Abstract
Background
The gastrointestinal tract (GIT) microbiota is essential to metabolic health, and the prevalence of the Western diet (WD) high in fat and sugar is increasing, with evidence highlighting a negative interaction between the GIT and WD, resulting in liver dysfunction. Additionally, an adverse in utero environment such as placental insufficiency resulting in low birth weight (LBW) offspring, contributes to an increased risk of metabolic diseases such as fatty liver infiltration and liver dysfunction in later life. We sought to understand the potential interactive effects of exposure to a WD upon growing LBW offspring. We postulated that LBW offspring when challenged with a poor postnatal diet, would display an altered microbiota and more severe liver metabolic dysfunction.
Methods
The fecal microbiota of normal birth weight (NBW) and LBW young guinea pig offspring, weaned onto either a control diet (CD) or WD was determined with 16S rRNA gene next generation sequencing at young adulthood following the early rapid growth phase after weaning. A liver blood chemistry profile was also performed.
Results
The life-long consumption of WD following weaning into young adulthood resulted in increased total cholesterol, triglycerides and alanine aminotransferase levels in association with an altered GIT microbiota when compared to offspring consuming CD. Neither birth weight nor sex were associated with any significant changes in microbiota alpha diversity, by measuring the Shannon’s diversity index. One hundred forty-eight operational taxonomic units were statistically distinct between the diet groups, independent of birth weight. In the WD group, significant decreases were detected in Barnesiella, Methanobrevibacter smithii and relatives of Oscillospira guillermondii, while Butyricimonas and Bacteroides spp. were increased.
Discussion
These results describe the GIT microbiota in a guinea pig model of LBW and WD associated metabolic syndrome and highlight several WD specific GIT alterations associated with human metabolic disease.
Keywords: Microbiome
Introduction
Metabolic diseases such as obesity and the related metabolic syndrome are now considered to be an epidemic and an increasing burden on health care systems (Mathers et al., 2001). The gastrointestinal tract (GIT) microbiota is essential to metabolic health, and a dysfunctional GIT is closely linked to the development of aspects of metabolic syndrome. The GIT microbiota utilizes indigestible components of our diets and some suggest it may influence calorie harvesting from food (Turnbaugh et al., 2006; Zeng et al., 2013). It also has an important role in homeostasis and the maintenance of epithelial barriers, which when degraded may contribute to inflammation leading to chronic diseases characterized by metabolic dysfunction such as non-alcoholic fatty liver disease (NAFLD) and diabetes (Bäckhed et al., 2004; Dunne et al., 2014).
Due to the divergent nutritional requirements of various bacteria residing in the gut, diet has been shown to shape the composition of the microbiota, which in turn may lead to adverse health outcomes such as metabolic syndrome (Turnbaugh et al., 2008; Turnbaugh et al., 2009). Specifically, the consumption of a typical “Western” diet (WD) high in fat and sugar has been shown by some groups to alter the microbial diversity and relative abundance of two main phyla in humans and mice, Bacteroidetes and Firmicutes (Turnbaugh et al., 2009). For these reasons, the gastrointestinal microbiota is considered one of the potential environmental factors that advance the host to a metabolically diseased state (Hildebrandt et al., 2009).
An emerging factor potentially regulating the GIT microbiota composition is early life conditioning through pregnancy and during early postnatal life. While it is not yet clear how an adverse in utero environment specifically impacts the new born microbiota, studies report that placental insufficiency outcomes are associated with an altered neonatal GIT and caecocolonic microbiota, an alteration that in some reports continues into later life (Trahair et al., 1997; Sangild, Fowden & Trahair, 2000; Fança-Berthon et al., 2010; Yan et al., 2011). This altered gut flora is associated in animal and human studies with failure of adequate postnatal growth (Trahair et al., 1997; Yan et al., 2011). In support of these observations, gut microbiota modulation by diet, prebiotics, or probiotics may modify the growth pattern of the offspring or prevent the development of adverse in utero environment-induced diseases (Luoto et al., 2010; Arrieta et al., 2014). In addition to modulating the new born gut composition, the in utero environment, resulting in a reduced fetal growth trajectory, plays a major role in setting the offspring’s risk of metabolic disease later in life (Browne, 1962; Barker et al., 1993; Barker, 2000; Yan et al., 2011). This is referred to as the “thrifty hypothesis”, whereby low birth weight (LBW) offspring experience permanent changes in their metabolic function in utero, which are determinant in later postnatal life when challenged with nutrient excess (Thorn et al., 2011). These metabolic abnormalities include fatty infiltration of the liver and liver dysfunction highlighted by elevated alanine aminotransferase (ALT) levels (Angulo et al., 1999; Hales & Barker, 2001).
Guinea pigs have been used interdependently in the study of in utero growth, fetal development, and the impact diet has on postnatal growth (Fernandez & Volek, 2006; Sarr et al., 2014; Sarr et al., 2015; Thompson et al., 2014). A limited number of studies have described the guinea pig intestinal microbiota and have highlighted an overlap of phyla present in both the guinea pig and human GIT (Yanabe et al., 2001; Takahashi et al., 2005; Hildebrand et al., 2012). The aims of the present pilot study were to determine whether an in utero environment resulting in LBW is a factor in the compositional development of the gut and hepatic manifestations of metabolic syndrome, specifically altered ALT, and to investigate how a WD may impact these outcomes in growing offspring.
Materials and Methods
Ethics statement
Animal care, maintenance, and surgeries were conducted in accordance with the standards set by the Canadian Council on Animal Care. The University of Western Ontario Animal Use Subcommittee approved all procedures (AUP # 2010-229).
Animals and diets
Time-mated pregnant Dunkin-Hartley guinea pigs (Charles River Laboratories, Wilmington, MA, USA) were housed in a temperature (20–22 °C) and humidity (30%) controlled environment with a 12 h light–dark cycle and had access to chow and tap water provided ad libitum.
