An improved understanding of the molecular causes and modifiers of pulmonary arterial hypertension (PAH) has emerged during the last several decades. However, the survival of patients with PAH remains poor, and there is no curative therapy (1–6). Further, specific molecular information to directly inform decision-making at the individual level regarding prognosis and therapeutic decision-making remains insufficient. The next decade and beyond should see major advances in our capacity to precisely define the factors involved in PAH, and all forms of pulmonary hypertension, using traditional and novel approaches to personalized care.
Traditional molecular medicine approaches have been invaluable in the expansion of our knowledge of PAH, including but not limited to the application of traditional genetic approaches, genetic epidemiology studies, molecular signaling and structural analyses, and the use of molecular pharmacology and gene modification techniques. But more recently, advanced experimental techniques and systems analyses have emerged to shed light on the genomic, transcriptomic, epigenomic, metabolomic, proteomic, and other domains of molecular activity, and their interactions, that contribute to PAH pathogenesis and progression (7). “Omics” and related advances are crucial, but must be integrated with alternative data sources. The resultant synergy of traditional and novel molecular approaches will facilitate the personalization of PAH risk, treatment, and cure.
We now know that in a minority of cases, a rare genetic variation (mutation) in a single gene creates a cellular and systemic milieu of PAH susceptibility. In fact, for prediction of PAH development, there is no greater risk than being the carrier of a mutation in one of the known “pulmonary hypertension (PH)–specific genes,” such as bone morphogenetic protein receptor type 2 (BMPR2). And, about 20% of subjects thought to have idiopathic PAH (IPAH) actually have disease associated with mutations in this gene (8). For some families with a known BMPR2 gene mutation, preimplantation genetic testing under the direction of experienced clinicians may be a viable option for future pregnancies, in women without PAH, to ensure that the mutation is not carried forward (9). In addition, for those individuals with a BMPR2 mutation, and perhaps those without a mutation but reduced downstream signaling, mechanisms to enhance BMPR2 function and/or signaling by novel targeted approaches such as nonsense mutation read-through, manipulation of gene expression by alternative splicing, amplification of wild-type protein signaling, or other mechanisms are viable preventive and therapeutic possibilities on the horizon (10–14).
Yet, the presence of a mutation in BMPR2 does not guarantee the development of clinical PAH as there is markedly decreased penetrance, suggesting that additional molecular and environmental factors modify the development and phenotypic expression, for example, age of onset of PAH among susceptible hosts (8). The factors modifying PAH penetrance are likely complex, incorporating multiple sources of variability including sex (females are threefold more likely to develop PAH than males [15]); wild-type expression of the nonmutated BMPR2 allele (16); common and rare variations in other genes (17, 18); and epigenetic (19), biochemical (20), or other factors. It is also known that mutations in BMPR2, as well as in the hereditary hemorrhagic telangiectasia-associated PAH genes endoglin (ENG) and activin A receptor type II-like 1 (ACVRL1, ALK1) cause more severe PAH with shorter survival than seen in IPAH cases (21, 22). But, stratification of severity within genes according to genotype has not occurred because of the rarity of these mutations overall. Although rare, the prevalence of these mutations is significantly higher than that of other genes associated with PAH, such as caveolin-1 (CAV1), potassium channel two pore domain subfamily K member 3 (KCNK3), and eukaryotic translation initiation factor 2α kinase 4 (EIF2AK4) (23–25).
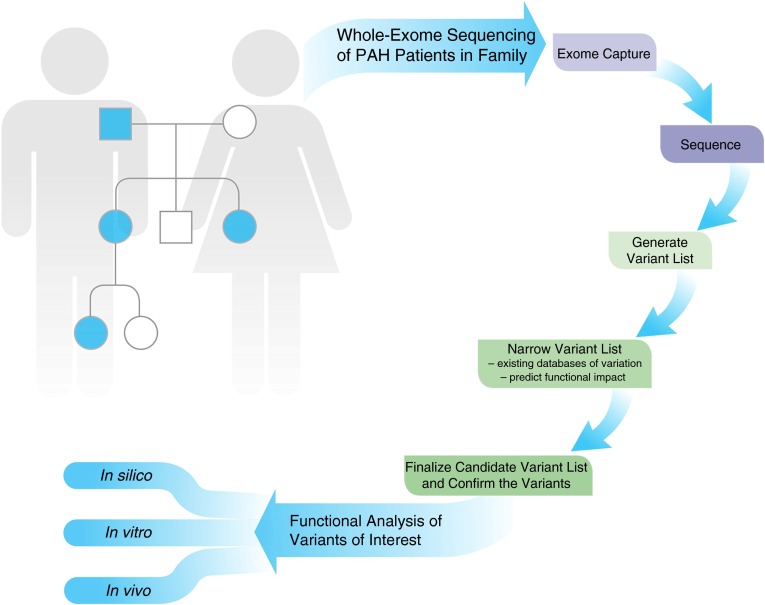
Mutations in CAV1, KCNK3, TopBP1, and EIF2AK4 were discovered in PAH by the use of whole-exome sequencing, which is one of several more recent approaches employing next-generation sequencing technology (23–27). Although it can be applied to unrelated subjects, whole-exome (and whole-genome) sequencing may be particularly effective in the setting of familial disease. For example, as shown in Figure 1, a family with PAH in multiple family members with available DNA may be analyzed, as performed with the CAV1 discovery. Briefly, relatively standardized approaches are employed to capture the exonic segments of each gene, which are then sequenced. Sequencing typically generates a raw sequence, which is subsequently evaluated and converted into a more mature final sequence of exons. This process is typically followed by careful analysis of the sequencing for common and rare genetic variations, which are differences from population databases of “normal” subjects, or subjects with other known diseases (e.g., Single Nucleotide Polymorphism Database, 1000 Genomes Project, and the human genome browser of University of California Santa Cruz). In most cases, when searching for rare variations (mutations), at this step common variants (e.g., more than 1% prevalence) are filtered out of the analysis. Careful analysis of the residual rare variants is then undertaken in a variety of ways, including confirmation of the prevalence in the population, using databases of variation (e.g., the Exome Variant Server [http://evs.gs.washington.edu/EVS/] and ExAC Browser [http://exac.broadinstitute.org/]); assessment of the various mutation types represented; and prediction of impact of the variations, using a variety of approaches (e.g., SIFT [http://sift.jcvi.org/] and PolyPhen-2 [http://genetics.bwh.harvard.edu/pph2/]).
Figure 1.
Whole-exome sequencing to identify rare variants relevant to pulmonary arterial hypertension (PAH) pathogenesis. A representative pedigree, similar to that employed for the discovery of a CAV1 mutation in a family with heritable PAH, is represented, with four patients with PAH in the modified family pedigree. Whole-exome sequencing is performed, which involves a number of complicated laboratory and bioanalytical steps, to determine the variants of interest for confirmation. Ultimately, confirmed candidate genes and variants require biological studies to determine the true functional impact of the changes discovered. Finally (data not shown), additional case subjects and control subjects should be genotyped for mutations in the candidate genes of interest.
