Abstract
In spite of its importance in cell function, targeting DNA is under-represented in the design of small molecules. A barrier to progress in this area is the lack of a variety of modules that recognize G•C base pairs (bp) in DNA sequences. To overcome this barrier an entirely new design concept for modules that can bind to mixed GC and AT sequences of DNA is reported. Because of their successes in biological applications, minor-groove-binding heterocyclic cations were selected as the platform for design. Binding to AT sequences requires H-bond donors while recognition of the G-NH2 requires an acceptor. The concept that we report here uses preorganized N-methylbenzimidazole (N-MeBI) thiophene modules for selective binding with mixed bp DNA sequences. The interaction between the thiophene sigma-hole (positive electrostatic potential) and electron donor nitrogen of N-MeBI preorganizes the conformation for accepting an H-bond from G-NH2. The compound-DNA interactions were evaluated with a powerful array of biophysical methods and the results show that N-MeBI-thiophene monomer compounds can strongly and selectively recognize single G•C bp sequences. Replacing the thiophene with other moieties significantly reduces binding affinity and specificity, as predicted by the design concept. These results show that the use of molecular features, such as sigma-hole, can lead to new approaches for small molecules in biomolecular interactions.
Keywords: Sigma-hole, biosensor-SPR, G•C bp recognition, N-methylbenzimidazole-thiophene, DNA minor groove
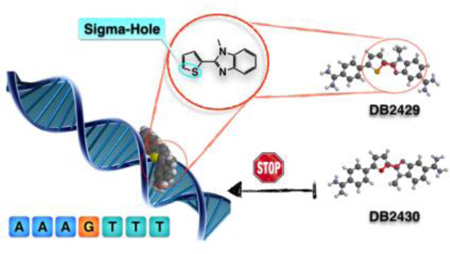
Graphical abstract
A new concept of Sigma-hole is applied in design of N-methylbenzimidazole-thiophene compounds to specifically recognize G•C base pair in the DNA minor groove.
Introduction
Design and preparation of new types of compounds that recognize mixed base pairs (bps) nucleic acid sequences is a major goal for the use of DNA as a cellular receptor for therapeutics and biotechnology applications as well as for development of gene specific probes.[1–8] Such compounds, for example, are being designed as transcription factor inhibitors by binding to a DNA promoter sequence rather than by targeting the transcription factor part of the complex.[9–14] There are almost unlimited numbers of applications of gene specific probes of this type. The current lack of a variety of small molecules that can bind strongly and sequence specifically to mixed A•T and G•C bp sequences of DNA is, however, a barrier to progress in this field. With the current increasing availability of both gene sequences and knowledge of their functions from bacteria to humans, there is clearly an unmet need for new sequence specific, small molecule DNA probes.[7,9,15] Significant variations in the cell uptake potential of molecular structures and pharmacokinetic differences will also require discovery and development of a diversity of different molecular systems for DNA recognition that do not now exist.[16–19]
Compounds: design concepts
We report here a new concept for the modular design of novel mixed sequence DNA-binding compounds that focuses on both the DNA minor groove structure and chemistry as well as on key chemical and structural features of the designed compounds.[3,20–22] The compounds, for example, should have appropriate curvature to fit the convex shape of the minor groove.[21–25] Although not essential, cationic groups have been attached at one or both ends of the new compounds to help solubility and increase the electrostatic interaction with negatively charged DNA backbone.[13,21,25–27] Amidine cationic groups that have been quite successful for targeting DNA, provide a H-bond donating, planar surface for optimum interactions with A•T bps as flanking sites of G•C bps.[24,28,29] An amidine attached to a six member ring, Figure 1, will also have a slight twist that helps the compound track the helical curvature of the minor groove. For the design of G•C bp binding capability, H-bond acceptor functions are needed to form a specific interaction with the G-NH2 groups that project into the minor groove.[15,30,31] Linked heterocyclic systems have been used in our design schemes because of their excellent cell uptake properties,[9,27] flexibility, and strong molecular interaction potential, particularly with the DNA minor groove.[26,30] The goal is to design modules that can be linked in a variety of ways to strongly and specifically recognize a range of target DNA sequences. Many well characterized A•T recognition units have been reported, such as benzimidazole/indole,[32] phenyl-amidine with numerous types of linking groups, such as -O-, -NH-, furan and others,[26,28,33,34] pyrrole-amide,[6,35–37] and alkylamines,[38] however, very few types of molecules and modules that specifically recognize one or more G•C bps have been reported.[15,34,37,39] Pyridine-based H-bond acceptor units (such as DB2120, Figure 1A),[30,34] and azabenzimidazole (azaBI) H-bond acceptors (DB2277, Figure 1A),[31,40] have recently been designed to recognize G•C bp and complement the polyamide imidazole group[4]. Dervan’s group and others have expanded the five-member hydrocycle used in polyamide. The amide-hydrocycle unit has also been replaced with BI or azaBI to recognize A·T or G·C bps.[4,41–44] Our understanding of G•C bp recognition by small molecule minor groove binders remains very limited, however, and additional molecular structures are needed to not only expand the use of the DNA as a cellular drug receptor but also the compound uptake potential in different organisms and cell types
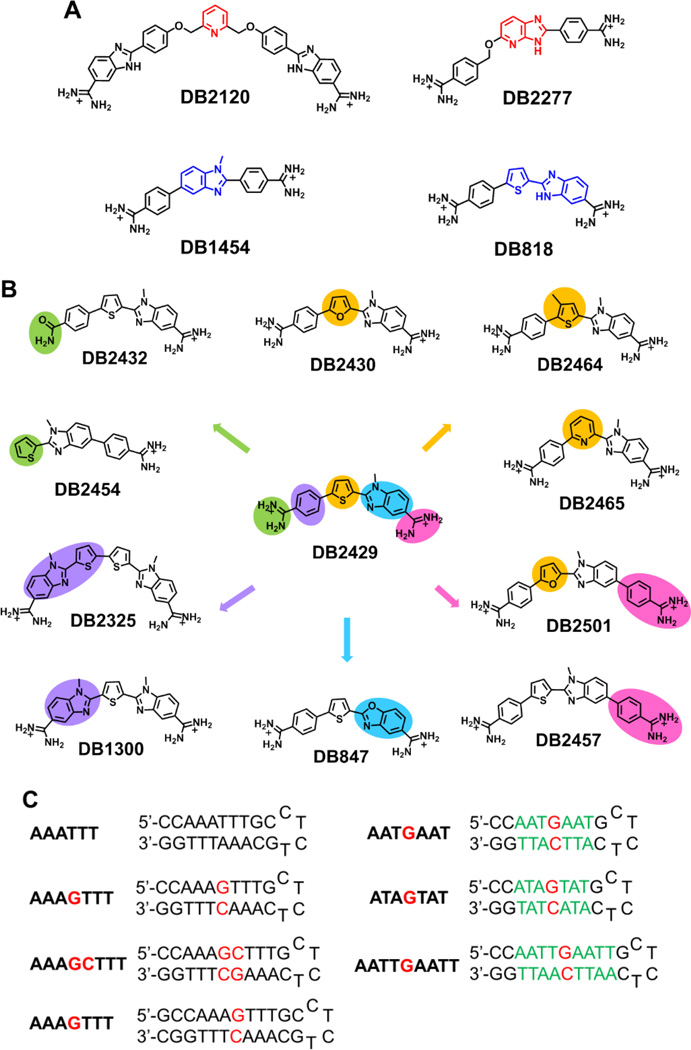
Figure 1.
