ABSTRACT
Much of our knowledge on adipogenesis comes from cell culture models of preadipocyte differentiation. Adipogenesis is induced by treating confluent preadipocytes with the adipogenic cocktail, which activates transcription factors (TFs) glucocorticoid receptor (GR) and CREB within minutes and increases expression of TFs C/EBPβ, C/EBPδ, KLF4, and Krox20 within hours. All of these TFs have been shown to be capable of promoting adipogenesis in culture when they are overexpressed. However, it has remained unclear whether endogenous KLF4 and Krox20 are required for adipogenesis in culture and in vivo. Using conditional knockout mice and derived white and brown preadipocytes, we show that endogenous KLF4 and Krox20 are dispensable for adipogenesis in culture and for brown adipose tissue development in mice. In contrast, the master adipogenic TF peroxisome proliferator-activated receptor γ (PPARγ) is essential. These results challenge the existing model on transcriptional regulation in the early phase of adipogenesis and highlight the need of studying adipogenesis in vivo.
KEYWORDS: KLF4, Krox20, PPARγ, adipogenesis
INTRODUCTION
Adipose tissues, including white adipose tissue (WAT) and brown adipose tissue (BAT), play critical roles in energy metabolism and homeostasis. Understanding molecular mechanisms underlying the development of adipose tissues is of potential importance for treating obesity and type II diabetes. Much of our knowledge on adipose tissue development comes from studies using cell culture model systems such as the mouse 3T3-L1 preadipocytes (1, 2). Adipogenesis of 3T3-L1 is induced by stimulating confluent cells with the adipogenic cocktail, which consists of synthetic glucocorticoid dexamethasone, insulin, the phosphodiesterase inhibitor isobutylmethylxanthine (IBMX), and fetal bovine serum (FBS) (3–5).
Studies on transcriptional regulation of adipogenesis have revealed a cascade of sequentially induced transcription factors (TFs) that promotes adipogenesis in culture (6, 7). The adipogenic cocktail activates TFs GR (also known as NR3C1) and CREB within minutes and induces expression of TFs C/EBPβ, C/EBPδ, KLF4, and Krox20 (also known as Egr2) within hours (8). C/EBPβ and C/EBPδ directly activate the expression of two principal adipogenic TFs, peroxisome proliferator-activated receptor γ (PPARγ) and C/EBPα, which cooperate to induce expression of thousands of adipocyte genes, resulting in differentiation toward mature adipocytes (9). C/EBPα, -β, and -δ and PPARγ have been shown to be essential for adipogenesis in mice (10, 11).
Besides C/EBPβ and C/EBPδ, KLF4 and Krox20 are also induced in the early phase of 3T3-L1 adipogenesis (12, 13). Krox20 and Klf4 are specifically induced by FBS and IBMX, respectively. Krox20 is induced within 1 h, while the Klf4 mRNA level peaks at 2 h after the addition of an adipogenic cocktail to confluent 3T3-L1 cells. Ectopic expression of Krox20 promotes adipogenesis in both 3T3-L1 preadipocytes and NIH 3T3 fibroblasts. Conversely, RNA interference (RNAi)-mediated knockdown of Krox20 or Klf4 has been shown to inhibit 3T3-L1 adipogenesis (12, 13). Ectopic KLF4 and Krox20 cooperate to promote C/EBPβ expression, thus providing a possible mechanism by which these two TFs positively regulate adipogenesis in culture (13). However, the roles of endogenous Krox20 and KLF4 in adipogenesis have remained unclear.
We have investigated the roles of adipogenic TFs in adipogenesis by employing conditional knockout (cKO) mice and derived white and brown preadipocytes. By deleting Klf4, Krox20, and Pparγ genes individually, we found that surprisingly, endogenous Krox20 and KLF4 are dispensable for adipogenesis of preadipocytes in culture and for BAT development in mice. In contrast, PPARγ is essential for adipogenesis both in culture and in mice.
RESULTS
KLF4 and Krox20 are dispensable for the induction of early adipogenic TFs.
To profile gene expression changes during adipogenesis, confluent 3T3-L1 white preadipocytes and Pparγf/f brown preadipocytes were induced to undergo differentiation. RNAs were extracted in the early phase (0 h, 1 h, 2 h, and 4 h) and later phase (day 2 and day 7) of adipogenesis for RNA sequencing (RNA-Seq) analysis. The results from both cell lines confirmed previous observations that Krox20 and Klf4 were induced transiently in the early phase of adipogenesis (Fig. 1A and B) (12, 13). Krox20 was induced to the maximum level at 1 h but dropped quickly at 4 h. Klf4 was induced to the maximum level at 2 h but started to drop at 4 h. Both Krox20 and Klf4 levels were very low at day 2 and day 7 of adipogenesis. Cebpβ and Cebpδ were also induced in the early phase but were expressed throughout the differentiation. In contrast, the master adipogenic TF Pparγ was induced at day 2 and reached the maximal level at day 7.
FIG 1.
KLF4 and Krox20 are dispensable for induction of early adipogenic TFs. (A, B) 3T3-L1 white preadipocytes (A) and Pparγf/f immortalized brown preadipocytes (B) were induced to undergo adipogenesis. Cells were collected at the indicated time points for RNA-Seq. RPKM values from RNA-Seq indicate gene expression levels. D2, day 2; D7, day 7. (C, D) Klf4f/f (C) and Krox20f/f (D) immortalized brown preadipocytes were infected with retroviral vector MSCVhygro expressing Cre. Adipogenesis was induced with the adipogenic cocktail, and cells were collected at indicated time points for qRT-PCR analysis of Klf4, Krox20, Cebpβ, Cebpδ, Klf5, and Klf9.