Chow-fed pregnant guinea pigs underwent uterine artery ablation (UAA) surgery at mid gestation (∼32 days, term 69 days) to generate normal and low birth weight offspring (NBW and LBW, respectively) due to chronic placental insufficiency as described previously (Turner & Trudinger, 2009; Sarr et al., 2014; Thompson et al., 2014). Sows delivered spontaneously at term (∼67 days) and birth weight was recorded. Guinea pig pups from a UAA pregnancy weighing less than 85 grams were defined as LBW, and pups weighing 90 grams or greater at birth were defined as NBW (Elias et al., 2015). Five days prior to weaning the postnatal control diet (CD, TD: 110240; Harlan Laboratories, Madison, WI, USA) was introduced to the pups through the maternal feeding tray. At 15 days of age the offspring were weaned, separated by sex, weighed, housed in individual cages, and randomized to either CD or a Western diet (WD, WD: 110239; Harlan Laboratories), as described previously (Thompson et al., 2014). Briefly, the diets differed in kilocalorie density (3.4 vs 4.2 kcal g−1), but were matched for protein and macronutrients. The percentage of kilocalories for CD and WD from protein was 21.6 and 21.4, from fat was 18.4 and 45.3, and from carbohydrates was 60 and 33.3. Additionally, the WD contained 2.5 g kg−1 cholesterol. To avoid litter effects, only one LBW/NBW animal per sex from a single litter was assigned to each diet. From the time of weaning, food intake was recorded daily until sacrifice by CO2 inhalation at young adulthood ∼150 days. At sacrifice, blood was collected to quantify total cholesterol and triglyceride levels, as well as to conduct a liver blood chemistry profile (ALB, ALP, ALT, BA, BUN, GGT, and TBIL) using a Vetscan VS2 (Abaxis, Union City, CA). Fecal samples were also collected at sacrifice by emptying colon contents into a sterile bag, then immediately stored at −80 °C until further analysis.
Fecal DNA extraction
The MoBio PowerSoil® 96-Well Soil DNA Isolation Kit (Mobio, Carlsbad, CA), was used according to the modified Earth Microbiome Project standard protocols (Earth Microbiome Project, 2016). Approximately 0.25 g of each fecal sample was transferred to each well using sterile pipette tips, and extracted DNA was stored sealed at −20 °C until PCR.
Fecal sample polymerase chain reaction
Fifty microlitres of the DNA template extract was transferred to a 96-well PCR plate (Axygen, Union City, CA). The BioMek® 3000 Laboratory Automation Workstation was used for automated PCR reagent set up. Amplifications of the V4 region of the 16S ribosomal RNA gene were carried out with the primers ACACTCTTTCCCTACACGACGCTCTTCCGATCTNNNNxxxxxxxxGTGCCAGCMGCCGCGGTAA and CGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATCTNNNNxxxxxxxxGGACTACHVGGGTWTCTAAT wherein xxxxxxxx is a sample specific nucleotide barcode and the preceding sequence is a portion of the Illumina adapter sequence for library construction. Ten microlitres (2.3 pmol/µl) each of a total of 32 primers, 16 left and right with unique barcodes were arrayed in 96 well plates. Using a BioMek 3000® (Beckman Coulter, Brea, CA, USA) 2µl of the DNA template was transferred into a plate containing 10 µl of each unique primer. Then 20 µl of Promega GoTaq® Colourless Master Mix (Promega, Maddison, WI, USA), containing the necessary dNTPs, PCR reaction buffer, MgCl2, and GoTaq® DNA Polymerase was added to the DNA template and primers. The final plate was firmly sealed with a foil PCR plate cover. This plate was placed in the Eppendorf Mastercycler® thermal cycler (Eppendorf, Mississauga, ON), where the lid was kept at 105 °C. An initial hot start temperature of 95 °C was used for two minutes to activate the GoTaq®. This was followed by 25 cycles of 95 °C for one minute, 50 °C for one minute, and 72 °C for one minute. After completion, the reaction was held at 4 °C until collection and then the amplicons were stored at −20 °C.
DNA sequencing and data analysis
Samples were sent to the London Regional Genomics Centre at Robarts Research Institute (Western University, London, ON, CAN), where the sample quantification, clean-up, and sequencing were also performed. Amplicons were quantified using Picogreen (Quant-It; Life Technologies, Burlington, ON, CAN) and pooled at equimolar concentrations before cleanup (QIAquick PCR clean up; Qiagen, Germantown, MD, USA). The final samples were sequenced using the MiSeq by Illumina® platform, with 2 × 300 bp paired-end chemistry. Obtained reads were quality filtered and overlapped using USEARCH including reads with one or fewer sequencing errors, and binned into OTUs based on 97% identity (Edgar, 2010). Statistical significance in animal characteristics and hematological analysis was determined using 2-way ANOVA (GraphPad Software, San Diego, CA, USA). Diversity analysis was performed using the R package Vegan (version 2.3-2), differential abundance analysis was performed using the R package ALDEx2 (version 1.4.0) and all additional analysis was performed in base R (version 3.2.2). Utilized scripts are provided in Data S7 and demultiplexed reads are available in the NCBI Sequence Read Archive: BioProject ID PRJNA344687 (Edgar, 2010; Fernandes et al., 2013; Fernandes et al., 2014; R Development Core Team, 2008).