(A) Chemical structure of reported single G•C bp binding molecules (top) and some stand molecules used in this study (below). (B) Chemical Structure of DB2429 and its analogues used in this study. Green: Mono-amidine; Orange: Furan, Pyridine, 3-methylthiophene; Pink: Extended Phenyl-amidine; Blue: Benzoxazole-thiophene module; Purple: bis-N-MeBI-thiophene modules. (C) The DNA sequences used in this study; DNA sequences with 5'-biotin labels were used for SPR studies.
The new design concept that we report here obeys the “rules” described above but is based on the hypothesis that an N-methylbenzimidazole (N-MeBI) module with the methyl pointed away from the minor groove floor would make an excellent new G-NH2 recognition unit. Unfortunately, none of our original designs, such as DB1454 in Figure 1A, gave significant strong and specific binding to G•C bps. Analysis of ab initio molecular models suggested that an N-MeBI-thiophene module could fit the minor groove shape[24] to successfully recognize G•C bps based on interactions between low-lying σ* orbitals (the “sigma-hole”) with positive electrostatic potential and electron donors such as the nitrogen atom of N-MeBI.[45–47] Such intramolecular interactions can favorably modulate the conformational preferences of a molecule.[48] The conformation that results from these intramolecular 1,4 N-S interactions of the thiophene and adjacent nitrogen heterocyclic ring pre-organizes the module to point the unsubstituted N of N-MeBI into the minor groove floor for accepting a H-bond from the minor groove G-NH2.
From extended compound designs (Figure 1B), synthetic studies, and biophysical experiments described here, we show that the N-MeBI-thiophene motif can provide excellent G•C recognition ability in an appropriate motif. Unlike classical heterocyclic thiophene derivatives such as DB818 (Figure 1A),[23] the new N-MeBI-thiophenes bind significantly more strongly to sequences with G•C bps than to pure AT sequences.
Extensions and tests of the Sigma-Hole design concept for DNA recognition
To test the key role of the thiophene in DB2429, furan (DB2430, DB2501), pyridine (DB2465) and phenyl (DB1454) analogs were prepared. The importance of the diamidines was investigated with the mono-amidines, DB2432 and DB2454. The significance of molecular shape and substituents was probed with DB2464, DB2325, DB1300 and DB2457. The role of the N-MeBI in DB2429 has been evaluated with benzoxazole DB847. Extensive biophysical analysis of the DNA interactions of these compounds show very clearly that the N-MeBI-thiophene module provides an essential basic recognition unit for mixed bps DNA sequence recognition in this compound set. The results validate the use of the sigma-hole concept in the design of DNA targeted agents. This type of compound preorganization to recognize target sites can significantly increase affinity and specificity independent of compound-receptor contacts and is a valuable new concept in DNA recognition modules. (Figure 1B)
Results and Discussion
Compound synthesis
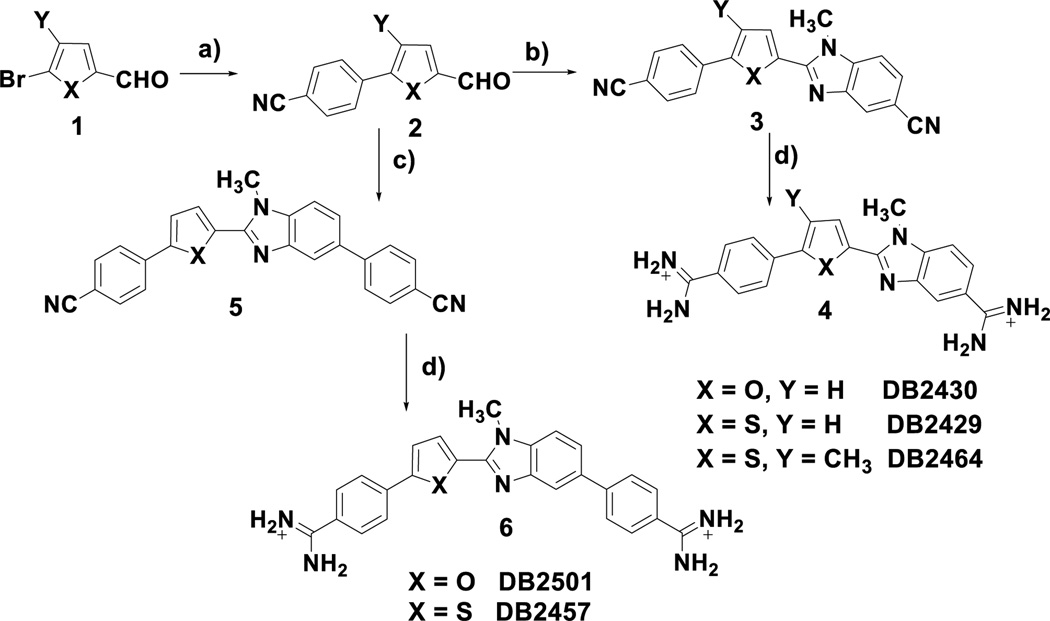
The syntheses of the key N-methylbenzimidazole heterocycles are outlined in Scheme 1. Reaction of the readily available bromoheteroaryl aldehydes (1) with 4-cyanophenylboronic acid under standard Suzuki coupling[49] conditions conveniently provides the 5-(4-cyanophenyl)-2-formyl 5-ring heterocycles (2) which can be used to prepare both the tri- and tetra-heteroaryl target molecules 4 and 6. Oxidative condensation and cyclization, mediated by sodium metabisulfite, of the aldehydes 2 with 3-amino-4-(methylamino) benzonitrile gives the bis-nitriles 3 in acceptable yields.[50] The bis-nitriles 3 are readily converted into the desired diamidines 4 by the action of LiN(TMS)2 in THF.[51] In a similar sequence the aldehydes 2 are allowed to react with 3'-amino-4'-(methylamino) [1,1'-biphenyl]- 4-carbonitrile, again mediated by sodium metabisulfite, to produce the bis-nitriles 5 which are converted to the diamidines 6 as previously described. The supplemental information contains the experimental details for the compounds described in Scheme 1 as well as the other novel compounds in Figure 1B used in this study.