To investigate whether endogenous KLF4 and Krox20 are required for the expression of early adipogenesis factors, primary brown preadipocytes were isolated from Klf4f/f or Krox20f/f BATs and were immortalized with simian virus 40 large T antigen (SV40T). The immortalized brown preadipocytes were infected with retroviral Cre to stably delete Klf4 or Krox20 gene (Fig. 1C and D). Consistent with RNA-Seq data, Klf4 and Krox20 were induced at early time points after induction of differentiation in control cells but not in KO cells. However, deletion of Klf4 or Krox20 from brown preadipocytes had little effect on the induction of early adipogenic TFs, including Cebpβ, Cebpδ, Klf5, Klf9, Klf4, and Krox20 by the adipogenic cocktail (Fig. 1C and D). These data indicate that KLF4 and Krox20 are dispensable for the induction of early adipogenic TFs.
KLF4 is dispensable for adipogenesis in culture.
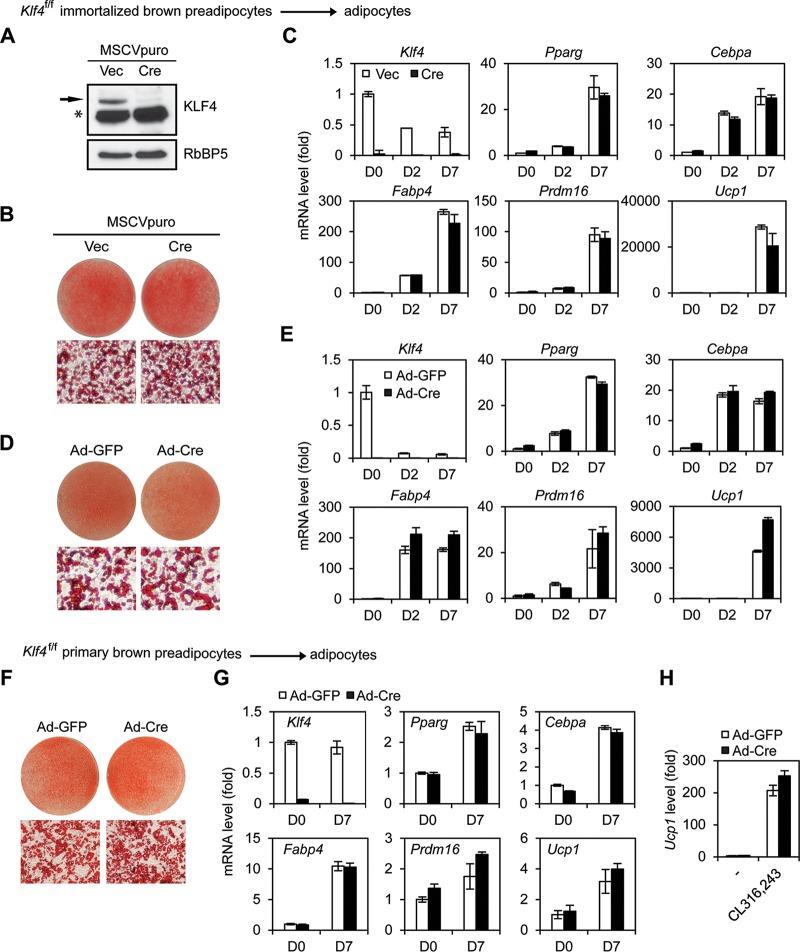
Next, we investigated whether KLF4 is required for adipogenesis. We induced adipogenesis in retroviral Cre-infected Klf4f/f immortalized brown preadipocytes. To our surprise, deletion of Klf4 had little effect on brown adipogenesis and induction of adipogenesis markers Pparγ, Cebpα, and Fabp4 and brown adipocyte markers Prdm16 and Ucp1 (Fig. 2A to C). To eliminate the possible involvement of any compensation mechanism during the 2-week process required for retroviral Cre-mediated stable gene deletion, we acutely infected Klf4f/f brown preadipocytes with adenoviral Cre. Acute deletion of Klf4 also had little effect on adipogenesis (Fig. 2D and E).
FIG 2.
KLF4 is dispensable for brown adipogenesis in culture. (A to C) Klf4f/f immortalized brown preadipocytes were infected with retroviral vector MSCVpuro expressing Cre. (A) Western blot analysis of KLF4 before differentiation. The asterisk indicates a nonspecific band. RbBP5 was used as a loading control. (B) Oil Red O staining at day 7 of adipogenesis. Upper panels, stained dishes; lower panels, representative fields under the microscope. (C) qRT-PCR analysis of Klf4, adipogenesis markers (Pparg, Cebpa, and Fabp4) and brown adipocyte markers (Prdm16 and Ucp1) at indicated time points of adipogenesis. D0, day 0. (D, E) Klf4f/f immortalized brown preadipocytes were infected with adenoviruses expressing GFP (Ad-GFP) or Cre (Ad-Cre). Two days later, cells were replated, followed by adipogenesis assays. (D) Oil Red O staining. (E) qRT-PCR analysis of gene expression. (F to H) Klf4f/f primary brown preadipocytes were infected with Ad-GFP or Ad-Cre, followed by adipogenesis assays. (F) Oil Red O staining. (G) qRT-PCR analysis of gene expression. (H) D7 mature brown adipocytes were treated with 100 nM CL-316,243 for 4 h, followed by qRT-PCR analysis of Ucp1 expression.