In the case of the CAV1 discovery (26), four patients with PAH in the same family were analyzed by whole-exome sequencing, providing the opportunity to search for shared heterozygous mutations in a family with autosomal dominant PAH. An initial detection of 54,540 genomic locations with variations was trimmed to 10,088 (18%) shared genomic locations, and then trimmed to 653 heterozygous variants shared by all four patients, and then trimmed to 52 shared mutations, which were ultimately narrowed to 16 shared mutations of interest after careful review of mutation type, functional impact, and conservation across species. Ultimately, 11 of the 16 candidate mutations were confirmed by standard sequencing (Sanger sequencing), and a fifth case of PAH in the family was used to filter the list down to three candidate mutations. From that list, careful analysis of the genes including tissue expression, conservation of the genetic site, mutation type, prediction of mutation impact, and other factors led to the identification of a CAV1 mutation as the primary target. As is typically employed, additional case subjects and control subjects were screened for CAV1 mutations to ultimately improve confidence of the finding.
Although the true prevalence of CAV1 mutations and mutations in other PH-specific genes has yet to be fully elucidated, at least two current programs underway are searching for common and rare variants (mutations) in large numbers of patients with PAH; therefore, the true prevalence of mutations in PH-specific genes will be elucidated soon, which may facilitate more detailed genotype–phenotype studies (National Biological Sample and Data Repository for PAH in the United States and BRIDGE-PAH [Biomedical Research Identification of Genetic Etiology of PAH] in the United Kingdom). In addition, in the near future, modifier genes that predispose to susceptibility to PAH or genes that promote resiliency may be known; these genes could be incorporated into a risk calculator to provide precise prediction as to which mutation carriers will develop disease and at what age. Concurrent with such a molecular breakthrough, novel approaches to detect early pulmonary vascular disease are needed to allow therapeutic intervention at the presymptomatic phase of disease (which is currently undetectable) (28).
Because mutations account for only a modest proportion of PAH cases, there is a critical need to identify the genetic and nongenetic factors involved in PAH pathogenesis, progression, and response to therapy, regardless of the presence or absence of a PH-specific gene mutation. Those same factors that appear to modify BMPR2 penetrance, as noted previously, may contribute in the setting of no PH-specific gene mutation, as well as additional variations such as common polymorphisms in the endostatin gene, endothelin-related genes, sex hormone–related genes, other mutations across the genome, or at the CBLN2 locus (27, 29–35). In addition, a growing body of work implicates complicated molecular perturbations, such as disruptions in energy metabolism and mitochondrial dynamics involving both the pulmonary vasculature and right ventricle (5, 19). No set of variations occurs in isolation. To date, however, most molecular discoveries have been made in relative isolation without integration of multiple gene variants or molecular features, due to the limitations of traditional variant studies.
The complexity of cellular derangements that contribute to PAH requires a broad scientific approach that can be ultimately narrowed to the individual level. Achievement of this lofty goal requires integration of data from multiple sources involving traditional and novel molecular platforms, and physicians and scientists with a broad range of expertise. The components include well-defined human subject cohorts monitored longitudinally, “omics”-level data from those same subjects across multiple cell types, and the clinical and scientific expertise to determine disease onset, response to therapy, and disease progression. These resources will provide the necessary foundation for scientific approaches to advance understanding of the molecular complexity of PAH. Next, we present some of the more recent data on precision medicine in PAH and then discuss how modern gene-editing technology can be used to understand current and future discoveries.
PAH and the Concept of Precision Medicine
Although some patients do well on current therapies for prolonged periods, PAH remains a deadly disease and a significant proportion of patients rapidly decline despite appropriate and aggressive therapy (2, 36). With the notable exception of calcium channel blocker therapy, our current treatment recommendations are based on clinical markers of disease severity, matching more severe disease to prostaglandin and combination therapy, while recommending treatment with oral therapies for patients with preserved right ventricular function and more mild disease (37). The current U.S. Food and Drug Administration (FDA)-approved PAH therapies offer many choices to tailor to patients’ preferences and disease severity, but also create uncertainty, as we cannot easily predict which patients may benefit from which treatment class. Clinicians at the bedside know that certain patients often have dramatic responses to one particular class of medications, while they have no response to another. The only way to identify these responses, presently, is by trial and error. Unfortunately, when ineffective medications are tried, disease often progresses, and thus valuable time is lost. Although modern molecular techniques and next-generation sequencing can and should be used to understand disease etiology, it can also be used to predict drug responsiveness. Given the persistent high mortality in PAH and the current inability to predict drug responsiveness, a key use of molecular investigations should be to match an individual patient to the drug most likely to be effective, called “precision medicine” (38).
The concept of precision medicine is not new in PAH, as we have long known that patients who meet specific criteria for hemodynamic response to inhaled nitric oxide during right heart catheterization have long-term favorable responses to calcium channel blocker therapy (39). Although detailed phenotypic assessments and classification are a major part of the broader PH field, analysis of differential treatment response according to PAH subtype remains an underused precision opportunity; for example, last year Rhee and colleagues used individual participant data from phase 3 placebo-controlled randomized controlled trials of therapies for PAH submitted to the FDA for drug approval to demonstrate that PAH treatment may be less effective in connective tissue disease–associated PAH compared with IPAH (40). Careful phenotyping offers great opportunity to fine-tune the selection of therapies for the appropriate subtype of patient.
In addition, “omics” studies may be employed to better understand disease processes, and ultimately support the development and selection of therapeutics for certain individuals. A growing body of literature supports the concept that irregularities in microRNA activity and related networks promote PAH pathophysiology, which creates exciting opportunities for precision therapeutics design (19, 41–44). Rhodes and colleagues analyzed pulmonary artery endothelial cells from control subjects and subjects with IPAH, using RNA sequencing to determine novel pathways perturbed in relation to reduced BMPR2 signaling (45). We identified a peripheral blood signature of RNA expression patterns that faithfully identifies patients with a PAH subphenotype responsive to calcium channel blocker therapy, suggesting that peripheral blood can be used to identify drug responsiveness in PAH (46). Using whole-exome sequencing, genetic variants that may underlie this subphenotype were also uncovered (47). In another example of precision pharmacogenomics, Benza and colleagues explored genetic variants in endothelin-1 (ET-1) signaling that predict good responses to ET-1 receptor antagonists (34). These different and equally successful approaches highlight potential ways forward for our field in using modern molecular medicine techniques to improve patient care in PAH. The traditional scientific approach used by Benza and colleagues tested the hypothesis that variants in ET-1 receptor or signaling will underlie different responses to endothelin receptor antagonist therapy. They used genome-wide association study data to generate a candidate list of variants in ET-1 signaling and then performed targeted genotyping for the candidates. One of their candidate variants (rs11157866) predicted response to endothelin receptor antagonism. Although successful in this study, this methodology may miss important molecular associations as one is limited by the current knowledge of molecular disease etiology.