Results
Physical and hematological analysis of animal groups
The characteristics of each animal feeding group are displayed in Table 1. Animals body weights at sacrifice were lower in the LBW group (p = 0.011), but were not significantly different between the diet groups, despite the differences in the diets’ nutritional compositions. Both daily caloric intake and liver triglycerides were significantly elevated in the WD group (p = 0.027 and p < 0.0001). Liver blood chemistry profiles revealed no difference in alkaline phosphatase, bile acids, or blood urea nitrogen. Diet, but not birth weight, was a significant factor where WD groups had higher albumin, alanine aminotransferase, and cholesterol than CD groups (p = 0.031, p = 0.011, p < 0.0001, respectively). There was a significant diet and birth weight interaction in total bilirubin (p = 0.024).
Table 1. Animal characteristics and metadata.
| Control diet | Western diet | Diet | Birth weight | Interaction | |||
|---|---|---|---|---|---|---|---|
| NBWa (n = 9) | LBWb (n = 10) | NBW (n = 6) | LBW (n = 8) | ||||
| Distribution of Sex | F:4/M:5 | F:5/M:5 | F:2/M:4 | F:5/M:3 | |||
| Body weight (g) | 752.13 ± 36.00 | 648.46 ± 42.15 | 704.45 ± 27.9 | 585.99 ± 46.59 | NSc | P < 0.05 | NS |
| Daily energy intake (kcal) | 149.94 ± 5.48 | 192.98 ± 22.16 | 213.92 ± 9.40 | 210.81 ± 19.17 | P < 0.05 | NS | NS |
| Liver Triglycerides | 3.92 ± 0.74 | 5.51 ± 1.08 | 68.40 ± 11.39 | 75.74 ± 14.20 | P < 0.0001 | NS | NS |
| Blood analysis | |||||||
| ALBd | 4.04 ± 0.10 | 3.50 ± 0.41 | 4.20 ± 0.17 | 4.33 ± 0.17 | P < 0.05 | NS | NS |
| ALPe | 62.40 ± 11.30 | 43.00 ± 8.50 | 65.14 ± 7.67 | 76.67 ± 7.56 | NS | NS | NS |
| ALTf | 49.20 ± 3.11 | 49.00 ± 5.58 | 94.57 ± 16.40 | 125.50 ± 32.57 | P < 0.05 | NS | NS |
| BAg | 58.80 ± 21.26 | 46.00 ± 19.09 | 59.43 ± 10.19 | 63.33 ± 16.67 | NS | NS | NS |
| BUNh | 27.80 ± 35.60 | 26.75 ± 4.77 | 33.57 ± 4.35 | 28.83 ± 2.01 | NS | NS | NS |
| TBILi | 0.05 ± 0.05 | 0.20 ± 0.00 | 0.23 ± 0.04 | 0.10 ± 0.06 | NS | NS | P < 0.05 |
| Cholesterol | 77.00 ± 14.08 | 72.50 ± 15.18 | 418.00 ± 41.07 | 449.67 ± 30.41 | P < 0.0001 | NS | NS |
Notes.
NBW, Normal birth weight, or >90 g.
LBW, Low birth weight, or <85 g.
NS, Not significant.
ALB, Albumin.
ALP, Alkaline Phosphatase.
ALT, Alanine Aminotransferase.
BA, Bile Acids.
BUN, Blood Urea Nitrogen.
TBIL, Total Bilirubin.
Values are represented as mean ± the standard error of the mean. Significance tests were performed using 2-way ANOVA.
16S rRNA gene sequence-based characterization of the guinea pig fecal microbiota
Bacterial DNA amplified from fecal samples was grouped by 97% sequence similarity and assigned to a taxonomy using the Ribosomal Database Project Classifier (Wang et al., 2007). The sequences were grouped into 11 phyla, 19 classes, 29 orders, 45 families, and 73 genera, after filtering for OTUs representing 0.1% in any sample (data in Data S1). The Shannon’s index was used to measure the diversity of the individual samples (Shannon, 1948; Haegeman et al., 2013) and, surprisingly, a 2-way ANOVA did not detect a statistically significant effect of diet or birth weight groups (Fig. S1A, Data S2). Similarly, there were no significant differences between the mean number of reads per sample (Fig. S1B).
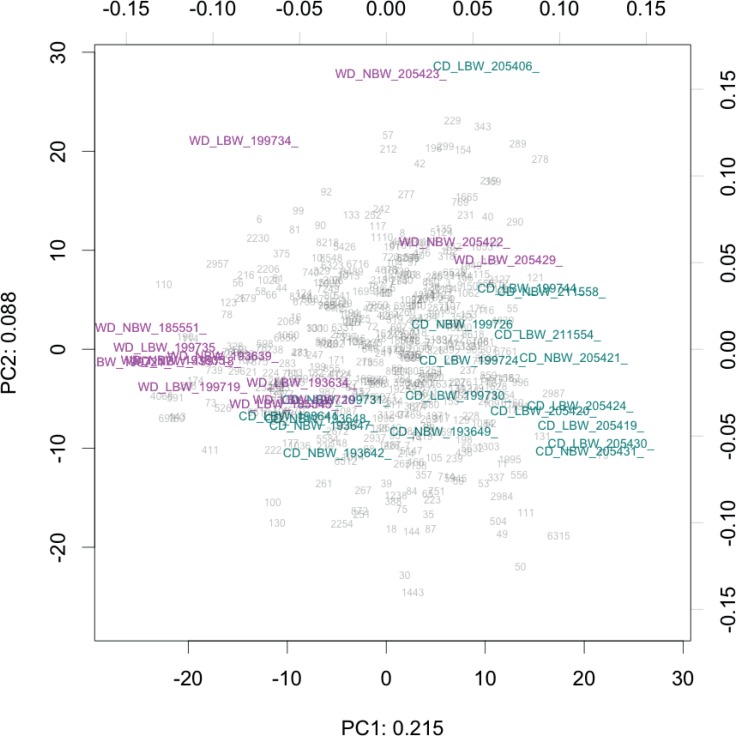
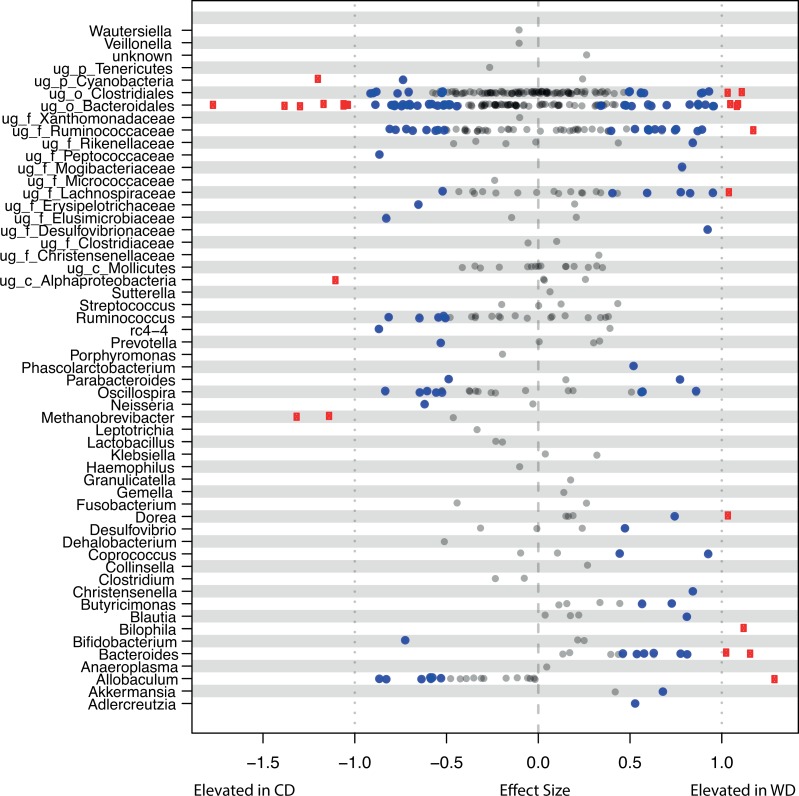
