Abstract
Rationale: The potential role of the airway microbiota in dictating immune responses and infection outcomes in HIV-associated pneumonia is largely unknown.
Objectives: To investigate whether microbiologically and immunologically distinct subsets of patients with HIV and pneumonia exist and are related to mortality.
Methods: Bronchoalveolar lavage samples from Ugandan patients with HIV and pneumonia (n = 182) were obtained at study enrollment (following antibiotic treatment); patient demographics including 8- and 70-day mortality were collected. Lower airway bacterial community composition was assessed via amplification and sequencing of the V4 region of the 16S ribosomal RNA gene. Host immune response gene expression profiles were generated by quantitative polymerase chain reaction using RNA extracted from bronchoalveolar lavage fluid. Liquid and gas chromatography mass spectrometry was used to profile serum metabolites.
Measurements and Main Results: Based on airway microbiome composition, most patients segregated into three distinct groups, each of which were predicted to encode metagenomes capable of producing metabolites characteristically enriched in paired serum samples from these patients. These three groups also exhibited differences in mortality; those with the highest rate had increased ceftriaxone administration and culturable Aspergillus, and demonstrated significantly increased induction of airway T-helper cell type 2 responses. The group with the lowest mortality was characterized by increased expression of T-cell immunoglobulin and mucin domain 3, which down-regulates T-helper cell type 1 proinflammatory responses and is associated with chronic viral infection.
Conclusions: These data provide evidence that compositionally and structurally distinct lower airway microbiomes are associated with discrete local host immune responses, peripheral metabolic reprogramming, and different rates of mortality.
Keywords: HIV, microbiota, pneumonia, immune response, mortality
At a Glance Commentary
Scientific Knowledge on the Subject
Patients with HIV and pneumonia exhibit greater lung microbial diversity than patients with pneumonia without HIV; however, it is unknown if distinct pathogenic lower airway microbiomes exist in patients with HIV and pneumonia and whether these relate to host immune response and mortality within this population.
What This Study Adds to the Field
Using a cohort of 182 patients with HIV and pneumonia, we identified three compositionally distinct microbial community states in the lower airways. Each exhibits unique metagenomic functional capacity, induces distinct lower airway immune responses, and is associated with a unique profile of circulating metabolites and with different rates of mortality. These results provide evidence that microbiologically and immunologically distinct subsets of patients with HIV and pneumonia exist and that these distinctions are related to clinical outcomes, thus arguing for the potential need to tailor therapy based on the specific microbiome dysbiosis and related immune and metabolic dysfunction exhibited by these patients.
Sub-Saharan Africa accounts for 71% of persons estimated to be living with HIV infection worldwide, with 63,000 AIDS-related deaths per year in Uganda alone (1). Pulmonary infections pose a common and frequently fatal comorbidity in patients with HIV in Africa; two of the most prevalent are tuberculosis (TB) and bacterial pneumonia, incident in approximately 80% of this patient population (2). Overall, TB is the leading cause of death in patients with HIV worldwide (3, 4). In HIV–TB coendemic areas, bacterial pneumonia is a common cause of hospital admission, with mortality rates over 30% even with antiretroviral and antibiotic treatments (5, 6).
Even in the absence of acute respiratory infection, patients with HIV exhibit a broader breadth of lower airway bacterial taxa compared with that detected in healthy subjects (7), indicating that HIV infection may present a risk factor for developing pulmonary infection. Despite high morbidity and mortality within this population, little is known about factors that influence heterogeneity in patient outcomes, and, specifically, whether variation in airway microbiota composition and immune response are related to patient survival. We demonstrated, in a non–HIV-infected, antimicrobial-treated pneumonia cohort, that following antimicrobial treatment, a precipitous decline in airway microbiome diversity and domination of the community by a distinct respiratory pathogen (e.g., Streptococcus pneumonia or Pseudomonas aeruginosa) is associated with increased 28-day mortality (8). In a study of 60 Ugandan patients with HIV and antimicrobial-treated pneumonia, patients with reduced airway bacterial microbiota richness and diversity exhibited higher bacterial burden and increased expression of proinflammatory tumor necrosis factor-α and matrix metalloprotinase-9 (9), thus providing the first evidence that HIV airway microbiota composition is related to immune response.
These observations led us to hypothesize that in the context of HIV-associated immune dysfunction and antimicrobial administration, acute pneumonia patient subsets can be identified based on their lower airway bacterial composition. We further rationalized that these compositionally distinct airway microbiota function as discrete pathogenic units that induce characteristic airway immune responses and are associated with mortality. To address this hypothesis, we examined clinical and demographic factors related to the bacterial airway microbiome, and relationships between community composition, host immune response, and patient outcomes in a large cohort of Ugandan patients with HIV and pneumonia. Some of the results of these studies have been previously reported in an abstract (10).
Methods
Subjects and Sample Collection
We enrolled subjects with HIV admitted to Mulago Hospital in Kampala, Uganda for acute pneumonia from October 2009 to December 2011 as part of the Lung MicroCHIP (Lung Microbiome in Cohorts of HIV-Infected Persons) study. Patients underwent two sputum acid-fast bacilli smear examinations to diagnose pulmonary TB. Acid-fast bacilli smear-negative patients underwent bronchoscopy with bronchoalveolar lavage (BAL) for clinical diagnosis, with 10 ml set aside for microbiome analysis (9). Bronchoscopy was performed a median of 3 days after hospital admission (interquartile range, 1–4 d). More than 98% of subjects had received antibiotics before bronchoscopy. Serum was collected on hospital Day 1, at enrollment. Clinical data were collected and diagnoses were assigned as previously described (9). Study endpoint was mortality follow-up at 70 days after bronchoscopy (see Methods in the online supplement).