One example of a different strategy for molecular prediction of drug responses is an unbiased, discovery-based approach to precision medicine in PAH. This methodology uses technology such as next-generation sequencing, RNA sequencing or microarray, and proteomics or metabolomics to measure thousands of chemicals, proteins, transcripts, or genes. Rather than having an a priori idea of which findings are important, those with the strongest association with disease are selected for further study, without the requirement for biologic plausibility in all cases. The challenge of this approach is that it may identify compounds unimportant to the disease or may identify so many potential targets that prioritization requires advanced bioinformatics and confirmation of functional consequences. However, if one wants solely to identify strong predictors of drug responsiveness, the unbiased discovery approach may offer the best hope for finding molecules or genes that fulfill this requirement. Considering the urgency of patients with progressive disease treated by ineffective therapy, the specific functions of these genes, proteins, or metabolites may matter little in the short term if they are strong predictors of response to a particular therapy. The hard work of figuring out how these molecules are relevant to disease could be sorted out after confirming their predictive value. Of course, this approach necessitates serial acquisition of samples in advance and over time from patients exposed to therapeutics.
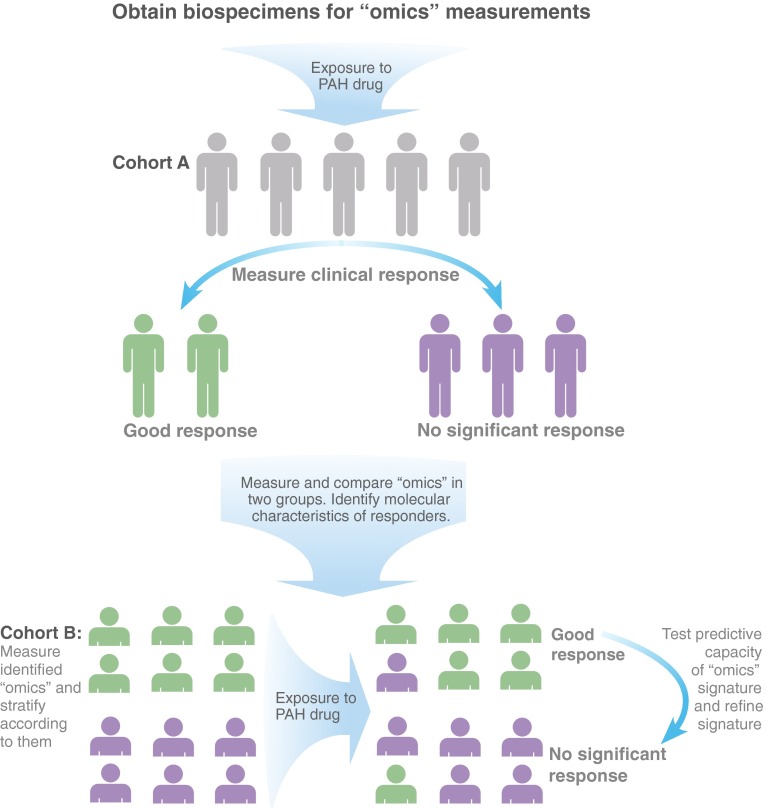
Regardless of methodology, identified molecules or genes of significance to PAH can be used for multiple purposes. Most urgently, they can be used to predict responses to already FDA-approved therapies in PAH. Next, they can potentially be used to improve clinical trial design in PAH (Figure 2). At present, data from clinical trials of new therapeutics are analyzed in aggregate, so that variability in individual patient response is generally not presented. If we find clinical “superresponder” patients in new drug trials, “omics” techniques could be applied to identify molecular markers of these patients. Future trials could enroll only patients with this excellent response, thereby saving money and time and potentially maximizing therapeutic benefit for enrollees (48). For instance, in the case of the drug imatinib, if superresponders were identified in clinical trials and specimens available for analyses (49), next-generation sequencing may have been able to find genetic predictors of this response to select patients in whom the risk-to-benefit ratio of this therapy is warranted. Preselecting patients for study trial participation has already proven successful in cystic fibrosis (50), and it should be forthcoming in PAH.
Figure 2.
The identification of genes, pathways, and molecules relevant to pulmonary arterial hypertension (PAH) can be used for multiple purposes, including the improvement of clinical trial design. Subjects exposed to a given therapeutic could be compared in a number of different ways, including according to therapeutic response. Comprehensive approaches to explore the shared variations among those who respond and those who do not respond may support the determination of a predictive signature of response to help refine patient selection for a given therapeutic.
Thus there are many potential ways to use the broad “omics” data that will be generated through application of modern molecular medicine in PAH. An additional critical step will be to take the identified genes, pathways, and molecules and use them to expand our understanding of disease etiology, which is critical to developing curative therapy. This will require the application of traditional and novel molecular techniques, such as gene editing.
Gene Editing
A set of powerful new technologies that allow precision editing of genomes has been developed. These have multiple potential applications in pulmonary hypertension research and therapy, from understanding the specific variants arising from “omics” studies to designing cells for targeted drug delivery applications, to the repair of mutations. Two relatively new technologies allow targeting of specific nucleotides: TALEN and CRISPR; each of these has multiple related technologies (51). TALEN, and the related technologies of zinc finger nucleases (52) and MegaTAL (53), use modular protein-based sequence recognition, whereas CRISPR uses RNA-guided sequence recognition. Although variations on these technologies are likely to develop over time, the core technologies are unlikely to change.
Transcription activator–like (TAL) effectors were derived from the plant pathogen Xanthomonas in 2007 (54), with a nuclease function for gene editing added in 2010 (55). The combination, TAL effector–like nucleases (TALENs), consist of a modular array of TAL recognition sequences fused to a FokI nuclease (56). These are inserted in pairs, one for each strand, and work as a dimer to create double-stranded breaks in specific DNA sequences.
The components of the other major method of making targeted cuts in the genome, CRISPR–Cas9, are also derived from bacteria and archaea, in which they are part of a viral defense system (57). It consists of clustered regularly interspaced short palindromic repeats (CRISPR), which bind a guide RNA and an associated endonuclease (Cas9). Binding specificity is thus dependent on RNA–DNA interaction strength. The main advantage of CRISPR–Cas9 over the TALEN-based technology is its speed of production and extremely low cost; its disadvantages, as discussed later, are a likely inherently lower specificity, and the legal challenges that have not been resolved as of this writing (58).
Fundamentally, TALEN and CRISPR both cause site-specific cleavage, which allows gene editing through two mechanisms: homologous recombination (HR) and nonhomologous end joining (NHEJ). When TALEN or CRISPR make the site-specific double-stranded break, DNA repair mechanisms are employed to repair the break. If a piece of DNA matching the sites flanking the cleavage site is available, the cell can use HR to repair the damage. If no such DNA is available, the cell will use NHEJ.
