groups were derived from the hierarchical clustering analysis shown in Fig. 2A. Normalized enrichment scores of the top 10 most enriched BTMs are shown for patients from cluster 1 (A), complete cluster 2 (B), cluster 2a (C) and cluster 2b (D). (E) Mean of week 5 to 0 BTM activity score changes were calculated for T and NK cell modules as well as myeloid cell and inflammation modules. Pearson correlation analysis was performed.
In contrast, the top 10 most enriched modules in patients belonging to cluster 2 were characterized by upregulation of BTMs associated with T cells, NK cell and B cells. In addition, there was also one cell cycle BTM within the top 15 enriched BTMs at week 5 in patients of cluster 2. In this group of patients, the myeloid-leading edge genes were temporarily downregulated at week 5 compared to baseline except for NFE2 (Fig. S2A). Based on the patient clustering results (Fig. 2A), we further sought to dissect patients within cluster 2 and divided them into subgroups 2a and 2b. Transcriptional upregulation of T and NK cell modules were found in both subclusters (Figs. 3C and D). However, upregulation of B cell modules was only found in patients belonging to cluster 2a, whereas cell cycle and mitosis modules were only enriched in cluster 2b patients.
The distinct patient clustering driven by myeloid or T and NK cell modules suggested that these transcriptional changes were mutually exclusive. To test this hypothesis, we first calculated the week 5 to 0 differences for each of these two sets of BTMs. We then determined the mean of all BTM week 5 to 0 differences contained in the myeloid cluster and the T and NK cell cluster. Correlation analysis revealed a highly significant, inverse relationship between these two means (Fig. 3E). These results therefore indicate that in our subjects, the upregulation of BTMs consistent with myeloid cells, or with T and NK cells is mutually exclusive. Thus, segregated patients were characterized by an enrichment of non-overlapping transcriptional modules post vaccination.
Prolonged progression-free and overall survival in patients with NK and T cell BTM enrichment at week 5
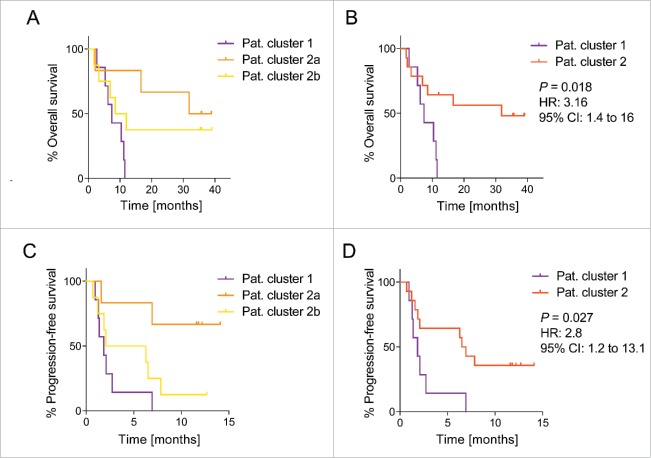
Increased expression of genes associated with monocytes and other myeloid cells could indicate rising levels of circulating myeloid-derived suppressor cells (MDSCs). These heterogeneous subsets of immature myeloid cells have been described in NSCLC patients and were linked with unfavorable clinical outcomes.34-36 Furthermore, transcriptional profiling studies performed in PBMCs derived from NSCLC patients indicated a positive correlation between survival and the expression of T cell-associated genes.37-39 We therefore hypothesized that patients belonging to cluster 2 with a higher activity of T and NK cell modules after vaccination should exhibit a better clinical outcome compared to patients in cluster 1. This was indeed the case as all patients in our cohort with a higher activity of myeloid cells at week 5 compared to week 0 were short-term survivors and died within less than a year (Figs. 4A and B). We found similar results when we compared the progression-free survival between cluster 1 and cluster 2 patients (Figs. 4C and D). In our cohort comprising 22 patients, there were seven patients with a prolonged survival exceeding 30 mo. All of these long-term survivors were part of patient cluster 2. We were thus able to identify a significant difference in overall and progression-free survival between patients belonging to cluster 1 and cluster 2 (Figs. 4B and D).
Figure 4.
Patients with NK and (T)cell BTM enrichment post CV9201 treatment are associated with a prolonged survival. Kaplan–Meier curves for overall survival (A) and progression-free survival (C) are shown for patients belonging to cluster 1, 2a and 2b. Overall survival (B) and progression-free survival rates (D) are shown for patients belonging to cluster 1 compared to patients belonging to cluster 2. Log-rank test was performed to calculate the hazard ratio and p value.
Changes in the transcriptional signature correlate with changes in lymphocyte subsets and neutrophil/lymphocyte ratio
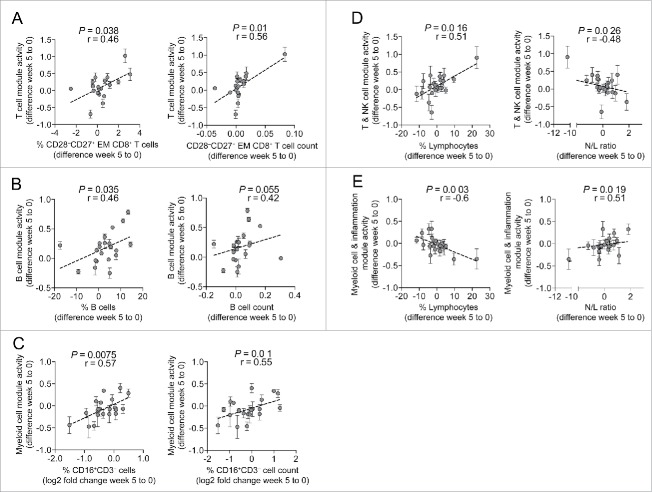
The transcriptional changes at week 5 post treatment could either be the result of changes in the cellular composition in peripheral blood or alternatively indicate transcriptional activation of distinct leukocyte subsets or a combination of both. To address this question, we performed correlation analyses between selected BTM activity scores and lymphocyte phenotyping data measured by flow cytometry. Differentiation states of CD4+ and CD8+ T cells were assessed based on their cell surface expression of CCR7 and CD45RA, and we defined effector memory T cells as CCR7−CD45RA− cells.40,41 We further dissected these effector memory cells based on their expression patterns of CD27 and CD28. Intriguingly, changes in T cell module activity scores derived from the top 15 most enriched BTMs in patient cluster 2a (Fig. 3C) were positively correlated with absolute and relative changes of CD27+CD28− effector memory CD8+ T cells (Fig. 5A). Consistent with this finding, a number of genes associated with T effector functions were enriched in cluster 2 patients at week 5 including CCL5, CTSW, the genes encoding for gramzymes A, B, H, M and K (GZMA, GZMB, GZMH, GZMM, GZMK), Perforin (PRF1), KLRF1 and the transcription factors T-bet (TBX21) and LEF1 (Figs. S2C and D). Similarly, there was a positive correlation between changes in B cell module activity scores and changes in relative and absolute B cell numbers (Fig. 5B). Although flow cytometric data were not obtained during the course of the clinical trial to allow precise identification of myeloid cells, CD16+CD3−CD 56− lymphocytes to a large degree consist of HLA-DR-expressing myeloid cells as well as some NK cells devoid of CD56 (personal observation). Using these criteria, we were able to identify a positive association between changes in myeloid BTM scores with relative and absolute changes in CD16+CD3−CD56− cells (Fig. 5C).
