SUMMARY
Applications of ESCs require faithful chromatin changes during differentiation but the fate of each X-chromosome-state in differentiating ESCs is unclear. Female human ESC-lines either carry two active X-chromosomes (XaXa), an Xa and inactive-X-chromosome with or without XIST-RNA-coating (XiXIST+Xa;XiXa), or an Xa and an eroded-Xi (XeXa) where the Xi no longer expresses XIST-RNA and has partially reactivated. Here, we established XiXa, XeXa, and XaXa ESC-lines and followed their X-chromosome-state during differentiation. Surprisingly, we found that the X-state pre-existing in primed ESCs is maintained in differentiated cells. Consequently, differentiated XeXa and XaXa cells lacked XIST, did not initiate X-inactivation, and displayed higher X-linked gene-expression than XiXa cells. These results demonstrate that X-chromosome-dosage-compensation is not required for ESC differentiation. Our data imply that XiXIST+Xa ESCs are most suited for downstream applications and show that all other X-states are abnormal byproducts of our ESC-derivation and propagation methods.
Graphical Abstract
INTRODUCTION
X-chromosome-inactivation (XCI) compensates for the difference in X-chromosome number between female and male placental mammals and is orchestrated by the long non-coding RNA Xist, which induces the transcriptional silencing of one X-chromosome in females in early embryonic development (Augui et al., 2011; Disteche, 2012). The silencing of the X-chromosome is accompanied by heterochromatin formation and stably maintained throughout the lifetime of the organism (Augui et al., 2011; Disteche, 2012). Genetic approaches have indicated that XCI is necessary for development and adult homeostasis (Schulz and Heard, 2013; Yang et al., 2016).
Given the importance of XCI in vivo, the X-chromosome state of human embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) has been addressed by a number of researchers, arriving at the conclusion that these cells have different XCI-states (Anguera et al., 2012; Barakat et al., 2015; Hall et al., 2008; Hoffman et al., 2005; Kim et al., 2014; Lengner et al., 2010; Mekhoubad et al., 2012; Nazor et al., 2012; Pomp et al., 2011; Shen et al., 2008; Silva et al., 2008; Tchieu et al., 2010; Tomoda et al., 2012; Vallot et al., 2015; Xie et al., 2016). At least four X-chromosome states have been described: (i) two active X-chromosomes without XIST expression (XaXa); (ii) an XIST RNA-coated Xi with an Xa (XiXIST+Xa); (iii) an Xi lacking XIST expression with an Xa (XiXa); and (iv) an Xi lacking XIST RNA with partial loss – erosion – of its silent state (eroded Xi, Xe) with an Xa (XeXa). The epigenetic state of the X-chromosome not only varies between pluripotent stem cell (PSC) lines, but also between subcultures of a given line and even within a cell population. In addition, it appears that modified culture conditions affect the epigenetic state of the X-chromosome. A consistent view on the basis and functional outcomes of each XCI state is currently lacking.
Human PSCs represent a focal point of regenerative medicine since they can differentiate into all germ layers and are a powerful research tool for understanding early human development and modeling human diseases (Thomson et al., 1998). Therefore it is important that PSCs faithfully recapitulate transcriptional and epigenetic changes of in vivo differentiation, such as achieving the XiXIST+Xa state in female cells. This is the case in mouse ESC lines as they are consistently XaXa without Xist expression in the undifferentiated state and stably inactivate one X-chromosome by expressing Xist during differentiation (Augui et al., 2011). Thus, it is important to understand whether human PSCs with different XCI-states are capable of establishing or maintaining an Xi, or whether the XCI state pre-existing in PSCs is maintained upon differentiation.
Currently, the fate of X-chromosomes upon differentiation of human PSCs is controversial. Some reports argued that XaXa ESCs and iPSCs undergo XCI upon differentiation (Hall et al., 2008; Lengner et al., 2010; Silva et al., 2008; Tomoda et al., 2012; Ware et al., 2009). This pattern would best mirror that of differentiating mouse ESCs, which has dogmatically made XaXa PSCs most sought-after. Similarly, XeXa cells have been described to undergo XCI upon differentiation (Vallot et al., 2015). Conversely, other reports described that the XeXa state is maintained during differentiation (Mekhoubad et al., 2012; Nazor et al., 2012). In many cases the human PSC lines analyzed have had heterogeneous XCI-states in the undifferentiated state, with both XiXIST+Xa/XiXa and XaXa/XeXa cells in the same culture. An increase in the fraction of XiXIST+Xa/XiXa cells during differentiation was interpreted as evidence of XCI (Hall et al., 2008; Hoffman et al., 2005; Silva et al., 2008; Tomoda et al., 2012; Vallot et al., 2015). However, this result can also be explained without XCI if differentiation gave a selective advantage to XiXIST+Xa/XiXa cells compared to XaXa and XeXa cells.
Here, we systematically characterized the XCI-state in primed female human ESCs and their differentiated progeny. Based on our concern that heterogeneity has made it difficult to assess changes in the XCI-state upon differentiation, we established ESC lines with homogeneous and well-defined XCI states. Our results show that in our cell culture system, the XaXa state arises during ESC derivation from blastocysts in addition to the faithfully inactivated XiXIST+Xa state. Both states can be stabilized and carried over into established ESC lines. The abnormal XiXa (without XIST) and XeXa states appear due to culture-induced erosion of the Xi during the propagation of established XiXIST+Xa ESCs, consistent with recent reports (Mekhoubad et al., 2012; Tchieu et al., 2010; Vallot et al., 2015). All lines regardless of XCI state could differentiate and the XCI state pre-existing in ESCs was maintained in differentiated cells, indicating that X-to-autosome dosage compensation is not required for in vitro differentiation. Our works provides a framework for the epigenetic changes of the X-chromosome during derivation and propagation of ESCs under conventional primed culture condition and the consequences for differentiation, and a model for how the blastocyst state of the X transitions towards XCI.
RESULTS
Characterization of the XCI-state in human ESC lines at the single cell level
To study the state of the X-chromosome in human ESCs upon differentiation, we evaluated undifferentiated ESC lines derived and maintained in conventional FGF2-containing primed culture media (Table S1). Specifically, we applied RNA fluorescent in situ hybridization (RNA FISH) to 10 newly derived female ESC lines (UCLA1, UCLA3, UCLA4, UCLA5, UCLA8, UCLA9, UCLA14, UCLA16, UCLA17, and UCLA18) at intermediate passage (P13–25) after at least one freeze-thaw cycle, and to three widely used female ESC lines from other institutions (H9, H7, and ESI03; at P21–38). We expected abnormal XCI states to be present at intermediate passage. For newly established lines, derivation was performed under both normoxic and hypoxic conditions, but oxygen levels did not influence the resulting XCI-state (Table S1). All new lines expressed the pluripotency transcription factors OCT4 and NANOG and surface markers SSEA4 and TRA1-81 and formed teratomas containing all three germ layers (Fig S1A, Table S1). DNA copy number and metaphase chromosome spread analyses confirmed that the ESC lines were karyotypically normal (Fig S1B, S2, Table S1).
We utilized RNA FISH to examine the expression and nuclear localization of XIST, the key regulator of XCI, as well as the nascent transcription of nine X-linked genes (MID1, CASK, AMMECR1, WDR44, THOC2, GPC3, MTMR1, ATRX, HUWE1) distributed along the entire X-chromosome (Fig S3A), all subject to silencing by XCI (Balaton et al., 2015). Human female fibroblasts, and male (H1) and female (H9) ESC lines were used to validate these assays (Fig S3B). We anticipated the analysis of these well-distributed X-linked genes to distinguish between the XaXa, XeXa, and XiXa states. In addition, RNA FISH for UTX, a gene known to escape XCI (Fig S3A/B) (Balaton et al., 2015), was used to verify that cells with two X-chromosomes were scored.