Scheme 1.
Reagents and conditions: a) 4-cyanophenylboronic acid, Pd(PPh3)4, Na2CO3/H2O, dioxane, reflux b) 3-amino-4-methylaminobenzonitrile, sodium metabisulfite, DMF, reflux c) 3'-amino-4'-methylamino[1,1'-biphenyl]-4-carbonitrile, sodium metabisulfite, DMF,110 °C d) i. LiN(TMS)2, THF, rt ii. HCl, ethanol.
Thermal melting (Tm): relative binding affinity
DNA thermal melting experiments provide a rapid qualitative evaluation of the relative binding affinities of compounds with selected DNA sequences.[52] Small molecules binding with DNA generally enhance the duplex stability and an increase in the DNA melting temperature upon the addition of ligands which can be used as a screen for relative binding affinity.[11] Hairpin DNA oligomers, which have monomolecular melting transition (Figure 1) were chosen as model sequences to study relative binding affinities. The benzimidazole-thiophene DB818 (Figure 1A) has very strong binding with pure A-tract sequences, as previously reported.[24] The ΔTm results of DB2429 and its analogues are shown in Table 1. DB2429, the parent N-MeBI-thiophene after replacing the benzimidazole of DB818 with the N-MeBI moiety, has a high ΔTm (11 °C) with the target single G•C bp containing AAAGTTT sequence but weaker interactions with AAATTT (ΔTm=5 °C) and AAAGCTTT (ΔTm = 5 °C). The insertion of a phenyl ring between the N-MeBI and an amidine group of DB2429 yields DB2457 with a slightly increased Tm value with AAAGTTT (ΔTm=12 °C). Interestingly, the addition of a 3-methyl group into DB2429 increased the sequence selectivity due to a much lower affinity with the AAATTT and AAAGCTTT sequences. Modification of the thiophene group to furan (DB2430) and pyridine (DB2465) resulted in compounds with much weaker binding to single G sequences. To evaluate the effects of the N-MeBI without an adjacent thiophene ring, DB1454 was compared with DB2429 and found to have very low binding with the three test sequences. From these results it is clear that the special molecular properties of the N-MeBI-thiophene moiety help to enhance DNA minor groove binding ability and specificity. The benzoxazole, DB847, was considered to be a possible additional way to use the thiophene sigma-hole-N interaction to provide single G•C bp binding but the compound gave relatively moduate Tm increase (AAAGTTT, ΔTm = 7 °C). Molecular modeling, described below, provides a possible explanation for these results. In order to potentially expand the application of the N-MeBI-thiophene motif, DB1300 and DB2325 with two N-MeBI-thiophene modules, were studied. The binding activities of the compounds with AAAGTTT are relatively low and there is no significant interaction with the two G•C bps containing sequences. It seems likely that the curvature and placement of H-bond acceptors in these compounds does not properly index with donors on bp at the floor of the minor groove.
Table 1.
Thermal melting studies (ΔTm, °C) of DB2429 and analogues with pure AT and mixed DNA sequences.[a]
| DNA | AAA TTT |
AAA G TTT |
AAA GC TTT |
DNA | AAA TTT |
AAA G TTT |
AAA GC TTT |
|---|---|---|---|---|---|---|---|
| Ligand | Ligand | ||||||
| DB2429 | 5 | 11 | 5 | DB1300 | 4 | 8 | 3 |
| DB2464 | 3 | 10 | 2 | DB2325 | 2 | 8 | <1 |
| DB847 | 5 | 7 | 2 | DB2457 | 6 | 12 | 5 |
| DB2501 | 3 | 3 | <1 | DB2430 | 8 | 6 | 3 |
| DB2432 | 2 | 3 | <1 | DB2465 | 5 | 4 | 3 |
| DB1454 | 2 | 2 | <1 | DB2454 | <1 | <1 | <1 |
ΔTm = Tm (the complex) − Tm (the free DNA). 3 µM DNA sequences were studied in Tris-HCl buffer (50 mM Tris-HCl, 100 mM NaCl, 1 mM EDTA, pH 7.4) with the ratio of 2:1 [ligand/DNA]. An average of two independent experiments with a reproducibility of ±0.5 °C. Full DNA sequences: AAATTT-5’-CCAAATTTGCCTCTGCAAATTTGG-3’; AAAGTTT-5’-CCAAAGTTTGCTCTCAAACTTTGG-3’; AAAGCTTT-5’-CCAAAGCTTTGCTCTCAAAGCTTTGG-3’.
Based on the excellent results for AAAGTTT binding with N-MeBI-thiophenes, DB2429 and DB2457 were evaluated with sequences AATGAAT, ATAGTAT and AATTGAATT (Supporting Information Table S1). For both compounds the binding with all three DNAs was weaker than with AAAGTTT. In summary, with the compounds in Figure 1B, only the mono-N-MeBI-thiophene sigma-hole diamidines give strong and specific binding to the AAAGTTT mixed bp DNA sequence.
Biosensor-surface plasmon resonance (SPR): binding affinity and stoichiometry
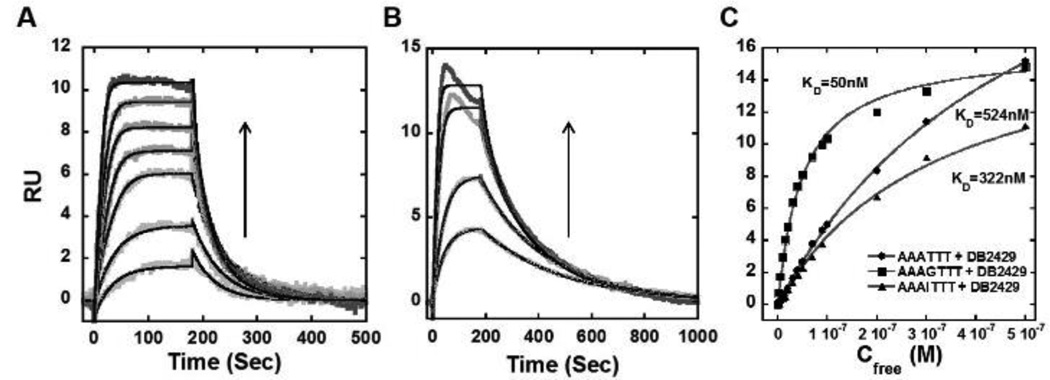
Biosensor-SPR methods provide an excellent way to quantitatively evaluate the interaction of small organic molecules with immobilized biomolecules.[53] SPR provides sensitive real-time progress of the binding reaction as well as the equilibrium binding affinity, kinetics, and stoichiometry of complex formation.[21,26] Based on the Tm results, the interactions of DB2429 with AAATTT, AAAGTTT and AAAITTT were evaluated by SPR (Figure 2). As can be seen in Figure 2A, DB2429 binds strongly with the single G•C bp containing sequence. Global kinetics fitting yielded a single binding site and an approximate KD of 50 nM for DB2429 with an AAAGTTT sequence. The reactions are relatively fast and the RU response in the plateau region was also plotted vs. the free compound concentration to determine the KD values for other sequences. The maximum value (RUmax) allows determination of stoichiometry.[53] The AAAITTT sequence was used to evaluate, in a more direct fashion, the influence of the G-NH2 group on binding in the minor groove. Based on the RUmax, both the single “G” and “I” sequences had only one binding site. The AAATTT sequence appeared to have a quite weaker second binding site that could not be saturated under our conditions. As with the Tm results, DB2429 has much stronger binding with AAAGTTT than AAATTT sequences. The KD value of AAAGTTT binding with DB2429 is quite strong (50 nM) compared to the KD values of AAATTT (524 nM) and AAAITTT (322 nM) (Figure 2C). This difference shows that DB2429 binds to AAAGTTT with excellent selectivity and that the G-NH2 is an essential component of the strong interaction.