Figure 5.
Changes in the transcriptional signature correlate with changes in lymphocyte subsets. (A) Changes in T cell module activity scores were correlated with changes in either relative or absolute numbers of CD28−CD27+ effector memory CD8+ T cells. (B) Changes in B cell module activity scores were correlated with changes in relative or absolute numbers of B cells. (C) Myeloid cell modules were correlated with changes in relative or absolute numbers of CD16+CD3−CD56− cells. (D) Changes in BTM cluster 2a were correlated with changes in lymphocyte frequencies or N/L ratio. (E) Changes in BTM cluster 1 were correlated with changes in lymphocyte frequencies or N/L ratio. Spearman's rank correlation test was performed for all analyses. Error bars indicate standard deviations of changes in BTM activity scores. EM – effector memory.
In addition, the cellular composition of peripheral blood, in particular lymphocyte and neutrophil frequencies, was routinely assessed during the clinical trial as safety parameters by differential blood count. We determined the neutrophil-to-lymphocyte (N/L) ratio, a previously described marker with prognostic value in various cancer indications, including NSCLC.42-46 Module activity changes in cluster 2a were linked to the changes in lymphocyte frequencies and inversely correlated with changes in the N/L ratio (Fig. 5D). Conversely, transcriptional changes in BTM cluster 1 were negatively associated with changes in lymphocyte percentages but positively correlated with changes in the N/L ratio (Fig. 5E). In summary, these data suggest that the detected transcriptional changes may be in part the result of changes in the cellular composition of peripheral blood cells. Furthermore, by demonstrating correlations with the N/L ratio, we show that our transcriptional signatures are consistent with previously established prognostic biomarkers in cancer.
Discussion
Active cancer immunotherapy based on CureVac's mRNA technology was previously shown to be safe and well tolerated and potently induced vaccine antigen-specific immune responses.26 Similar observations were made in the clinical trial of CV9201 in NSCLC described here (manuscript in preparation). In a subset of this NSCLC trial, we sought to identify transcriptional differences post vaccination in 22 stage IV lung cancer patients. The aim of the study was to generate hypotheses regarding underlying biological processes associated with protection detectable in peripheral blood and to identify potential biomarker candidates for mRNA-based immunotherapies in NSCLC.
Whole-genome transcriptional profiling is a widely used approach to identify molecular signatures correlating with vaccine-induced immune protection and to generate hypotheses for biomarkers in clinical cancer trials.47 Since transcriptome profiling by microarrays was introduced approximately 15 y ago, many different analysis tools have been developed to process the data.48,49 Traditional supervised approaches to analyze transcriptional data frequently involve identifying differentially expressed genes followed by pathway and annotation enrichment analyses.50 Although pathway analyses for gene expression data have been helpful to elucidate the biological context in many experimental settings, we were confronted with a number of limitations when applying this approach to our PBMC transcriptome data. PBMC transcriptomes contain expression signals from a variety of cells, which limits the sensitivity of pathway analyses. Also, due to the heterogeneous nature of NSCLC disease,1 we expected and indeed encountered a substantial degree of additional heterogeneity in our samples. As pointed out by Li et al., a substantial part of the knowledge contained in pathway databases are of limited value to assess PBMC transcriptomes, due to the sometimes artificial experimental conditions, in which pathways had been identified, e.g., cell lines, knock-out models, etc. Furthermore, not all genes in a given pathway are necessarily regulated at the level of transcription.32
One of the inherent challenges in transcriptome data analysis is to reduce the dimensionality and the complexity of the data set in a meaningful way without losing critical biological information. Summarizing genes into transcriptional modules has become a powerful tool to assess gene networks.51 For our analyses, we used the BTMs published by Li et al.32 These modules were constructed by integrating large amounts of publically available gene expression data generated on peripheral blood cells. Because these modules were generated from PBMC expression data, we found the annotation to be more informative than other available modules and libraries. In addition, these modules offered sufficient plasticity to be applied to a disease setting as heterogeneous as lung cancer, while still enabling us to obtain immunologically relevant information. The use of BTMs, especially in conjunction with GSEA, is a powerful and sensitive approach, which yielded novel biological insights despite our relatively small cohort size.
Using unsupervised clustering based on BTM activity score changes over time, we identified a segregation of vaccinated NSCLC patients into two distinct groups, each with a discrete molecular signature. We demonstrated that these transcriptional changes after vaccination correlated with our flow cytometric phenotyping data as well as the N/L ratio. These findings suggest that the gene expression differences could be a result of changes in the cellular composition of PBMCs although transcriptional activation of leukocyte subsets cannot be ruled out.