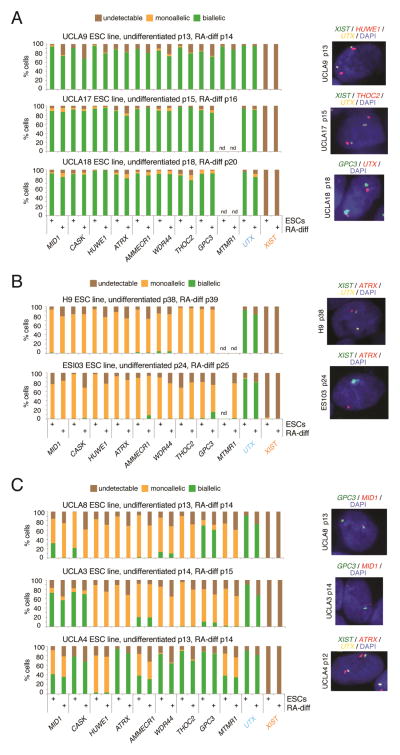
Applying the multi-gene RNA FISH approach, we found dramatic differences between ESC lines. UCLA9, UCLA17, and UCLA18 displayed bi-allelic expression for all assayed X-linked genes and lacked XIST expression in all cells (Fig 1A), which led us to classify them as homogeneously XaXa. A few cells displayed no or mono-allelic nascent transcription of the X-linked genes, which is likely due to transcriptional bursts. The pluripotency specific lncRNA XACT (Vallot et al., 2015) was also expressed from both X-chromosomes (Fig S3C). H9 and ESI03 displayed mono-allelic expression for all tested X-linked genes (Fig 1B). XIST expression was completely lacking in H9 cells and present in only 3% of ESI03 cells (Fig 1B); hence, we classified these cell lines as XiXa. Eight ESC lines (UCLA1, UCLA3, UCLA4, UCLA5, UCLA8, UCLA14, UCLA16, and H7) lacked XIST expression and displayed mono- and bi-allelic expression of at least one of the examined X-linked genes (Fig 1C, S3D), suggesting that these lines carried an Xe that arose from the Xi with XIST expression (Anguera et al., 2012; Mekhoubad et al., 2012; Nazor et al., 2012; Shen et al., 2008; Vallot et al., 2015). To test this idea, we analyzed the XCI-state of these lines at earlier passage and found that UCLA1, UCLA4, UCLA14, UCLA16, and UCLA8 had an XIST-coated X-chromosome or an accumulation of H3K27me3 indicative of the XIST-coated Xi (Fig S3E–H; see below for UCLA8; the other three ESC were not available at earlier passage). This result was consistent with erosion of the XiXIST+Xa state accompanied by XIST loss and partial Xi reactivation upon propagation of these cells, and ruled out the possibility that these lines underwent only partial XCI in the first place. In some XeXa ESC lines, only one or two genes were bi-allelically expressed (GPC3 in UCLA8; AMMECR and GPC3 in UCLA1; CASK and MID1 in UCLA3) indicating slight Xi-erosion (Fig 1C, S3D). Conversely, UCLA4 showed bi-allelic expression for eight out of nine genes, indicating substantial Xi-erosion (Fig 1C). Moreover, our RNA FISH analysis suggested that some of the genes tested were more susceptible to erosion than others (for instance MID1, CASK, GPC3, AMMECR vs. HUWE1) (Fig 1C, S3D). HUWE1 was the only gene tested with mono-allelic expression across all XeXa ESC lines studied here (Fig 1A/C).
Figure 1. X-chromosome analysis in undifferentiated human ESCs and upon RA-induced differentiation by multi-gene RNA FISH.
A) Quantification of RNA FISH results in XaXa ESC lines for the nascent transcription foci of nine X-linked genes normally subject to XCI, of the X-linked gene UTX that escapes XCI, and of XIST for different ESC lines in the undifferentiated state after the cells went through at least one freeze/thaw cycle since derivation, and after 10 days of retinoic acid (RA)-induced differentiation, at the indicated passage of ESCs. Representative RNA FISH images of ESCs are given.
B) As in (A), except for XiXa ESC lines.
C) As in (A), except for XeXa ESC lines with different degrees of Xi-erosion.
See also Figures S1, S2, S3 and S5.
We conclude that our ESC lines, at intermediate passage, are all XIST-negative and can be classified as XiXa, XeXa, and XaXa based on RNA FISH for nine X-linked genes. The XiXa and XaXa ESC lines were essentially homogeneous for their XCI state, whereas XeXa lines expressed certain X-linked genes mono-allelically in some cells and bi-allelically in the others.
Comprehensive analysis of XCI state by DNA methylation profiling and RNA-seq
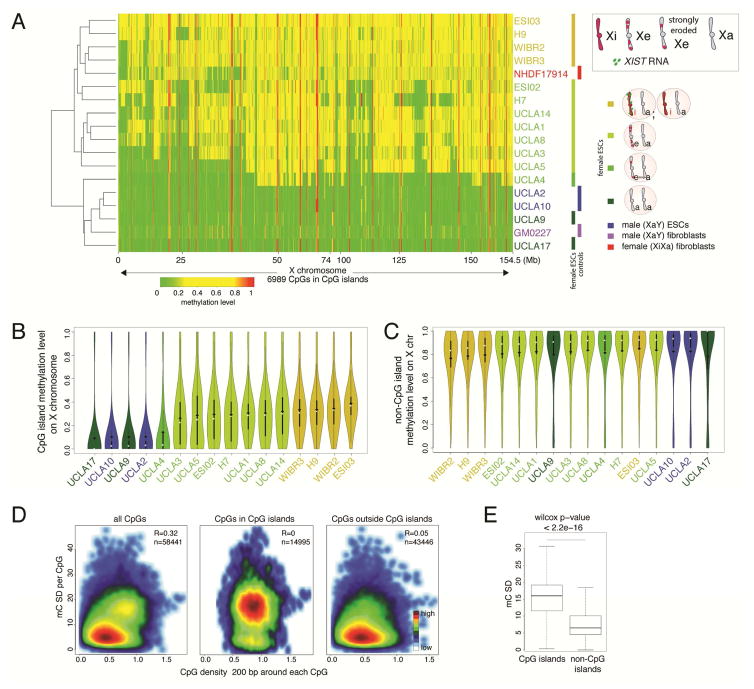
Despite single cell resolution, RNA FISH is limited to only a small number of genes. Therefore, we examined DNA methylation levels of CpG islands (CGIs) on the X-chromosome in 11 of the aforementioned female ESC lines (at the same passage as for RNA FISH analysis) and three additional female lines (ESI02, WIBR2, and WIBR3) by conducting reduced representation bisulfite sequencing (RRBS) (Meissner et al., 2005) (Fig S4A). This was particularly important for XaXa and XiXa lines to ensure that these states extended across the entire X-chromosome. Analysis of CGIs associated with X-linked genes examined by RNA FISH above demonstrated that these genes lacked methylation when they were bi-allelically expressed and were intermediately methylated when mono-allelically expressed as a result of methylation on the Xi and absence thereof on the Xa (Fig S4B). This result validated the use of methylation as faithful marker of the XCI state (Nazor et al., 2012; Sharp et al., 2011).
Methylation analysis across the X demonstrated that UCLA9 and UCLA17 nearly completely lacked CGI methylation and closely mirrored the methylation pattern of male ESC lines UCLA2 and UCLA10 and male fibroblasts with their solitary Xa, indicating that the XaXa state was an X-chromosome-wide phenomenon (Fig 2A/B, S4C). H9, ESI03, WIBR2, and WIBR3 showed a uniform pattern of intermediate methylation across the X similar to female XiXIST+Xa fibroblasts, consistent with their chromosome-wide classification as XiXa (Fig 2A/B, S4C). UCLA1, UCLA3, UCLA4, UCLA5, UCLA8, UCLA14, H7, and ESI02 lacked CGI methylation in parts of the X-chromosome in agreement with their XeXa state (Fig 2A/B, S4C). UCLA4 showed comparably little CGI methylation highlighting the extensive erosion of the Xi in this line.
Figure 2. CGI methylation distinguishes XeXa, XaXa, and XiXa ESCs.
A) Heatmap of unsupervised hierarchical clustering of RRBS-based methylation levels for CpGs within X-linked CGIs in indicated female and male ESC lines and control fibroblasts. Methylation level of 1 indicates 100% methylation and 0 absence of methylation. CpGs with constitutively low (<0.15) and high (>0.85) methylation were excluded to highlight Xi-linked intermediate methylation.
B) Violin plots of the distribution of methylation levels of CpGs in X-linked CGIs in indicated cell lines. White square=median, black diamond=mean.
C) As in (B), except for methylation levels of X-linked CpGs outside CGIs.
D) Plot of standard deviation (SD) of methylation of all X-linked CpGs and the subsets within and outside of CGIs, respectively, relative to CpG density in the 200-nucleotide window around each CpG considered, in female ESC lines with different XCI states. The number of CpGs analyzed is given.
E) Boxplot of SD values defined in (D) for X-linked CpGs within and outside of CGIs and significance testing of the difference.
See also Figure S4.