Principal component analysis (PCA) of centered log-transformed distances highlighted differences in the microbiota samples between the CD and WD fed animals (Fig. 1) (Van den Boogaart & Tolosana-Delgado, 2013). Distance on this plot represents overall dissimilarity between the microbiota profiles of the samples using the Aitchison distance which is appropriate for compositional data (Van den Boogaart & Tolosana-Delgado, 2013). The distance of each OTU from the centre of the plot is proportional to the standard deviation in the dataset (up to the limit of the projection displayed). Comparing Aitchison distance, microbiota profiles did not cluster on the plot by sex or birth weights of the animals, but did cluster distinctly when the animals were grouped by diet type (Fig. 1, Fig. S2). Diet groups were significantly different (p < 0.1) for 148 OTUs, when tested using the R package ALDEx2 with the non-parametric Wilcoxon rank-sum test with false discovery rate correction (Data S3, Fig. S3). The most significant of the 148 is OTU 22, and is most closely related to the genus Barnesiella from the family Porphyromonadaceae (p = 1.08 × 10−4) which was decreased in the WD animals. Other notable OTUs include the Archaea Methanobrevibacter (OTU 11 and 6160) and Oscillospira (OTU 77) which were reduced in the WD animals, while the genera Bacteroides (OTU 26 and 51), Bilophila (OTU 26), Coprococcus (OTU 165), and Desulfovibrio (OTU 76) were enriched in the same group. Figure 2 illustrates the differential OTUs between the diet groups and shows how individual OTUs from the same lineage behave. OTUs are plotted by effect size (the median of the ratio of the between diet group difference and the largest of the variance within groups) and Benjamini–Hochberg corrected Wilcoxon rank-sum test.
Figure 1. Compositional biplot of all samples.
Samples are coloured according to diet, OTUs are shown as grey numbers. Approximately 30% of the variance is explained in the first two components. The biplot is drawn to show the relationship between the samples (scale = 0), as opposed to OTUs.
Figure 2. Stripchart of differential OTUs between diet groups.
OTUs with a Benjamini–Hochberg corrected p-value from Wilcoxon rank-sum test <0.1 are plotted in blue. OTUs with p < 0.1 and an absolute effect size >1 are red. OTUs are summarized to genus. If genus is unknown (ug_), the lowest known taxonomic rank is stated (f_Clostridiaceae).
The most abundant phyla in all samples were Bacteroidetes and Firmicutes. Diet had a suggestive but not significant effect increasing the relative proportion of Bacteroidetes (p = 0.0832) in the WD group. The relative proportion of Firmicutes was significantly lower in the WD group compared to control (p = 0.0049). Birth weight had no effect on the relative proportions of either phylum.
Discussion
This was a pilot study investigating the gut microbiota in an established guinea pig model of metabolic syndrome. The model utilizes a combination of uterine artery ablation to induce LBW offspring, with a postnatal diet high in total fat and sugar, and produces a non-overweight phenotype with impaired vascular function, increased visceral adiposity, and liver fibrosis with fatty infiltration of the liver, hallmarks of metabolic disease (Sarr et al., 2014; Sarr et al., 2015; Thompson et al., 2014).