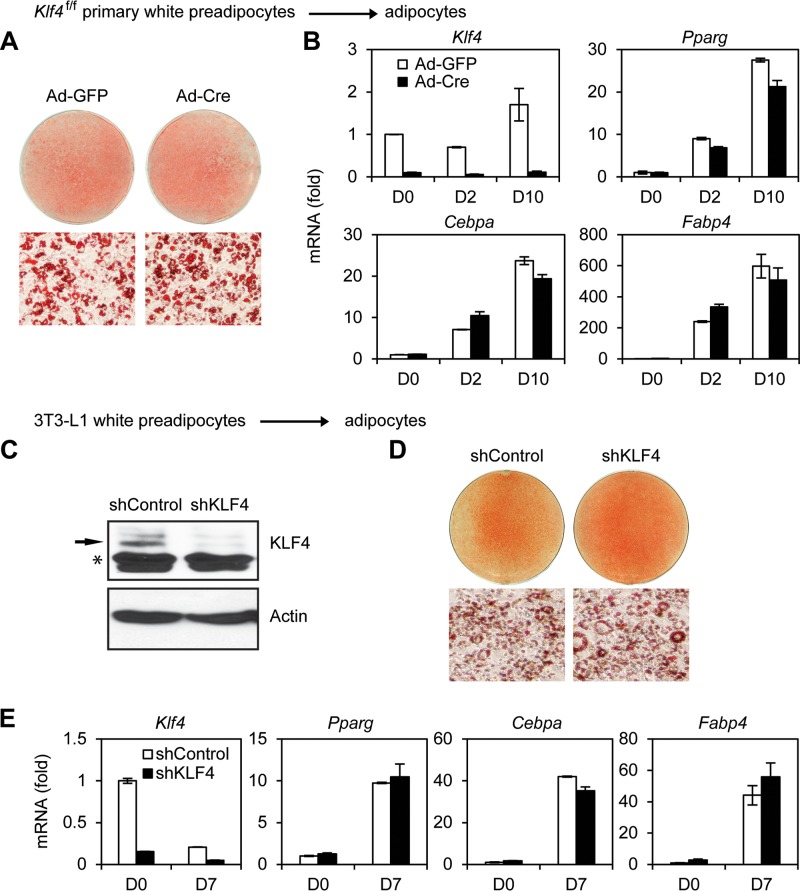
Deletion of Klf4 also had little effect on adipogenesis of primary brown preadipocytes (Fig. 2F and G). After primary preadipocytes differentiated to mature brown adipocytes, cells were treated by CL316,243, a selective adrenergic-β3 receptor agonist. Deletion of Klf4 had little effect on CL316,243-induced expression of Ucp1, a critical thermogenic gene, in brown adipocytes (Fig. 2H). Consistent with our observations in brown adipogenesis, deletion of Klf4 in primary white preadipocytes or knockdown of Klf4 in immortalized 3T3-L1 cells had little effect on adipogenesis (Fig. 3). Together, these data indicate that KLF4 is dispensable for adipogenesis in culture.
FIG 3.
KLF4 is dispensable for white adipogenesis in culture. (A, B) Klf4f/f primary white preadipocytes were infected with Ad-GFP or Ad-Cre, followed by adipogenesis assays. (A) Oil Red O staining of differentiated cells at D10 (day 10). (B) qRT-PCR analysis of gene expression. (C to E) 3T3-L1 white preadipocytes were infected with lentiviral vector expressing control (shControl) or KLF4 knockdown shRNA (shKLF4), followed by adipogenesis assays. (C) Western blot analysis of KLF4 in preadipocytes. The asterisk indicates a nonspecific band. (D) Oil Red O staining of differentiated cells at D7. (E) qRT-PCR analysis of gene expression.
Krox20 is dispensable for adipogenesis in culture.
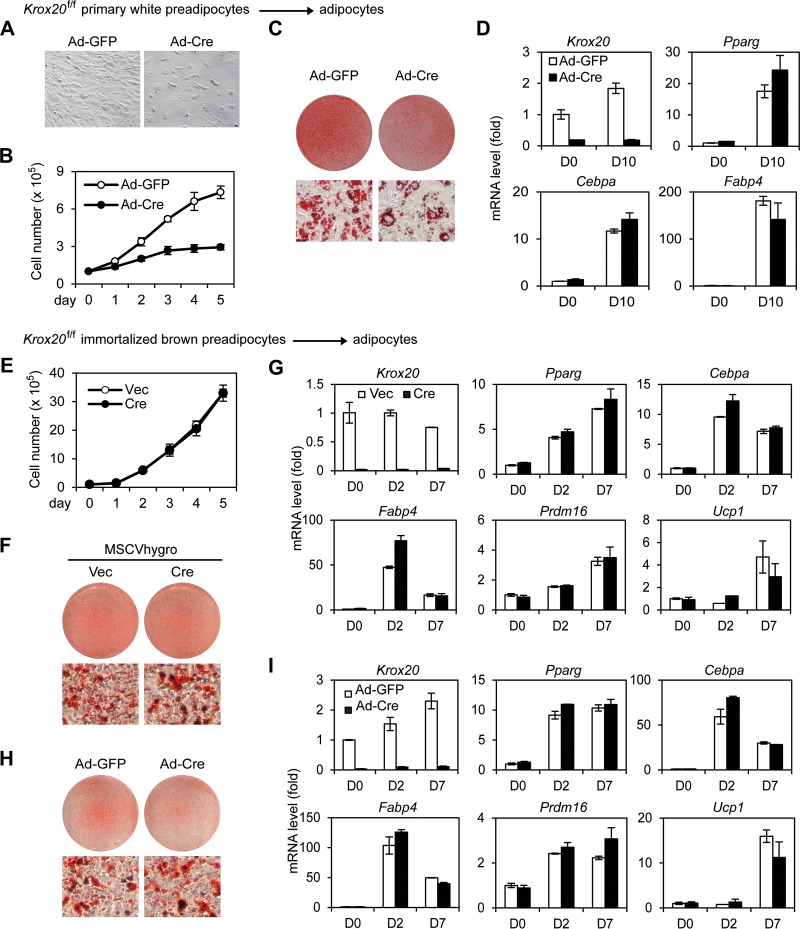
To investigate whether endogenous Krox20 is required for adipogenesis, primary white preadipocytes were isolated from Krox20f/f mice and were infected with adenoviral Cre. Deletion of Krox20 in primary white preadipocytes led to a growth defect (Fig. 4A and B). After cells reached confluence, adipogenesis was induced. Although Cre-infected cells showed reduced lipid droplet accumulation after 10 days of differentiation (Fig. 4C), reverse transcription-quantitative PCR (qRT-PCR) revealed that deletion of Krox20 in preadipocytes had little effect on the induction of adipogenesis markers (Fig. 4D). To distinguish the roles of Krox20 in cell differentiation versus proliferation, we turned to Krox20f/f immortalized brown preadipocytes. Deletion of Krox20 had little effect on the growth of immortalized brown preadipocytes (Fig. 4E). We found that either stable deletion by retroviral Cre or acute deletion by adenoviral Cre of Krox20 in preadipocytes did not affect brown adipogenesis and induction of adipogenesis markers (Fig. 4F to I), suggesting that Krox20 is dispensable for adipogenesis in culture.