Figure 2.
Representative SPR sensorgrams for (A) DB2429 and (B) DB2457 in the presence of AAAGTTT hairpin DNA, concentrations of DB2429 from bottom to top are 5, 10, 20, 30, 40, 50, and 70 nM, and for DB2457 from bottom to top are 5, 10, 20 and 30 nM; (C) Comparison of steady-state binding plots for DB2429 with AAAGTTT, AAATTT and AAAITTT sequences. The data are fitted to a steady state binding function using a 1:1 model to determine equilibrium binding constants. In (A) and (B) the solid black lines are best fit values for global kinetic fitting of the results with a single site function.
Based on the results with DB2429, several additional analogues were evaluated with AAATTT, AAAGTTT, and AAAGCTTT binding sites by using biosensor-SPR (Table 2). DB2457, the extended N-MeBI-phenyl analogue, shows 10 times stronger binding affinity than DB2429 with AAAGTTT (Table 2). The differences in binding affinity are most clearly seen in the global kinetic fitting of the dissociation phase of the sensorgrams (Figure 2). With minor groove binders that are structurally related, differences in affinity can be related to changes in either the on or off rate or some combination. With DB2429 and DB2457 very similar on-rates (ka = 1.3 ± 0.25×106 M−1 s−1) are observed while the off-rate kd for DB2457 is 10-fold lower (0.51 ± 1.3×10−2 S−1) than for DB2429 (kd = 6.5 ± 1.8×10−2 S−1). As a result of this slow off-rate, DB2457 is the strongest binder for single G•C bp containing sequences in this set of molecules. The extra phenyl group not only facilitated the higher binding ability for AAAGTTT but also increased the sequence selectivity of 50 times over the pure AT sequence (KD = 222 nM for AAATTT). DB2464, the 3-methylthiophene compound, however, has lower binding affinity (KD = 70 nM) with AAAGTTT in agreement with the Tm results. The three related N-MeBI compounds showed much lower affinity for the “I” containing sequence (Table 2). All of these results indicate that the N-MeBI moiety is playing a crucial role in G-NH2 recognition and DNA sequence selectivity.
Table 2.
Summary of binding affinity (KD, nM) for the interaction of all test compounds with biotin labeled DNA sequences using biosensor-SPR method [a]
| DB 2429 |
DB 2464 |
DB 2432 |
DB 847 |
DB 1300 |
DB 2457 |
DB 2430 |
DB 2465 |
|
|---|---|---|---|---|---|---|---|---|
| AAA TTT |
524 | 1160 | NB | 731 | 277 | 222 | 74 | 330 |
| AAA G TTT |
50 | 70 | 303 | 559 | 196 | 4 | 180 | 690 |
| AAA GC TTT |
1086 | 1060 | NB | NB | 708 | 192 | 360 | 1400 |
| AAA I TTT |
322 | 883 | -- | -- | -- | 322 | -- | -- |
| ATA G TAT |
432 | 590 | -- | -- | -- | 297 | -- | -- |
| AATT G AATT |
983 | 1493 | -- | -- | -- | 226 | -- | -- |
All the results in this table were investigated in Tris-HCl buffer (50 mM Tris-HCl, 100 mM NaCl, 1 mM EDTA, 0.05% P20, pH 7.4) at a 100 µL/min flow rate. “--” experiment not done, “NB” no binding. The listed binding affinities are an average of two independent experiments carried out with two different sensor chips and the values are reproducible within 10% experimental errors.
The binding of DB2429, DB2464, and DB2457 were also determined with a single G•C bp and other AT flanking sequences (ATAGTAT, AATTGAATT). Interestingly, relative to AAAGTTT, the compounds show weaker binding towards all other sequences (Table 2). Other compounds were also tested by SPR experiments with AAATTT, AAAGTTT, and AAAGCTTT. In agreement with the Tm results, DB2432, DB847 and DB1300 all have weak to no binding with AAAGTTT and other sequences. Surprisingly, DB2430 with an N-MeBI-furan motif shows higher binding affinity for AAATTT (KD = 74 nM) over AAAGTTT (KD = 180 nM). DB2465 with an N-MeBI-pyridine motif has quite weak binding to all test sequences (Table 2). The SPR results are all correlated with the Tm experiments.
In summary, an N-MeBI adjacent to a phenyl, a pyridine or a furan ring has very poor binding with the target AAAGTTT sequence. Our optimum thiophene sigma-hole compound, DB2457, however, has a KD with AAAGTTT of 4 nM and a KD with AAATTT of >200 nM. Clearly the N-MeBI-thiophene is an effective new module for strong, single G•C bp specific recognition.
Fluorescence emission spectroscopy: sequence dependent fluorescence differences
Fluorescence emission spectroscopy is an effective and sensitive method to study the binding between DNA and fluorescent small molecules[54] and to monitor the cellular location of compounds in culture.[10,28,55] The fluorescence titration results of DB2429 and DB2430 with sequences AAATTT, AAAGTTT and AAAGCTTT are shown in supporting information (Figure S3). The fluorescence intensity (F.I.) of DB2429 was quenched by all three test sequences but to different extents. The AAAGTTT sequence showed the highest level of F.I. decrease and the spectra reached the saturation state near a 1:1 [ligand/DNA] ratio, indicating monomer complex formation in agreement with SPR results. The intensity of the furan, DB2430, was quenched by AAAGTTT and AAAGCTTT to a smaller extent than DB2429. In summary, adding a G•C bp to the AAATTT sequence causes a marked decrease in fluorescence for all test compounds.