In the current study, birth weight was not significantly associated with an altered GIT microbiota. However, a change in GIT microbiota was observed as a function of the animals’ diet, an effect strong enough to possibly overshadow any potential influence of birth weight on the microbiota. Alterations in the relative abundance of specific OTUs in the guinea pig GIT are in agreement with both human studies and other animal models. For example, the genus Bacteroides was significantly higher in the WD group, and is observed to be elevated in overweight women, while the genus Methanobrevibacter and relatives of Oscillospira guillermondii have been associated with low BMI in humans and were both comparatively decreased in the WD group (Collado et al., 2008; Million et al., 2011; Million et al., 2013). OTU 22 is most closely related to the genus Barnesiella, and was the most significantly different between our diet groups. Barnesiella has previously been shown to be increased in a non-obese diabetic rat model, but was decreased in our WD group (Zened et al., 2012; Marietta et al., 2013). Interestingly, this organism has been shown in rodents to be a marker of health as it assists in the clearance of less desirable bacterial colonization following antibiotic use, and is important in microbiome restoration (Ubeda et al., 2013). Similarly, this bacterial group may be outcompeted in our WD group but act as a marker of health in the CD animals.
When analyzing the data at the phylum level, it was observed that the guinea pig fecal microbiota is dominated by Bacteroidetes and Firmicutes in all diet and birth weight groups, similar to other rodent reports (Eckburg et al., 2005; Hildebrand et al., 2012). It was interesting to note that the prototypical decrease in relative abundance of Bacteroidetes accompanied by an increase in Firmicutes captured through a “B∕F” ratio as observed by some in obese or metabolic syndrome studies, was not observed in our WD group (Ley et al., 2006; Turnbaugh et al., 2006; Furet et al., 2010). Diet and birth weight did not have any effect on the relative proportion of Bacteroidetes, but the proportion of Firmicutes was decreased in the WD group compared to the CD group. This is not the first study unable to replicate the “stereotypical” shift observed between B/F phyla. Indeed, a recent meta analysis concluded that the ratio of Firmicutes and Bacteroidetes is not a consistent feature when comparing human obese and lean gut microbiota (Walters, Xu & Knight, 2014). Other factors contributing to the current study’s lack of B∕F change may be that guinea pigs are herbivorous and undergo hindgut fermentation, or that changes in B/F present at a later life stage than what was investigated herein (Duncan et al., 2008; Ley et al., 2008; Schwiertz et al., 2009; Walters, Xu & Knight, 2014). We caution that the convention of using the ratio of B∕F as a marker of the microbiome in metabolic disease may not be suitable for all animal models, especially in studies investigating the animal’s native microbiota as opposed to animals colonized by the human microbiota. Functional metagenomics, or reporting changes in particular genera and species, are likely to provide more insight (Hildebrand et al., 2012).
Alanine aminotransferase is one of the most commonly used markers in screening for liver disease, and levels were elevated in our WD fed animals independent of birth weight (Miyake et al., 2012). It is known that the GIT microbiota is a major factor in shifting the host to a metabolically diseased state, and the WD fed groups not surprisingly displayed elevated ALT levels and altered microbial markers, many of which are observed in humans with metabolic syndrome (Ley et al., 2006; Tims et al., 2012; Zened et al., 2012; Marietta et al., 2013; Ubeda et al., 2013). If diet is predominantly shaping the GIT microbiota, undesirable microbial products associated with WD may be translocating to the liver via the portal vein, impacting the liver function (Moore et al., 1991; Ilan, 2012; Hu et al., 2016). This can further induce hepatic tissue injury via activation of the inflammasome and chemokine release, creating a vicious circle of liver dysfunction (Ilan, 2012).
This study reports the WD-related changes to the gut microbiota of non-overweight young-adult guinea pigs with signs of early metabolic dysfunction (Sarr et al., 2014). The in utero environment resulting in low birth weight and metabolic disease at young adulthood appeared to have no impact upon the GIT microbiota, contrary to other reports in rats (Fança-Berthon et al., 2010). This lack of birth weight associated GIT changes at the age studied was in contrast to the driving role that diet appeared to play. A large number of OTUs identified by partial 16S rRNA gene sequence analysis were significantly different based on diet. Since changes were not largely detected in response to birth weight, if the microbiome does have a role here it may be occurring on a subtler basis rather than a global microbial shift. It is also possible the dietary effect on the microbiota was overshadowing any birth-weight related effects. An increase in the relative proportion of Firmicutes and decrease in Bacteroidetes was not observed in our WD group, and microbial diversity was largely unchanged. Despite this, several differential OTUs reported in the guinea pig associated with elevated ALT are also reported to occur in association with human metabolic disease. These observations highlight the potential usefulness of the guinea pig in understanding the negative impact of a diet high in saturated fats and sugar upon the GIT and its possible contribution to the development of metabolic disease.
Supplemental Information
OTUs were filtered to 0.1% in any sample.
Shannon’s diversity and read counts are not different between birth weight or diet groups.
Output from diversity function in R (Vegan 2.3-2) performed on OTU table.
The samples are coloured according to diet and birth weight groups. The biplot is drawn to show the relationship between the OTUs [scale = 1].
Output from ALDEx2 test for significantly different OTUs between diet groups.
OTUs divergent between diet groups by Wilcoxon rank-sum test are shown. Colour corresponds to the log2 of the OTUs’ abundance.
R scripts are provided here to generate figures and data 1, 2, S2, S3, S4, S5, S6 from the provided OTU table (S1)
Acknowledgments
We wish to thank Brad Matushewski for his assistance with animal surgeries and Dr. Yves Bureau for assistance with animal group statistics.
Funding Statement
Funding was provided by the Canadian Institutes of Health Research (#MOP-209113) and the W. Garfield Weston Foundation. OS was supported by the Whaley Postdoctoral Fellowship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Kait Al conceived and designed the experiments, performed the experiments, analyzed the data, wrote the paper, prepared figures and/or tables, reviewed drafts of the paper.
Ousseynou Sarr performed the experiments, analyzed the data, reviewed drafts of the paper.
Kristyn Dunlop reviewed drafts of the paper.
Gregory B. Gloor analyzed the data, reviewed drafts of the paper.
Gregor Reid contributed reagents/materials/analysis tools, reviewed drafts of the paper.