FIG 4.
Krox20 is dispensable for adipogenesis in culture. (A, B) Krox20 knockout (KO) reduces growth of primary white preadipocytes. Krox20f/f primary white preadipocytes were infected with Ad-GFP or Ad-Cre. (A) Cell morphology of Krox20 KO cells under the microscope. (B) Cell growth rates: 1× 105 cells were plated, and the cumulative cell numbers were determined every day for 5 days. (C, D) Ad-GFP or Ad-Cre infected Krox20f/f primary white preadipocytes were cultured until confluence. Two days later, cells were treated with the adipogenic cocktail to induce adipogenesis. (C) Oil Red O staining at D10. (D) qRT-PCR analysis of gene expression. (E to G) Krox20f/f immortalized brown preadipocytes were infected with MSCVhygro expressing Cre. (E) Cell growth rates. (F) Oil Red O staining at day 7 of adipogenesis. (G) qRT-PCR analysis of Krox20 and adipogenesis markers at indicated time points of adipogenesis. (H, I) Krox20f/f immortalized brown preadipocytes were infected with Ad-GFP or Ad-Cre. Two days later, cells were replated, followed by adipogenesis assays. (H) Oil Red O staining at day 7 of adipogenesis. (I) qRT-PCR analysis of gene expression.
KLF4 is dispensable for BAT development.
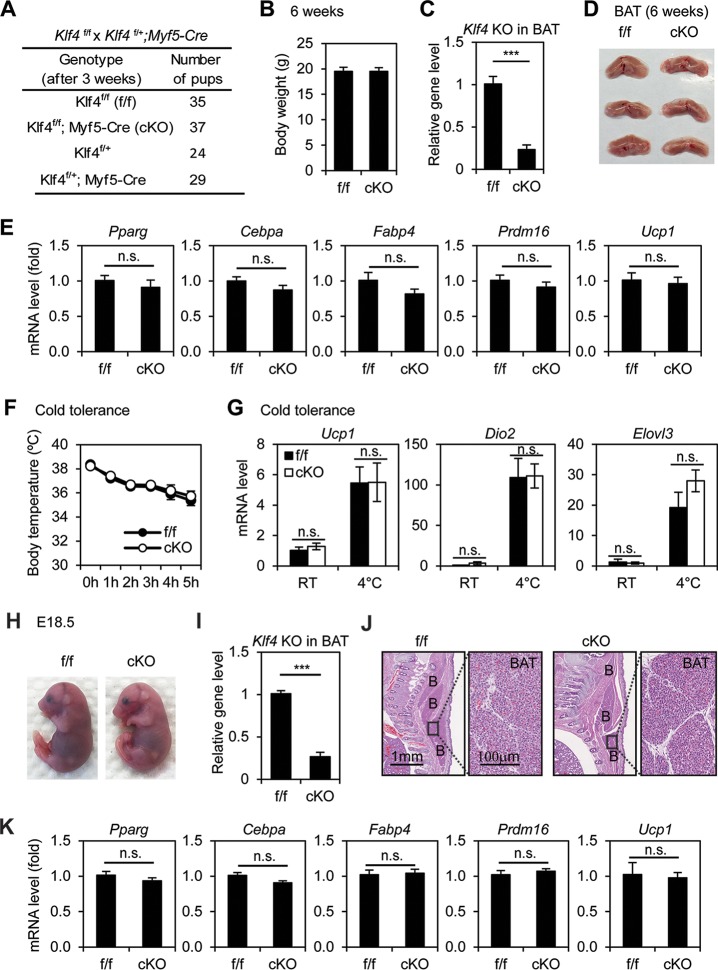
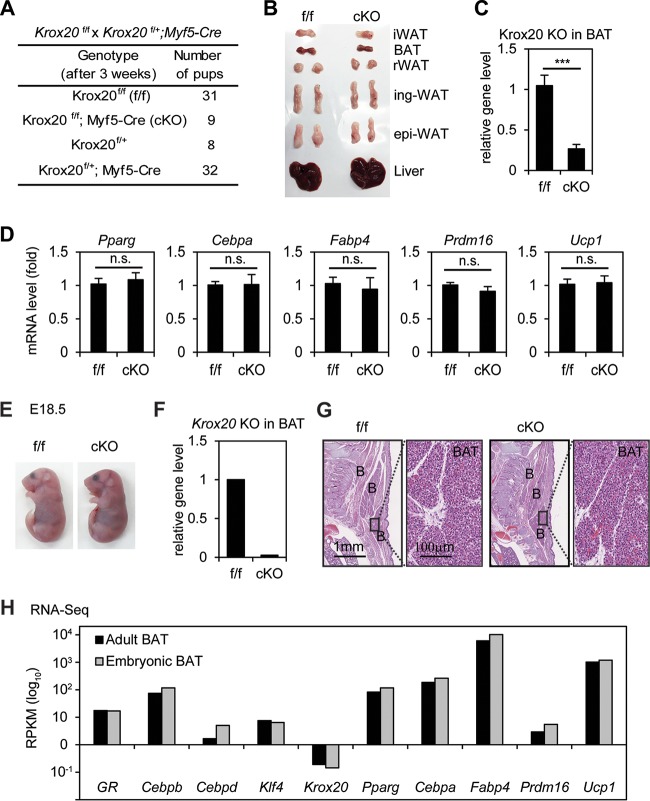
To find out whether KLF4 is required for BAT development, we generated Klf4f/f; Myf5-Cre mice. The Myf5-Cre transgene specifically deletes floxed genes in precursor cells of both BAT and muscles in the back (14). Littermate Klf4f/f mice were used as the control. Klf4f/f; Myf5-Cre pups survived well beyond 6 weeks after birth and showed body weight similar to that of control mice, although the Klf4 gene level was reduced by over 70% in the BAT of KO mice (Fig. 5A to C). Consistent with the adipogenesis data in cell culture, deletion of Klf4 by Myf5-Cre in Klf4f/f mice did not affect the development of BAT and associated expression of adipogenesis and BAT markers (Fig. 5D and E). Consistently, Klf4f/f; Myf5-Cre mice maintained normal body temperatures, were cold tolerant, and displayed behavior similar to that of control mice in the cold tolerance test (Fig. 5F and G). Similar results obtained from E18.5 embryos confirmed that KLF4 is dispensable for BAT development (Fig. 5H to K). While a full physiological characterization of Klf4f/f; Myf5-Cre mice needs to be done in the future, these data indicate that KLF4 is dispensable for BAT development in mice.