The analysis of DNA methylation not only confirmed our RNA FISH-based classification of the XCI states in our ESC lines, but also led to additional insights. The XaXa state in female mouse ESCs is associated with global DNA hypomethylation compared to male ESCs (Zvetkova et al., 2005). We found no evidence of autosomal hypomethylation in female human XaXa lines compared to male and female XiXa ESCs, indicating that the presence of two active X-chromosomes, unlike in mouse, is not associated with dramatic changes in autosomal methylation in human ESCs (Fig S4D/E). Furthermore, we addressed whether methylation changes on the Xe extended beyond CGIs. While CGI methylation differed strongly between XCI-states, methylation outside these regions was consistently high and did not differ between XCI states (Fig 2B/C, S4F). Consistent with this finding, the analysis of methylation variance along the X-chromosome in XaXa, XiXa, and XeXa ESC lines and CpG density demonstrated that regions with high CpG content, such as CGIs, were specifically prone for erosion (Fig 2D/E). Thus, Xi-erosion is largely confined to CGIs, suggesting that the erosion process specifically acts on gene-regulatory elements. Lastly, we determined the methylation level of imprint control regions and found no correlation between the epigenetic state of the X and imprint methylation (Fig S4G), indicating that these two mechanisms of mono-allelic expression regulation are distinctly affected in human ESCs (Nazor et al., 2012).
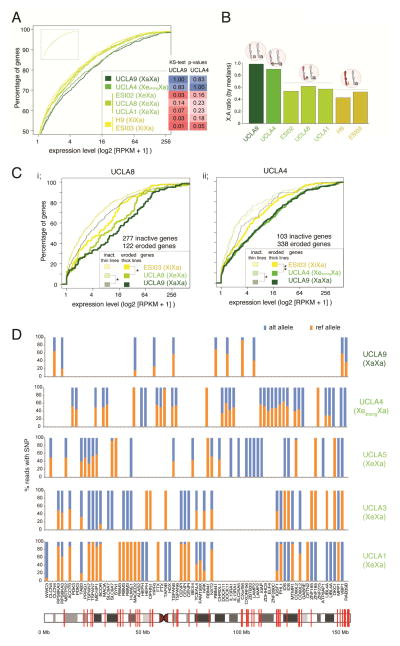
To determine whether the XeXa and XaXa ESCs displayed higher expression of X-linked genes compared to XiXa lines, we generated RNA-seq data for the XiXa lines H9 and ESI03, the weakly eroded XeXa lines ESI02, UCLA1, and UCLA8, the strongly eroded line UCLA4, and the XaXa line UCLA9. We observed a significant increase in X-linked gene expression levels in XaXa and strongly eroded ESCs relative to XiXa lines (Fig 3A). Moreover, we found a higher median X-linked relative to autosomal gene expression in UCLA9 and UCLA4 (Fig 3B) indicating that X-to-autosome dosage compensation did not occur in these two lines and, therefore, is not critical for the self-renewal of human ESCs. Moreover, based on the presence or absence of CGI methylation within promoter regions, we defined X-linked genes as putative active and inactive, respectively (Table S2). For the slightly eroded line UCLA8 and the strongly eroded line UCLA4, genes with demethylated CGIs (i.e. putatively active due Xi-erosion) were expressed at a level similar to the XaXa ESC line UCLA9, while genes with intermediary CGI methylation (i.e. putatively inactive) mirrored the level of the XaXi ESC line ESI03 (Fig 3C). Thus, Xi-erosion leads to the loss of methylation and an increase of the dose of affected X-linked genes, although cause and consequence remain unclear.
Figure 3. X-linked gene expression is higher in XeXa and XaXa than XiXa ESCs.
A) Cumulative distribution functions (CDFs) of X-linked gene expression for indicated female ESC lines based on RNA-seq data. Inset gives the CDFs of autosomal gene expression for the same cell lines (same XY-scales). ks-test p-values are given for the comparison of X-linked gene expression distributions between the XaXa line UCLA9 and the strongly eroded XeXa line UCLA4, respectively, and all other lines.
B) X-to-autosome (X:A) ratio of median gene expression in indicated female ESC lines based on RNA-seq data.
C) (i) CDFs of expression values of eroded (thick lines) and inactive (thin lines) genes on the X in the XeXa line UCLA8 defined based on the methylation level of promoter-associated CGIs. The expression of the same two gene sets in the XaXa line UCLA9 and the XiXa line ESI03 was also plotted for comparison. *=ks-test p-value ≤ 0.05 in comparisons of eroded and inactive genes between UCLA8 and the two ESC lines. (ii) As in (i) except for inactive and eroded X-linked genes for the strongly eroded line UCLA4. **=p-value < 0.005.
D) Allelic expression analysis based on RNA-seq reads. X-axis lists genes subject to XCI with informative SNPs covered by five or more RNA-seq reads in the indicated female ESC lines, and the Y-axis the proportion of reads originating from the reference (ref) or alternate (alt) allele. If more than one such SNP was present in the same gene, the gene appears more than once. If the given SNP was not informative or did not have ≥5 reads in a given ESC line, it appears as a blank on the graph. Red lines show SNP locations in the X-chromosome image.
Since XCI is non-random in most human ESC lines, likely due to clonal expansion during derivation (Shen et al., 2008), we also analyzed RNA-seq data for the allelic expression state of X-linked genes based on single nucleotide polymorphisms (SNPs) (Fig 3D). Consistent with non-random XCI, many genes with informative SNPs expressed solely either the reference or the alternate allele and few genes expressed both alleles in UCLA1, UCLA3 and UCLA5 due to Xi-erosion. The reverse was true for UCLA4 in agreement with its more extensive Xi-erosion. In UCLA9, we did not find any informative gene with mono-allelic expression, agreeing with it being XaXa rather than XeXa.
The XCI-state of ESCs is maintained during differentiation
The unambiguous classification of the XCI-state in our ESC lines allowed us to test how the XaXa, XeXa, and XiXa states fare during differentiation. This analysis excluded the XiXIST+Xa cells since this XCI-state is labile and quickly disappears during ESC propagation (see below) (Shen et al., 2008). Despite differences in XCI state, all ESC lines formed teratomas containing differentiated cells of all three germ layers (Fig S1A, Table S1) suggesting that the starting XCI-state does not affect pluripotency. We used retinoic acid (RA) to drive in vitro differentiation of XaXa (UCLA9, 17,18), XeXa (UCLA1, 3, 4, 5, 8, 14, 16, and H7), and XiXa (H9 and ESI03) ESC lines for 10 days, and determined whether these lines were capable of inducing XIST and de novo XCI by multi-gene RNA FISH (Fig S5A). Quantification was restricted to only OCT4-negative differentiated cells. Since all ESC lines were XIST-negative in the undifferentiated state, de novo XCI would easily be detectable by the upregulation of XIST. Upon differentiation, the proportion of X-linked genes with mono- and bi-allelic expression was nearly identical to that in the pluripotent cell of origin and XIST was undetectable (Fig 1A–C, S3D). An Xi-like enrichment of H3K27me3, a hallmark of the XIST-expressing Xi (Plath et al., 2003), was also absent in RA-differentiated cells (Fig S5B). Similarly, spontaneously differentiating ESCs did not exhibit XIST expression (Fig S5C). Thus, neither XIST nor XCI were induced de novo during spontaneous and RA-induced differentiation of XIST-negative XeXa, XaXa, and XiXa ESC lines.
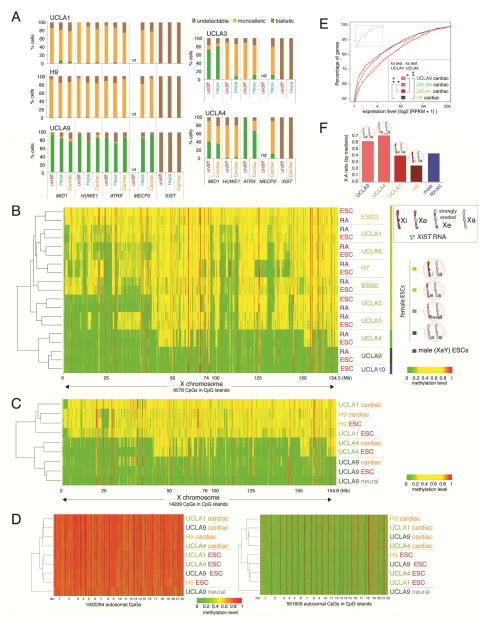
To validate our findings further, we determined the XCI-state in ESC-derived differentiated cells of defined character. We differentiated UCLA9 (XaXa), UCLA1 (weak XeXa), UCLA4 (strong XeXa), and H9 (XaXi) towards cardiomyocytes, and UCLA9, UCLA1, UCLA3 (XeXa), and H9 into hepatocyte-like cells. More than 90% of the cells successfully differentiated into cardiomyocytes or hepatocytes. Hepatocyte-like identity was confirmed by immunostaining for ALBUMIN and AFP, and measurement of ALBUMIN secretion (Fig S5D–F). Differentiated cardiomyocytes displayed expected characteristics including spontaneous beating, appropriate calcium transients, an increase in beating frequency in response to beta-adrenergic stimulation, expression of cardiomyocyte markers, and a striated pattern of cardiac TROPONIN I (Fig S5G–J). RNA FISH indicated that TROPONIN I-positive cardiomyocytes and ALBUMIN-positive hepatocyte-like cells maintained the bi- or mono-allelic expression pattern of X-linked genes that was present in the respective undifferentiated ESC line (Fig 4A, S5E). Moreover, there was no evidence for expression of XIST or induction of the XIST-dependent H3K27me3 accumulation in differentiated cells (Fig 4A, S5D/H). Similar results were obtained for the XaXa ESC line UCLA9 upon differentiation into TUJ1-positive neurons (Fig S5K).