In NHEJ, although the DNA is rejoined, there is often a deletion or insertion of a small number of nucleotides. If the cleavage site was in the middle of a coding sequence or essential regulatory element, this will serve to destroy gene expression or function. Without addition of a homologous recombination template, then, TALEN and CRISPR are capable only of truncation or knockout, not precision edits. However, the efficiency is relatively high—successfully transfected cells will have DNA insertions or deletions at the target site in 10–60% of cells (59).
Homologous recombination is much more powerful. In HR, the cell is provided with a DNA template to repair the damage caused by TALEN or CRISPR, which precisely matches the surrounding area, aside from the specific gene edits desired. Historically, this technique has been used for decades to create precision gene-edited mice. However, without use of TALEN or CRISPR to drive local DNA repair mechanisms, the efficiency was extraordinarily low (less than 1 in 1,000,000 cells) and even then required DNA arms that precisely matched the surrounding sequence with total lengths approaching 10,000 bp. This was possible only in specific strains of clonal mice for which libraries of DNA were available. With TALEN and CRISPR, the efficiency is dramatically higher (between 1 in 100 and 1 in 1,000 cells), and the size of the homology arms can be much smaller (fewer than 1,000 bp on each side) (60).
In PAH research, the usefulness of these techniques is clear. In the past, we were limited to viral or plasmid-based expression of mutations, in which regulation and expression levels were generally completely nonphysiological. With these new techniques, we can easily and rapidly create cell lines with the precise mutations arising from our “omics,” sequencing, and genome-wide association study approaches, to allow testing of the effects individually and in combination with the exact alterations identified in our studies. Moreover, because all of these variations can be created on the same background line, they are perfectly controlled.
Further, there are several therapeutic possibilities derived from gene editing. These make use of the strong evidence that several varieties of circulating cells, including endothelial progenitor cells and a variety of bone marrow–derived cells, are found in the vicinity of the pulmonary vascular lesions and may play a role in disease etiology or progression (61). Closest to the clinic is use of cell-based therapies for drug delivery. In the PHACeT (Endothelial Nitric Oxide Synthase Gene–Enhanced Progenitor Cell Therapy for PAH) trial, endothelial progenitor cells were transfected with endothelial nitric oxide synthase (eNOS) and introduced into patients (62). Short-term hemodynamic benefits were not sustained at 3 months but there were persistent improvements in 6-minute walk test distance. Most importantly, the transient transfection strategy resulted in detectable eNOS expression for only about 1 week. One might imagine that using gene-editing approaches would result in longer term delivery of eNOS, and would be safer than other stable gene therapy approaches such as viral integration, because potentially dangerous random integration effects are removed. The safety and some efficacy of this initial trial provide promise for future targeted delivery of eNOS, or potentially other proteins.
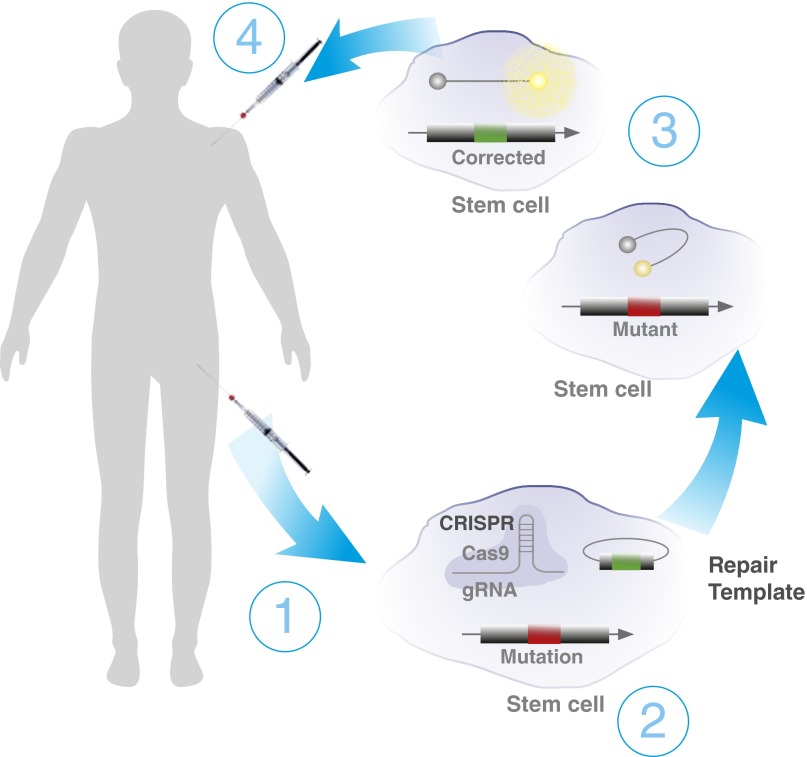
Another possibility, however, is correction of known deleterious mutations. There is evidence from several groups that bone marrow cells alone are sufficient to drive PAH, including our finding that control bone marrow–derived stem cells can reduce the PAH phenotype when transplanted into lethally irradiated Bmpr2 mutant mice (63, 64). This suggests the future possibility of autologous bone marrow transplantation after edited correction of stem cells as a therapeutic possibility; that is, correction of the mutation from the stem cells of a BMPR2 mutation carrier with PAH could be a therapeutic approach (Figure 3) (64). However, this is technically challenging for a variety of reasons, including the need for high efficiency, the need to avoid off-target effects, and the need for high speed to avoid genomic instability or terminal differentiation of stem cells.
Figure 3.
Schematic of the use of gene-editing techniques to repair a known gene mutation as a therapeutic approach, in this case demonstrating the manipulation of a patient’s own stem cells. (1) CRISPR–Cas9 can be used with homologous recombination to repair a bone morphogenetic protein receptor type 2 (BMPR2) mutation for autologous transplant in patient bone marrow. (2) Bone marrow is extracted from the patient and sorted for stem cells. Stem cells are transfected with CRISPR–Cas9 guided to the mutation site, with homologous recombination DNA containing corrected sequences. (3) Small-molecule nonhomologous end-joining blockers can be used to increase the efficiency of homologous recombination. (4) Molecular probes that fluoresce only when bound to the corrected RNA can be used to sort for cells that have had their mutations corrected. Corrected stem cells are reintroduced into the patient. This process is still a hypothetical approach to correct stem cells with a BMPR2 mutation that may promote pulmonary arterial hypertension; there are a number of technical barriers that must be overcome as described in text. Cas9 = CRISPR-associated protein 9; CRISPR = clustered regularly interspaced short palindromic repeats; gRNA = guide RNA.
The need for high efficiency is a particularly difficult problem as homologous recombination has an efficiency of at best 1%. Some sorting or selection technology must be employed to identify the cells that have been correctly edited, ideally without leaving any sorting markers embedded in the cell. Transposase-based approaches such as Piggybac have been used for successful footprint-free gene editing in, for instance, correction of cystic fibrosis mutations in human induced pluripotent stem cells (65), and so the problem is solvable, but becomes difficult when combined with the need for rapid editing.