FIG 5.
KLF4 is dispensable for BAT development and function. Klf4f/f mice were crossed with Klf4f/+; Myf5-Cre mice to obtain Klf4f/f; Myf5-Cre (conditional KO [cKO]) mice and littermate control (Klf4f/f [f/f]) mice. (A to E) KLF4 is dispensable for brown adipose tissue (BAT) development. Six-week-old cKO and f/f mice were characterized. (A) Genotyping results. The expected ratios of the four genotypes are 1:1:1:1. (B) Body weight of f/f (n = 8) or cKO (n = 13) male mice. (C) Confirmation of Klf4 deletion in BAT by qPCR analysis of genomic DNA. ***, P < 0.001. (D) Pictures of BATs isolated from f/f and cKO mice (n = 3 per group). (E) RNA was extracted from BAT of f/f (n = 8) or cKO (n = 13) mice for qRT-PCR analysis of adipogenesis markers Pparγ, Cebpα, and Fabp4 and BAT markers Prdm16 and Ucp1. Data are presented as means ± SEM. n.s., no significance. (F, G) Cold tolerance test. cKO mice and their littermate controls (n = 5 per group) were acutely exposed to 4°C for 5 h. (F) Body temperatures were measured every hour. (G) qRT-PCR analysis of genes involved in thermogenesis in BAT. RT, room temperature. Data are presented as means ± SEM. (H to K) Characterization of E18.5 embryos. (H) Representative pictures of E18.5 embryos. (I) Confirmation of Klf4 deletion in E18.5 BAT by qPCR analysis of genomic DNA. (J) E18.5 embryos were sagittally sectioned along the midline. The sections of the interscapular area were stained with hematoxylin and eosin (H&E). B, BAT. (K) RNAs were extracted from E18.5 BAT of f/f (n = 7) or cKO (n = 9) embryos for qRT-PCR analysis.
Krox20 is dispensable for BAT development.
To investigate whether Krox20 is required for BAT development, we tried to generate Krox20f/f; Myf5-Cre mice. However, since the Krox20 gene (chromosome 10 [Chr10], bp 67534453 to 67547493) is located on the same chromosome as the Myf5-Cre transgene (Chr10, bp 107482908 to 107486927), the recombination frequency is 1/20 according to the distance between two loci (40 Mb) (15). Among 80 mice, we obtained 9 Krox20f/f; Myf5-Cre adult mice (Fig. 6A). Krox20f/f; Myf5-Cre pups survived well beyond 8 weeks after birth without any obvious developmental problems. Consistent with the adipogenesis data in cell culture, deletion of Krox20 did not affect BAT development and expression of adipogenesis and BAT markers, although the Krox20 gene level was reduced by over 90% in the BAT of KO mice (Fig. 6B to D). At the E18.5 stage, we obtained only 2 Krox20f/f; Myf5-Cre embryos of 37, which appeared grossly normal compared to the littermate Krox20f/f embryos (Fig. 6E). One embryo was used for extracting RNA to confirm Krox20 deletion (Fig. 6F). The other was subjected to histological analysis. The results revealed similar morphology of BATs and muscles for Krox20f/f; Myf5-Cre and Krox20f/f embryos (Fig. 6G). Furthermore, RNA-Seq analysis of interscapular BAT from wild-type adult (8-week-old) mice and embryos (E18.5) showed that compared with other early adipogenic TFs (GR, Cebpβ, Cebpδ, and Klf4) or adipogenesis markers (Pparγ, Cebpα, Fabp4, Prdm16, and Ucp1), the expression level of Krox20 was very low (RPKM < 0.1) in BAT (Fig. 6H), suggesting that Krox20 is unlikely to play a major role in BAT function. Together, while a full physiological characterization of Krox20f/f; Myf5-Cre mice may need to be done, these data indicate that Krox20 is dispensable for BAT development in mice.
FIG 6.
Krox20 is dispensable for BAT development. Krox20f/f were crossed with Krox20f/+; Myf5-Cre to obtain Krox20f/f; Myf5-Cre (conditional KO [cKO]) and littermate control (Krox20f/f [f/f]) mice. (A to D) Krox20 is dispensable for BAT development. Eight-week-old cKO and f/f mice were characterized. (A) Genotyping results. The expected ratios of the four genotypes are 4:1:1:4. (B) Isolated adipose tissues. iWAT, interscapular WAT; epi-WAT, epididymal WAT; ing-WAT, inguinal WAT; rWAT, retroperitoneal WAT. (C) Confirmation of Krox20 deletion in BAT by qPCR analysis of genomic DNA. (D) RNA was extracted from BAT of f/f and cKO mice for qRT-PCR analysis of adipogenesis markers Pparγ, Cebpα, and Fabp4 and BAT markers Prdm16 and Ucp1. (E to G) Characterization of E18.5 embryos. (E) Representative pictures of E18.5 embryos. (F) Krox20 deletion in BAT was confirmed by qRT-PCR of genomic DNA. (G) H&E staining of the interscapular area. B, BAT. (H) Krox20 mRNA is largely absent in BAT. The mRNA levels of early (GR, Cebpβ, Cebpδ, Klf4, and Krox20) or late (Pparγ, Cebpα, Fabp4, Prdm16, and Ucp1) adipogenesis genes in BATs isolated from adult (8-week-old) mice or embryos (E18.5) were determined by RNA-Seq. RPKM values from RNA-Seq indicate gene expression levels (log10 scale). ***, P < 0.001; n.s., no significance.