Jeremy Burton conceived and designed the experiments, contributed reagents/materials/analysis tools, wrote the paper, reviewed drafts of the paper.
Timothy R.H. Regnault conceived and designed the experiments, performed the experiments, contributed reagents/materials/analysis tools, wrote the paper, reviewed drafts of the paper.
Animal Ethics
The following information was supplied relating to ethical approvals (i.e., approving body and any reference numbers):
The University of Western Ontario Animal Use Subcommittee approved all procedures, AUP Number: 2010-229. Animal care, maintenance, and surgeries were conducted in accordance with the standards set by the Canadian Council on Animal Care.
Data Availability
The following information was supplied regarding data availability:
The raw data has been supplied as a Supplemental File.
References
- Angulo et al. (1999).Angulo P, Keach JC, Batts KP, Lindor KD. Independent predictors of liver fibrosis in patients with nonalcoholic steatohepatitis. Hepatology. 1999;30:1356–1362. doi: 10.1002/hep.510300604. [DOI] [PubMed] [Google Scholar]
- Arrieta et al. (2014).Arrieta M-C, Stiemsma L, Amenyogbe N, Brown E, Finlay B. The intestinal microbiome in early life: health and disease. Frontiers in Immunology. 2014;5 doi: 10.3389/fimmu.2014.00427. Article 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed et al. (2004).Bäckhed F, Ding H, Wang T, Hooper LV, Koh G, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker (2000).Barker DJP. In utero programming of cardiovascular disease. Theriogenology. 2000;53:555–574. doi: 10.1016/S0093-691X(99)00258-7. [DOI] [PubMed] [Google Scholar]
- Barker et al. (1993).Barker DJP, Hales CN, Fall CHD, Osmond C, Phipps K, Clark PMS. Type 2 (non-insulin-dependent) diabetes mellitus, hypertension and hyperlipidaemia (syndrome X): relation to reduced fetal growth. Diabetologia. 1993;36:62–67. doi: 10.1007/BF00399095. [DOI] [PubMed] [Google Scholar]
- Browne (1962).Browne J. Placental insufficiency. Postgraduate Medical Journal. 1962;38:225–228. doi: 10.1136/pgmj.38.438.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado et al. (2008).Collado M, Isolauri E, Laitinen K, Salminen S. Distinct composition of gut microbiota during pregnancy in overweight and normal-weight women. The American Journal of Clinical Nutrition. 2008;88:894–899. doi: 10.1093/ajcn/88.4.894. [DOI] [PubMed] [Google Scholar]
- Duncan et al. (2008).Duncan SH, Lobley GE, Holtrop G, Ince J, Johnstone AM, Louis P, Flint HJ. Human colonic microbiota associated with diet, obesity and weight loss. International Journal of Obesity. 2008;32:1720–1724. doi: 10.1038/ijo.2008.155. [DOI] [PubMed] [Google Scholar]
- Dunne et al. (2014).Dunne JL, Triplett EW, Gevers D, Xavier R, Insel R, Danska J, Atkinson MA. The intestinal microbiome in type 1 diabetes. Clinical & Experimental Immunology. 2014;177:30–37. doi: 10.1111/cei.12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earth Microbiome Project (2016).Earth Microbiome Project EMP protocols and standards: DNA extraction protocol. http://www.earthmicrobiome.org/emp-standard-protocols/dna-extraction-protocol/ [7 July 2016];2016
- Eckburg et al. (2005).Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar (2010).Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Elias et al. (2015).Elias AA, Ghaly A, Matushewski B, Regnault TR, Richardson BS. Maternal nutrient restriction in guinea pigs as an animal model for inducing fetal growth restriction. Reproductive Sciences. 2015;23:219–227. doi: 10.1177/1933719115602773. [DOI] [PubMed] [Google Scholar]
- Fança-Berthon et al. (2010).Fança-Berthon P, Hoebler C, Mouzet E, David A, Michel C. Intrauterine growth restriction not only modifies the cecocolonic microbiota in neonatal rats but also affects its activity in young adult rats. Journal of Pediatric Gastroenterology and Nutrition. 2010;51:402–413. doi: 10.1097/MPG.0b013e3181d75d52. [DOI] [PubMed] [Google Scholar]
- Fernandes et al. (2013).Fernandes AD, Macklaim JM, Linn TG, Reid G, Gloor GB. ANOVA-like differential expression (ALDEx) analysis for mixed population RNA-seq. PLoS ONE. 2013;8:e67019–15. doi: 10.1371/journal.pone.0067019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes et al. (2014).Fernandes AD, Reid JN, Macklaim JM, McMurrough TA, Edgell DR, Gloor GB. Unifying the analysis of high-throughput sequencing datasets: characterizing RNA-seq, 16S rRNA gene sequencing and selective growth experiments by compositional data analysis. Microbiome. 2014;2:1–13. doi: 10.1186/2049-2618-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez & Volek (2006).Fernandez M, Volek J. Guinea pigs: a suitable animal model to study lipoprotein metabolism, atherosclerosis and inflammation. Nutrition & Metabolism. 2006;3:1–6. doi: 10.1186/1743-7075-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furet et al. (2010).Furet J-P, Kong L-C, Tap J, Poitou C, Basdevant A, Bouillot J-L, Mariat D, Corthier G, Dore J, Henegar C, Rizkalla S, Clément K. Differential adaptation of human gut microbiota to bariatric surgery–induced weight loss links with metabolic and low-grade inflammation markers. Diabetes. 2010;59:3049–3057. doi: 10.2337/db10-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haegeman et al. (2013).Haegeman B, Hamelin J, Moriarty J, Neal P, Dushoff J, Weitz J. Robust estimation of microbial diversity in theory and in practice. The ISME Journal. 2013;7:1092–1101. doi: 10.1038/ismej.2013.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hales & Barker (2001).Hales NC, Barker DJ. The thrifty phenotype hypothesis. British Medical Bulletin. 2001;60:5–20. doi: 10.1093/bmb/60.1.5. [DOI] [PubMed] [Google Scholar]