Because CRISPR–Cas9 uses RNA–DNA interactions for its specificity, it inherently has difficulty with off-target effects (66). Numerous methodologies to reduce these off-target effects have been proposed, and can reduce these effects under carefully controlled circumstances (67). Nonetheless, in actual practice, there are substantial fidelity and specificity issues, as demonstrated by the detailed analysis done in one study in which CRISPR was used to edit human preimplantation embryos (68). Although TALENs theoretically could have off-target cleavage, this is much less of an issue for current-generation TALENs. Several studies using TALEN for gene editing have failed to find any evidence of off-target mutations (69, 70).
Finally, gene editing must be done in such a way as to preserve the primitive, long-term repopulating cell populations in hematopoietic stem and progenitor cells, which will require either improved understanding of the signaling factors needed to preserve the stem state, or greatly increased speed (at present, editing by homologous recombination takes months). A number of groups are working on these issues, for instance, by delivering the gene-editing components through viruses (71). This could also hold promise for the targeted correction of somatic lung cells with spontaneous mutations due to increased mutagen sensitivity, which may be a feature of PAH (72–74). The use of gene editing has yet to reach the bedside, but this rapidly evolving field may provide great opportunities for therapeutic approaches in the near future.
Conclusion
It is an exciting new era for PH-focused clinicians, researchers, and patients. PH-specific mutations identify individuals with substantial risk of disease, and provide an opportunity to provide more in-depth information about disease severity and progression a priori. However, much work remains to broaden our understanding of how rare variations in specific genes associate with PAH in some but not all cases. For example, the clinical value of a mutation in a PH-specific gene is currently limited by the reduced penetrance and broad age at onset across a lifetime—there is a critical need to identify the additional susceptibility and resiliency factors that modify disease penetrance and expression.
It is likely that those factors will be involved in other forms of pulmonary hypertension, in terms not only of pathogenesis, but also sensitivity to therapeutics, performance of the right ventricle under stress, and other functions. Traditional and novel technologies must be employed and merged to facilitate a broader understanding of these molecular and other factors that coalesce to promote and perpetuate the PAH state. Also, we must recognize that a “one size fits all” approach to therapeutics must be abandoned in favor of a more detailed molecular understanding that facilitates the precise selection of treatment approaches for each individual. This will require investigators to incorporate sample collection into future clinical trials consistent with the need to determine molecular or clinical characteristics that segregate responders and nonresponders. Exciting new approaches are on the horizon, such as gene-editing techniques, with tremendous potential to accelerate progress at the bench, and bedside, with roles such as cell-based drug delivery and mutation correction. The era of precision medicine is truly here (38); it is time for prevention and treatment strategies that make individual variability the priority in PH.
Footnotes
Supported by NIH grants P01 HL108800 (E.D.A., J.W., J.E.L., and A.R.H.) and 1U01 HL125212-01 (A.R.H.).
Originally Published in Press as DOI: 10.1164/rccm.201605-0905PP on July 11, 2016
References
- 1.D’Alonzo GE, Barst RJ, Ayres SM, Bergofsky EH, Brundage BH, Detre KM, Fishman AP, Goldring RM, Groves BM, Kernis JT, et al. Survival in patients with primary pulmonary hypertension: results from a national prospective registry. Ann Intern Med. 1991;115:343–349. doi: 10.7326/0003-4819-115-5-343. [DOI] [PubMed] [Google Scholar]
- 2.Humbert M, Sitbon O, Chaouat A, Bertocchi M, Habib G, Gressin V, Yaïci A, Weitzenblum E, Cordier JF, Chabot F, et al. Survival in patients with idiopathic, familial, and anorexigen-associated pulmonary arterial hypertension in the modern management era. Circulation. 2010;122:156–163. doi: 10.1161/CIRCULATIONAHA.109.911818. [DOI] [PubMed] [Google Scholar]
- 3.Badesch DB, Raskob GE, Elliott CG, Krichman AM, Farber HW, Frost AE, Barst RJ, Benza RL, Liou TG, Turner M, et al. Pulmonary arterial hypertension: baseline characteristics from the REVEAL Registry. Chest. 2010;137:376–387. doi: 10.1378/chest.09-1140. [DOI] [PubMed] [Google Scholar]
- 4.Maron BA, Leopold JA. Emerging concepts in the molecular basis of pulmonary arterial hypertension. II. Neurohormonal signaling contributes to the pulmonary vascular and right ventricular pathophenotype of pulmonary arterial hypertension. Circulation. 2015;131:2079–2091. doi: 10.1161/CIRCULATIONAHA.114.006980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ryan JJ, Archer SL. Emerging concepts in the molecular basis of pulmonary arterial hypertension. I. Metabolic plasticity and mitochondrial dynamics in the pulmonary circulation and right ventricle in pulmonary arterial hypertension. Circulation. 2015;131:1691–1702. doi: 10.1161/CIRCULATIONAHA.114.006979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tuder RM, Archer SL, Dorfmüller P, Erzurum SC, Guignabert C, Michelakis E, Rabinovitch M, Schermuly R, Stenmark KR, Morrell NW. Relevant issues in the pathology and pathobiology of pulmonary hypertension. J Am Coll Cardiol. 2013;62(25) Suppl:D4–D12. doi: 10.1016/j.jacc.2013.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang RS, Maron BA, Loscalzo J. Systems medicine: evolution of systems biology from bench to bedside. Wiley Interdiscip Rev Syst Biol Med. 2015;7:141–161. doi: 10.1002/wsbm.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soubrier F, Chung WK, Machado R, Grünig E, Aldred M, Geraci M, Loyd JE, Elliott CG, Trembath RC, Newman JH, et al. Genetics and genomics of pulmonary arterial hypertension. J Am Coll Cardiol. 2013;62(25) Suppl:D13–D21. doi: 10.1016/j.jacc.2013.10.035. [DOI] [PubMed] [Google Scholar]
- 9.Frydman N, Steffann J, Girerd B, Frydman R, Munnich A, Simonneau G, Humbert M. Pre-implantation genetic diagnosis in pulmonary arterial hypertension due to BMPR2 mutation. Eur Respir J. 2012;39:1534–1535. doi: 10.1183/09031936.00185011. [DOI] [PubMed] [Google Scholar]