PPARγ is essential for adipogenesis and BAT development.
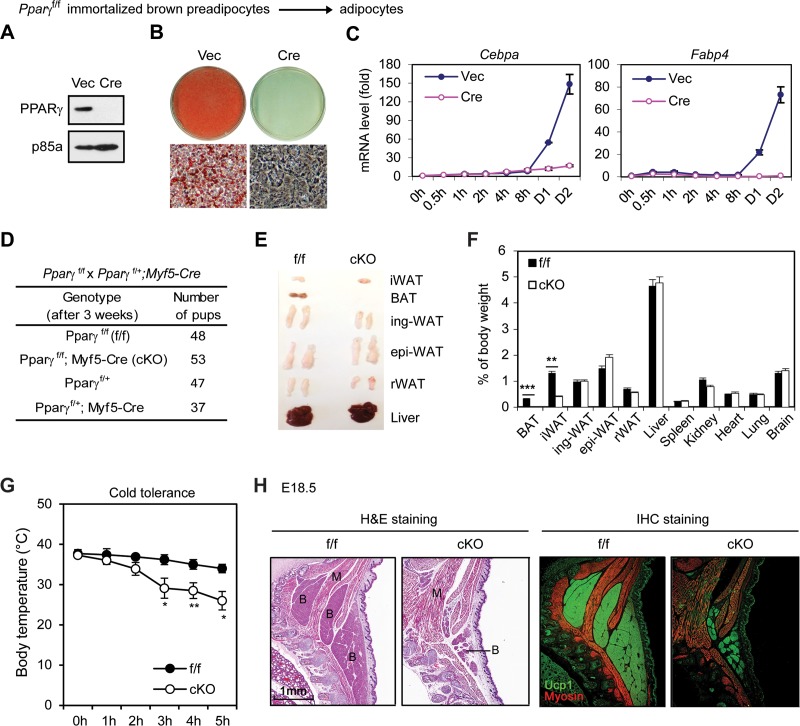
As a positive control, we infected Pparγf/f immortalized brown preadipocytes with retroviral Cre. Deletion of Pparγ gene in preadipocytes prevented adipogenesis and induction of adipogenesis markers such as Cebpα and Fabp4 (Fig. 7A to C). This result is consistent with the known essential role of PPARγ for adipogenesis.
FIG 7.
PPARγ is essential for adipogenesis and BAT development. (A to C) Pparγf/f immortalized brown preadipocytes were infected with MSCVhygro expressing Cre. (A) Western blot analysis of PPARγ. p85a was used as a loading control. (B) Seven days after induction of adipogenesis, cells were stained with Oil Red O. (C) Time course qRT-PCR analysis of adipogenic markers Cebpα and Fabp4. (D to G) Characterization of 6-week-old Pparγf/f; Myf5-Cre mice. (D) Genotyping results. (E) Representative pictures of isolated adipose tissues and livers. (F) The relative weight of each tissue and organ is shown as a percentage of body weight. (G) Pparγf/f; Myf5-Cre mice are cold intolerant. Six-week-old mice were housed at 4°C, and their body temperatures were measured every hour (n = 6). *, P < 0.05; **, P < 0.01, ***, P < 0.001. (H) E18.5 embryos were sagittally sectioned along the midline. The sections of the interscapular area were stained with H&E (left panels) or with antibodies recognizing BAT marker UCP1 (green) and skeletal muscle marker myosin (red) (right panels). IHC, immunohistochemistry.
We also generated Pparγf/f; Myf5-Cre mice by crossing Pparγf/f with Myf5-Cre mice to define the role of PPARγ in BAT development in mice. Pparγf/f; Myf5-Cre mice were obtained close to the expected Mendelian ratio, and all survived well beyond 6 weeks after birth without any obvious developmental defects (Fig. 7D). Compared to littermate controls, Pparγf/f; Myf5-Cre mice showed near-complete absence of BAT and marked decreases of interscapular WAT mass (Fig. 7E and F). Consistent with the lack of BAT, Pparγf/f; Myf5-Cre mice were unable to maintain body temperatures and were cold intolerant when exposed to environmental cold (Fig. 7G). Immunohistochemistry analysis of E18.5 embryos using antibodies against the BAT marker UCP1 and the muscle marker myosin confirmed the severe defect in BAT but not in muscle development in Pparγf/f; Myf5-Cre embryos (Fig. 7H). Since Myf5-Cre is selectively expressed in precursor cells giving rise to BAT and muscles in the back (14), these results indicate that PPARγ is required for BAT but not muscle development and that BAT is dispensable for the survival of mice in the laboratory setting.
DISCUSSION
We show that endogenous KLF4 and Krox20 are dispensable for white and brown adipogenesis in culture. Consistent with these observations, KLF4 and Krox20 are dispensable, while PPARγ is essential, for BAT development in mice. These results indicate that while KLF4 and Krox20 are transiently induced by the adipogenic cocktail in the early phase of preadipocyte differentiation in culture, they do not play major roles in adipogenesis. Among components of the adipogenic cocktail, IBMX and FBS induce the expression of Klf4 and Krox20, respectively (12, 13). The unexpected findings presented here suggest that the adipogenic cocktail accelerates adipogenesis independent of KLF4 and Krox20.