Ethics Statement
The Makerere University School of Medicine Research Ethics Committee, the Mulago Hospital Research and Ethics Committee, the Uganda National Council for Science and Technology, and the University of California San Francisco Committee on Human Research approved the protocol. Subjects provided written, informed consent.
DNA and RNA Extraction
Total DNA and RNA were extracted from whole BAL in parallel using an AllPrep DNA/RNA extraction kit (Qiagen, Hilden, Germany) (see Methods in the online supplement) (11). RNA quality and purity were assessed as previously described (9).
16S Ribosomal RNA Gene Amplification and Sequencing
The V4 region of bacterial 16S ribosomal RNA (rRNA) gene was amplified using primers with multiplex sequencing barcodes (see Table E1 and Methods in the online supplement). A mock community was used to monitor for contamination and standardize across runs. Sequencing was performed using a MiSeq platform and MiSeq Control Software version 2.2.0 (Illumina, San Diego, California). Raw sequencing data are available via the SRA database under SRP077299.
Microbiome Data Processing
A total of 251-bp paired-end sequence reads were assembled using FLASh (12), and quality-trimmed using QIIME (see Methods section in the online supplement) (13). Chimeras were removed using ChimeraSlayer (14). Each sample was rarefied 100 times to 100,000 reads in the R environment (15); the centroid of each sample distribution was subsequently used for analysis (n = 182). Greengenes database May 2013 (16) was used to classify taxa; singleton operational taxonomical units were removed.
Immune Gene Expression
Total RNA (0.5 μg) was reverse transcribed; cDNA gene expression was assayed using real-time polymerase chain reaction and analyzed using the delta-delta CT method to normalize gene expression (see Methods in the online supplement) (17).
Metabolic Profiling
Metabolic profiles were generated from 100 μl of patient serum (n = 30) by ultrahigh-performance liquid and gas chromatography-tandem mass spectrometry at Metabolon according to a standard protocol.
Microbial and Statistical Analyses
Microbial analyses were performed using QIIME software (13). Results were visualized using Emperor (18). Metagenomic predictions were generated using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) (19). Procrustes (“transform_coordinate_matrices.py” script, “-r 1000”) and Mantel tests (“compare_distance_matrices.py” script) were performed in QIIME using Bray Curtis dissimilarity (compositional dissimilarity based on taxon relative abundance) (20). Statistical analyses (e.g., one-way analysis of variance [ANOVA], Kruskal-Wallis) were performed in the R environment. Dirichlet multinomial mixtures (DMM) and log-rank test were performed using the DirichletMultinomial (21) and survival packages, respectively. Permutational multivariate analysis of variance (PERMANOVA [22], vegan version 2.3.0, 1,000 permutations) and principal coordinate analysis (PCoA) were performed using weighted UniFrac (23, 24) and Canberra dissimilarity measurements. PERMANOVA independently considers each factor (e.g., age, sex) against bacterial community β-diversity variance, permuting data independently, and thus does not require false-discovery correction. The resulting R2 provides the proportion of variation explained (e.g., a factor that has a R2 = 0.021, explains 2% of the variation in community composition).
Results
Lower Airway Microbiota Composition Is Associated with Demographic, Clinical, and Microbiologic Factors
Lower airway bacterial microbiota profiles of 190 Ugandan patients with HIV with acute pneumonia were generated by 16S rRNA amplicon sequencing of whole BAL fluid (see Figure E1A: read depth). Overall, 182 samples with sufficient sequence reads and adequate bacterial community coverage were used for all microbiota analyses (see Figure E1B). A total of 6,915 operational taxonomical units (>97% 16S rRNA V4-sequence similarity; range, 124–869; median, 335.5 taxa per sample) were identified indicating robust bacterial presence.
Demographic and clinical data (see Table E2) were used in PERMANOVA analysis (22). PERMANOVA allows for the identification of factors related to observed variation in bacterial β-diversity (intersample bacterial compositional differences); we measured β-diversity using a weighted UniFrac dissimilarity matrix, which considers phylogenetic relatedness and species abundance in distance calculations (24). Sex (R2 = 0.021; P < 0.017) (see Figure E2A), consumption of alcohol ever (R2 = 0.015; P < 0.045) (see Figure E2B), the presence of culturable Aspergillus in BAL (R2 = 0.038; P < 0.004) (see Figure E2C), BAL or sputum culture positivity for Mycobacterium (R2 = 0.027; P < 0.021) (see Figure E2D), and ceftriaxone administration within the last 2 weeks, or at the time of bronchoscopy (R2 = 0.016; P < 0.040 and R2 = 0.061; P < 0.001, respectively ) (Table 1, Figure 1A) were significantly related to airway bacterial community composition. Seventy-day mortality trended strongly toward a relationship with airway microbiota composition (Canberra [β-diversity distance based on taxa presence/absence]; R2 = 0.0061; P < 0.053). Mortality trended strongly towards significance using a Canberra but not a weighted UniFrac dissimilarity matrix, suggesting that presence (or absence) of particular taxa in airway communities is related to mortality, rather than relative abundance or phylogenetic relatedness of community members present.
Table 1.