The first patient group exhibited an upregulation of myeloid cell-associated and inflammation-related modules, which could indicate an accumulation of monocytes and other myeloid cells (e.g., neutrophils) in the periphery. A possible explanation for this phenomenon could be an enrichment of a heterogeneous set of immuno-suppressive cells known as MDSCs. MDSCs are immature myeloid cells that accumulate due to an abnormal differentiation process in disease settings.52 Expansion of MDSCs was observed in patients of virtually all cancer types53 including NSCLC.34,54 In general, the frequency of MDSCs in peripheral blood depends on the tumor burden and elevated numbers of these cells correlate with disease progression and overall poor prognosis.55 In addition, the upregulation of myeloid cells was accompanied by an enrichment of inflammation and immune activation modules (Fig. 3A). Cancer-related inflammation has long been recognized to be one of the immunological hallmarks of disease and one of the key drivers of tumor promotion.56,57 Circulating inflammation markers were described to have prognostic capacity in lung cancer.58,59 In accordance with these observations, we found that all patients with a transcriptional enrichment of myeloid and immune activation modules at week 5 died within 1 y. In contrast, patients belonging to cluster 2 exhibited an upregulation of modules containing genes reflecting T, B and NK cell biology. All long-term survivors belonged to patient cluster 2. Increased activity of these modules was associated with a general increase of lymphocyte frequencies in peripheral blood. Notably, enriched T cell modules were associated with a distinct effector memory CD8+ T cell subset with a CD27+CD28− phenotype. Effector memory CD8+ T cells are heterogeneous and further subsets can be delineated based on the surface expression of CD27 and CD28.60,61 Effector memory CD8+ T cells lacking surface expression of the B7-receptor CD28 are more likely to constitutively produce effector molecules, including granzyme B, perforin and IFNγ compared to their CD28+ counterpart.60 In support of this notion, we observed an upregulation of a number of genes associated with T effector functions in cluster 2 patients at week 5 compared to week 0. These genes included CCL5, CTSW, GZMA, GZMB, GZMH, GZMM, GZMK, PRF1, KLRF1, TBX21 and LEF1. Importantly, CD28, though part of several enriched T cell modules in patient cluster 2, was not among the enriched leading edge genes, which is in accordance with the finding that alterations in the T cell module activity scores correlated with the frequency of CD27+CD28− effector memory CD8+ T cells. Although long-term survivors were characterized by increased T cell module activity scores in comparison with short-term survivors, they further exhibited an upregulation of effector cell modules (predominantly T and NK cell modules) after treatment.
Our findings are corroborated by previously published studies reporting transcriptional analyses of peripheral blood samples in NSCLC.37,38,62 These studies indicate that lung cancer induces tumor-specific molecular signatures, which can be detected in the periphery. Analysis of prognostic genes, which are immune cell type-specific revealed a significant association of myeloid cells and T cells with survival.38,63 All of the 21 myeloid cell-associated genes with prognostic value were exclusively negatively correlated with survival. Among 23 prognostic T cell genes, eight genes were associated with better survival, including LEF1 and GZMK, which were also enriched in our cluster 2 patient group.38 In contrast to our longitudinal study, these results were generated in a cross-sectional study. In addition, the study by Kossenkov et al. analyzed NSCLC patients with clinical stages I–IIIA prior to surgical resection, whereas our study only included advanced stage IV patients. In spite of this discrepancy, analogous immune cell-specific gene signatures could be identified indicating similar biological processes in NSCLC patients at various disease stages. These data are in accordance with our finding that long-term survivors compared with short-term survivors displayed elevated of T and NK cell transcriptional module activity at baseline and subsequent time points. We furthermore showed that changes in activity scores of myeloid cell modules and T cell modules correlated with the N/L ratio, a well-established biomarker in various cancer indications.42-46 Our data, however, are more informative than the N/L ratio. For example, we show that patients in cluster 2 with increases in T and NK cell module activity scores at week 5 are heterogeneous and can further be dissected based on expression changes of cell cycle and mitosis genes. Further studies are required to confirm whether these signatures can be reproduced in independent study cohorts and whether changes in cell cycle module activities correlate with differential clinical outcomes.
Changes in gene expression at week 5 compared to baseline could reflect effects of RNActive® treatment, treatment-independent differential disease progression or other unknown variables. Notably, most direct transcriptional perturbations in peripheral blood induced by vaccinations, such as immunizations against flu antigens, seem to occur rapidly after treatment (e.g., after 24 h) and are very transient in nature.64,65 In contrast, our post vaccine blood samples were obtained 2 weeks after the third or fifth vaccination, respectively. The transcriptional changes over time in our study therefore seem to be the result of effects lasting in the range of weeks instead of days. It is tempting to speculate that the increase of T and NK cell modules at week 5 compared to week 0 could be a result of CV9201 immunotherapy and thus represent a candidate biomarker for clinical benefit post treatment. In support of this hypothesis, we found that all long-term survivors were part of patient cluster 2 displaying a further increase of T and NK cell module activity at week 5 compared to baseline. Conversely, the increase of myeloid cell- and inflammation-related genes in some patients could reflect lung cancer disease progression since MDSC expansion correlates with tumor burden.
In the absence of an untreated control cohort, we cannot address the question of whether mRNA-based immunotherapy with CV9201 can increase T cell gene expression and reduce myeloid module activity and whether it induces clinical benefit. In addition, our data set is limited by a relatively small cohort size as well as the absence of an independent testing cohort, thus precluding extensive cross-validation or analysis of the robustness of these molecular signatures. Our findings certainly warrant further investigation in randomized controlled clinical trials. Placebo-controlled clinical trials in NSCLC patients addressing the clinical efficacy of CV9201 treatment in combination with chemo- and radiotherapy are currently in preparation. These studies will allow formal testing of the hypotheses generated with these initial results, with the aim to identify reliable, immunological correlates and biomarkers with predictive value in cancer immunotherapy.
Material and methods
Two-component mRNA cancer immunotherapy CV9201
CureVac AG proprietary technology has generated mRNA molecules with increased stability and translatability (patents EP1392341B1, EP1604688B1). All mRNA vaccines used in the present study were produced in accordance with this technology and Good Manufacturing Practice guidelines (CureVac AG, Tübingen, Germany). The RNActive®-based vaccine CV9201 consisted of a mixture of 50% free mRNA (component 1) and 50% mRNA complexed with protamine (MEDA Pharma) at a weight ratio of 2:1 (component 2). First, mRNA was complexed by the addition of protamine-Ringer lactate solution and, after stable complexation, free mRNA was added.19 CV9201 consists of RNActive®-based mRNA components encoding the NSCLC-specific antigens NY-ESO1, MAGE-C1, MAGE-C2, Survivin, and 5T4. The five antigens were formulated separately.