- 10.Long L, Ormiston ML, Yang X, Southwood M, Gräf S, Machado RD, Mueller M, Kinzel B, Yung LM, Wilkinson JM, et al. Selective enhancement of endothelial BMPR-II with BMP9 reverses pulmonary arterial hypertension. Nat Med. 2015;21:777–785. doi: 10.1038/nm.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunmore BJ, Drake KM, Upton PD, Toshner MR, Aldred MA, Morrell NW. The lysosomal inhibitor, chloroquine, increases cell surface BMPR-II levels and restores BMP9 signalling in endothelial cells harbouring BMPR-II mutations. Hum Mol Genet. 2013;22:3667–3679. doi: 10.1093/hmg/ddt216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spiekerkoetter E, Tian X, Cai J, Hopper RK, Sudheendra D, Li CG, El-Bizri N, Sawada H, Haghighat R, Chan R, et al. FK506 activates BMPR2, rescues endothelial dysfunction, and reverses pulmonary hypertension. J Clin Invest. 2013;123:3600–3613. doi: 10.1172/JCI65592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drake KM, Zygmunt D, Mavrakis L, Harbor P, Wang L, Comhair SA, Erzurum SC, Aldred MA. Altered microRNA processing in heritable pulmonary arterial hypertension: an important role for Smad-8. Am J Respir Crit Care Med. 2011;184:1400–1408. doi: 10.1164/rccm.201106-1130OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cogan J, Austin E, Hedges L, Womack B, West J, Loyd J, Hamid R. Role of BMPR2 alternative splicing in heritable pulmonary arterial hypertension penetrance. Circulation. 2012;126:1907–1916. doi: 10.1161/CIRCULATIONAHA.112.106245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larkin EK, Newman JH, Austin ED, Hemnes AR, Wheeler L, Robbins IM, West JD, Phillips JA, III, Hamid R, Loyd JE. Longitudinal analysis casts doubt on the presence of genetic anticipation in heritable pulmonary arterial hypertension. Am J Respir Crit Care Med. 2012;186:892–896. doi: 10.1164/rccm.201205-0886OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamid R, Cogan JD, Hedges LK, Austin E, Phillips JA, III, Newman JH, Loyd JE. Penetrance of pulmonary arterial hypertension is modulated by the expression of normal BMPR2 allele. Hum Mutat. 2009;30:649–654. doi: 10.1002/humu.20922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodriguez-Murillo L, Subaran R, Stewart WC, Pramanik S, Marathe S, Barst RJ, Chung WK, Greenberg DA. Novel loci interacting epistatically with bone morphogenetic protein receptor 2 cause familial pulmonary arterial hypertension. J Heart Lung Transplant. 2010;29:174–180. doi: 10.1016/j.healun.2009.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phillips JA, III, Poling JS, Phillips CA, Stanton KC, Austin ED, Cogan JD, Wheeler L, Yu C, Newman JH, Dietz HC, et al. Synergistic heterozygosity for TGFβ1 SNPs and BMPR2 mutations modulates the age at diagnosis and penetrance of familial pulmonary arterial hypertension. Genet Med. 2008;10:359–365. doi: 10.1097/GIM.0b013e318172dcdf. [DOI] [PubMed] [Google Scholar]
- 19.Chen X, Talati M, Fessel JP, Hemnes AR, Gladson S, French J, Shay S, Trammell A, Phillips JA, Hamid R, et al. Estrogen metabolite 16α-hydroxyestrone exacerbates bone morphogenetic protein receptor type II–associated pulmonary arterial hypertension through microRNA-29–mediated modulation of cellular metabolism. Circulation. 2016;133:82–97. doi: 10.1161/CIRCULATIONAHA.115.016133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Austin ED, Cogan JD, West JD, Hedges LK, Hamid R, Dawson EP, Wheeler LA, Parl FF, Loyd JE, Phillips JA., III Alterations in oestrogen metabolism: implications for higher penetrance of familial pulmonary arterial hypertension in females. Eur Respir J. 2009;34:1093–1099. doi: 10.1183/09031936.00010409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Evans JD, Girerd B, Montani D, Wang XJ, Galiè N, Austin ED, Elliott G, Asano K, Grünig E, Yan Y, et al. BMPR2 mutations and survival in pulmonary arterial hypertension: an individual participant data meta-analysis. Lancet Respir Med. 2016;4:129–137. doi: 10.1016/S2213-2600(15)00544-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Girerd B, Montani D, Coulet F, Sztrymf B, Yaici A, Jaïs X, Tregouet D, Reis A, Drouin-Garraud V, Fraisse A, et al. Clinical outcomes of pulmonary arterial hypertension in patients carrying an ACVRL1 (ALK1) mutation. Am J Respir Crit Care Med. 2010;181:851–861. doi: 10.1164/rccm.200908-1284OC. [DOI] [PubMed] [Google Scholar]
- 23.Best DH, Sumner KL, Austin ED, Chung WK, Brown LM, Borczuk AC, Rosenzweig EB, Bayrak-Toydemir P, Mao R, Cahill BC, et al. EIF2AK4 mutations in pulmonary capillary hemangiomatosis. Chest. 2014;145:231–236. doi: 10.1378/chest.13-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eyries M, Montani D, Girerd B, Perret C, Leroy A, Lonjou C, Chelghoum N, Coulet F, Bonnet D, Dorfmüller P, et al. EIF2AK4 mutations cause pulmonary veno-occlusive disease, a recessive form of pulmonary hypertension. Nat Genet. 2014;46:65–69. doi: 10.1038/ng.2844. [DOI] [PubMed] [Google Scholar]
- 25.Ma L, Roman-Campos D, Austin ED, Eyries M, Sampson KS, Soubrier F, Germain M, Trégouët DA, Borczuk A, Rosenzweig EB, et al. A novel channelopathy in pulmonary arterial hypertension. N Engl J Med. 2013;369:351–361. doi: 10.1056/NEJMoa1211097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Austin ED, Ma L, LeDuc C, Berman Rosenzweig E, Borczuk A, Phillips JA, III, Palomero T, Sumazin P, Kim HR, Talati MH, et al. Whole exome sequencing to identify a novel gene (caveolin-1) associated with human pulmonary arterial hypertension. Circ Cardiovasc Genet. 2012;5:336–343. doi: 10.1161/CIRCGENETICS.111.961888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Jesus Perez VA, Yuan K, Lyuksyutova MA, Dewey F, Orcholski ME, Shuffle EM, Mathur M, Yancy L, Jr, Rojas V, Li CG, et al. Whole-exome sequencing reveals TopBP1 as a novel gene in idiopathic pulmonary arterial hypertension. Am J Respir Crit Care Med. 2014;189:1260–1272. doi: 10.1164/rccm.201310-1749OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Austin ED, Kawut SM, Gladwin MT, Abman SH. Pulmonary hypertension: NHLBI Workshop on the Primary Prevention of Chronic Lung Diseases. Ann Am Thorac Soc. 2014;11:S178–S185. doi: 10.1513/AnnalsATS.201312-443LD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roberts KE, Fallon MB, Krowka MJ, Benza RL, Knowles JA, Badesch DB, Brown RS, Jr, Taichman DB, Trotter J, Zacks S, et al. Pulmonary Vascular Complications of Liver Disease Study Group. Serotonin transporter polymorphisms in patients with portopulmonary hypertension. Chest. 