Figure 4. The XCI state of ESCs is maintained in differentiated cells.
A) Quantification of X-linked gene RNA FISH pattern and XIST in ESC lines and ESC-derived cardiomyocytes and hepatocytes.
B) Heatmap of unsupervised hierarchical clustering of RRBS-based methylation levels for CpGs within X-linked CGIs in indicated female and male ESC lines and their day 10 RA-induced differentiation products. CpGs with constitutive low (<0.15) and high (>0.85) methylation levels were not included to highlight the intermediate methylation due to XCI.
C) As in (B), except for indicated female ESC lines in the undifferentiated state and upon directed differentiation into neurons and cardiomyocytes.
D) Heatmap of unsupervised hierarchical clustering of RRBS-based methylation levels for all autosomal CpGs (left) and only those within CGIs (right) for the same cell lines/conditions shown in (C). The numbers below indicate chromosomes, which are separated by dashed lines.
E) CDFs of X-linked gene expression for ESC-derived cardiomyocytes. Inset shows normalized autosomal expression data for the same cell lines at the same XY-scales. p-values of the ks-test for the difference of expression distributions between UCLA9 and UCLA4, respectively, and all other lines are given (*≤0.05; **≤0.0005).
F) X:A ratio of median gene expression in cardiomyocytes derived from the indicated ESC lines based on RNA-seq.
See also Figures S3 and S5.
To extend these findings from a few X-linked genes to the entire X-chromosome, we measured DNA methylation in cardiomyocytes, neurons, and day 10 RA-differentiation cultures derived from XaXa, XeXa and XiXa ESC lines. Unsupervised hierarchical clustering revealed that X-linked CGI methylation in differentiated cells remained highly similar to that in starting undifferentiated ESC and differed between cell lines (Fig 4B/C). This was not true for autosomal methylation, which clustered by differentiation state and not by cell line (Fig 4D). Thus, differentiation is not associated with a change in the epigenetic state of the X-chromosome, regardless of the pre-existing XCI-state in ESCs. In support of this observation, X-linked gene expression was significantly higher in cardiomyocytes derived from XaXa ESCs (UCLA9) and the strongly eroded XeXa ESCs (UCLA4) compared to XiXa ESCs (H9) and slightly eroded XeXa ESCs (UCLA1) (Fig 4E, S5L). Similarly, the median expression level of X-linked genes relative to autosomal genes was increased (Fig 4F), indicating that in vitro differentiation can tolerate improper X-to-autosome dosage compensation. The X-to-autosome ratios in cardiomyocytes were generally lower than those for ESCs (Fig 4F vs 3B), but the reason is unclear. Cardiomyocytes derived from UCLA1, UCLA4, and UCLA9 displayed no changes in X-chromosome copy number, confirming that X-linked gene expression differences arose from epigenetic, not genetic differences (Fig S2). We conclude that de novo XCI and X-to-autosome dosage compensation are not required for differentiation of female human ESCs. Our data reveal that in addition to the XIST-negative XeXa and XiXa states, the XaXa state in our ESC lines – widely sought after in human PSCs for its putative ability to undergo XCI – is in fact an abnormal epigenetic state of the X-chromosome as it lacks the ability to induce XIST and XCI upon differentiation.
X-linked silencing correlates with differentiation propensity
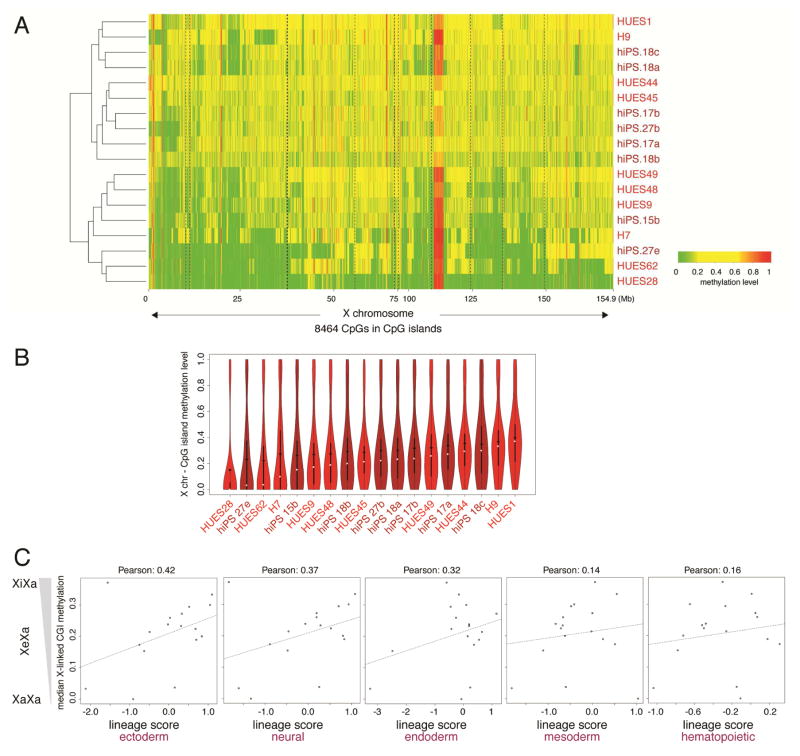
Human ESCs and iPSCs are often variable in their differentiation propensity (Bock et al., 2011), and the contribution of XCI-state to this phenomenon is unknown. To examine this, we took advantage of a previously reported estimation of differentiation propensity towards ectoderm, mesoderm, and endoderm, coined ‘lineage score’, for 18 human female ESC and iPSC lines (Bock et al., 2011). We used RRBS data provided by the same study to derive methylation values for X-linked CGIs. As expected, CGI methylation varied widely between the 18 lines (Fig 5A). A subset of lines displayed intermediate CGI methylation along the entire X, most closely matching the XiXa state, while others were eroded to different degree. One ESC line (HUES28) nearly lacked CGI methylation, mirroring the pattern of male PSCs, and hence was considered XaXa (Fig 5A, S6A). Next, we correlated the median CGI methylation level as a measure of the XCI state of these PSC lines (Fig 5B, Fig S6B) with their lineage score. This analysis revealed a positive correlation between XCI status and lineage score for all germ layers (Fig 5C). The relationship was strongest for ectoderm and neural lineage (Fig 5C). This unbiased approach therefore suggested that PSC lines with extensive erosion (no XCI) differentiate less efficiently than those with a proper Xi or slight Xi-erosion (with XCI). Thus, lack of XCI may be one of the factors that contribute to differentiation biases of PSCs.
Figure 5. X-chromosome silencing correlates with differentiation propensity.
A) Heatmap of unsupervised hierarchical clustering of RRBS-based methylation levels for CpGs within X-linked CGIs in indicated female ESC (red) and iPSC (dark red) lines obtained from Bock et al (2011). CpGs with constitutive low (<0.2) and high (>0.6) methylation were not included to emphasize intermediate methylation due to XCI.
B) Violin plots of methylation levels of CpGs in X-linked CGIs for cell lines shown in (A). White square=median, black diamond=mean.
C) Scatterplot of the median methylation level of X-linked CGIs for each cell lines shown in (A) (low median methylation indicates presence of an Xa or strongly eroded Xe and high methylation indicates an Xi or weakly eroded Xe) versus the lineage scores for differentiation propensity for the same line.
See also Figure S6.
XaXa and XiXIST+Xa cells emerge during ESC derivation, yet XiXa and XeXa arise during ESC propagation
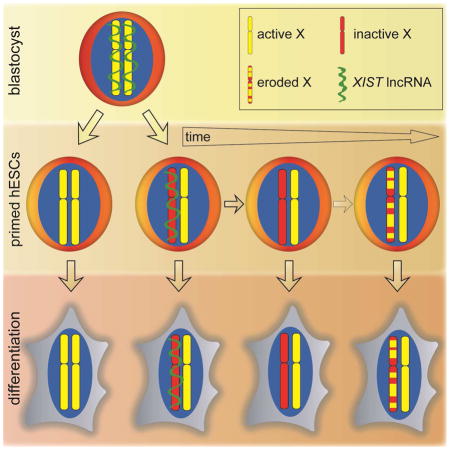
None of the XCI states observed in primed human ESCs captured that of the pre-implantation blastocyst, where XIST is expressed from both X-chromosomes yet the X-chromosomes are active (XaXIST+XaXIST+) (Okamoto et al., 2011). This disparity is likely best explained by the transition from naïve to primed pluripotency when ESCs are derived in FGF2-containing media (Davidson et al., 2015). Regardless, we wanted to understand how the different XCI-states of primed human ESCs arise during derivation and propagation.
