Abstract
Nucleotide sequences of the gapA gene, encoding the glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase, were determined for 16 strains of Salmonella and 13 strains of Escherichia coli recovered from natural populations. Pairs of sequences from strains representing the eight serovar groups of Salmonella differed, on average, at 3.8% of nucleotide sites and 1.1% of inferred amino acids, and comparable values for E. coli were an order of magnitude smaller (0.2% and 0.1%, respectively). The rate of substitution at synonymous sites was significantly higher for codons specifying the catalytic domain of the enzyme than for those encoding the NAD(+)-binding domain, but the nonsynonymous substitution rate showed the opposite relationship. For Salmonella, statistical tests for nonrandom clustering of polymorphic sites failed to provide evidence that intragenic recombination or gene conversion has contributed to the generation of allelic diversity. The topology of a tree constructed from the gapA sequences was generally similar to that of phylogenetic trees of the strains based on multilocus enzyme electrophoresis, but the level of divergence of gapA in Salmonella group V from other Salmonella and E. coli strains is much greater than that indicated by DNA hybridization for the genome as a whole.
Full text
PDF

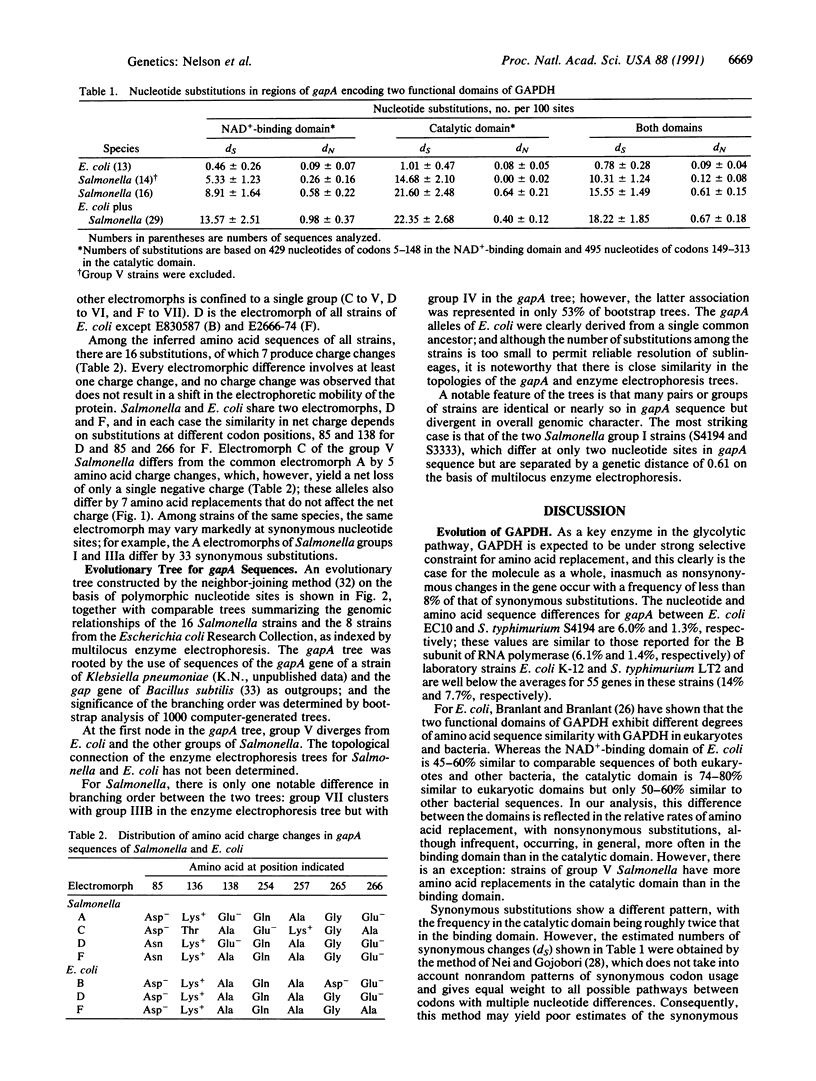
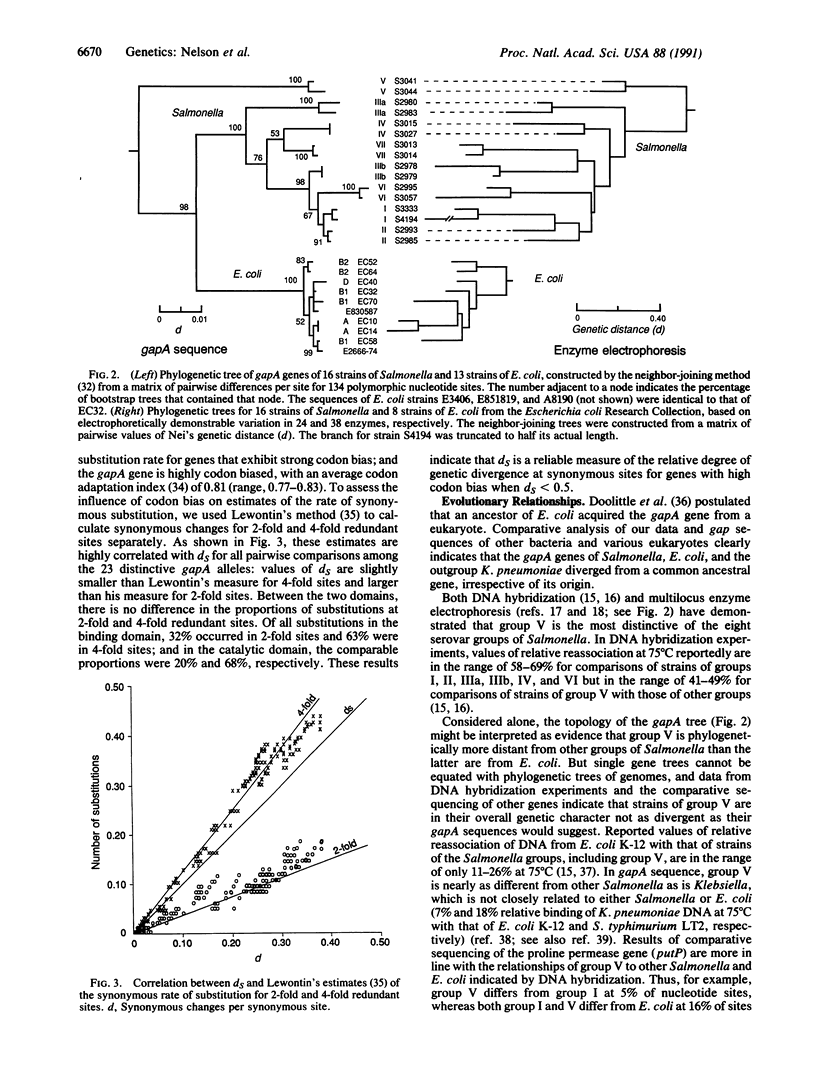

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alefounder P. R., Perham R. N. Identification, molecular cloning and sequence analysis of a gene cluster encoding the class II fructose 1,6-bisphosphate aldolase, 3-phosphoglycerate kinase and a putative second glyceraldehyde 3-phosphate dehydrogenase of Escherichia coli. Mol Microbiol. 1989 Jun;3(6):723–732. doi: 10.1111/j.1365-2958.1989.tb00221.x. [DOI] [PubMed] [Google Scholar]
- Barcak G. J., Wolf R. E., Jr Comparative nucleotide sequence analysis of growth-rate-regulated gnd alleles from natural isolates of Escherichia coli and from Salmonella typhimurium LT-2. J Bacteriol. 1988 Jan;170(1):372–379. doi: 10.1128/jb.170.1.372-379.