2009;135:1470–1475. doi: 10.1378/chest.08-1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roberts KE, Fallon MB, Krowka MJ, Brown RS, Trotter JF, Peter I, Tighiouart H, Knowles JA, Rabinowitz D, Benza RL, et al. Pulmonary Vascular Complications of Liver Disease Study Group. Genetic risk factors for portopulmonary hypertension in patients with advanced liver disease. Am J Respir Crit Care Med. 2009;179:835–842. doi: 10.1164/rccm.200809-1472OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Germain M, Eyries M, Montani D, Poirier O, Girerd B, Dorfmüller P, Coulet F, Nadaud S, Maugenre S, Guignabert C, et al. Genome-wide association analysis identifies a susceptibility locus for pulmonary arterial hypertension. Nat Genet. 2013;45:518–521. doi: 10.1038/ng.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Willers ED, Newman JH, Loyd JE, Robbins IM, Wheeler LA, Prince MA, Stanton KC, Cogan JA, Runo JR, Byrne D, et al. Serotonin transporter polymorphisms in familial and idiopathic pulmonary arterial hypertension. Am J Respir Crit Care Med. 2006;173:798–802. doi: 10.1164/rccm.200509-1361OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Damico R, Kolb TM, Valera L, Wang L, Housten T, Tedford RJ, Kass DA, Rafaels N, Gao L, Barnes KC, et al. Serum endostatin is a genetically determined predictor of survival in pulmonary arterial hypertension. Am J Respir Crit Care Med. 2015;191:208–218. doi: 10.1164/rccm.201409-1742OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benza RL, Gomberg-Maitland M, Demarco T, Frost AE, Torbicki A, Langleben D, Pulido T, Correa-Jaque P, Passineau MJ, Wiener HW, et al. Endothelin-1 pathway polymorphisms and outcomes in pulmonary arterial hypertension. Am J Respir Crit Care Med. 2015;192:1345–1354. doi: 10.1164/rccm.201501-0196OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kerstjens-Frederikse WS, Bongers EM, Roofthooft MT, Leter EM, Douwes JM, Van Dijk A, Vonk-Noordegraaf A, Dijk-Bos KK, Hoefsloot LH, Hoendermis ES, et al. TBX4 mutations (small patella syndrome) are associated with childhood-onset pulmonary arterial hypertension. J Med Genet. 2013;50:500–506. doi: 10.1136/jmedgenet-2012-101152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benza RL, Miller DP, Gomberg-Maitland M, Frantz RP, Foreman AJ, Coffey CS, Frost A, Barst RJ, Badesch DB, Elliott CG, et al. Predicting survival in pulmonary arterial hypertension: insights from the Registry to Evaluate Early and Long-Term Pulmonary Arterial Hypertension Disease Management (REVEAL) Circulation. 2010;122:164–172. doi: 10.1161/CIRCULATIONAHA.109.898122. [DOI] [PubMed] [Google Scholar]
- 37.Galiè N, Humbert M, Vachiery JL, Gibbs S, Lang I, Torbicki A, Simonneau G, Peacock A, Vonk Noordegraaf A, Beghetti M, et al. 2015 ESC/ERS guidelines for the diagnosis and treatment of pulmonary hypertension. Rev Esp Cardiol (Engl Ed) 2016;69:177. doi: 10.1016/j.rec.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 38.Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;372:793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rich S, Brundage BH. High-dose calcium channel–blocking therapy for primary pulmonary hypertension: evidence for long-term reduction in pulmonary arterial pressure and regression of right ventricular hypertrophy. Circulation. 1987;76:135–141. doi: 10.1161/01.cir.76.1.135. [DOI] [PubMed] [Google Scholar]
- 40.Rhee RL, Gabler NB, Sangani S, Praestgaard A, Merkel PA, Kawut SM. Comparison of treatment response in idiopathic and connective tissue disease–associated pulmonary arterial hypertension. Am J Respir Crit Care Med. 2015;192:1111–1117. doi: 10.1164/rccm.201507-1456OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wallace E, Morrell NW, Yang XD, Long L, Stevens H, Nilsen M, Loughlin L, Mair KM, Baker AH, MacLean MR. A sex-specific microRNA-96/5–hydroxytryptamine 1B axis influences development of pulmonary hypertension. Am J Respir Crit Care Med. 2015;191:1432–1442. doi: 10.1164/rccm.201412-2148OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen T, Zhou G, Zhou Q, Tang H, Ibe JC, Cheng H, Gou D, Chen J, Yuan JX, Raj JU. Loss of microRNA-17–92 in smooth muscle cells attenuates experimental pulmonary hypertension via induction of PDZ and LIM domain 5. Am J Respir Crit Care Med. 2015;191:678–692. doi: 10.1164/rccm.201405-0941OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schlosser K, White RJ, Stewart DJ. miR-26a linked to pulmonary hypertension by global assessment of circulating extracellular microRNAs. Am J Respir Crit Care Med. 2013;188:1472–1475. doi: 10.1164/rccm.201308-1403LE. [DOI] [PubMed] [Google Scholar]
- 44.Rhodes CJ, Wharton J, Boon RA, Roexe T, Tsang H, Wojciak-Stothard B, Chakrabarti A, Howard LS, Gibbs JS, Lawrie A, et al. Reduced microRNA-150 is associated with poor survival in pulmonary arterial hypertension. Am J Respir Crit Care Med. 2013;187:294–302. doi: 10.1164/rccm.201205-0839OC. [DOI] [PubMed] [Google Scholar]
- 45.Rhodes CJ, Im H, Cao A, Hennigs JK, Wang L, Sa S, Chen PI, Nickel NP, Miyagawa K, Hopper RK, et al. RNA sequencing analysis detection of a novel pathway of endothelial dysfunction in pulmonary arterial hypertension. Am J Respir Crit Care Med. 2015;192:356–366. doi: 10.1164/rccm.201408-1528OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hemnes AR, Trammell AW, Archer SL, Rich S, Yu C, Nian H, Penner N, Funke M, Wheeler L, Robbins IM, et al. Peripheral blood signature of vasodilator-responsive pulmonary arterial hypertension. Circulation. 2015;131:401–409. [Discussion, 409. doi: 10.1161/CIRCULATIONAHA.114.013317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hemnes AR, Zhao M, West J, Newman JH, Rich S, Archer SL, Robbins IM, Blackwell TS, Cogan J, Loyd JE, et al. Critical genomic networks and vasoreactive variants in idiopathic pulmonary arterial hypertension. Am J Respir Crit Care Med. 2016;194:464–475. doi: 10.1164/rccm.201508-1678OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pencina MJ, Peterson ED. Moving from clinical trials to precision medicine: the role for predictive modeling. JAMA. 2016;315:1713–1714. doi: 10.1001/jama.2016.4839. [DOI] [PubMed] [Google Scholar]