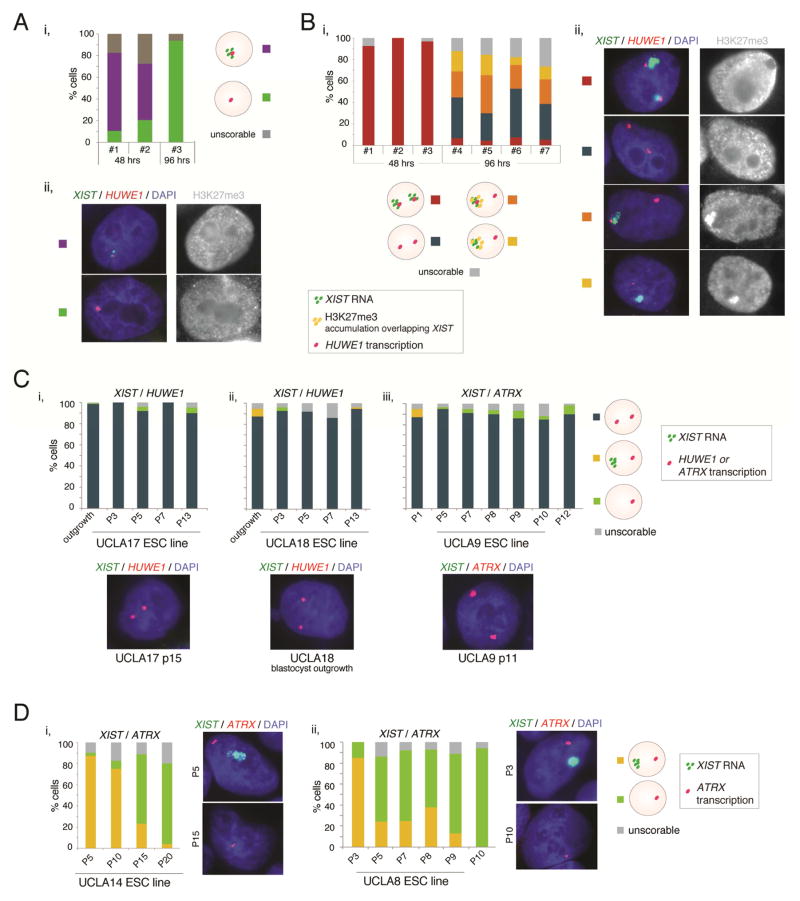
We confirmed the XCI-state of human blastocysts by RNA FISH for XIST and nascent transcription foci of HUWE1, which distinguished the Xa from the Xe in our ESC lines (Fig 1, S3D). All blastocysts analyzed either expressed single or double signals of XIST and HUWE1 (Fig S7A/B), indicative of male XaXIST+Y and female XaXIST+XaXIST+ blastocysts, as previously described (Okamoto et al., 2011). Next, we examined how XIST expression and the transcriptional state of HUWE1 changed during ESC derivation by performing RNA FISH in combination with H3K27me3 immunostaining on blastocysts 48 and 96 hours (h) after plating onto feeders in FGF2-containing culture media. In blastocysts, H3K27me3 accumulation of the X does not accompany XIST expression (Okamoto et al., 2011). At 48h, approximately 24h after blastocyst attachment, the majority of cells maintained the state of the blastocyst such that male and female cells still displayed one and two XIST-expressing active X-chromosomes without H3K27me3 accumulation, respectively (Fig 6A/B). At 96h, when a larger number of cells had grown out, we did not detect XIST expression in male cells (Fig 6A), indicating that XIST became silenced on the single X-chromosome. Conversely, female embryos, detected based on the number of sites with XIST/HUWE1 expression per cell, showed various X-chromosome states at 96h (Fig 6B). On average, 20% of scorable outgrowing cells from four female blastocysts displayed the prototypical post-XCI pattern with a strong XIST cloud and overlapping H3K27me3 enrichment on one X and expression of HUWE1 from the other X (XiXIST+/H3K27me3+Xa). The remaining 80% of cells displayed bi-allelic expression of HUWE1 and therefore carried two active X-chromosomes. Of these, less than 10% showed the pattern of the blastocyst with XIST expression and absence of H3K27me3 accumulation (XaXIST+XaXIST+), approximately 50% lacked an XIST signal and H3K27me3 accumulation (XaXa), and around 40% carried an accumulation of both XIST and H3K27me3 on one of the two X-chromosomes (XaXIST+/H3K27me3+Xa), a pattern that is normally seen in mouse ESCs soon after Xist coats the X but before silencing is initiated. In a second derivation experiment with four additional female embryos, the blastocyst XCI-state was already lost at 48h after plating. In this case, both XaXa and XiXIST+Xa states were equally present at 48h, and the majority of cells from an additional embryo displayed the XiXIST+Xa state at 96h (Fig S7C/D). These data indicated that the pre-XCI pattern of the female blastocyst is quickly lost in primed ESC culture conditions.
Figure 6. The XaXa and XiXIST+Xa states are stabilized during ESC derivation.
A) (i) Quantification of RNA FISH patterns for XIST and X-linked gene HUWE1 and co-immunostaining for H3K27me3, in cells of male blastocysts at 48hrs and 96hrs after plating in ESC culture media. No enrichment of H3K27me3 was observed. “Male” classification was based on the predominant mono-allelic HUWE1 and/or XIST pattern. (ii) Representative images of the XIST/HUWE1 RNA FISH (left) and the H3K27me3 co-staining (right) for the two main patterns observed in (i). The XIST signal was typically weak.
B) As in (A), except for seven female pre-implantation blastocysts, classified based on two distinct nuclear regions of XIST and/or HUWE1 RNA FISH signal.
C) Quantification of the XCI patterns and representative RNA FISH images of XIST, HUWE1 and ATRX, in the blastocyst outgrowths and throughout early passages (p) for the XaXa ESC lines UCLA17 (i), UCLA18 (ii), and UCLA9 (iii) without any freeze/thaw cycle.
D) As in (C), but for the early passages of UCLA14 (i) and UCLA8 (ii), which were XiXIST+Xa at the earliest passage. Representative RNA FISH images capture the XIST-positive and XIST-negative states, while mono-allelic ATRX expression is maintained.
See also Figure S6.
To understand which XCI-states persist in derivation cultures over time, we analyzed three blastocyst outgrowths at three weeks after plating by RNA FISH for XIST and HUWE1. In two blastocyst outgrowths, the vast majority of cells were XaXa as they expressed HUWE1 bi-allelically without XIST (Fig 6Ci/ii). Interestingly, in one of those two, 7% of cells displayed the post-XCI XiXIST+Xa state and clustered together within the large pool of XaXa cells implying that the XiXIST+Xa cells arose from cells that had faithfully undergone XCI soon after plating (Fig S7E). The third blastocyst outgrowth displayed XIST expression in the majority of cells indicative of the XiXIST+Xa state (S7F). Together, these data indicated that XaXa and XiXIST+Xa states can both be stabilized under our derivation cultures.
Since pluripotent cells may represent a minority in blastocyst outgrowths, we confirmed these findings in established ESC lines by following their XCI-state through early passages. We established new ESC lines by isolating a small piece of the blastocyst outgrowth. ESC lines derived from blastocyst outgrowths that were largely XaXa (UCLA17 and UCLA18) displayed the XIST-negative XaXa state at passage 3 (P3) and all subsequent passages (with or without freezing) (Fig 6Ci,ii). This finding was consistent with the XaXa state of UCLA17 and UCLA18 at intermediate passage (Fig 1A/2A). The third ESC lines classified as XaXa at intermediate passage (Fig 1A/2A) was also in this state at passage 1 (Fig 6Ciii). Thus, all three XaXa ESC lines described in this study carried this state already at the earliest passage and stably maintained it during ESC propagation, consistent with the notion that the XaXa state arises during ESC derivation.
Two other ESC lines followed through early passages, UCLA8 and UCLA14, displayed XIST expression and mono-allelic expression of the X-linked gene ATRX at early passage indicative of the XiXIST+Xa state (Fig 6D). After only a few passages (without freezing of the cells), XIST expression was gradually lost in both lines such that by passage 20 almost no XIST-positive cells remained (Fig 6D). While the loss of XIST in UCLA8 and UCLA14 did not coincide with the activation of ATRX (Fig 6D), RNA-seq data and RNA FISH for XACT demonstrated that the transition from the XIST-positive to the XIST-negate state coincided with Xi-erosion (Fig S7G–I), in agreement with prior reports (Mekhoubad et al., 2012; Nazor et al., 2012; Shen et al., 2008; Vallot et al., 2015). Thus, in addition to XaXa cells, the XiXIST+Xa state can be stably maintained in blastocyst outgrowths and carried over into established ESC lines. However, in contrast to the XaXa state, the XiXIST+Xa state is unstable as it loses XIST during propagation and in many cases undergoes Xi-erosion.