- Hildebrand et al. (2012).Hildebrand F, Ebersbach T, Nielsen H, Li X, Sonne S, Bertalan M, Dimitrov P, Madsen L, Qin J, Wang J, Raes J, Kristiansen K, Licht T. A comparative analysis of the intestinal metagenomes present in guinea pigs (Cavia porcellus) and humans (Homo sapiens) BMC Genomics. 2012;13:514. doi: 10.1186/1471-2164-13-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt et al. (2009).Hildebrandt MA, Hoffmann C, Sherrill-mix SA, Keilbaugh SA, Hamady M, Chen Y, Knight R, Ahima RS, Bushman F, Wu GD. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology. 2009;137:1716–1724. doi: 10.1053/j.gastro.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu et al. (2016).Hu Y, Zhang H, Li J, Cong X, Chen Y, He G, Chi Y, Liu Y. Gut-derived lymphocyte recruitment to liver and induce liver injury in non-alcoholic fatty liver disease mouse model. Journal of Gastroenterology and Hepatology. 2016;31:676–684. doi: 10.1111/jgh.13183. [DOI] [PubMed] [Google Scholar]
- Ilan (2012).Ilan Y. Leaky gut and the liver: a role for bacterial translocation in nonalcoholic steatohepatitis. World Journal of Gastroenterology. 2012;18:2609–2610. doi: 10.3748/wjg.v18.i21.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley et al. (2008).Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey R, Bircher SJ, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, Gordon JI. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley et al. (2006).Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- Luoto et al. (2010).Luoto R, Kalliomaki M, Laitinen K, Isolauri E. The impact of perinatal probiotic intervention on the development of overweight and obesity: follow-up study from birth to 10 years. International Journal of Obesity. 2010;34:1531–1537. doi: 10.1038/ijo.2010.50. [DOI] [PubMed] [Google Scholar]
- Marietta et al. (2013).Marietta EV, Gomez AM, Yeoman C, Tilahun AY, Clark CR, Luckey DH, Murray JA, White BA, Kudva YC, Rajagopalan G. Low incidence of spontaneous Type 1 diabetes in non-obese diabetic mice raised on gluten-free diets is associated with changes in the intestinal microbiome. PLoS ONE. 2013;8:e78687–9. doi: 10.1371/journal.pone.0078687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathers et al. (2001).Mathers CD, Vos T, Lopez AD, Salomon J, Ezzati M. National burden of disease studies: a pratical guide. World Health Organization; Geneva: 2001. [Google Scholar]
- Million et al. (2013).Million M, Angelakis E, Maraninchi M, Henry M, Giorgi R, Valero R, Vialettes B, Raoult D. Correlation between body mass index and gut concentrations of Lactobacillus reuteri, Bifidobacterium animalis, Methanobrevibacter smithii and Escherichia coli. International Journal of Obesity. 2013;37:1460–1466. doi: 10.1038/ijo.2013.20. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Million et al. (2011).Million M, Maraninchi M, Henry M, Armougom F, Richet H, Carrieri P, Valero R, Raccah D, Vialettes B, Raoult D. Obesity-associated gut microbiota is enriched in Lactobacillus reuteri and depleted in Bifidobacterium animalis and Methanobrevibacter smithii. International Journal of Obesity. 2011;36:817–825. doi: 10.1038/ijo.2011.153. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Miyake et al. (2012).Miyake T, Kumagi T, Hirooka M, Koizumi M, Furukawa S, Ueda T, Tokumoto Y, Ikeda Y, Abe M, Kitai K, Hiasa Y, Matsuura B, Onji M. Metabolic markers and ALT cutoff level for diagnosing nonalcoholic fatty liver disease: a community-based cross-sectional study. Journal of Gastroenterology. 2012;47:696–703. doi: 10.1007/s00535-012-0534-y. [DOI] [PubMed] [Google Scholar]
- Moore et al. (1991).Moore FA, Moore EE, Poggetti R, McAnena OJ, Peterson VM, Abernathy CM, Parsons PE. Gut bacterial translocation via the portal vein: a clinical perspective with major torso trauma. Journal of Trauma. 1991;31:629–636. doi: 10.1097/00005373-199105000-00006. [DOI] [PubMed] [Google Scholar]
- R Development Core Team (2008).R Development Core Team . R Foundation for Statistical Computing; Vienna: 2008. [Google Scholar]
- Sangild, Fowden & Trahair (2000).Sangild P, Fowden A, Trahair J. How does the foetal gastrointestinal tract develop in preparation for enteral nutrition after birth? Livestock Production Science. 2000;66:141–150. doi: 10.1016/S0301-6226(00)00221-9. [DOI] [Google Scholar]
- Sarr et al. (2015).Sarr O, Blake A, Thompson JA, Zhao L, Rabicki K, Walsh JC, Welch I, Regnault TR. The differential effects of low birth weight and western diet consumption upon early life hepatic fibrosis development in guinea pig. The Journal of Physiology. 2015;594:1753–1772. doi: 10.1113/JP271777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarr et al. (2014).Sarr O, Thompson JA, Zhao L, Lee T-Y, Regnault TR. Low birth weight male guinea pig offspring display increased visceral adiposity in early adulthood. PLoS ONE. 2014;9:e98433–13. doi: 10.1371/journal.pone.0098433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwiertz et al. (2009).Schwiertz A, Taras D, Schäfer K, Beijer S, Bos NA, Donus C, Hardt PD. Microbiota and SCFA in lean and overweight healthy subjects. Obesity. 2009;18:190–195. doi: 10.1038/oby.2009.167. [DOI] [PubMed] [Google Scholar]
- Shannon (1948).Shannon CE. A mathematical theory of communication. The Bell System Technical Journal. 1948;27:379–423. [Google Scholar]