Patients and study design
Study participants were at least 18 y of age with advanced, stage IIIB or IV NSCLC (UICC V6.0 criteria) with stable disease or objective response according to Response Evaluation Criteria in Solid Tumors (RECIST), version 1.0, after first-line treatment (chemotherapy, chemo-radiotherapy) for advanced, unresectable or metastatic disease.66 Patients must have had a life expectancy of >6 mo as assessed by the investigator. Eligibility criteria further included an Eastern Cooperative Oncology Group (ECOG) performance status of ≤ 1 and adequate renal, hepatic, cardiac and bone marrow function. Informed consent was obtained from all patients prior to any study-related procedures. The study was conducted in accordance with the Declaration of Helsinki and Good Clinical Practice guidelines and was registered at the European Clinical Trials Database with EudraCT number 2008-007785-39. The protocol was approved by ethics committees/review boards of participating study sites. The primary objective of the clinical trial was assessment of safety and secondary objectives were evaluation of induction of immune responses.
Patients were vaccinated at weeks 1, 2, 3, 5 and 7 with 320 μg mRNA of each antigen for a total dose of 1,600 μg mRNA per vaccination. Two intradermal injections of each antigen were applied to two injections sites, one into the thigh and one into the upper arm of the same body half. Injection sites of each antigen were rotated clockwise at different vaccination days. After end or discontinuation of vaccination, further antitumor therapy was at the discretion of the investigator. Peripheral blood sampling was performed at day 0 (baseline), week 5 (2 weeks post third vaccination) and week 9 (2 weeks post 5th vaccination). Transcriptome profiling analysis was restricted to stage IV NSCLC patients treated in the phase IIa part of the study and subjects were chosen based on sample availability. Demographic patient data is provided in Table 1 as well as Table S1.
Preparation of peripheral blood mononuclear cells (PBMCs)
To accommodate the challenges underlying a multi-centric clinical trial and to consistently ensure a high quality of cells, PBMC preparation from blood samples was required to be performed within 6 h after venipuncture. A study-specific and standardized PBMC preparation kit (Interlab GmbH, Germany) containing all necessary materials, buffers and media was used and PBMC purification were carried out by trained personnel in validated central laboratories according to standard operating procedures (SOPs). Briefly, blood was transferred from heparin tubes to Leucosep tubes and centrifuged. PBMCs were harvested from the interphase and washed twice. Viability was assessed by Trypan Blue staining, yield and red blood cell contamination was assessed by Tuerk's solution for cell counting. After cell counting, PBMCs were slowly frozen in serum-free Cryo SFM media (Promocell, Heidelberg, Germany) using Mr. Frosty freezing devices (VWR, Darmstadt, Germany) at −80° for 24 h before transferring frozen cells to liquid nitrogen. Blood neutrophil and lymphocyte counts were determined from peripheral blood using validated diagnostic assays at a central laboratory provider (InterLab Services worldwide GmbH, Munich, Germany).
Gene expression profiling
RNA was isolated from re-thawed PBMCs frozen in PBS using RNEasy (Qiagen) according to the manufacturer's instructions. To ensure that re-thawing of PBMC samples did not affect any down-stream analyses, RNA quality control was performed on a 2100 Bioanalyzer (Agilent Technologies) and only samples with a RIN value ≥6.5 were further processed. Cyanine-3-labeling of cRNA and hybridization using the Agilent SurePrint G3 Human Gene Expression 8×60K v2 Microarray platform was performed by IMGM, Munich, Germany according to manufacturer's manuals (Agilent Technologies). Raw data was processed using Feature Extraction 10.7.3.1 and GeneSpring GX 12.6.1. Quantile normalization was performed and log2-transformed data was filtered for probes detectable in at least one sample resulting in gene expression data for 48,605 probes.
Calculation of blood transcriptional module (BTM) activity scores and gene set enrichment analysis (GSEA)
To address the expected heterogeneity of the samples derived from individual patients, we sought to apply a modular approach to gene expression analysis developed by Li et al.32 These 346 BTMs represent sets of genes, which are transcriptionally coordinately expressed. Most of these BTMs reflect sets of genes involved in the regulation of an immunological process, such as signaling pathways or represent sets of genes specifically enriched in immune cells, such as T or B cells. BTM activity scores were determined, which represent the mean expression of all genes contained in a module. Following the example of Li et al. probes with the highest average expression across all samples were chosen for genes targeted by multiple probes.32
GSEA was performed as described previously.67 Ranking of gene lists was performed using unpaired Student's t-test for cross-sectional comparisons. For longitudinal contrasts, pre-ranked gene lists were generated based on t scores obtained from paired Student's t-test analyses.
Phenotyping of lymphocyte subsets
After thawing PBMCs in PBS containing the endonuclease benzonase (Novagen/Merck Milipore Schwalbach, Germany), cells were directly stained in PBEA buffer (PBS, 0.5% BSA, 2 mM EDTA and 0.01% NaN3 ) for 30 min on 4°C. An overview of the B cell-, T cell- and NK cell/regulatory T cell (Treg)-phenotyping panels including fluorochromes is provided in Table S2. Cells were washed with PBEA buffer and fixed in PBS containing 1% formaldehyde. The following monoclonal antibodies were used after initial testing and titration: anti-CD3 Alexa-eFluor 780, anti-CD4 PE-Cy7, anti-CD19 Pe-Cy7, anti-CD27 APC, anti-CD28 FITC, and anti-CCR7 PE (eBioscience, SanDiego, USA), anti-CD4 PerCP-Cy5.5, anti-CD8+ PerCP-Cy5.5, anti-CD16 PerCP-Cy5.5, anti-CD20 FITC, anti-CD25 V450, anti-CD38 PerCP-Cy5.5, anti-CD45RA HV450, anti-CD69 FITC, anti-CD86 V450, anti-IFN-g APC, anti-IgD PE, anti-TNF-a FITC (Becton Dickenson GmbH, Heidelberg, Germany), anti-IL-2 PacificBlue (BioLegend,); anti-CD56 PE (Miltenyi, Bergisch Gladbach, Germany).