Clinical, Demographic, and Microbiologic Features Are Significantly Associated with Airway Bacterial β-Diversity in Patients with HIV and Pneumonia
| Variable | Sample (n) | Yes/No* | Min–Max (Median) | PERMANOVA |
|
|---|---|---|---|---|---|
| R2 | P Value | ||||
| Clinical and demographic | |
||||
| 70-day mortality† | 182 | 143/39 (live/dead) | 0.006 | 0.053 | |
| Alcohol ever consumed | 182 | 113/69 | 0.015 | 0.045 | |
| Ceftriaxone at bronchoscopy | 174 | 54/120 | 0.061 | 0.001 | |
| Ceftriaxone within last 2 wk | 178 | 137/41 | 0.016 | 0.040 | |
| Culture identified Aspergillus | 157 | 15/142 | 0.038 | 0.004 | |
| Sex | 182 | 110/72 (F/M) | 0.021 | 0.017 | |
| TB-positive by culture | 182 | 40/1/141 (positive/scanty/negative) | 0.027 | 0.021 | |
| Microbiologic | |
||||
| Chao1 | 182 | 170–1,326 (484.1) | 0.080 | 0.001 | |
| Faith’s phylogenetic diversity | 182 | 8.792–45.83 (21.97) | 0.091 | 0.001 | |
| Observed species | 182 | 39–865 (340.5) | 0.076 | 0.001 | |
| Shannon diversity | 182 | 0.642–6.427 (4.011) | 0.169 | 0.001 | |
| Simpson diversity | 182 | 0.112–0.977 (0.867) | 0.154 | 0.001 | |
Definition of abbreviations: Max = maximum; Min = minimum; PERMANOVA = permutational multivariate analysis of variance; TB = tuberculosis.
Unless otherwise noted.
PERMANOVA value calculated using a Canberra distance matrix.
Figure 1.
Antibiotic administration, α-diversity, and probabilistic modeling differentiate bacterial community types within the lower airways of patients with HIV and pneumonia. Principal coordinate analysis of n = 182 lower airway bronchoalveolar lavage bacterial community profiles of Ugandan patients with HIV and pneumonia illustrates that (A) ceftriaxone (in yellow vs. no ceftriaxone in purple), a third-generation cephalosporin, administered at time of bronchoscopy is significantly associated with community composition (PERMANOVA, R2 = 0.061, P < 0.001), as is (B) Shannon diversity (PERMANOVA, R2 = 0.17, P < 0.001, scaled from high [red] to low [blue]). (C) Based on Laplace approximation, for which a lower value indicates a better model fit, Dirichlet multinomial mixtures (DMM) identified two compositionally distinct bacterial microbiota (n = 136 and n = 46) in the lower airways of patients with HIV and pneumonia. (D) Principal coordinate analysis illustrates that DMM-defined lower airway bacterial communities are compositionally distinct (PERMANOVA, R2 = 0.246, P < 0.001). PC = principal coordinate; PERMANOVA = permutational multivariate analysis of variance.
Because microbes engage in interspecies cell-cell communication that dictates abundance and behavior of other microorganisms in their environment (25, 26), we rationalized that interspecies interactions also occur in complex multispecies bacterial microbiota, resulting in deterministic community structures. Indeed, Shannon diversity index (which considers abundance and richness; PERMANOVA: R2 = 0.17, P < 0.001) (Figure 1B), Faith’s phylogenetic diversity (phylogenetic variation, R2 = 0.09, P < 0.001), Chao1 index (species richness estimator, R2 = 0.08, P < 0.001), and observed species richness (total species, R2 = 0.08, P < 0.001), were all significantly associated with airway bacterial β-diversity. These α-diversity indices (measurements of variation within samples) explained a greater degree of microbial community variability (8–17%) than clinical or demographic features (reflected in the strength of PCoA groupings in Figures 1 and E1), suggesting that microbiologic influences seem to play a larger role in defining airway taxonomic content than clinical-demographic features.
Patients with HIV and Pneumonia Stratify into Two Groups Based on Bacterial Community Composition
DMM (21) modeling examines taxa frequencies and determines how many “metacommunities” or microbial community states (MCS) exist within a dataset. Application of DMM to our cohort identified two significantly distinct MCS (n = 46 and n = 136; R2 = 0.246; P < 0.001) using a Laplace approximation (Figures 1C and 1D), which evaluates model fit (lowest Laplace value corresponds to the number of metacommunities that best fit the model) (Figure 1C). Specific cooccurring bacterial families were characteristically enriched in these two groups; MCS1 microbiota was characteristically dominated by Pseudomonadaceae, which typically cooccurred with Sphingomonadaceae and Prevotellaceae. The second, larger group, exhibited a reciprocal gradient of Streptococcaceae or Prevotellaceae domination, which we designated MCS2A and MCS2B, respectively. Streptococcaceae-dominated MCS2A communities coassociated with Prevotellaceae and Veillonellaceae, and Prevotellaceae-dominated MCS2B assemblages with Veillonellaceae and Streptococcaceae (Figure 2A). These distinct microbial states exhibited significant differences in diversity, with MCS1 exhibiting the lowest mean diversity compared with MCS2A or MCS2B communities (Faith’s phylogenetic diversity; one-way ANOVA, P < 0.001) (Figure 2B).
Figure 2.
Two compositionally distinct lower airway microbial states exist in patients with HIV and pneumonia. (A) Mean community composition of each state at the family level. (B) Lower airway phylogenetic diversity differs significantly across microbial states (one-way analysis of variance [ANOVA], P < 0.001). (C) Principal coordinate analysis plot illustrating weighted UniFrac distances permits visualization of MCS1 (green) and the sister states MCS2A (blue) and MCS2B (red), which collectively explain a significant proportion of bacterial community variation (PERMANOVA, R2 = 0.67, P < 0.001) within the lower airways of this patient population. Patient lower airway communities that do not fit one of these three mean community compositions are depicted in gray. MCS = microbial community state; PC = principal coordinate; PERMANOVA = permutational multivariate ANOVA.