- Takahashi et al. (2005).Takahashi T, Karita S, Yahaya M, Goto M. Radial and axial variations of bacteria within the cecum and proximal colon of guinea pigs revealed by PCR-DGGE. Bioscience, Biotechnology, and Biochemistry. 2005;69:1790–1792. doi: 10.1271/bbb.69.1790. [DOI] [PubMed] [Google Scholar]
- Thompson et al. (2014).Thompson JA, Sarr O, Piorkowska K, Gros R, Regnault TR. Low birth weight followed by postnatal over-nutrition in the guinea pig exposes a predominant player in the development of vascular dysfunction. The Journal of Physiology. 2014;592:5429–5443. doi: 10.1113/jphysiol.2014.275016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorn et al. (2011).Thorn S, Rozance P, Brown L, Hay W. The intrauterine growth restriction phenotype: fetal adaptations and potential implications for later life insulin resistance and diabetes. Seminars in Reproductive Medicine. 2011;29:225–236. doi: 10.1055/s-0031-1275516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tims et al. (2012).Tims S, Derom C, Jonkers D, Vlietinck R, Saris W, Kleerebezem M, Vos W, Zoetendal E. Microbiota conservation and BMI signatures in adult monozygotic twins. The ISME Journal. 2012;7:707–717. doi: 10.1038/ismej.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trahair et al. (1997).Trahair JF, DeBarro TM, Robinson JS, Owens JA. Restriction of nutrition in utero selectively inhibits gastrointestinal growth in fetal sheep. The Journal of Nutrition. 1997;127:637–641. doi: 10.1093/jn/127.4.637. [DOI] [PubMed] [Google Scholar]
- Turnbaugh et al. (2008).Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host & Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh et al. (2006).Turnbaugh P, Ley R, Mahowald M, Magrini V, Mardis E, Gordon J. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1131. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Turnbaugh et al. (2009).Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Science Translational Medicine. 2009;1:1–12. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner & Trudinger (2009).Turner AJ, Trudinger BJ. A modification of the uterine artery restriction technique in the guinea pig fetus produces asymmetrical ultrasound growth. Placenta. 2009;30:236–240. doi: 10.1016/j.placenta.2008.11.023. [DOI] [PubMed] [Google Scholar]
- Ubeda et al. (2013).Ubeda C, Bucci V, Caballero S, Djukovic A, Toussaint NC, Equinda M, Lipuma L, Ling L, Gobourne A, No D, Taur Y, Jenq RR, Van den Brink MR, Xavier JB, Pamer EG. Intestinal microbiota containing Barnesiella species cures vancomycin-resistant Enterococcus faecium colonization. Infection and Immunity. 2013;81:965–973. doi: 10.1128/IAI.01197-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Boogaart & Tolosana-Delgado (2013).Van den Boogaart GK, Tolosana-Delgado R. Analyzing compositional data with R. Springer-Verlag; Berlin Heidelberg: 2013. [Google Scholar]
- Walters, Xu & Knight (2014).Walters W, Xu Z, Knight R. Meta-analyses of human gut microbes associated with obesity and IBD. FEBS Letters. 2014;588:4223–4233. doi: 10.1016/j.febslet.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2007).Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and Environmental Microbiology. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan et al. (2011).Yan X, Huang Y, Wang H, Du M, Hess B, Ford S, Nathanielsz P, Zhu M. Maternal obesity induces sustained inflammation in both fetal and offspring large intestine of sheep. Inflammatory Bowel Diseases. 2011;17:1513–1522. doi: 10.1002/ibd.21539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanabe et al. (2001).Yanabe M, Shibuya M, Gonda T, Asai H, Tanaka T, Sudou K, Narita T, Matsui T, Itoh K. Establishment of specific pathogen-free guinea-pig colonies using limited-flora guinea-pigs associated with conventional guinea-pig flora, and monitoring of their cecal flora. Experimental Animals/Japanese Association for Laboratory Animal Science. 2001;50:105–113. doi: 10.1538/expanim.50.105. [DOI] [PubMed] [Google Scholar]
- Zened et al. (2012).Zened A, Combes S, Cauquil L, Mariette J, Klopp C, Bouchez O, Troegeler-Meynadier A, Enjalbert F. Microbial ecology of the rumen evaluated by 454 GS FLX pyrosequencing is affected by starch and oil supplementation of diets. FEMS Microbiology Ecology. 2012;83:504–514. doi: 10.1111/1574-6941.12011. [DOI] [PubMed] [Google Scholar]
- Zeng et al. (2013).Zeng H, Liu J, Jackson MI, Zhao FQ, Yan L, Combs GF. Fatty liver accompanies an increase in Lactobacillus species in the hind gut of C57BL/6 mice fed a high-fat diet. Journal of Nutrition. 2013;143:627–631. doi: 10.3945/jn.112.172460. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
OTUs were filtered to 0.1% in any sample.
Shannon’s diversity and read counts are not different between birth weight or diet groups.
Output from diversity function in R (Vegan 2.3-2) performed on OTU table.
The samples are coloured according to diet and birth weight groups. The biplot is drawn to show the relationship between the OTUs [scale = 1].
Output from ALDEx2 test for significantly different OTUs between diet groups.
OTUs divergent between diet groups by Wilcoxon rank-sum test are shown. Colour corresponds to the log2 of the OTUs’ abundance.
R scripts are provided here to generate figures and data 1, 2, S2, S3, S4, S5, S6 from the provided OTU table (S1)
Data Availability Statement
The following information was supplied regarding data availability:
The raw data has been supplied as a Supplemental File.