- 49.Hoeper MM, Barst RJ, Bourge RC, Feldman J, Frost AE, Galié N, Gómez-Sánchez MA, Grimminger F, Grünig E, Hassoun PM, et al. Imatinib mesylate as add-on therapy for pulmonary arterial hypertension: results of the randomized IMPRES study. Circulation. 2013;127:1128–1138. doi: 10.1161/CIRCULATIONAHA.112.000765. [DOI] [PubMed] [Google Scholar]
- 50.Accurso FJ, Rowe SM, Clancy JP, Boyle MP, Dunitz JM, Durie PR, Sagel SD, Hornick DB, Konstan MW, Donaldson SH, et al. Effect of VX-770 in persons with cystic fibrosis and the G551D-CFTR mutation. N Engl J Med. 2010;363:1991–2003. doi: 10.1056/NEJMoa0909825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.West J, Gill WW. Genome editing in large animals. J Equine Vet Sci. 2016;41:1–6. doi: 10.1016/j.jevs.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Isalan M, Choo Y, Klug A. Synergy between adjacent zinc fingers in sequence-specific DNA recognition. Proc Natl Acad Sci USA. 1997;94:5617–5621. doi: 10.1073/pnas.94.11.5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boissel S, Jarjour J, Astrakhan A, Adey A, Gouble A, Duchateau P, Shendure J, Stoddard BL, Certo MT, Baker D, et al. megaTALs: a rare-cleaving nuclease architecture for therapeutic genome engineering. Nucleic Acids Res. 2014;42:2591–2601. doi: 10.1093/nar/gkt1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sugio A, Yang B, Zhu T, White FF. Two type III effector genes of Xanthomonas oryzae pv. oryzae control the induction of the host genes OsTFIIAγ1 and OsTFX1 during bacterial blight of rice. Proc Natl Acad Sci USA. 2007;104:10720–10725. doi: 10.1073/pnas.0701742104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Certo MT, Morgan RA. Salient features of endonuclease platforms for therapeutic genome editing. Mol Ther. 2016;24:422–429. doi: 10.1038/mt.2016.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rath D, Amlinger L, Rath A, Lundgren M. The CRISPR–Cas immune system: biology, mechanisms and applications. Biochimie. 2015;117:119–128. doi: 10.1016/j.biochi.2015.03.025. [DOI] [PubMed] [Google Scholar]
- 58.Sherkow JS. Law, history and lessons in the CRISPR patent conflict. Nat Biotechnol. 2015;33:256–257. doi: 10.1038/nbt.3160. [DOI] [PubMed] [Google Scholar]
- 59.Nemudryi AA, Valetdinova KR, Medvedev SP, Zakian SM. TALEN and CRISPR/Cas genome editing systems: tools of discovery. Acta Naturae. 2014;6:19–40. [PMC free article] [PubMed] [Google Scholar]
- 60.Whitelaw CB, Sheets TP, Lillico SG, Telugu BP. Engineering large animal models of human disease. J Pathol. 2016;238:247–256. doi: 10.1002/path.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hu J, Xu Q, McTiernan C, Lai YC, Osei-Hwedieh D, Gladwin M. Novel targets of drug treatment for pulmonary hypertension. Am J Cardiovasc Drugs. 2015;15:225–234. doi: 10.1007/s40256-015-0125-4. [DOI] [PubMed] [Google Scholar]
- 62.Granton J, Langleben D, Kutryk MB, Camack N, Galipeau J, Courtman DW, Stewart DJ. Endothelial NO-synthase gene–enhanced progenitor cell therapy for pulmonary arterial hypertension: the PHACeT trial. Circ Res. 2015;117:645–654. doi: 10.1161/CIRCRESAHA.114.305951. [DOI] [PubMed] [Google Scholar]
- 63.Asosingh K, Farha S, Lichtin A, Graham B, George D, Aldred M, Hazen SL, Loyd J, Tuder R, Erzurum SC. Pulmonary vascular disease in mice xenografted with human BM progenitors from patients with pulmonary arterial hypertension. Blood. 2012;120:1218–1227. doi: 10.1182/blood-2012-03-419275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yan L, Chen X, Talati M, Nunley BW, Gladson S, Blackwell T, Cogan J, Austin E, Wheeler F, Loyd J, et al. Bone marrow–derived cells contribute to the pathogenesis of pulmonary arterial hypertension. Am J Respir Crit Care Med. 2016;193:898–909. doi: 10.1164/rccm.201502-0407OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Camarasa MV, Gálvez VM. Robust method for TALEN-edited correction of pF508del in patient-specific induced pluripotent stem cells. Stem Cell Res Ther. 2016;7:26. doi: 10.1186/s13287-016-0275-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 66.Cradick TJ, Fine EJ, Antico CJ, Bao G. CRISPR/Cas9 systems targeting β-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 2013;41:9584–9592. doi: 10.1093/nar/gkt714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR–Cas9. Nat Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liang P, Xu Y, Zhang X, Ding C, Huang R, Zhang Z, Lv J, Xie X, Chen Y, Li Y, et al. CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein Cell. 2015;6:363–372. doi: 10.1007/s13238-015-0153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dreyer AK, Hoffmann D, Lachmann N, Ackermann M, Steinemann D, Timm B, Siler U, Reichenbach J, Grez M, Moritz T, et al. TALEN-mediated functional correction of X-linked chronic granulomatous disease in patient-derived induced pluripotent stem cells. Biomaterials. 2015;69:191–200. doi: 10.1016/j.biomaterials.2015.07.057. [DOI] [PubMed] [Google Scholar]
- 70.Menger L, Gouble A, Marzolini MA, Pachnio A, Bergerhoff K, Henry JY, Smith J, Pule M, Moss P, Riddell SR, et al. TALEN-mediated genetic inactivation of the glucocorticoid receptor in cytomegalovirus-specific T cells. Blood. 2015;126:2781–2789. doi: 10.1182/blood-2015-08-664755. [DOI] [PubMed] [Google Scholar]
- 71.Wang J, Exline CM, DeClercq JJ, Llewellyn GN, Hayward SB, Li PW, Shivak DA, Surosky RT, Gregory PD, Holmes MC, et al. Homology-driven genome editing in hematopoietic stem and progenitor cells using ZFN mRNA and AAV6 donors. Nat Biotechnol. 2015;33:1256–1263. doi: 10.1038/nbt.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Federici C, Drake KM, Rigelsky CM, McNelly LN, Meade SL, Comhair SA, Erzurum SC, Aldred MA. Increased mutagen sensitivity and DNA damage in pulmonary arterial hypertension. Am J Respir Crit Care Med. 2015;192:219–228. doi: 10.1164/rccm.201411-2128OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Aldred MA, Comhair SA, Varella-Garcia M, Asosingh K, Xu W, Noon GP, Thistlethwaite PA, Tuder RM, Erzurum SC, Geraci MW, et al. Somatic chromosome abnormalities in the lungs of patients with pulmonary arterial hypertension. Am J Respir Crit Care Med. 2010;182:1153–1160. doi: 10.1164/rccm.201003-0491OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Drake KM, Comhair SA, Erzurum SC, Tuder RM, Aldred MA. Endothelial chromosome 13 deletion in congenital heart disease–associated pulmonary arterial hypertension dysregulates SMAD9 signaling. Am J Respir Crit Care Med. 2015;191:850–854. doi: 10.1164/rccm.201411-1985LE. [DOI] [PMC free article] [PubMed] [Google Scholar]