The Xi of propagating iPSCs undergoes erosion instead of reactivation
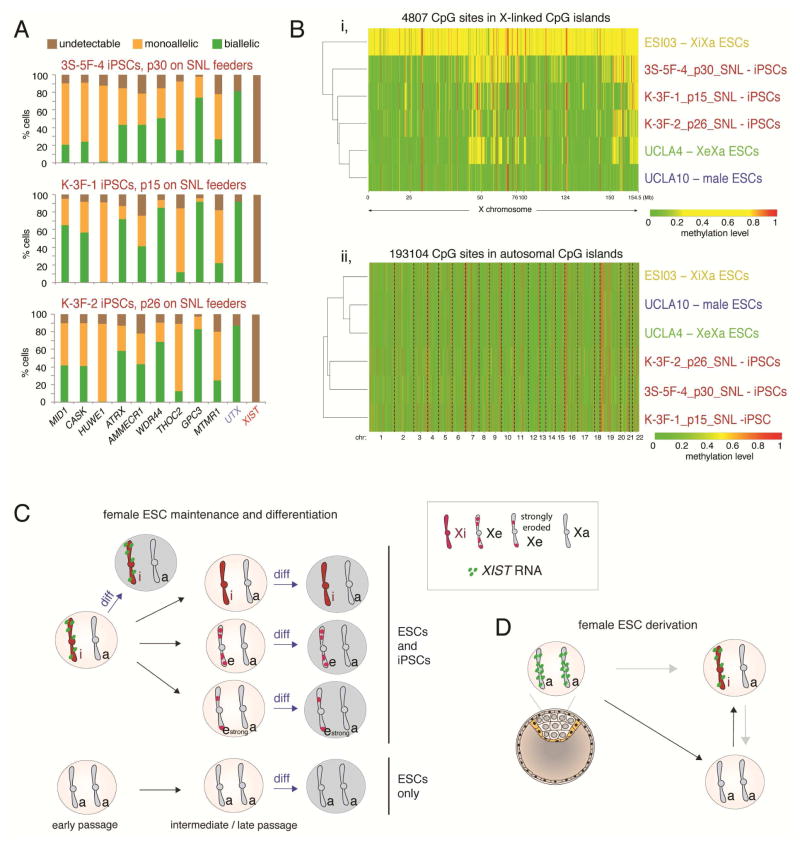
Our finding that the XaXa state arises during ESC derivation and not during propagation suggested that iPSCs that are in the XiXIST+Xa when first derived (Mekhoubad et al., 2012; Tchieu et al., 2010) should erode, as seen in XiXIST+Xa ESCs, but not transition to the XaXa state in the primed state. Supporting this idea only one ESC line, but no iPSC line, exhibited the XaXa state in Figure 5A. We made a similar observation when inspecting published X-linked CGI-methylation data for a large number of female ESCs and iPSCs (Figure 5 in (Nazor et al., 2012)), where five female ESC lines, but no iPSC line, were XaXa. However, contrary to our idea, a recent report indicated that established female XiXa iPSCs can be converted to the XaXa state upon transfer onto SNL-fibroblasts, which are immortalized mouse fibroblasts that secret human LIF (Tomoda et al., 2012), based on the measurement of bi-allelic expression of only a few X-linked genes by RNA FISH and increased X-linked gene expression, which may not distinguish between strong erosion and the XaXa state. Therefore, we re-investigated the XCI state of three SNL-treated iPSC lines (Tomoda et al., 2012). Multi-gene RNA FISH revealed that all three SNL-iPSC lines carried a severely eroded Xi that expressed HUWE1 mono-allelically and most other tested X-linked genes bi-allelically (Fig 7A). X-linked CGI methylation of these iPSC lines was similar to our most severely eroded ESC line UCLA4 (Fig 7B). These data suggest that XiXIST+Xa iPSCs, like XiXIST+Xa ESCs, can be driven into extensive Xi-erosion through specific culture conditions, but never reactivate the Xi completely (no XaXa state) when the primed pluripotent state is maintained. Thus, we propose that the XaXa state is specific to ESCs, established during derivation from the blastocyst, and not relevant for primed in iPSCs (Fig 7C).
Figure 7. Summary of XCI states observed during derivation, propagation and upon differentiation of ESCs, and comparison to iPSCs.
A) Quantification of the RNA FISH pattern of indicated X-linked genes normally subject to XCI, of the X-linked gene UTX that escapes XCI, and of XIST in three SNL-converted iPSC lines (Tomoda et al., 2012).
B) (i) Heatmap of unsupervised hierarchical clustering of RRBS-based methylation levels for CpGs within X-linked CGIs in female iPSCs described in (A), and indicated ESC lines for comparison. Only CpGs that are not constitutively lowly (<0.15) and highly (>0.85) methylated were plotted to better capture the intermediate methylation due to XCI. (ii) As in (i), expect that CpGs in autosomal CGIs were analyzed, revealing clustering by culture condition.
C) Scheme summarizing changes in XCI-state during ESC propagation and differentiation.
D) As in (C), but during ESC derivation.
See also Figure S7.
DISCUSSION
In female mammals, X-to-autosome dosage compensation is achieved by X-chromosome inactivation and current evidence indicates that this process is essential for female embryonic development and adult homeostasis (Schulz and Heard, 2013; Yang et al., 2016). The prevailing model therefore suggests that human primed female ESCs and iPSCs undergo XCI during differentiation, but the literature on the state of the X in undifferentiated PSCs and upon differentiation has remained controversial (Anguera et al., 2012; Barakat et al., 2015; Hall et al., 2008; Hoffman et al., 2005; Kim et al., 2014; Lengner et al., 2010; Mekhoubad et al., 2012; Nazor et al., 2012; Pomp et al., 2011; Shen et al., 2008; Silva et al., 2008; Tchieu et al., 2010; Tomoda et al., 2012; Vallot et al., 2015; Xie et al., 2016). We applied genomics approaches and multi-gene RNA FISH to define the XCI-state in ESCs at both the single cell and population level (summarized in Table S1). Moreover, we derived new ESC lines from a small area of the blastocyst outgrowth rather than dissociation of the entire outgrowth, which led to relatively homogeneous XCI patterns in most lines, enabling us to draw robust conclusions about any changes of the XCI-state upon differentiation.
We found that neither XaXa nor XeXa ESC lines, regardless of the degree of Xi-erosion, are competent for XIST upregulation and de novo initiation of XCI upon induction of differentiation, resulting in the maintenance of the epigenetic state that pre-existed in the ESC population (Fig 7C). While we did not analyze the fate of the XiXIST+Xa state in differentiating ESCs, prior studies have shown that this state is also maintained upon differentiation (Fig 7C) (Mekhoubad et al., 2012). Similarly, XiXa ESCs do not induce XIST upon differentiation (Fig 7C). Differentiated XeXa and XaXa cells express X-linked genes more highly than XiXa cells, indicating the absence of an alternative XIST-independent dosage compensation process during in vitro differentiation. Since ESC lines can undergo differentiation into all three germ layers regardless of XCI-state, we conclude that an imbalance of the dosage between X-chromosomes and autosomes is compatible not only with the pluripotent state but also with differentiated cell identities in vitro. Thus, while XCI is a mandatory process in vivo, it is not required in vitro.
Our work demonstrates that the pre-XCI state of the blastocyst (XaXIST+XaXIST+) quickly transitions into the post-XCI state with an XIST RNA-coated Xi (XiXIST+Xa) or the XaXa state without XIST expression, revealing an unexpectedly heterogeneity early in the ESC derivation process that can be maintained in cultures over time (Fig 7D, S7J). The occurrence of a large number of XaXa cells in ESC derivation is intriguing and may indicate that cells normally proceed from the XaXIST+XaXIST+ pre-XCI state to the XiXIST+Xa post-XCI state via the XaXa state, i.e. the silencing of XIST on both X-chromosomes and subsequent XIST upregulation on one X, which would then lead to initiation of silencing (Fig 7D, black arrows). In this model, the XaXa state would be a transient intermediate of the usual sequence towards XCI. Under derivation conditions, there may only be a short time window in which XIST can be re-induced in XaXa cells to mediate XCI. For instance, genome-wide methylation changes occurring during ESC derivation (Smith et al., 2014) may permanently silence XIST. In support of this idea, several studies have demonstrated that the absence of XIST in established ESCs correlates with DNA methylation of the XIST promoter region (Shen et al., 2008; Tchieu et al., 2010). Missing the window of opportunity for XIST re-expression would result in the maintenance of XaXa cells.