Using dominant family to classify samples, PCoA-ordination of weighted UniFrac distance matrices confirmed a strong and significant relationship between MCS class and bacterial β-diversity (PERMANOVA, R2 = 0.670, P < 0.001) (Figure 2C), corroborating the existence of compositionally distinct microbial states. Removal of the dominant family reads and reapplication of DMM to the remaining data yielded the same two groups (n = 46, n = 136), indicating that dominant family is not the sole defining feature of these airway microbiota.
Lower Airway States Are Related to Clinical and Microbiologic Factors
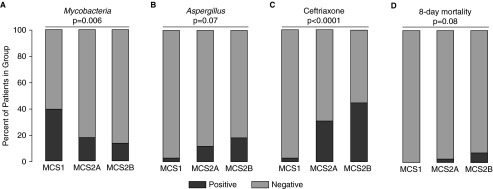
We next asked whether the specific factors that explained the variation in bacterial β-diversity were differentially associated with microbial state (see Table E2). Neither sex nor alcohol consumption significantly differed across groups; however, MCS1 communities had significantly higher Mycobacterium detection (chi-square, P = 0.006) (Figure 3A), whereas MCS2B communities exhibited increased culturable Aspergillus (chi-square, P = 0.07) (Figure 3B). In parallel, the MCS2B group had significantly increased ceftriaxone administered (n = 29/65), whereas MCS1 patients were almost never treated with ceftriaxone (n = 1/36, chi-square, P < 0.0001) (Figure 3C). Because ceftriaxone administration may be reflective of infection severity, we compared variables associated with disease severity on study enrollment (e.g., fever, sputum production, chest pain) between ceftriaxone-treated and -untreated patients; however, no statistically significant differences were observed.
Figure 3.
Culture positivity for Mycobacterium or Aspergillus, and antibiotic administration and mortality, differ between microbial community states (MCS). (A) Mycobacterium (chi-square test, P = 0.006) or (B) Aspergillus (P = 0.07) culture positivity, (C) ceftriaxone administration at bronchoscopy (P < 0.0001), and (D) mortality after 1 week of enrollment (P = 0.08) differ among microbial states (positive in black, negative in gray).
Mortality was tracked from enrollment through 70 days after bronchoscopy. MCS2B patients exhibited the most deaths at 1 week after enrollment (n = 5/67), whereas MCS1 patients all survived (P = 0.08) (Figure 3D). At 70 days, MCS2B patients still had the highest mortality (22%), followed by MCS2A (16%) and MCS1 patients (13%), although this trend did not reach significance (log-rank test, P > 0.05) (see Figure E3).
Lower Airway Microbial States Are Predicted to Encode Functionally Distinct Metagenomes
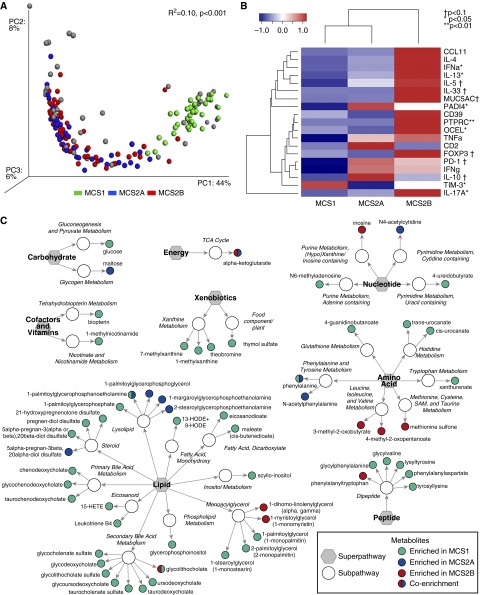
We next predicted the metagenomic content of each microbial state using the PICRUSt (19) package. Each microbial state was predicted to encode significantly distinct metagenomes and enriched for a characteristic set of gene pathways (PERMANOVA, R2 = 0.10, P < 0.001) (Figure 4A). A total of 238 Kyoto Encyclopedia of Genes and Genomes (27, 28) pathways differed significantly between the three groups (Kruskal-Wallis, 329 pathways tested, q < 0.05) (see Table E3). Despite decreased community diversity, MCS1 communities were predicted to encode a greater range of functional pathways compared with the other groups (Kruskal-Wallis, pairwise, P < 0.001, q < 0.05). This group was predicted to be significantly enriched for a broad range of pathways involved in β-lactam, linoleic, and arachidonic acid, and tryptophan metabolism, most of which (69%) were encoded by Pseudomonadaceae in these communities. MCS2A bacterial communities were enriched for pathways involved in biosynthesis of flavonols and ion channels, whereas MCS2B bacterial communities encoded glycan metabolism and glycosphingolipid biosynthesis pathways and lacked type II polyketide biosynthesis. Few pathways were predicted to be significantly enriched in MCS2B, indicating that associated increased mortality risk may be either driven by differential expression of pathways shared across compositionally distinct communities, or that nonbacterial species, such as Aspergillus, detected with greater frequency in this microbial state, contribute substantially to their associated pathogenesis.
Figure 4.
Airway microbial states are predicted to encode distinct metagenomes, and each is shown to induce different lower airway immunologic responses and is associated with significantly different serum metabolomes. (A) Principal coordinate analysis of in silico metagenomic predictions (generated using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States) indicates that the variation in predicted metagenomic content is significantly explained by microbial community states (MCS) designation (PERMANOVA, R2 = 0.10, P < 0.001). Predicted metagenomes of non-MCS samples (gray) share functional similarity with MCS samples. (B) Quantitative real-time polymerase chain reaction array assessing immune-associated gene expression within the lower airways of patients identifies specific cytokines that differ significantly in relative expression across MCS. Pathway significance: *P < 0.05; †P < 0.1; **P < 0.01. (C) Comparative liquid and gas chromatography–tandem mass spectrometry metabolomic analysis of paired patient serum identified 60 metabolites that differed significantly among all three groups (Kruskal-Wallis, P < 0.05). Enriched in MCS1 versus MCS 2A and 2B, green; MCS2A versus MCS1 and 2B, blue; MCS2B versus MCS1 and 2A, red; bi-colored circles indicate enrichment in both MCS versus the non-depicted MCS. HETE = hydroxyeicosatetraenoic acid; HODE = hydroxyoctadecadienoic acid; PC = principal coordinate; PERMANOVA = permutational multivariate analysis of variance; SAM = S-adenosyl methionine; TCA = tricarboxylic acid cycle.