Flow cytometry
All flow cytometric measurements were performed on a calibrated FACSCanto II (4-2-2) cytometer (Becton Dickenson, Heidelberg, Germany). Acquisition of data including compensation was performed using BD FACSDiva 6.1 and use of compensation beads. Accumulated data was analyzed using FlowJo versions 7.5.2 and 10.0.7. (Treestar Inc., Oregon, USA). Tethered and non-viable cells were excluded from analysis. Absolute cell counts from lymphocyte subpopulations were determined by normalizing data to lymphocyte counts derived from differential blood tests. All laboratory procedures were performed according to approved SOPs.
Statistical analysis
Differentially expressed genes between groups were determined based on false discovery rate (FDR)-corrected p-values in combination with fold change (FC) thresholds. In detail, p values were generated by the Welch's approximate t-test followed by the Benjamini and Hochberg correction for multiple testing using GeneSpring G x 12.6.1. FC thresholds of ranging from 1.5 to 3 were applied in our analysis. Hierarchical clustering, calculation of t values of paired t-tests and PCA were performed using MeV version 4.9.0. Unsupervised hierarchical clustering of BTM activity scores was performed using Euclidean distance and average linkage clustering. Survival analyses including Log-rank test were performed with GraphPad Prism version 6.0.5. GraphPad Prism was also used to plot all graphs except for heatmaps and to perform Pearson and Spearman's rank correlation analyses.
Supplementary Material
Disclosure of potential conflicts of interest
HSH, SDK, BS, UGV, AS, KJK, VW, IH, MFM are or were employed at CureVac AG. HSH, SDK and MFM are named as inventors on a patent application for a gene expression-based biomarker filed by CureVac AG.
Acknowledgments
We thank all participating medical doctors (Martin Sebastian, Lotta v. Boehmer, Alfred Zippelius, Frank Mayer, Martin Reck, Djordje Atanackovic, Michael Thomas, Folker Schneller, Jan Stoehlmacher, Eray Goekkurt, Helga Bernhard, Andreas Gröschel, Robert Bals, Susanne Schmidt, Elke Jäger and Alexander Knuth) and staff of the participating hospitals and importantly all participating patients. Helen Dietrich and Simone Eppler provided excellent technical support with phenotypic analyses by flow cytometry. We acknowledge Fatma Doener and Madeleine Hipp for critical review of our manuscript and for helpful discussions.
Author contributions
HSH, SDK, MF conceived the study; HSH, LD, OK, JBM analyzed the data; BS, UGK, AS, KJK, VW, IH were responsible for the clinical trial and provided clinical material and data; HSH, SDK and JBM wrote the manuscript; all authors read and discussed the manuscript.
References
- 1.Chen Z, Fillmore CM, Hammerman PS, Kim CF, Wong KK. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer 2014; 14:535-46; PMID:25056707; http://dx.doi.org/ 10.1038/nrc3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Owonikoko TK, Ragin CC, Belani CP, Oton AB, Gooding WE, Taioli E, Ramalingam SS. Lung cancer in elderly patients: an analysis of the surveillance, epidemiology, and end results database. J Clin Oncol 2007; 25:5570-7; PMID:18065729; http://dx.doi.org/ 10.1200/JCO.2007.12.5435 [DOI] [PubMed] [Google Scholar]
- 3.Reck M. What future opportunities may immuno-oncology provide for improving the treatment of patients with lung cancer? Ann Oncol 2012; 23 Suppl 8:viii28-34; PMID:22918925; http://dx.doi.org/ 10.1093/annonc/mds260 [DOI] [PubMed] [Google Scholar]
- 4.Mellman I, Coukos G, Dranoff G. Cancer immunotherapy comes of age. Nature 2011; 480:480-9; PMID:22193102; http://dx.doi.org/ 10.1038/nature10673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity 2013; 39:1-10; PMID:23890059; http://dx.doi.org/ 10.1016/j.immuni.2013.07.012 [DOI] [PubMed] [Google Scholar]
- 6.Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, Patnaik A, Aggarwal C, Gubens M, Horn L et al.. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med 2015; 372:2018-28; PMID:25891174; http://dx.doi.org/ 10.1056/NEJMoa1501824 [DOI] [PubMed] [Google Scholar]
- 7.Chen L, Han X. Anti-PD-1/PD-L1 therapy of human cancer: past, present, and future. J Clin Invest 2015; 125:3384-91; PMID:26325035; http://dx.doi.org/ 10.1172/JCI80011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Topalian SL, Sharpe AH. Balance and imbalance in the immune system: life on the edge. Immunity 2014; 41:682-4; PMID:25517610; http://dx.doi.org/ 10.1016/j.immuni.2014.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Cowey CL, Lao CD, Schadendorf D, Dummer R, Smylie M, Rutkowski P et al.. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N Engl J Med 2015; 373:23-34; PMID:26027431; http://dx.doi.org/ 10.1056/NEJMoa1504030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wing K, Onishi Y, Prieto-Martin P, Yamaguchi T, Miyara M, Fehervari Z, Nomura T, Sakaguchi S. CTLA-4 control over Foxp3+ regulatory T cell function. Science 2008; 322:271-5; PMID:18845758; http://dx.doi.org/ 10.1126/science.1160062 [DOI] [PubMed] [Google Scholar]
- 11.Boon T, Coulie PG, Van den Eynde BJ, van der Bruggen P. Human T cell responses against melanoma. Annu Rev Immunol 2006; 24:175-208; PMID:16551247; http://dx.doi.org/ 10.1146/annurev.immunol.24.021605.090733 [DOI] [PubMed] [Google Scholar]