In an alternative model, it is possible that XaXIST+XaXIST+ cells induce XIST silencing on only one X, whereas XIST on the other X is enabled to induce XCI (Fig 7D, top gray arrow). In this scenario, the XaXa state would be reflective of a state not within the normal sequence towards XCI in human development, potentially due to aberrant silencing of XIST on both X-chromosomes instead on one. Lastly, it is conceivable that XaXa cells are derived from XiXIST+Xa cells due to silencing of XIST immediately after initiation of XCI when XCI is still in a reversible state (Fig 7D, right gray arrow). However, given that at 48h of blastocyst plating in primed ESC culture media the XaXa state was predominant, we favor the first model of the XaXa state being an intermediate towards the post-XCI XiXIST+Xa state. Regardless, both the XaXa and XiXIST+Xa states can be manifested in early passage ESCs. The proportion of the XiXIST+Xa or XaXa cells in a blastocyst outgrowth may depend on exact culture conditions, thawing process and genomic background, and/or exact developmental stage of the blastocyst, which remains to be elucidated in the future. The XiXIST+Xa pattern becomes prevalent more often than the XaXa state, such that the majority of early passage female ESC lines are XiXIST+Xa (Nazor et al., 2012; O'Leary et al., 2012; Silva et al., 2008).
In agreement with published data (Mekhoubad et al., 2012; Nazor et al., 2012; Shen et al., 2008; Vallot et al., 2015), we observed that established XiXIST+Xa ESCs predictably lose XIST and spontaneously re-express Xi-linked genes yielding XeXa cells. Notably, we found that all ESCs that initially displayed XIST expression and were XiXIST+Xa eroded but none of them induced complete X-chromosome-wide re-activation as we see in XaXa cells. Hence, XaXa cells arise specifically during derivation from the blastocyst and not from the Xi by erosion (Fig 7C/D). We propose the primed iPSCs should therefore also not normally be in the XaXa state (Fig 7C). Our data are most consistent with the notion that the intrinsic state of ESCs cultured in classic primed media conditions is XiXIST+Xa because all other XCI states, including the XaXa state, display abnormalities upon differentiation such as lack of XIST expression, lack of XCI, increased X-linked gene expression, and/or altered DNA methylation patterns. The XiXIST+Xa state is consistent with the post-XCI state and intermediary DNA methylation of X-linked CGIs in mouse epiblast stem cells (Pasque et al., 2011), which, based on their morphology, signaling requirement, and gene expression are in a similar developmental state as primed human ESCs (Davidson et al., 2015).
Several studies concluded that XaXa ESCs can induce XCI upon induction of differentiation (Hall et al., 2008; Lengner et al., 2010; Silva et al., 2008; Tomoda et al., 2012; Ware et al., 2009). While most of these studies examined PSC lines heterogeneous for the XCI-state such that skewing of cell populations may have masked the absence of XCI, one study proposed that XaXa ESCs can be derived under low oxygen and undergo XCI in the self-renewing state upon transfer to normoxic conditions (Lengner et al., 2010). This may suggest that the XaXa state we describe here is specific for the particular derivation condition that we used. However, based on available genomics data, we analyzed independent ESC lines from other labs derived and cultured under various conditions (Bock et al., 2011; Nazor et al., 2012) and found several XaXa ESC lines, supporting our conclusion that the XaXa state can be stably maintained in ESCs grown in normoxic conditions. While it remains possible that hypoxic culture conditions during derivation may stabilize the XaXa state in a window of opportunity that enables XIST upregulation and XCI during the propagation of established ESC lines, other studies have now shown that the derivation under hypoxic conditions yields XiXIST+Xa ESCs and that hypoxia enhances XIST loss in XiXIST+Xa ESCs (O'Leary et al., 2012; Xie et al., 2016), which is consistent with our findings.
Given that PSCs are used for understanding early human development, regenerative medicine, and disease-in-a-dish studies, it is critical to consider that abnormal XCI states may interfere with the intended application or confound biological interpretation (Mekhoubad et al., 2012). Thus, a careful analysis of the epigenetic status of the X needs to be performed before using female PSCs. Additionally, it is important to consider paths to clearing the abnormalities of the X-chromosome in female PSCs. For instance, it may be possible to develop methods that enable the re-induction of XIST because XIST has the capability to initiate silencing in human PSCs when expressed ectopically (Jiang et al., 2013). Alternatively, our recent study (Sahakyan et al, 2016, in press) indicates that newly developed culture methods that enable reprogramming of human PSCs to developmentally earlier pluripotent states induce the pre-XCI state of the human blastocyst and reset epigenetic abnormalities of the X-chromosome in primed PSCs, enabling faithful and XIST-mediated XCI upon differentiation.
EXPERIMENTAL PROCEDURES
Cell lines, pre-implantation embryos, ESC derivation and culture
Two male and 10 female ESC lines used in this study were derived in primed culture conditions at UCLA (Table S1). H7, H9, ESI02, and ESI03 ESCs were obtained from the WiCell Research Institute’s WISC Bank, WIBR2 and WIBR3 ESCs from the Whitehead Institute (Lengner et al., 2010), and the iPSC lines 3S-5F-4, K-3F-1, K-3F-2 under SNL/LIF culture conditions from the Gladstone Institute (Tomoda et al., 2012). For ESC derivation and blastocyst outgrowth analyses, surplus embryos were donated from couples that underwent in vitro fertilization treatment under informed consent. Table S1 summarizes derivation conditions and analyses performed for each ESC line. For more details see Supplemental Experimental Procedures. Human embryo studies presented here had the approval of the UCLA Institutional Review Board (IRB#11-002027) and Embryonic Stem Cell Research Oversight (ESCRO) Committee (2008–015 and 2007–009).
RNA FISH
RNA FISH, immunofluorescence, and the acquisition of images were performed as described (Tchieu et al., 2010). Briefly, for RNA FISH, we used fluorescently labeled DNA probes generated by random priming from BACs or FOSMIDS. To quantify staining patterns, at least 200 single cells distributed over 10 fields of view were inspected under the microscope for each gene and cell line. Experiments were performed at least in triplicate for RNA FISH and immunostainings of undifferentiated ESC and retinoic acid differentiation, and in duplicate for the directed differentiations. For more details see Supplemental Experimental Procedures.
Expression, methylation, and karyotype analysis
To prepare ESCs for analysis by RNA FISH, RRBS, RNA-seq and CNV arrays, cells were feeder depleted, except for the WIBR2/WIBR3 and Gladstone iPSCs. Libraries were built according to standard protocols. For more details see Supplemental Experimental Procedures.
Data deposition
The accession number for the genomics data is GSE88933.
Supplementary Material
Acknowledgments
We are grateful to Drs. S. Yamanaka, B. Panning, K. Tomoda, R. Jaenisch, and M. Mitalipova for sharing cell lines. SP was supported by CIRM; G.B. by Philip Whitcome, Dissertation Year and Quantitative and Computational Biosciences Training Fellowships at UCLA; AS by the Ruth L. Kirschstein NRSA F31, Philip Whitcome, and Mangasar M. Mangasarian Fellowships; RK by the UCLA Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research (BSCRC) and CIRM; CC by CIRM and a Leukemia and Lymphoma Research Visiting Fellowship (10040); S.F. by a QCB Collaboratory Fellowship; ATC by NIH HD079546; WL by NIH P01 GM099134; and KP by the UCLA BSCRC and David Geffen School of Medicine, the Iris Cantor-UCLA Women’s Health Center and CTSI (UL1TR000124), CIRM, and NIH P01 GM099134. The authors have no conflict of interest to declare. Funds for human embryo banking and ESC derivation were provided by BSCRC and CIRM.