Microbial Community States Induce Distinct and Characteristic Lower Airway Immune Responses
RNA extracted from a subset of compositionally representative BAL samples (n = 10/MCS) was used to analyze expression of a diverse panel of immune markers, chosen for their known associations with HIV, chronic bacterial infections, or airway inflammatory responses. mRNA expression levels (glyceraldehyde phosphate dehydrogenase–normalized) were used to generate a multivariate profile of host immune response. PERMANOVA analysis, which in this case was used to assess whether profiles of immune gene expression were related to MCS, indicated that a significant relationship existed (PERMANOVA, R2 = 0.168, P < 0.005). Specific immune responses were significantly enriched in particular MCS (one-way ANOVA, P < 0.05) (Figure 4B). For example, MCS1 patients exhibited significantly higher expression of T-cell immunoglobulin and mucin domain (TIM)-3, a glycoprotein expressed by T and innate cells, that down-regulates T-helper cell type (Th) 1 activity and proinflammatory responses (29), and plays a key role in T-cell dysfunction that occurs during chronic viral infection (30, 31).
By contrast, MCS2A demonstrated the lowest TIM-3 expression and significantly increased expression of protein-arginine deiminase type-4 (PADI4), which converts arginine to citrulline, an α-amino acid post-translationally incorporated into histones, filaggrin, and proteins involved in myelination (32). Additionally, this MCS trended toward significantly higher levels of IL-10 (antiinflammatory cytokine) and programmed cell death protein 1 (T-cell negative regulator and exhaustion marker), and lower levels of forkhead box P3 (master regulator of regulatory T cells) expression. MCS2B subjects displayed IFN-α, which characteristically protects against infection in immunocompetent subjects, but is associated with rapid disease progression in HIV infection (33). IL-13 (mediator of Th2 cell function), occludin/ELL domain-containing protein 1 (maintains and regulates tight junctions), and protein tyrosine phosphatase receptor type C (expressed on microvesicles produced by HIV-infected cells) were also significantly increased. These patients also trended toward increased expression of IL-5 (mediator of Th2 cell function, which stimulates B-cell growth), MUC5AC (the primary airway mucin), programmed cell death protein 1, and IL-33 (proinflammatory cytokine that induces Th2 responses), indicating a significant Th2 skew.
Lower Airway Microbial States Are Associated with Distinct Circulating Metabolites
Paired serum from patients for whom BAL immune profiles were generated (n = 30) were examined using liquid and gas chromatography–mass spectrometry to determine whether distinct systemic metabolic profiles were associated with airway MCS. A total of 60 metabolites differed significantly between all three groups (Kruskal-Wallis, P < 0.05) (Figure 4C; see Table E4). As the in silico metagenomic analysis predicted, MCS1 patients were characterized by significant enrichment of xanthurenate (a tryptophan metabolite) and arachidonic acid metabolites, including the eicosanoids leukotriene B4, a proinflammatory lipid-mediator, and 15-hydroxyeicosatetraenoic acid, which induces pulmonary vasoconstriction and edema (34). In addition, MCS1 patients were significantly enriched for multiple products of primary and secondary bile acid metabolism (e.g., chenodeoxycholate, glycodeoxycholate, taurochenodeoxycholate, and ursodeoxycholate), several of which have been shown to up-regulate inflammatory responses along with LPS (35), indicating that the activities of the gastrointestinal microbiome may also contribute to the tone of host inflammation in MCS1.
As predicted, MCS2B patients were characterized by significantly reduced relative levels of circulating metabolites compared with MCS1 patients. However, significant increases in amino acid metabolites 3-methyl-2-oxobutyrate and 4-methyl-2-oxopentanoate (both involved in valine and leucine metabolism), monoacylglycerols associated with lipid metabolism (1-dihomo-linolenylglycerol and 1-myristoylglycerol), and inosine (purine metabolism) were significantly enriched. MCS2A patients exhibited a mixture of metabolites identified in the other MCS but at lower concentrations, with a unique increase in lysolipid and pyrimidine metabolism and a decrease in monoacylglycerols. Thus, products of several of the biosynthetic or metabolic pathways predicted to discriminate between patients with specific airway MCS were significantly and differentially enriched in their circulation.
To verify that specific MCS, their predicted metagenomes, local airway immune responses, and serum metabolites were interrelated, we applied Procrustes (13, 36) and Mantel (37) analyses. Both confirmed a strong and significant correlation between each of these data matrices. We examined the correlation between bacterial community composition and (1) PICRUSt metagenomic prediction (Procrustes: r2 = 0.513, P < 0.001; Mantel: r2 = 0.674, P < 0.001), (2) airway immune expression (Procrustes: r2 = 0.147, P = 0.031; Mantel: r2 = 0.122, P = 0.067), or (3) serum metabolites (Procrustes: r2 = 0.414, P < 0.001; Mantel r2 = 0.286, P < 0.001). This result indicates that our patients with HIV and pneumonia who possess distinct airway MCS exhibit corresponding features of immune dysfunction and a characteristic peripheral metabolome.