- 12.Melief CJ, van Hall T, Arens R, Ossendorp F, van der Burg SH. Therapeutic cancer vaccines. J Clin Invest 2015; 125:3401-12; PMID:26214521; http://dx.doi.org/ 10.1172/JCI80009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Melero I, Gaudernack G, Gerritsen W, Huber C, Parmiani G, Scholl S, Thatcher N, Wagstaff J, Zielinski C, Faulkner I et al.. Therapeutic vaccines for cancer: an overview of clinical trials. Nat Rev Clin Oncol 2014; 11:509-24; PMID:25001465; http://dx.doi.org/ 10.1038/nrclinonc.2014.111 [DOI] [PubMed] [Google Scholar]
- 14.Sahin U, Kariko K, Tureci O. mRNA-based therapeutics–developing a new class of drugs. Nat Rev Drug Discov 2014; 13:759-80; PMID:25233993; http://dx.doi.org/ 10.1038/nrd4278 [DOI] [PubMed] [Google Scholar]
- 15.Kallen KJ, Heidenreich R, Schnee M, Petsch B, Schlake T, Thess A, Baumhof P, Scheel B, Koch SD, Fotin-Mleczek M. A novel, disruptive vaccination technology: self-adjuvanted RNActive((R)) vaccines. Hum Vaccin Immunother 2013; 9:2263-76; PMID:23921513; http://dx.doi.org/ 10.4161/hv.25181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H, Bauer S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science 2004; 303:1526-9; PMID:14976262; http://dx.doi.org/ 10.1126/science.1093620 [DOI] [PubMed] [Google Scholar]
- 17.Scheel B, Braedel S, Probst J, Carralot JP, Wagner H, Schild H, Jung G, Rammensee HG, Pascolo S. Immunostimulating capacities of stabilized RNA molecules. Eur J Immunol 2004; 34:537-47; PMID:14768059; http://dx.doi.org/ 10.1002/eji.200324198 [DOI] [PubMed] [Google Scholar]
- 18.Fotin-Mleczek M, Zanzinger K, Heidenreich R, Lorenz C, Thess A, Duchardt KM, Kallen KJ. Highly potent mRNA based cancer vaccines represent an attractive platform for combination therapies supporting an improved therapeutic effect. J Gene Med 2012; 14:428-39; PMID:22262664; http://dx.doi.org/ 10.1002/jgm.2605 [DOI] [PubMed] [Google Scholar]
- 19.Fotin-Mleczek M, Duchardt KM, Lorenz C, Pfeiffer R, Ojkic-Zrna S, Probst J, Kallen KJ. Messenger RNA-based vaccines with dual activity induce balanced TLR-7 dependent adaptive immune responses and provide antitumor activity. J Immunother 2011; 34:1-15; PMID:21150709; http://dx.doi.org/ 10.1097/CJI.0b013e3181f7dbe8 [DOI] [PubMed] [Google Scholar]
- 20.Probst J, Weide B, Scheel B, Pichler BJ, Hoerr I, Rammensee HG, Pascolo S. Spontaneous cellular uptake of exogenous messenger RNA in vivo is nucleic acid-specific, saturable and ion dependent. Gene Ther 2007; 14:1175-80; PMID:17476302; http://dx.doi.org/ 10.1038/sj.gt.3302964 [DOI] [PubMed] [Google Scholar]
- 21.Scheel B, Aulwurm S, Probst J, Stitz L, Hoerr I, Rammensee HG, Weller M, Pascolo S. Therapeutic anti-tumor immunity triggered by injections of immunostimulating single-stranded RNA. Eur J Immunol 2006; 36:2807-16; PMID:17013976; http://dx.doi.org/ 10.1002/eji.200635910 [DOI] [PubMed] [Google Scholar]
- 22.Carralot JP, Probst J, Hoerr I, Scheel B, Teufel R, Jung G, Rammensee HG, Pascolo S. Polarization of immunity induced by direct injection of naked sequence-stabilized mRNA vaccines. Cell Mol Life Sci 2004; 61:2418-24; PMID:15378210; http://dx.doi.org/ 10.1007/s00018-004-4255-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pascolo S. Messenger RNA-based vaccines. Expert Opin Biol Ther 2004; 4:1285-94; PMID:15268662; http://dx.doi.org/ 10.1517/14712598.4.8.1285 [DOI] [PubMed] [Google Scholar]
- 24.Hoerr I, Obst R, Rammensee HG, Jung G. In vivo application of RNA leads to induction of specific cytotoxic T lymphocytes and antibodies. Eur J Immunol 2000; 30:1-7; PMID:10602021; http://dx.doi.org/ 10.1002/1521-4141(200001)30:1%3c1::AID-IMMU1%3e3.0.CO;2- [DOI] [PubMed] [Google Scholar]
- 25.Heidenreich R, Jasny E, Kowalczyk A, Lutz J, Probst J, Baumhof P, Scheel B, Voss S, Kallen KJ, Fotin-Mleczek M. A novel RNA-based adjuvant combines strong immunostimulatory capacities with a favorable safety profile. Int J Cancer 2015; 137(2):372-84; PMID:25530186; http://dx.doi.org/26082837 10.1002/ijc.29402 [DOI] [PubMed] [Google Scholar]
- 26.Kubler H, Scheel B, Gnad-Vogt U, Miller K, Schultze-Seemann W, Vom Dorp F, Parmiani G, Hampel C, Wedel S, Trojan L et al.. Self-adjuvanted mRNA vaccination in advanced prostate cancer patients: a first-in-man phase I/IIa study. J Immunother Cancer 2015; 3:26; PMID:26082837; http://dx.doi.org/ 10.1186/s40425-015-0068-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chaussabel D, Pascual V, Banchereau J. Assessing the human immune system through blood transcriptomics. BMC Biol 2010; 8:84; PMID:20619006; http://dx.doi.org/ 10.1186/1741-7007-8-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Querec TD, Akondy RS, Lee EK, Cao W, Nakaya HI, Teuwen D, Pirani A, Gernert K, Deng J, Marzolf B et al.. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol 2009; 10:116-25; PMID:19029902; http://dx.doi.org/ 10.1038/ni.1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Furman D, Jojic V, Kidd B, Shen-Orr S, Price J, Jarrell J, Tse T, Huang H, Lund P, Maecker HT et al.. Apoptosis and other immune biomarkers predict influenza vaccine responsiveness. Mol Syst Biol 2013; 9:659; PMID:23591775; http://dx.doi.org/ 10.1038/msb.2013.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaucher D, Therrien R, Kettaf N, Angermann BR, Boucher G, Filali-Mouhim A, Moser JM, Mehta RS, Drake DR 3rd, Castro E et al.. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J Exp Med 2008; 205:3119-31; PMID:19047440; http://dx.doi.org/ 10.1084/jem.20082292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakaya HI, Wrammert J, Lee EK, Racioppi L, Marie-Kunze S, Haining WN, Means AR, Kasturi SP, Khan N, Li GM et al.. Systems biology of vaccination for seasonal influenza in humans. Nat Immunol 2011; 12:786-95; PMID:21743478; http://dx.doi.org/ 10.1038/ni.2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li S, Rouphael N, Duraisingham S, Romero-Steiner S, Presnell S, Davis C, Schmidt DS, Johnson SE, Milton A, Rajam G et al.. Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nat Immunol 2014; 15:195-204; PMID:24336226; http://dx.doi.org/ 10.1038/ni.2789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chaussabel D, Baldwin N. Democratizing systems immunology with modular transcriptional repertoire analyses. Nat Rev Immunol 2014; 14:271-80; PMID:24662387; http://dx.doi.org/ 10.1038/nri3642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heuvers ME, Muskens F, Bezemer K, Lambers M, Dingemans AM, Groen HJ, Smit EF, Hoogsteden HC, Hegmans JP, Aerts JG. Arginase-1 mRNA expression correlates with myeloid-derived suppressor cell levels in peripheral blood of NSCLC patients. Lung Cancer 2013; 81:468-74; PMID:23850196; http://dx.doi.org/ 10.1016/j.lungcan.2013.06.005 [DOI] [PubMed] [Google Scholar]
- 35.Vetsika EK, Koinis F, Gioulbasani M, Aggouraki D, Koutoulaki A, Skalidaki E, Mavroudis D, Georgoulias V, Kotsakis A. A circulating subpopulation of monocytic myeloid-derived suppressor cells as an independent prognostic/predictive factor in untreated non-small lung cancer patients. J Immunol Res 2014; 2014:659294; PMID:25436215; http://dx.doi.org/ 10.1155/2014/659294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schneider T, Sevko A, Heussel CP, Umansky L, Beckhove P, Dienemann H, Safi S, Utikal J, Hoffmann H, Umansky V. Serum inflammatory factors and circulating immunosuppressive cells are predictive markers for efficacy of radiofrequency ablation in non-small-cell lung cancer. Clin Exp Immunol 2015; 180(3):467-74; PMID:25644608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Showe MK, Vachani A, Kossenkov AV, Yousef M, Nichols C, Nikonova EV, Chang C, Kucharczuk J, Tran B, Wakeam E et al.. Gene expression profiles in peripheral blood mononuclear cells can distinguish patients with non-small cell lung cancer from patients with nonmalignant lung disease. Cancer Res 2009; 69:9202-10; PMID:19951989; http://dx.doi.org/ 10.1158/0008-5472.CAN-09-1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kossenkov AV, Dawany N, Evans TL, Kucharczuk JC, Albelda SM, Showe LC, Showe MK, Vachani A. Peripheral immune cell gene expression predicts survival of patients with non-small cell lung cancer. PLoS One 2012; 7:e34392; PMID:22479623; http://dx.doi.org/ 10.1371/journal.pone.0034392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou C, Licciulli S, Avila JL, Cho M, Troutman S, Jiang P, Kossenkov AV, Showe LC, Liu Q, Vachani A et al.. The Rac1 splice form Rac1b promotes K-ras-induced lung tumorigenesis. Oncogene 2013; 32:903-9; PMID:22430205; http://dx.doi.org/ 10.1038/onc.2012.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 1999; 401:708-12; PMID:10537110; http://dx.doi.org/ 10.1038/44385 [DOI] [PubMed] [Google Scholar]
- 41.Hamann D, Baars PA, Rep MH, Hooibrink B, Kerkhof-Garde SR, Klein MR, van Lier RA. Phenotypic and functional separation of memory and effector human CD8+ T cells. J Exp Med 1997; 186:1407-18; PMID:9348298; http://dx.doi.org/ 10.1084/jem.186.9.1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomita M, Shimizu T, Ayabe T, Yonei A, Onitsuka T. Preoperative neutrophil to lymphocyte ratio as a prognostic predictor after curative resection for non-small cell lung cancer. Anticancer Res 2011; 31:2995-8; PMID:21868550 [PubMed] [Google Scholar]
- 43.Kishi Y, Kopetz S, Chun YS, Palavecino M, Abdalla EK, Vauthey JN. Blood neutrophil-to-lymphocyte ratio predicts survival in patients with colorectal liver metastases treated with systemic chemotherapy. Ann Surg Oncol 2009; 16:614-22; PMID:19130139; http://dx.doi.org/ 10.1245/s10434-008-0267-6 [DOI] [PubMed] [Google Scholar]
- 44.Chua W, Charles KA, Baracos VE, Clarke SJ. Neutrophil/lymphocyte ratio predicts chemotherapy outcomes in patients with advanced colorectal cancer. Br J Cancer 2011; 104:1288-95; PMID:21448173; http://dx.doi.org/ 10.1038/bjc.2011.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li J, Jiang R, Liu WS, Liu Q, Xu M, Feng QS, Chen LZ, Bei JX, Chen MY, Zeng YX. A large cohort study reveals the association of elevated peripheral blood lymphocyte-to-monocyte ratio with favorable prognosis in nasopharyngeal carcinoma. PLoS One 2013; 8:e83069; PMID:24386144; http://dx.doi.org/ 10.1371/journal.pone.0083069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stotz M, Gerger A, Eisner F, Szkandera J, Loibner H, Ress AL, Kornprat P, AlZoughbi W, Seggewies FS, Lackner C et al.. Increased neutrophil-lymphocyte ratio is a poor prognostic factor in patients with primary operable and inoperable pancreatic cancer. Br J Cancer 2013; 109:416-21; PMID:23799847; http://dx.doi.org/ 10.1038/bjc.2013.332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pulendran B, Li S, Nakaya HI. Systems vaccinology. Immunity 2010; 33:516-29; PMID:21029962; http://dx.doi.org/ 10.1016/j.immuni.2010.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quackenbush J. Computational analysis of microarray data. Nat Rev Genet 2001; 2:418-27; PMID:11389458; http://dx.doi.org/ 10.1038/35076576 [DOI] [PubMed] [Google Scholar]