Circular dichroism (CD): probing the binding mode and ratio
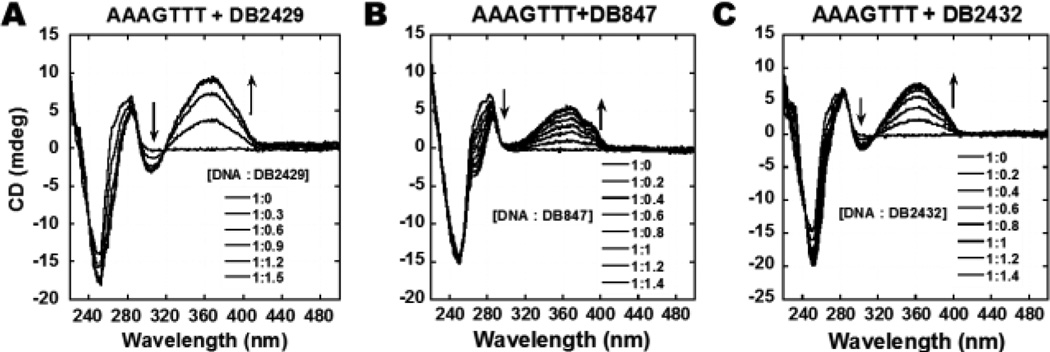
CD titration experiments as a function of compound concentration were evaluated to monitor the binding mode and the saturation limit for compound binding with DNA sequences (Figure 3). CD spectra monitor the asymmetric environment of the compounds binding to DNA and therefore can be used to obtain information on the binding mode.[56,57] There are no CD signals for the free compounds but on the addition of the compounds into DNA, substantial positive induced CD signals (ICD) arise in the compound absorption region between 300 and 450 nm wavelength. These positive ICD signals indicate a minor groove binding mode by these molecules as expected from their structures. A monotonic increase of ICD signals at the range of 320 to 440 nm wavelength was observed by incremental addition of DB2429, DB847, and DB2432 to the AAAGTTT sequence. As can be seen from Figure 3A and 3B, DB2429 and DB847 are binding in the minor groove of the AAAGTTT sequences with 1:1 stoichiometry, in agreement with SPR results. In summary, the CD titration results confirm a minor groove binding mode for the compounds in Figure 1.
Figure 3.
Circular dichroism spectra of the titration of representative compounds, (A) DB2429, (B) DB847, and (C) DB2432 with a 5 µM AAAGTTT sequence in the Tm buffer at 25 °C. Arrows indicate the changes.
Competition electrospray ionization mass spectrometry (ESI–MS): binding stoichiometry and relative binding affinity
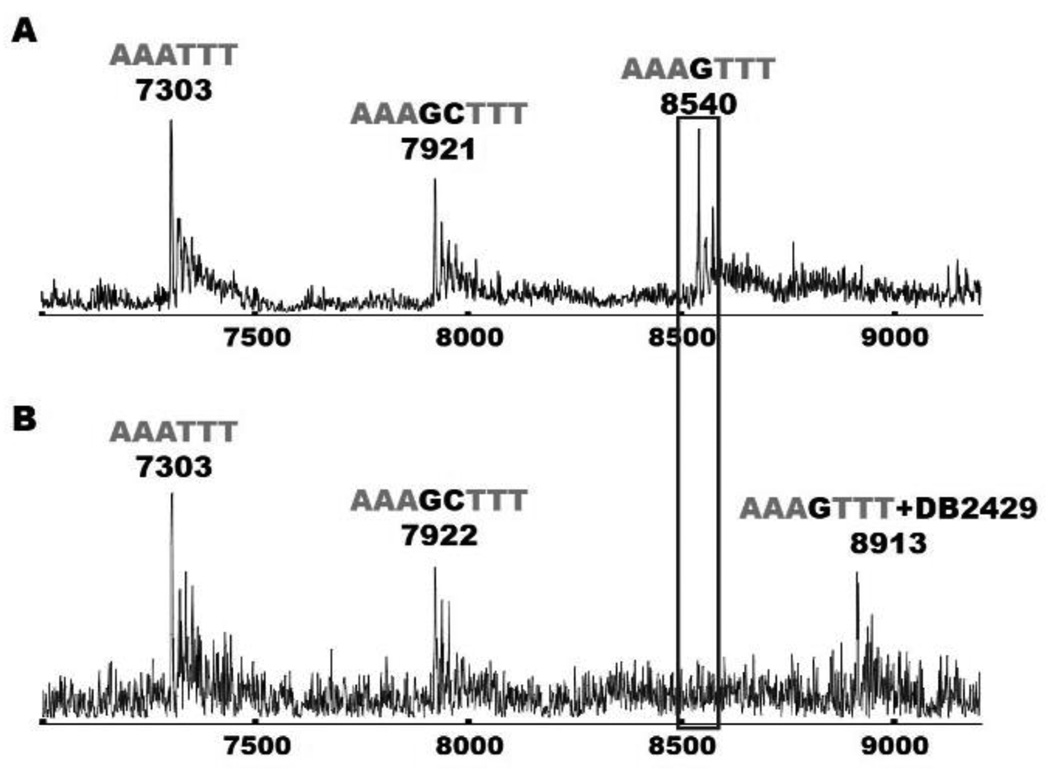
Mass spectrometry is an excellent method for the evaluation of relative binding affinity,[26,30,40] stoichiometry and the validation of experimental results obtained from other methods, such as SPR, where macromolecule-ligand stoichiometry is obtained by fitting the signal at different concentrations.[50,58] In this case the N-MeBI- thiophene compounds are far from ideal and a considerable amount of the compounds are adsorbed on the surface of the injector tube. Satisfactory results were obtained with the DB2429 complex to evaluate stoichiometry with the AAAGTTT sequence (Supporting Information Figure S4). The AAAGTTT sequence has a molecular weight of 8539 g/mol and this is observed as a single peak in the mass spectrometry results. With the addition of the compound DB2429 (mass of 374.5 g/mol) a new peak was observed at m/z = 8913 (difference of 374) indicating a 1:1 [ligand/DNA] complex formation which agrees with SPR and other results. For understanding the sequence specificity and binding stoichiometry of DB2429 with additional sequences, a competition ESI-MS experiment was performed (Figure 4).[58] Figure 4A represents the signal of three free DNA sequences, AAATTT, AAAGTTT, and AAAGCTTT at their molecular mass positions. With the addition of DB2429 the peak of AAAGTTT disappeared with the simultaneous appearance of a new peak at m/z = 8914 as a 1:1 DB2429-AAAGTTT complex. There was no signal of other ligand-DNA complex peaks observed which indicates excellent specificity of DB2429 with the AAAGTTT sequence. The signal to noise in these experiments is less than usual due to the problem of thiophene compound adsorption to the injector tube. It is clear, however, in agreement with the Tm and SPR results, that ESI-MS also shows that DB2429 can selectively recognize single G•C bp sequences with 1:1 stoichiometry.