Footnotes
AUTHOR CONTRIBUTION
Conceptualization, K.P.; Methodology, S.P., G.B., K.P.; Formal Analysis, G.B., A.S., C.C., J.L., S.F.G.; Investigation, S.P., G.B., R.K., C.C., A.S., L.R., R.J.P.S.; Visualization, S.P., G.B., A.S., K.P.; Data Curation, G.B.; Writing – Original Draft, K.P.; Writing – Review and Editing, S.P., G.B., K.P.; Funding Acquisition, K.P.; Supervision, R.A., M.P., B.L., A.T.C., and K.P; Project Administration, K.P.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anguera MC, Sadreyev R, Zhang Z, Szanto A, Payer B, Sheridan SD, Kwok S, Haggarty SJ, Sur M, Alvarez J, et al. Molecular signatures of human induced pluripotent stem cells highlight sex differences and cancer genes. Cell Stem Cell. 2012;11:75–90. doi: 10.1016/j.stem.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augui S, Nora EP, Heard E. Regulation of X-chromosome inactivation by the X-inactivation centre. Nature reviews. 2011;12:429–442. doi: 10.1038/nrg2987. [DOI] [PubMed] [Google Scholar]
- Balaton BP, Cotton AM, Brown CJ. Derivation of consensus inactivation status for X-linked genes from genome-wide studies. Biol Sex Differ. 2015;6:35. doi: 10.1186/s13293-015-0053-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barakat TS, Ghazvini M, de Hoon B, Li T, Eussen B, Douben H, van der Linden R, van der Stap N, Boter M, Laven JS, et al. Stable X chromosome reactivation in female human induced pluripotent stem cells. Stem Cell Reports. 2015;4:199–208. doi: 10.1016/j.stemcr.2014.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock C, Kiskinis E, Verstappen G, Gu H, Boulting G, Smith ZD, Ziller M, Croft GF, Amoroso MW, Oakley DH, et al. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson KC, Mason EA, Pera MF. The pluripotent state in mouse and human. Development (Cambridge, England) 2015;142:3090–3099. doi: 10.1242/dev.116061. [DOI] [PubMed] [Google Scholar]
- Disteche CM. Dosage compensation of the sex chromosomes. Annual review of genetics. 2012;46:537–560. doi: 10.1146/annurev-genet-110711-155454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall LL, Byron M, Butler J, Becker KA, Nelson A, Amit M, Itskovitz-Eldor J, Stein J, Stein G, Ware C, et al. X-inactivation reveals epigenetic anomalies in most hESC but identifies sublines that initiate as expected. J Cell Physiol. 2008;216:445–452. doi: 10.1002/jcp.21411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman LM, Hall L, Batten JL, Young H, Pardasani D, Baetge EE, Lawrence J, Carpenter MK. X-inactivation status varies in human embryonic stem cell lines. Stem cells (Dayton, Ohio) 2005;23:1468–1478. doi: 10.1634/stemcells.2004-0371. [DOI] [PubMed] [Google Scholar]
- Jiang J, Jing Y, Cost GJ, Chiang JC, Kolpa HJ, Cotton AM, Carone DM, Carone BR, Shivak DA, Guschin DY, et al. Translating dosage compensation to trisomy 21. Nature. 2013;500:296–300. doi: 10.1038/nature12394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KY, Hysolli E, Tanaka Y, Wang B, Jung YW, Pan X, Weissman SM, Park IH. X Chromosome of female cells shows dynamic changes in status during human somatic cell reprogramming. Stem Cell Reports. 2014;2:896–909. doi: 10.1016/j.stemcr.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengner CJ, Gimelbrant AA, Erwin JA, Cheng AW, Guenther MG, Welstead GG, Alagappan R, Frampton GM, Xu P, Muffat J, et al. Derivation of pre-X inactivation human embryonic stem cells under physiological oxygen concentrations. Cell. 2010;141:872–883. doi: 10.1016/j.cell.2010.04.010. [DOI] [PubMed] [Google Scholar]
- Meissner A, Gnirke A, Bell GW, Ramsahoye B, Lander ES, Jaenisch R. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic acids research. 2005;33:5868–5877. doi: 10.1093/nar/gki901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhoubad S, Bock C, de Boer AS, Kiskinis E, Meissner A, Eggan K. Erosion of dosage compensation impacts human iPSC disease modeling. Cell Stem Cell. 2012;10:595–609. doi: 10.1016/j.stem.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazor KL, Altun G, Lynch C, Tran H, Harness JV, Slavin I, Garitaonandia I, Muller FJ, Wang YC, Boscolo FS, et al. Recurrent variations in DNA methylation in human pluripotent stem cells and their differentiated derivatives. Cell Stem Cell. 2012;10:620–634. doi: 10.1016/j.stem.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Leary T, Heindryckx B, Lierman S, van Bruggen D, Goeman JJ, Vandewoestyne M, Deforce D, de Sousa Lopes SM, De Sutter P. Tracking the progression of the human inner cell mass during embryonic stem cell derivation. Nature biotechnology. 2012;30:278–282. doi: 10.1038/nbt.2135. [DOI] [PubMed] [Google Scholar]
- Okamoto I, Patrat C, Thepot D, Peynot N, Fauque P, Daniel N, Diabangouaya P, Wolf JP, Renard JP, Duranthon V, et al. Eutherian mammals use diverse strategies to initiate X-chromosome inactivation during development. Nature. 2011;472:370–374. doi: 10.1038/nature09872. [DOI] [PubMed] [Google Scholar]
- Pasque V, Gillich A, Garrett N, Gurdon JB. Histone variant macroH2A confers resistance to nuclear reprogramming. The EMBO journal. 2011;30:2373–2387. doi: 10.1038/emboj.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science (New York, NY. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- Pomp O, Dreesen O, Leong DF, Meller-Pomp O, Tan TT, Zhou F, Colman A. Unexpected X chromosome skewing during culture and reprogramming of human somatic cells can be alleviated by exogenous telomerase. Cell Stem Cell. 2011;9:156–165. doi: 10.1016/j.stem.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Sahakyan A, Kim R, Chronis C, Sabri S, Bonora G, Theunissen TW, Kuoy E, Langerman J, Clark AT, Jaenisch R, Plath K. Human naïve pluripotent stem cells model X-chromosome dampening and X-inactviation. Cell Stem Cell. 2016 doi: 10.1016/j.stem.2016.10.006. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz EG, Heard E. Role and control of X chromosome dosage in mammalian development. Current opinion in genetics & development. 2013;23:109–115. doi: 10.1016/j.gde.2013.01.008. [DOI] [PubMed] [Google Scholar]
- Sharp AJ, Stathaki E, Migliavacca E, Brahmachary M, Montgomery SB, Dupre Y, Antonarakis SE. DNA methylation profiles of human active and inactive X chromosomes. Genome Res. 2011;21:1592–1600. doi: 10.1101/gr.112680.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Matsuno Y, Fouse SD, Rao N, Root S, Xu R, Pellegrini M, Riggs AD, Fan G. X-inactivation in female human embryonic stem cells is in a nonrandom pattern and prone to epigenetic alterations. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:4709–4714. doi: 10.1073/pnas.0712018105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva SS, Rowntree RK, Mekhoubad S, Lee JT. X-chromosome inactivation and epigenetic fluidity in human embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:4820–4825. doi: 10.1073/pnas.0712136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Chan MM, Humm KC, Karnik R, Mekhoubad S, Regev A, Eggan K, Meissner A. DNA methylation dynamics of the human preimplantation embryo. Nature. 2014;511:611–615. doi: 10.1038/nature13581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchieu J, Kuoy E, Chin MH, Trinh H, Patterson M, Sherman SP, Aimiuwu O, Lindgren A, Hakimian S, Zack JA, et al. Female human iPSCs retain an inactive X chromosome. Cell Stem Cell. 2010;7:329–342. doi: 10.1016/j.stem.2010.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science (New York, NY. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Tomoda K, Takahashi K, Leung K, Okada A, Narita M, Yamada NA, Eilertson KE, Tsang P, Baba S, White MP, et al. Derivation conditions impact X-inactivation status in female human induced pluripotent stem cells. Cell Stem Cell. 2012;11:91–99. doi: 10.1016/j.stem.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallot C, Ouimette JF, Makhlouf M, Feraud O, Pontis J, Come J, Martinat C, Bennaceur-Griscelli A, Lalande M, Rougeulle C. Erosion of X Chromosome Inactivation in Human Pluripotent Cells Initiates with XACT Coating and Depends on a Specific Heterochromatin Landscape. Cell Stem Cell. 2015;16:533–546. doi: 10.1016/j.stem.2015.03.016. [DOI] [PubMed] [Google Scholar]
- Ware CB, Wang L, Mecham BH, Shen L, Nelson AM, Bar M, Lamba DA, Dauphin DS, Buckingham B, Askari B, et al. Histone deacetylase inhibition elicits an evolutionarily conserved self-renewal program in embryonic stem cells. Cell Stem Cell. 2009;4:359–369. doi: 10.1016/j.stem.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie P, Ouyang Q, Leng L, Hu L, Cheng D, Tan Y, Lu G, Lin G. The dynamic changes of X chromosome inactivation during early culture of human embryonic stem cells. Stem Cell Res. 2016;17:84–92. doi: 10.1016/j.scr.2016.05.011. [DOI] [PubMed] [Google Scholar]
- Yang L, Kirby JE, Sunwoo H, Lee JT. Female mice lacking Xist RNA show partial dosage compensation and survive to term. Genes & development. 2016;30:1747–1760. doi: 10.1101/gad.281162.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zvetkova I, Apedaile A, Ramsahoye B, Mermoud JE, Crompton LA, John R, Feil R, Brockdorff N. Global hypomethylation of the genome in XX embryonic stem cells. Nature genetics. 2005;37:1274–1279. doi: 10.1038/ng1663. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.