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran P., Musser J. M., Helmuth R., Farmer J. J., 3rd, Frerichs W. M., Wachsmuth I. K., Ferris K., McWhorter A. C., Wells J. G., Cravioto A. Toward a population genetic analysis of Salmonella: genetic diversity and relationships among strains of serotypes S. choleraesuis, S. derby, S. dublin, S. enteritidis, S. heidelberg, S. infantis, S. newport, and S. typhimurium. Proc Natl Acad Sci U S A. 1988 Oct;85(20):7753–7757. doi: 10.1073/pnas.85.20.7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branlant G., Branlant C. Nucleotide sequence of the Escherichia coli gap gene. Different evolutionary behavior of the NAD+-binding domain and of the catalytic domain of D-glyceraldehyde-3-phosphate dehydrogenase. Eur J Biochem. 1985 Jul 1;150(1):61–66. doi: 10.1111/j.1432-1033.1985.tb08988.x. [DOI] [PubMed] [Google Scholar]
- Crosa J. H., Brenner D. J., Ewing W. H., Falkow S. Molecular relationships among the Salmonelleae. J Bacteriol. 1973 Jul;115(1):307–315. doi: 10.1128/jb.115.1.307-315.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle R. F., Feng D. F., Anderson K. L., Alberro M. R. A naturally occurring horizontal gene transfer from a eukaryote to a prokaryote. J Mol Evol. 1990 Nov;31(5):383–388. doi: 10.1007/BF02106053. [DOI] [PubMed] [Google Scholar]
- DuBose R. F., Dykhuizen D. E., Hartl D. L. Genetic exchange among natural isolates of bacteria: recombination within the phoA gene of Escherichia coli. Proc Natl Acad Sci U S A. 1988 Sep;85(18):7036–7040. doi: 10.1073/pnas.85.18.7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goullet P., Picard B. Comparative electrophoretic polymorphism of esterases and other enzymes in Escherichia coli. J Gen Microbiol. 1989 Jan;135(1):135–143. doi: 10.1099/00221287-135-1-135. [DOI] [PubMed] [Google Scholar]
- Herzer P. J., Inouye S., Inouye M., Whittam T. S. Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J Bacteriol. 1990 Nov;172(11):6175–6181. doi: 10.1128/jb.172.11.6175-6181.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi R. G., Ochman H. Production of single-stranded DNA templates by exonuclease digestion following the polymerase chain reaction. Nucleic Acids Res. 1989 Jul 25;17(14):5865–5865. doi: 10.1093/nar/17.14.5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori H., Osawa S. Evolution of ribosomal proteins in Enterobacteriaceae. J Bacteriol. 1978 Mar;133(3):1089–1095. doi: 10.1128/jb.133.3.1089-1095.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll J. S., Moxon E. R. Capsulation in distantly related strains of Haemophilus influenzae type b: genetic drift and gene transfer at the capsulation locus. J Bacteriol. 1990 Mar;172(3):1374–1379. doi: 10.1128/jb.172.3.1374-1379.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Minor L., Popoff M. Y., Laurent B., Hermant D. Individualisation d'une septième sous-espèce de Salmonella: S. choleraesuis subsp. indica subsp. nov. Ann Inst Pasteur Microbiol. 1986 Sep-Oct;137B(2):211–217. [PubMed] [Google Scholar]