Figure 4.
ESI-MS negative mode spectra of the competition binding of sequences AAATTT, AAAGTTT and AAAGCTTT (10 µM each) with 40 µM DB2429 in ammonium acetate buffer (150 mM ammonium acetate with 5% methanol (v/v), pH 6.8). (A) The ESI-MS spectra of free DNA mixture. (B) The ESI-MS spectra of DNA mixture with DB2429. The ESI-MS results shown here are deconvoluted spectra and molecular weights are shown with each peak.
Structural calculations and molecular docking: explanations for binding modes and mechanism
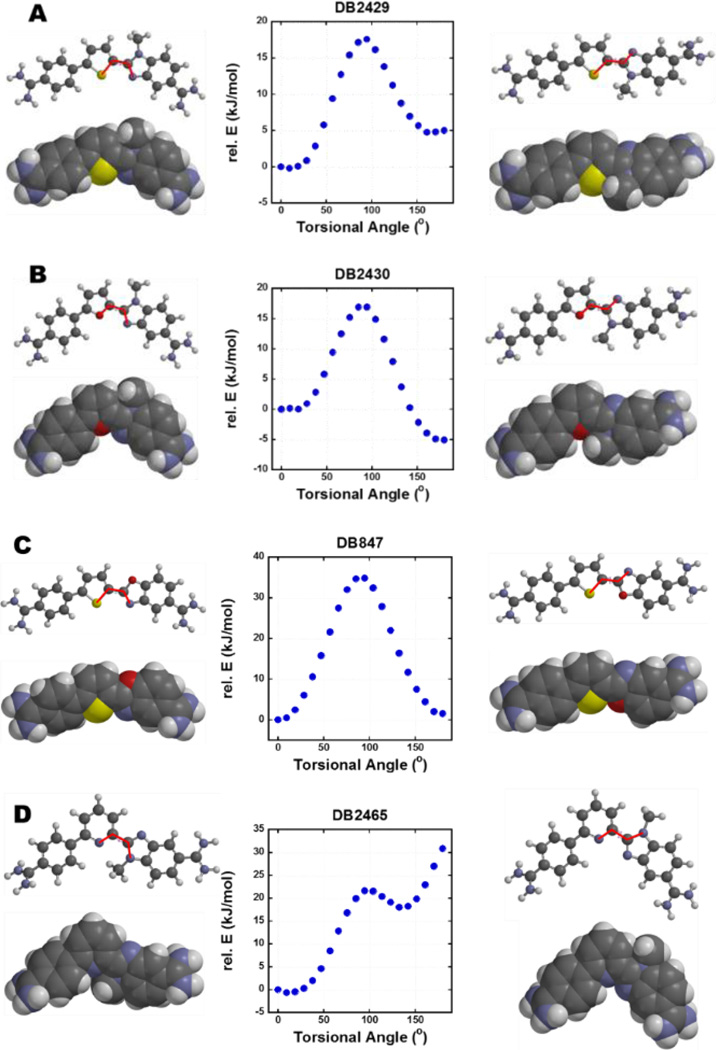
A specific question for the computational methods is: what is the low energy conformation of the test compounds and how well does this conformation match the minor groove? To help answer these, torsional angle maps and molecular conformations of DB2429, DB2430, DB847, and DB2465 were calculated (Figure 5). DB2429 has the lowest energy conformation at a torsional angle of 9° and this most stable structure of DB2429 has an appropriate shape and the G-NH2-N (H to N) H-bonding ability for the DNA minor groove (Figure 5A). Interestingly, DB2430, the furan derivative, reached the low energy state around a 180° torsional angle, the reverse orientation with the N-methyl group directed in the same orientation as the –O– of furan (Figure 5B). The N-methyl group is a steric block in this orientation and will not allow a hydrogen bond between the N-MeBI with G-NH2 to form. Results from SPR and Tm experiments show that DB2429 has a much stronger affinity with the AAAGTTT sequence than DB2430 in agreement with the observed structures. These results for the thiophene and furan compounds are also in complete agreement with the sigma-hole concept.[45–48] The torsional angle map of DB847 (Figure 5C), with the benzoxazole group, revealed that an orientation at 180° has a similar energy to 0°. As the SPR and Tm results show, DB847 exhibits intermediate binding affinity with the AAAGTTT sequence in agreement with the mixed orientational results. As with the furan derivative, the same side orientation of the N-methyl group and the “N” of pyridine in DB2465 represents the most stable structure (Figure 5D). The energy of the opposite orientation is quite high, due to the repulsive van der Waals interaction between the N-methyl group and a pyridine C-H. In agreement with the conformational predictions, DB2465 has the weakest binding ability with the AAAGTTT sequence among all the test compounds in this set (Table 2).
Figure 5.
Torsional angle maps for DB2429, DB2430, DB847, and DB2465; dihedral plots for (A) S-C of thiophene and C=N of N-MeBI. (B) O-C of furan and C=N of N-MeBI. (C) S-C of thiophene and C=N of benzoxazole. (D) N=C of pyridine and C=N of N-MeBI. All calculations are performed at the B3LYP/6–31G* level of theory. The scanned dihedral is depicted in red bold line.