Using 3T3-L1 white preadipocytes and immortalized brown preadipocytes, we confirmed the previous observations that Klf4 and Krox20 are transiently induced in the early phase of adipogenesis in culture (12, 13). Surprisingly, deletion of Klf4 or Krox20 did not affect the induction of other early adipogenic TFs, including C/EBPβ and C/EBPδ, which is consistent with our data that KLF4 and Krox20 are dispensable for adipogenesis. Our results do not exclude the possibility that overexpression of KLF4 and Krox20 stimulates adipogenesis in culture.
It was shown previously that small interfering RNA (siRNA)-mediated knockdown of Klf4 inhibited 3T3-L1 adipogenesis (13). In our hands, however, Klf4 knockdown by short hairpin RNA (shRNA) did not affect adipogenesis in 3T3-L1 cells (Fig. 3). Further, we demonstrated that KLF4 is dispensable for adipogenesis by deleting Klf4 gene in white and brown preadipocytes as well as in mice. It was also shown that siRNA-mediated knockdown of Krox20 inhibited 3T3-L1 adipogenesis (12). However, by deleting Krox20 gene in white and brown preadipocytes as well as in mice, we demonstrated that Krox20 is dispensable for adipogenesis.
Studies using cell culture model systems, especially 3T3-L1 adipogenesis, have enabled the discoveries of major adipogenic TFs, including C/EBPα, C/EBPβ, C/EBPδ, and PPARγ (6, 7). Our data showing that endogenous KLF4 and Krox20 are dispensable for adipogenesis in culture and BAT development in mice suggest that it is also important to study adipogenesis in vivo, a process that is poorly understood (16).
MATERIALS AND METHODS
Plasmids and antibodies.
Retroviral plasmids MSCVhygro-Cre and MSCVpuro-Cre have been described previously (17–19). The lentiviral shRNA plasmid pLKO.1 targeting Klf4 (clone ID TRCN0000238251) and shRNA control plasmid were from Sigma. Anti-KLF4 antibody (catalog number 4038) was from Cell Signaling. Anti-PPARγ (sc-7196X) and anti-p85α (sc-1637) antibodies were from Santa Cruz. Anti-RbBP5 (A300-109A) antibody was from Bethyl Laboratories.
Mouse experiments.
Klf4f/f mice (20), Krox20f/f mice (21), and Pparγf/f mice (22) (Jackson no. 004584) were crossed with Myf5-Cre mice (Jackson no. 007893). Histology and immunohistochemistry analyses of E18.5 embryos were done as described previously (18). For the cold tolerance test, individual mice were housed at 4°C as described previously (17). Body temperature was measured every hour for 5 h using a mouse rectal probe (Thermalert TH-5). At the end of experiments, mice were euthanized and BATs were collected. Data were presented as means ± standard errors of the means (SEM). Differences were analyzed with Student's t test. All mouse work was approved by the Animal Care and Use Committee of NIDDK, NIH.
Isolation of primary preadipocytes, immortalization, virus infection, and adipogenesis assay.
Primary brown preadipocytes were isolated from interscapular BAT of newborn Klf4f/f or Krox20f/f pups. Primary white adipocytes were isolated from inguinal WAT of Klf4f/f or Krox20f/f adult mice. Immortalization of primary brown preadipocytes by retroviral vectors expressing SV40T was done as described previously (23). Adipogenesis of primary white preadipocytes and the 3T3-L1 cell line was carried out as described previously (19). For brown adipogenesis assays, preadipocytes were plated at a density of 1 × 105 in each well of 6-well plates in growth medium (Dulbecco's modified Eagle medium [DMEM] plus 10% FBS) 4 days before induction of adipogenesis. At day 0, cells were fully confluent and were treated with differentiation medium (DMEM plus 10% FBS, 0.1 μM insulin, and 1 nM T3) supplemented with 0.5 mM IBMX, 1 μM dexamethasone, and 0.125 mM indomethacin. Two days later, cells were changed to the differentiation medium. The medium was replenished at 2-day intervals. Fully differentiated cells were either stained with Oil Red O or subjected to gene expression analysis by qRT-PCR.
qRT-PCR.
Total RNA was extracted using TRIzol (Invitrogen) and reverse transcribed using ProtoScript II first-strand cDNA synthesis kit (NEB), following the manufacturers' instructions. qRT-PCR was done using the following SYBR green primers: Klf4, forward, 5′-GTGCCCCGACTAACCGTTG-3′, and reverse, 5′-GTCGTTGAACTCCTCGGTCT-3′; Krox20, forward, 5′-TGACTATTGTGGCCGCAAGTT-3′, and reverse, 5′-TTCTGCCGAAGGTGGATCTT-3′. SYBR green primers for other genes were described previously (24).
RNA-Seq.
RNA-Seq experiments were performed during adipogenesis of 3T3-L1 white preadipocytes and Pparγf/f immortalized brown preadipocytes. For RNA-Seq analysis of BAT, interscapular BATs were isolated from wild-type adult (8-week-old) mice or E18.5 embryos. RNA-Seq library preparation was done following the protocol described previously (18). The expression levels of each gene were indicated by units of reads per kilobase per million (RPKM).
Statistical analyses.
Quantitative data in all figures are expressed as means ± standard errors. To compare between two groups, the statistical significance was calculated using the two-tailed unpaired t test under two experimental conditions. A P value of less than 0.05 was considered statistically significant.
Accession number.
All data sets described in this paper have been deposited in NCBI Gene Expression Omnibus under accession number GSE87113.
ACKNOWLEDGMENTS
We thank Klaus Kaestner for Klf4f/f mice, Patrick Charnay for Krox20f/f mice, Ronald Evans for Pparγf/f mice, Philippe Soriano for Myf5-Cre mice, and NIDDK Genomics Core for sequencing.
This work was supported by the Intramural Research Program of NIDDK, NIH, to K.G.
Footnotes
For a companion article on this topic, see https://doi.org/10.1128/MCB.00260-16.