Discussion
Factors that influence pneumonia outcomes in patients with HIV are poorly defined. We hypothesized that the airway microbiome may influence these outcomes. Three distinct microbial states were identified in this study; they exhibited significant differences in α-diversity, culturable Aspergillus or Mycobacterium, ceftriaxone administration, immune responses, and metabolic signatures, and trended toward differences in mortality. Recent work by Cribbs and colleagues (38) demonstrated patients with HIV are enriched for pneumonia-associated bacteria, including Streptococcus, even in the absence of airway infection, and exhibit a distinct metabolic microenvironment compared with healthy subjects. Together with this study, our work suggests that specific lower airway microbial states may lead to functionally relevant metabolic shifts that relate to distinct pathways of disease pathogenesis in individuals with HIV.
Segal and colleagues (39) have shown that healthy individuals who have an enrichment of oral taxa, including Prevotella, within their lower airways, exhibit increased inflammatory cytokines and Th17 cells. This corroborates our findings that Prevotellaceae-dominated airway microbiota promote inflammation within the lower airways, including IL-17A expression. Recent studies have demonstrated that composition of the airway microbiota influences susceptibility to Aspergillus infection (40), and that HIV-associated airway disease is related to fungal community alterations, including Aspergillus enrichment (41). Our data support these findings and suggest that Aspergillus prospers in a Prevotellaceae-dominated microbiota in the context of a Th2-skewed airway immune response.
Several recent studies have confirmed the capacity of multiple Aspergillus species to induce Th2 responses (42), particularly in early stage airway infection (43), suggesting that this species may not simply cocolonize MCS2B airways, but may actively define immunologic responses characteristic of this patient subgroup. Patients with this airway microbiota state were more likely to have been administered ceftriaxone and exhibited the highest mortality rates; one possible conclusion from these observations is that ceftriaxone administration selectively enriches for an MCS2B microbial community, and that their interkingdom microbial activities elicit a host immune response that increases mortality risk. However, the paucity of preantibiotic bronchoscopic samples, which are both ethically and logistically difficult to obtain, precludes definitive conclusions on whether ceftriaxone administration is responsible for the presence of this more severe MCS, or whether MCS2B assemblages preexisted in these patients’ airways before hospitalization.
MCS1 patients, who exhibited the lowest levels of profiled immune activation markers, were predicted to be enriched for pathways involved in linoleic and arachidonic acid metabolism. Leukotriene B4, a product of arachidonic acid metabolism typically produced by leukocytes in response to inflammatory mediators, was detected in significantly increased concentrations in these patients’ serum. Although circulating leukotriene B4 in MCS1 patients is likely produced by leukocytes, our data suggest that microbial metabolism of arachidonic acid may contribute to their circulating leukotriene B4 and that microbial-derived lipid inflammation may underlie their immune dysfunction. Mycobacterium was more prevalent in MCS1 patients, who also exhibited a significant increase in TIM-3 expression. This is consistent with the findings of Sada-Ovalle and colleagues (44) who demonstrated in vivo surface expression of TIM-3 on macrophages infected with Mycobacterium tuberculosis.
MCS2A seems to represent an intermediate microbial state, between the MCS2B and MCS1 groups in terms of clinical associations, α-diversity, composition, metabolites, and immune expression. This raises the possibility that airway microbiota may be dynamic and transition through distinct microbiologic states, particularly under antimicrobial selective pressure; however, large longitudinal studies are necessary to address this possibility. Nonetheless, patients with MCS2A were uniquely characterized by increased lower airways PADI4 expression. Extracellular bronchial PADI4 has been shown to citrullinate the innate immune defensin human cathelicidin LL-37/human cationic antimicrobial protein-18, rendering it less efficient at neutralizing lipopolysaccharide. PADI4 is detected in the airways of patients with chronic obstructive pulmonary disease, who also exhibit impaired antibacterial response against Streptococcus (45), indicating that MCS2A patients, who exhibit expansion of Streptococcaceae and induction of PADI4, may also have diminished capacity to respond to the dominant bacterial family present in their airways.
Although lower airway colonization is considered uniformly detrimental to patients, we show that specific, repeated airway microbiome states, discriminated on the basis of microbial composition, function, host immune response, and clinical outcomes, exist in subsets of patients with HIV and pneumonia. Although most patients fall into the three microbial states described, we recognize that not all patients belong to these groupings, which is not surprising in light of recent work by Twigg and colleagues (7) showing far greater variation across lower airway communities in patients with advanced HIV than healthy individuals. Though from a taxonomic standpoint, these samples did not fall into a specific MCS, their predicted metagenomes were consistent with those of MCS samples. This indicates that although currently economically challenging for large sample sets, metagenomic function may offer a preferred approach to define community states. In addition, larger cohorts of patients are necessary to sufficiently power studies examining other rarer microbial states and their immunologic and clinical implications. This cohort provides insight into Ugandan patients with HIV and pneumonia; however, these results may have limited applicability to patients in Western countries because of differences in patient demographics, laboratory testing, and antibiotic availability, and high HIV–TB coprevalence in Uganda. Furthermore, although BAL allows identification of general microbiota patterns within the lower airways, examining spatial-specific microbiota and their interactions with the host requires lung biopsies or brushings, which are beyond the scope of this study.
Although this study did not use paired oral-BAL samples to control for oral contamination, we have previously shown that oral and lower airway microbiota within patients with HIV and pneumonia display niche specificity (11). Our data trend toward a significant relationship between mortality and airway MCS. We calculated that to achieve an 80% likelihood of detecting a significant difference in mortality (power, 0.8), 100 patients per MCS would have to be studied, underscoring the need and utility of larger cohorts. Despite these limitations, this study identified several factors that shape microbial community composition in the lower airways of patients with HIV and pneumonia. Moreover, it identifies distinct bacterial microbiota states that repeat over large numbers of patients and builds an argument that pneumonia patient heterogeneity, with respect to both immunologic and clinical outcomes, may be related to compositional and functional differences in airway microbiomes.