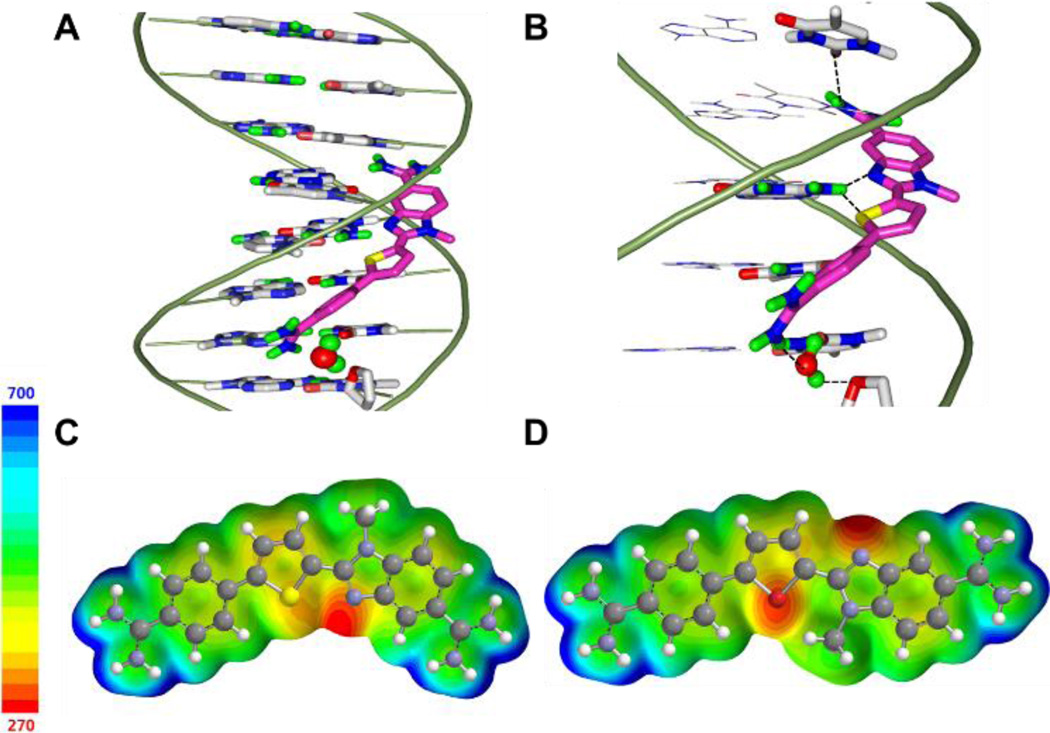
To evaluate ideas for how DB2429 and DB2457 are able to effectively bind to the AAAGTTT sequence, a molecular docking study was conducted with ds-[(5'-CCAAAGTTTG-3') (5'-CAAACTTTGG-3')] sequence using the Autodock software 4.2 package.[59] Low energy binding complexes were obtained for both compounds with the unsubstituted “N” of N-MeBI and “S” of thiophene forming bifurcated H-bonds to the exocyclic minor groove G-NH2 group (Figure 6B, S6B). The top amidine in Figure 6 and S6 participates in an H-bond with a thymine (T) carbonyl group at the complementary strand of DNA (the C-strand of the G•C bp) with 2.4 Å bonding distances for both DB2429 and DB2457. Interestingly, given this strong binding it is impossible to obtain simultaneous H-bonds by both amidine groups with A•T bps at the floor of the groove. The strong binding affinity and sequence selectivity of these two compounds can be explained by two possibilities: (i) a conformational change of the local DNA structure to allow the amidine and the N-MeBI to form an H-bond, (ii) if the DNA cannot undergo such a change due to the energy cost, a water molecule could be incorporated into the complex. In this case the non-H-bonded amidine is linked to base pair acceptor groups at the floor of the groove by the water molecule.
Figure 6.
Minor groove views of docked conformations of (A) DB2429 with the AAAGTTT sequence. The compounds are shown as stick model and colored by atom type (magenta for carbon, blue for nitrogen, yellow for thiophene sulfur, and green for amidine hydrogen). The DNA backbone is represented as a tube form in light green, DNA nucleobases are represented by sticks colored by atom type (gray for carbon, blue for nitrogen, red for oxygen, green for polar hydrogen). A water molecule (red sphere) is shown at the bottom amidine of each compound. (B) Important H-bond interactions between compounds and DNA nucleobases (shown with a black dashed line), the thiophene S and N-MeBI N form bifurcated hydrogen bonds with the exocyclic G6-NH in the minor groove, the bottom amidine group forms H-bonds with the thymine (T) carbonyl group through the water molecule, and the top amidine group forms a direct hydrogen bond with the thymine (T) carbonyl group. (C) (D) Electrostatic potential map of DB2429 and DB2430 with calculation performed at the B3LYP 6–31+G* level of theory (the energy range is negative to positive from red to blue color shown in a ladder).
We are unable to model the duplex DNA conformation in model (i) above, but we were able to construct a stable complex by including a water molecule at the floor of the minor groove. A bound water has been observed for linking the biphenyl benzimidazole diamidine, DB921, to the floor of the minor groove to form a strong complex.[32] One amidine group can form a direct H-bond to the DNA while the other is too far from the minor groove. An X-ray crystal structure clearly showed a water molecule completing the link between that amidine and N3 of Adenine (A) at the floor of the minor groove. Given this observation, we suggest that complexes with DB2429 and DB2457 involve a bridging water molecule between the compound and the floor of the minor groove (Figure 6, S6). Future work on NMR and molecular dynamics (MD) studies will be applied to address this issue.
The thiophene and furan ab initio models in Figure 6C, 6D illustrate the reversed equilibrium conformation of these systems as in Figure 5. The electrostatic potential maps provide an explanation for this observation. The thiophene C-S bond results in two electron deficient σ* orbitals on S. The N-MeBI unsubstituted N is in the close proximity with S and increases electron density from the lone pair electron of N to S. This can be seen in the diagram by the asymmetric electron density potential map on S. The N-MeBI side is significantly more electronegative than the phenyl side. The same interaction is not possible in the furan compound. In this case, the dominant inter-ring interaction is the negative potential on both the unsubstituted N and the furan O. These are rotated 180° apart to minimize the repulsive interaction. For the thiophene, the S-N σ-hole interaction provide the appropriate conformation for binding in the minor groove of DNA, while for the furan the O-N repulsion give an unfavorable conformation.
Conclusions
New design concept for molecular structures with quite different properties that can recognize mixed AT and GC DNA sequences will help to deal with the problem of a lack of a variety of compounds that can specifically target DNA. In this report we have used an entirely new concept to design and prepare compounds that can recognize G•C bps. By using the orienting interaction of a “S” of thiophene sigma-hole with the unsubstituted “N” of N-MeBI, a pre-organized module for the DNA minor groove is obtained that can accept an H-bond from the G–NH2. SPR, ESI-MS, fluorescence emission spectrometry and CD experiments all indicate that these N-MeBI-thiophene compounds bind specifically to a G•C bp with flanking A•T bps in a strong 1:1 minor groove complex. When the thiophene is replaced by a furan or pyridine moiety, the reverse orientation of the “O” in furan or “N” in pyridine and unsubstituted “N” in N-MeBI is favored and there is an energy barrier to convert the module to the H-bonding conformation with G-NH2. In agreement with SPR results, the furan and pyridine compounds bind more weakly to single G•C bp containing sequences than the thiophenes.
The rationally designed molecules, DB2429 and DB2457, are an important discovery in the design process for sequence specific DNA recognition. The large decrease in sequence specificity and affinity by either replacement of the thiophene or N-MeBI moiety demonstrates the importance of the N-MeBI-thiophene module in G•C bp recognition. The decreases in binding affinity for DB2429 and DB2457, after replacement of “G” by “I” in the minor groove of the DNA sequence are clear indications of a strong H-bond interaction between G-NH2 and the central N-MeBI-thiophene moiety. This new type of G•C bp specific recognition provides ideas on expansion of modules for sequence specific DNA recognition and in due course new therapeutics and biotechnology reagents. With these available units, we can now combine them with different linkers for recognition of expanded sequences with additional G•C base pairs.