REFERENCES
- 1.Green H, Kehinde O. 1975. An established preadipose cell line and its differentiation in culture. II. Factors affecting the adipose conversion. Cell 5:19–27. [DOI] [PubMed] [Google Scholar]
- 2.Green H, Kehinde O. 1976. Spontaneous heritable changes leading to increased adipose conversion in 3T3 cells. Cell 7:105–113. doi: 10.1016/0092-8674(76)90260-9. [DOI] [PubMed] [Google Scholar]
- 3.Gregoire FM, Smas CM, Sul HS. 1998. Understanding adipocyte differentiation. Physiol Rev 78:783–809. [DOI] [PubMed] [Google Scholar]
- 4.Rubin CS, Hirsch A, Fung C, Rosen OM. 1978. Development of hormone receptors and hormonal responsiveness in vitro. Insulin receptors and insulin sensitivity in the preadipocyte and adipocyte forms of 3T3-L1 cells. J Biol Chem 253:7570–7578. [PubMed] [Google Scholar]
- 5.Student AK, Hsu RY, Lane MD. 1980. Induction of fatty acid synthetase synthesis in differentiating 3T3-L1 preadipocytes. J Biol Chem 255:4745–4750. [PubMed] [Google Scholar]
- 6.Rosen ED, MacDougald OA. 2006. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol 7:885–896. doi: 10.1038/nrm2066. [DOI] [PubMed] [Google Scholar]
- 7.Farmer SR. 2006. Transcriptional control of adipocyte formation. Cell Metab 4:263–273. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ge K. 2012. Epigenetic regulation of adipogenesis by histone methylation. Biochim Biophys Acta 1819:727–732. doi: 10.1016/j.bbagrm.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cristancho AG, Lazar MA. 2011. Forming functional fat: a growing understanding of adipocyte differentiation. Nat Rev Mol Cell Biol 12:722–734. doi: 10.1038/nrm3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanaka T, Yoshida N, Kishimoto T, Akira S. 1997. Defective adipocyte differentiation in mice lacking the C/EBP[beta] and/or C/EBP[delta] gene. EMBO J 16:7432. doi: 10.1093/emboj/16.24.7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosen ED, Sarraf P, Troy AE, Bradwin G, Moore K, Milstone DS, Spiegelman BM, Mortensen RM. 1999. PPAR[gamma] is required for the differentiation of adipose tissue in vivo and in vitro. Mol Cell 4:611. doi: 10.1016/S1097-2765(00)80211-7. [DOI] [PubMed] [Google Scholar]
- 12.Chen Z, Torrens JI, Anand A, Spiegelman BM, Friedman JM. 2005. Krox20 stimulates adipogenesis via C/EBPbeta-dependent and -independent mechanisms. Cell Metab 1:93–106. doi: 10.1016/j.cmet.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 13.Birsoy K, Chen Z, Friedman J. 2008. Transcriptional regulation of adipogenesis by KLF4. Cell Metab 7:339–347. doi: 10.1016/j.cmet.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scime A, Devarakonda S, Conroe HM, Erdjument-Bromage H, Tempst P, Rudnicki MA, Beier DR, Spiegelman BM. 2008. PRDM16 controls a brown fat/skeletal muscle switch. Nature 454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dumont BL, Payseur BA. 2008. Evolution of the genomic rate of recombination in mammals. Evolution 62:276–294. doi: 10.1111/j.1558-5646.2007.00278.x. [DOI] [PubMed] [Google Scholar]
- 16.Rosen ED, Spiegelman BM. 2014. What we talk about when we talk about fat. Cell 156:20–44. doi: 10.1016/j.cell.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho YW, Hong S, Jin Q, Wang L, Lee JE, Gavrilova O, Ge K. 2009. Histone methylation regulator PTIP is required for PPARgamma and C/EBPalpha expression and adipogenesis. Cell Metab 10:27–39. doi: 10.1016/j.cmet.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee JE, Wang C, Xu S, Cho YW, Wang L, Feng X, Baldridge A, Sartorelli V, Zhuang L, Peng W, Ge K. 2013. H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. eLife 2:e01503. doi: 10.7554/eLife.01503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L, Xu S, Lee J-E, Baldridge A, Grullon S, Peng W, Ge K. 2013. Histone H3K9 methyltransferase G9a represses PPAR[gamma] expression and adipogenesis. EMBO J 32:45–59. doi: 10.1038/emboj.2012.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katz JP, Perreault N, Goldstein BG, Lee CS, Labosky PA, Yang VW, Kaestner KH. 2002. The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development 129:2619–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taillebourg E, Buart S, Charnay P. 2002. Conditional, floxed allele of the Krox20 gene. Genesis 32:112–113. doi: 10.1002/gene.10062. [DOI] [PubMed] [Google Scholar]
- 22.He W, Barak Y, Hevener A, Olson P, Liao D, Le J, Nelson M, Ong E, Olefsky JM, Evans RM. 2003. Adipose-specific peroxisome proliferator-activated receptor gamma knockout causes insulin resistance in fat and liver but not in muscle. Proc Natl Acad Sci U S A 100:15712–15717. doi: 10.1073/pnas.2536828100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klein J, Fasshauer M, Ito M, Lowell BB, Benito M, Kahn CR. 1999. beta 3-Adrenergic stimulation differentially inhibits insulin signaling and decreases insulin-induced glucose uptake in brown adipocytes. J Biol Chem 274:34795–34802. doi: 10.1074/jbc.274.49.34795. [DOI] [PubMed] [Google Scholar]
- 24.Jin Q, Wang C, Kuang X, Feng X, Sartorelli V, Ying H, Ge K, Dent SY. 2014. Gcn5 and PCAF regulate PPARgamma and Prdm16 expression to facilitate brown adipogenesis. Mol Cell Biol 34:3746–3753. doi: 10.1128/MCB.00622-14. [DOI] [PMC free article] [PubMed] [Google Scholar]