- Le Minor L., Véron M., Popoff M. Taxonomie des Salmonella. Ann Microbiol (Paris) 1982 Sep-Oct;133(2):223–243. [PubMed] [Google Scholar]
- Lewontin R. C. Inferring the number of evolutionary events from DNA coding sequence differences. Mol Biol Evol. 1989 Jan;6(1):15–32. doi: 10.1093/oxfordjournals.molbev.a040532. [DOI] [PubMed] [Google Scholar]
- Milkman R., Bridges M. M. Molecular evolution of the Escherichia coli chromosome. III. Clonal frames. Genetics. 1990 Nov;126(3):505–517. doi: 10.1093/genetics/126.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkman R., Crawford I. P. Clustered third-base substitutions among wild strains of Escherichia coli. Science. 1983 Jul 22;221(4608):378–380. doi: 10.1126/science.6346486. [DOI] [PubMed] [Google Scholar]
- Nei M., Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986 Sep;3(5):418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Nei M., Jin L. Variances of the average numbers of nucleotide substitutions within and between populations. Mol Biol Evol. 1989 May;6(3):290–300. doi: 10.1093/oxfordjournals.molbev.a040547. [DOI] [PubMed] [Google Scholar]
- Ochman H., Selander R. K. Standard reference strains of Escherichia coli from natural populations. J Bacteriol. 1984 Feb;157(2):690–693. doi: 10.1128/jb.157.2.690-693.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H., Wilson A. C. Evolution in bacteria: evidence for a universal substitution rate in cellular genomes. J Mol Evol. 1987;26(1-2):74–86. doi: 10.1007/BF02111283. [DOI] [PubMed] [Google Scholar]
- Pesole G., Bozzetti M. P., Lanave C., Preparata G., Saccone C. Glutamine synthetase gene evolution: a good molecular clock. Proc Natl Acad Sci U S A. 1991 Jan 15;88(2):522–526. doi: 10.1073/pnas.88.2.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves M. W., Evins G. M., Heiba A. A., Plikaytis B. D., Farmer J. J., 3rd Clonal nature of Salmonella typhi and its genetic relatedness to other salmonellae as shown by multilocus enzyme electrophoresis, and proposal of Salmonella bongori comb. nov. J Clin Microbiol. 1989 Feb;27(2):313–320. doi: 10.1128/jcm.27.2.313-320.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley M., Anilionis A. Conservation and variation of nucleotide sequences within related bacterial genomes: enterobacteria. J Bacteriol. 