- 49.Allison DB, Cui X, Page GP, Sabripour M. Microarray data analysis: from disarray to consolidation and consensus. Nat Rev Genet 2006; 7:55-65; PMID:16369572; http://dx.doi.org/ 10.1038/nrg1749 [DOI] [PubMed] [Google Scholar]
- 50.Khatri P, Sirota M, Butte AJ. Ten years of pathway analysis: current approaches and outstanding challenges. PLoS Comput Biol 2012; 8:e1002375; PMID:22383865; http://dx.doi.org/ 10.1371/journal.pcbi.1002375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Friedman N. Inferring cellular networks using probabilistic graphical models. Science 2004; 303:799-805; PMID:14764868; http://dx.doi.org/ 10.1126/science.1094068 [DOI] [PubMed] [Google Scholar]
- 52.Talmadge JE, Gabrilovich DI. History of myeloid-derived suppressor cells. Nat Rev Cancer 2013; 13:739-52; PMID:24060865; http://dx.doi.org/ 10.1038/nrc3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Condamine T, Ramachandran I, Youn JI, Gabrilovich DI. Regulation of tumor metastasis by myeloid-derived suppressor cells. Annu Rev Med 2015; 66:97-110; PMID:25341012; http://dx.doi.org/ 10.1146/annurev-med-051013-052304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Goeje PL, Bezemer K, Heuvers ME, Dingemans AC, Groen HJ, Smit EF, Hoogsteden HC, Hendriks RW, Aerts JG, Hegmans JP. Immunoglobulin-like transcript 3 is expressed by myeloid-derived suppressor cells and correlates with survival in patients with non-small cell lung cancer. Oncoimmunology 2015; 4:e1014242; PMID:26140237; http://dx.doi.org/ 10.1080/2162402X.2015.1014242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Diaz-Montero CM, Salem ML, Nishimura MI, Garrett-Mayer E, Cole DJ, Montero AJ. Increased circulating myeloid-derived suppressor cells correlate with clinical cancer stage, metastatic tumor burden, and doxorubicin-cyclophosphamide chemotherapy. Cancer Immunol Immunother 2009; 58:49-59; PMID:18446337; http://dx.doi.org/ 10.1007/s00262-008-0523-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144:646-74; PMID:21376230; http://dx.doi.org/ 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 57.Crusz SM, Balkwill FR. Inflammation and cancer: advances and new agents. Nat Rev Clin Oncol 2015; 12(10):584-96; PMID:26122183 [DOI] [PubMed] [Google Scholar]
- 58.Jiang AG, Chen HL, Lu HY. The relationship between glasgow prognostic score and serum tumor markers in patients with advanced non-small cell lung cancer. BMC Cancer 2015; 15:386; PMID:25956656; http://dx.doi.org/ 10.1186/s12885-015-1403-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alifano M, Mansuet-Lupo A, Lococo F, Roche N, Bobbio A, Canny E, Schussler O, Dermine H, Régnard JF, Burroni B et al.. Systemic inflammation, nutritional status and tumor immune microenvironment determine outcome of resected non-small cell lung cancer. PLoS One 2014; 9:e106914; PMID:25238252; http://dx.doi.org/ 10.1371/journal.pone.0106914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Romero P, Zippelius A, Kurth I, Pittet MJ, Touvrey C, Iancu EM, Corthesy P, Devevre E, Speiser DE, Rufer N. Four functionally distinct populations of human effector-memory CD8+ T lymphocytes. J Immunol 2007; 178:4112-9; PMID:17371966; http://dx.doi.org/ 10.4049/jimmunol.178.7.4112 [DOI] [PubMed] [Google Scholar]
- 61.Koch S, Larbi A, Derhovanessian E, Ozcelik D, Naumova E, Pawelec G. Multiparameter flow cytometric analysis of CD4 and CD8 T cell subsets in young and old people. Immun Ageing 2008; 5:6; PMID:18657274; http://dx.doi.org/ 10.1186/1742-4933-5-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kossenkov AV, Vachani A, Chang C, Nichols C, Billouin S, Horng W, Rom WN, Albelda SM, Showe MK, Showe LC. Resection of non-small cell lung cancers reverses tumor-induced gene expression changes in the peripheral immune system. Clin Cancer Res 2011; 17:5867-77; PMID:21807633; http://dx.doi.org/ 10.1158/1078-0432.CCR-11-0737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Showe MK, Kossenkov AV, Showe LC. The peripheral immune response and lung cancer prognosis. Oncoimmunology 2012; 1:1414-6; PMID:23243612; http://dx.doi.org/ 10.4161/onci.21096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sobolev O, Binda E, O'Farrell S, Lorenc A, Pradines J, Huang Y, Duffner J, Schulz R, Cason J, Zambon M et al.. Adjuvanted influenza-H1N1 vaccination reveals lymphoid signatures of age-dependent early responses and of clinical adverse events. Nat Immunol 2016; 17:204-13; PMID:26726811; http://dx.doi.org/ 10.1038/ni.3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nakaya HI, Hagan T, Duraisingham SS, Lee EK, Kwissa M, Rouphael N, Frasca D, Gersten M, Mehta AK, Gaujoux R et al.. Systems analysis of immunity to influenza vaccination across multiple years and in diverse populations reveals shared molecular signatures. Immunity 2015; 43:1186-98; PMID:26682988; http://dx.doi.org/ 10.1016/j.immuni.2015.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, Verweij J, Van Glabbeke M, van Oosterom AT, Christian MC et al.. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst 2000; 92:205-16; PMID:10655437; http://dx.doi.org/ 10.1093/jnci/92.3.205 [DOI] [PubMed] [Google Scholar]
- 67.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES et al.. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005; 102:15545-50; PMID:16199517; http://dx.doi.org/ 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.