Acknowledgments
Acknowledgment
The authors thank the patients and staff of Mulago Hospital for their contributions to the study. They also thank the research team who enrolled the patients who participated in this study. They thank Emily K. Cope and Kei E. Fujimura for assistance with sequencing analysis and figure design.
Footnotes
Supported by National Institutes of Health/NHLBI awards U01 HL098964, K24 HL087713, and R01 HL090335.
Author Contributions: L.H. and S.V.L. designed the study. W.W., I.A., P.B., S.K., S.F., S.S., E.C., J.L.D., and L.H. trained personnel in Uganda, enrolled patients, collected and shipped bronchoalveolar lavage and serum samples, and obtained demographic and clinical data. S.I. performed DNA extraction, amplification, 16S rRNA sequencing, and initial processing. D.L.L. profiled host immune responses and prepared samples for metabolomics analysis. M.K.S. analyzed clinical, microbiota, immune response, and metabolomic data. A.A.F. and M.R.S. provided statistical guidance. M.K.S. and S.V.L. wrote the manuscript. All authors reviewed and approved the final manuscript.
Originally Published in Press as DOI: 10.1164/rccm.201603-0523OC on July 22, 2016
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.World Health Organization. The gap report. Geneva, Switzerland: Joint United Nations Programme on HIV/AIDS; 2014.
- 2.World Health Organization. UNAIDS report on the global AIDS epidemic 2013. Geneva, Switzerland: Joint United Nations Programme on HIV/AIDS; 2013.
- 3.Badri M, Ehrlich R, Wood R, Pulerwitz T, Maartens G. Association between tuberculosis and HIV disease progression in a high tuberculosis prevalence area. Int J Tuberc Lung Dis. 2001;5:225–232. [PubMed] [Google Scholar]
- 4.Chaisson RE, Martinson NA. Tuberculosis in Africa: combating an HIV-driven crisis. N Engl J Med. 2008;358:1089–1092. doi: 10.1056/NEJMp0800809. [DOI] [PubMed] [Google Scholar]
- 5.French N, Gordon SB, Mwalukomo T, White SA, Mwafulirwa G, Longwe H, Mwaiponya M, Zijlstra EE, Molyneux ME, Gilks CF. A trial of a 7-valent pneumococcal conjugate vaccine in HIV-infected adults. N Engl J Med. 2010;362:812–822. doi: 10.1056/NEJMoa0903029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marshall CS, Curtis AJ, Spelman T, O’Brien DP, Greig J, Shanks L, du Cros P, Casas EC, da Fonseca MS, Athan E, et al. Impact of HIV-associated conditions on mortality in people commencing anti-retroviral therapy in resource limited settings. PLoS One. 2013;8:e68445. doi: 10.1371/journal.pone.0068445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Twigg HL, III, Knox KS, Zhou J, Crothers KA, Nelson DE, Toh E, Day RB, Lin H, Gao X, Dong Q, et al. Effect of advanced HIV infection on the respiratory microbiome. Am J Respir Crit Care Med. 2016;194:226–235. doi: 10.1164/rccm.201509-1875OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flanagan JL, Brodie EL, Weng L, Lynch SV, Garcia O, Brown R, Hugenholtz P, DeSantis TZ, Andersen GL, Wiener-Kronish JP, et al. Loss of bacterial diversity during antibiotic treatment of intubated patients colonized with Pseudomonas aeruginosa. J Clin Microbiol. 2007;45:1954–1962. doi: 10.1128/JCM.02187-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iwai S, Huang D, Fong S, Jarlsberg LG, Worodria W, Yoo S, Cattamanchi A, Davis JL, Kaswabuli S, Segal M, et al. The lung microbiome of Ugandan HIV-infected pneumonia patients is compositionally and functionally distinct from that of San Franciscan patients. PLoS One. 2014;9:e95726. doi: 10.1371/journal.pone.0095726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shenoy M, Iwai S, Lin D, Worodria W, Ayakaka I, Byanyima P, Kaswabuli S, Fong S, Stone SA, Chang E, et al. Functionally and immunologically distinct lung microbiomes of HIV-infected pneumonia patients are significantly related to mortality outcomes [abstract] Am J Respir Crit Care Med. 2015;191:A4699. doi: 10.1164/rccm.201603-0523OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwai S, Fei M, Huang D, Fong S, Subramanian A, Grieco K, Lynch SV, Huang L. Oral and airway microbiota in HIV-infected pneumonia patients. J Clin Microbiol. 2012;50:2995–3002. doi: 10.1128/JCM.00278-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, et al. Human Microbiome Consortium. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2008. [Google Scholar]
- 16.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 18.Vázquez-Baeza Y, Pirrung M, Gonzalez A, Knight R. EMPeror: a tool for visualizing high-throughput microbial community data. Gigascience. 2013;2:16. doi: 10.1186/2047-217X-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bray JR, Curtis JT. An ordination of upland forest communities of southern Wisconsin. Ecol Monogr. 1957;27:325–349. [Google Scholar]
- 21.Holmes I, Harris K, Quince C. Dirichlet multinomial mixtures: generative models for microbial metagenomics. PLoS One. 2012;7:e30126. doi: 10.1371/journal.pone.0030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46. [Google Scholar]