1980 Jul;143(1):366–376. doi: 10.1128/jb.143.1.366-376.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987 Jul;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sawyer S. A., Dykhuizen D. E., Hartl D. L. Confidence interval for the number of selectively neutral amino acid polymorphisms. Proc Natl Acad Sci U S A. 1987 Sep;84(17):6225–6228. doi: 10.1073/pnas.84.17.6225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer S. Statistical tests for detecting gene conversion. Mol Biol Evol. 1989 Sep;6(5):526–538. doi: 10.1093/oxfordjournals.molbev.a040567. [DOI] [PubMed] [Google Scholar]
- Selander R. K., Caugant D. A., Ochman H., Musser J. M., Gilmour M. N., Whittam T. S. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl Environ Microbiol. 1986 May;51(5):873–884. doi: 10.1128/aem.51.5.873-884.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987 Feb 11;15(3):1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith N. H., Beltran P., Selander R. K. Recombination of Salmonella phase 1 flagellin genes generates new serovars. J Bacteriol. 1990 May;172(5):2209–2216. doi: 10.1128/jb.172.5.2209-2216.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith N. H., Selander R. K. Molecular genetic basis for complex flagellar antigen expression in a triphasic serovar of Salmonella. Proc Natl Acad Sci U S A. 1991 Feb 1;88(3):956–960. doi: 10.1073/pnas.88.3.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens J. C. Statistical methods of DNA sequence analysis: detection of intragenic recombination or gene conversion. Mol Biol Evol. 1985 Nov;2(6):539–556. doi: 10.1093/oxfordjournals.molbev.a040371. [DOI] [PubMed] [Google Scholar]
- Stoltzfus A., Leslie J. F., Milkman R. Molecular evolution of the Escherichia coli chromosome. I. Analysis of structure and natural variation in a previously uncharacterized region between trp and tonB. Genetics. 1988 Oct;120(2):345–358. doi: 10.1093/genetics/120.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viaene A., Dhaese P. Sequence of the glyceraldehyde-3-phosphate dehydrogenase gene from Bacillus subtilis. Nucleic Acids Res. 1989 Feb 11;17(3):1251–1251. doi: 10.1093/nar/17.3.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittam T. S., Ochman H., Selander R. K. Multilocus genetic structure in natural populations of Escherichia coli. Proc Natl Acad Sci U S A. 1983 Mar;80(6):1751–1755. doi: 10.1073/pnas.80.6.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]