- 23.Lozupone C, Hamady M, Knight R. UniFrac: an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rutherford ST, Bassler BL. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med. 2012;2:a012427. doi: 10.1101/cshperspect.a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Antonic V, Stojadinovic A, Zhang B, Izadjoo MJ, Alavi M. Pseudomonas aeruginosa induces pigment production and enhances virulence in a white phenotypic variant of Staphylococcus aureus. Infect Drug Resist. 2013;6:175–186. doi: 10.2147/IDR.S49039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rahman AN, Clayton K, Mujib S, Fong IW, Ostrowski MA. TIM-3 and its immunoregulatory role in HIV infection. J Clin Cell Immunol. 2012;S7:007. [Google Scholar]
- 30.Jones RB, Ndhlovu LC, Barbour JD, Sheth PM, Jha AR, Long BR, Wong JC, Satkunarajah M, Schweneker M, Chapman JM, et al. Tim-3 expression defines a novel population of dysfunctional T cells with highly elevated frequencies in progressive HIV-1 infection. J Exp Med. 2008;205:2763–2779. doi: 10.1084/jem.20081398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Golden-Mason L, Palmer BE, Kassam N, Townshend-Bulson L, Livingston S, McMahon BJ, Castelblanco N, Kuchroo V, Gretch DR, Rosen HR. Negative immune regulator Tim-3 is overexpressed on T cells in hepatitis C virus infection and its blockade rescues dysfunctional CD4+ and CD8+ T cells. J Virol. 2009;83:9122–9130. doi: 10.1128/JVI.00639-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muller S, Radic M. Citrullinated autoantigens: from diagnostic markers to pathogenetic mechanisms. Clin Rev Allergy Immunol. 2015;49:232–239. doi: 10.1007/s12016-014-8459-2. [DOI] [PubMed] [Google Scholar]
- 33.Cha L, Berry CM, Nolan D, Castley A, Fernandez S, French MA. Interferon-alpha, immune activation and immune dysfunction in treated HIV infection. Clin Transl Immunology. 2014;3:e10. doi: 10.1038/cti.2014.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burhop KE, Selig WM, Malik AB. Monohydroxyeicosatetraenoic acids (5-HETE and 15-HETE) induce pulmonary vasoconstriction and edema. Circ Res. 1988;62:687–698. doi: 10.1161/01.res.62.4.687. [DOI] [PubMed] [Google Scholar]
- 35.Mobraten K, Haugbro T, Karlstrom E, Kleiveland CR, Lea T. Activation of the bile acid receptor TGR5 enhances LPS-induced inflammatory responses in a human monocytic cell line. J Recept Signal Transduct Res. 2015;35:402–409. doi: 10.3109/10799893.2014.986744. [DOI] [PubMed] [Google Scholar]
- 36.Gower JC. Generalized procrustes analysis. Psychometrika. 1975;40:33–51. [Google Scholar]
- 37.Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27:209–220. [PubMed] [Google Scholar]
- 38.Cribbs SK, Uppal K, Li S, Jones DP, Huang L, Tipton L, Fitch A, Greenblatt RM, Kingsley L, Guidot DM, et al. Correlation of the lung microbiota with metabolic profiles in bronchoalveolar lavage fluid in HIV infection. Microbiome. 2016;4:3. doi: 10.1186/s40168-016-0147-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Segal LN, Clemente JC, Tsay J-CJ, Koralov SB, Keller BC, Wu BG, Li Y, Shen N, Ghedin E, Morris A, et al. Enrichment of the lung microbiome with oral taxa is associated with lung inflammation of a Th17 phenotype. Nature Microbiology. 2016;1:16031. doi: 10.1038/nmicrobiol.2016.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kolwijck E, van de Veerdonk FL. The potential impact of the pulmonary microbiome on immunopathogenesis of Aspergillus-related lung disease. Eur J Immunol. 2014;44:3156–3165. doi: 10.1002/eji.201344404. [DOI] [PubMed] [Google Scholar]
- 41.Cui L, Lucht L, Tipton L, Rogers MB, Fitch A, Kessinger C, Camp D, Kingsley L, Leo N, Greenblatt RM, et al. Topographic diversity of the respiratory tract mycobiome and alteration in HIV and lung disease. Am J Respir Crit Care Med. 2015;191:932–942. doi: 10.1164/rccm.201409-1583OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Homma T, Kato A, Bhushan B, Norton JE, Suh LA, Carter RG, Gupta DS, Schleimer RP. Role of Aspergillus fumigatus in triggering protease-activated receptor-2 in airway epithelial cells and skewing the cells toward a T-helper 2 bias. Am J Respir Cell Mol Biol. 2015;54:60–70. doi: 10.1165/rcmb.2015-0062OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Urb M, Snarr BD, Wojewodka G, Lehoux M, Lee MJ, Ralph B, Divangahi M, King IL, McGovern TK, Martin JG, et al. Evolution of the immune response to chronic airway colonization with Aspergillus fumigatus hyphae. Infect Immun. 2015;83:3590–3600. doi: 10.1128/IAI.00359-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sada-Ovalle I, Chávez-Galán L, Torre-Bouscoulet L, Nava-Gamiño L, Barrera L, Jayaraman P, Torres-Rojas M, Salazar-Lezama MA, Behar SM. The Tim3-galectin 9 pathway induces antibacterial activity in human macrophages infected with Mycobacterium tuberculosis. J Immunol. 2012;189:5896–5902. doi: 10.4049/jimmunol.1200990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kilsgård O, Andersson P, Malmsten M, Nordin SL, Linge HM, Eliasson M, Sörenson E, Erjefält JS, Bylund J, Olin AI, et al. Peptidylarginine deiminases present in the airways during tobacco smoking and inflammation can citrullinate the host defense peptide LL-37, resulting in altered activities. Am J Respir Cell Mol Biol. 2012;46:240–248. doi: 10.1165/rcmb.2010-0500OC. [DOI] [PubMed] [Google Scholar]