Experimental Section
Biophysical experimental details, compound synthesis and characterisation data of the new compounds used in this article can be found in the Supporting Information.
Supplementary Material
Acknowledgments
This work was supported by the US NIH [Grant No. GM111749 to W.D.W and D.W.B]. The authors thank Carol Wilson for manuscript assistance and Sarah Laughlin-Toth for ESI-MS assistance.
References
- 1.Gottesfeld JM, Neely L, Trauger JW, Baird EE, Dervan PB. Nature. 1997;387:202–205. doi: 10.1038/387202a0. [DOI] [PubMed] [Google Scholar]
- 2.Dervan PB, Poulin-Kerstien AT, Fechter EJ. In: DNA Binders and Related Subjects. Waring MJ, Chaires JB, editors. Vol. 253. Berlin Heidelberg: Springer; 2005. pp. 1–31. [Google Scholar]
- 3.Neidle S. Nat. Prod. Rep. 2001;18:291–309. doi: 10.1039/a705982e. [DOI] [PubMed] [Google Scholar]
- 4.Satam V, Babu B, Patil P, Brien KA, Olson K, Savagian M, Lee M, Mepham A, Jobe LB, Bingham JP, Pett L, Wang S, Ferrara M, Bruce CD, Wilson WD, Lee M, Hartley JA, Kiakos K. Bioorg. Med. Chem. Lett. 2015;25:3681–3685. doi: 10.1016/j.bmcl.2015.06.055. [DOI] [PubMed] [Google Scholar]
- 5.Edwards TG, Vidmar TJ, Koeller K, Bashkin JK, Fisher C. PLoS ONE. 2013;8:e75406. doi: 10.1371/journal.pone.0075406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blackledge MS, Melander C. Bioorg. Med. Chem. 2013;21:6101–6114. doi: 10.1016/j.bmc.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tidwell RR, Boykin DW. In: Small Molecule DNA and RNA Binders: From Synthesis to Nucleic Acid Complexes. Demeunynck M, Bailly C, Wilson WD, editors. Weinheim, FRG: Wiley-VCH Verlag GmbH & Co. KGaA; 2002. pp. 414–460. [Google Scholar]
- 8.Pazos E, Mosquera J, Vázquez ME, Mascareñas JL. Chembiochem. 2011;12:1958–1973. doi: 10.1002/cbic.201100247. [DOI] [PubMed] [Google Scholar]
- 9.Koehler AN. Curr. Opin. Chem. Biol. 2010;14:331–340. doi: 10.1016/j.cbpa.2010.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Munde M, Wang S, Kumar A, Stephens CE, Farahat AA, Boykin DW, Wilson WD, Poon GMK. Nucleic Acids Res. 2014;42:1379–1390. doi: 10.1093/nar/gkt955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Munde M, Kumar A, Peixoto P, Depauw S, Ismail MA, Farahat AA, Paul A, Say MV, David-Cordonnier M-H, Boykin DW, Wilson WD. Biochemistry. 2014;53:1218–1227. doi: 10.1021/bi401582t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodríguez J, Mosquera J, García-Fandiño R, Vázquez ME, Mascareñas JL. Chem. Sci. 2016;7:3298–3303. doi: 10.1039/c6sc00045b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodríguez J, Mosquera J, Couceiro JR, Vázquez ME, Mascareñas JL. Chem. Sci. 2015;6:4767–4771. doi: 10.1039/c5sc01415h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Asamitsu S, Kawamoto Y, Hashiya F, Hashiya K, Yamamoto M, Kizaki S, Bando T, Sugiyama H. Bioorg. Med. Chem. 2014;22:4646–4657. doi: 10.1016/j.bmc.2014.07.019. [DOI] [PubMed] [Google Scholar]
- 15.Kielkopf CL, Baird EE, Dervan PB, Rees DC. Nat. Struct. Biol. 1998;5:104–109. doi: 10.1038/nsb0298-104. [DOI] [PubMed] [Google Scholar]
- 16.Cominetti MMD, Goffin SA, Raffel E, Turner KD, Ramoutar JC, O'Connell MA, Howell LA, Searcey M. Bioorg. Med. Chem. Lett. 2015;25:4878–4880. doi: 10.1016/j.bmcl.2015.06.014. [DOI] [PubMed] [Google Scholar]
- 17.Snyder RD, Holt PA, Maguire JM, Trent JO. Environ. Mol. Mutagen. 2013;54:668–681. doi: 10.1002/em.21796. [DOI] [PubMed] [Google Scholar]
- 18.Kumar S, Spano MN, Arya DP. Bioorg. Med. Chem. 2015;23:3105–3109. doi: 10.1016/j.bmc.2015.04.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie X, Choi B, Largy E, Guillot R, Granzhan A, Teulade-Fichou M-P. Chem. Eur. J. 2013;19:1214–1226. doi: 10.1002/chem.201203710. [DOI] [PubMed] [Google Scholar]
- 20.Bishop EP, Rohs R, Parker SCJ, West SM, Liu P, Mann RS, Honig B, Tullius TD. ACS Chem. Biol. 2011;6:1314–1320. doi: 10.1021/cb200155t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Munde M, Lee M, Neidle S, Arafa R, Boykin DW, Liu Y, Bailly C, Wilson WD. J. Am. Chem. Soc. 2007;129:5688–5698. doi: 10.1021/ja069003n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.White S, Szewczyk JW, Turner JM, Baird EE, Dervan PB. Nature. 1998;391:468–471. doi: 10.1038/35106. [DOI] [PubMed] [Google Scholar]
- 23.Rohs R, West SM, Sosinsky A, Liu P, Mann RS, Honig B. Nature. 2009;461:1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mallena S, Lee MPH, Bailly C, Neidle S, Kumar A, Boykin DW, Wilson WD. J. Am. Chem. Soc. 2004;126:13659–13669. doi: 10.1021/ja048175m. [DOI] [PubMed] [Google Scholar]
- 25.Vázquez O, Vázquez ME, Blanco JB, Castedo L, Mascareñas JL. Angew. Chem. Int. Ed. Engl. 2007;46:6886–6890. doi: 10.1002/anie.200702345. [DOI] [PubMed] [Google Scholar]
- 26.Liu Y, Chai Y, Kumar A, Tidwell RR, Boykin DW, Wilson WD. J. Am. Chem. Soc. 2012;134:5290–5299. doi: 10.1021/ja211628j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hahn L, Buurma NJ, Gade LH. Chem. Eur. J. 2016;22:6314–6322. doi: 10.1002/chem.201504934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson WD, Tanious FA, Mathis A, Tevis D, Hall JE, Boykin DW. Biochimie. 2008;90:999–1014. doi: 10.1016/j.biochi.2008.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nanjunda R, Wilson WD. Current Protocols in Nucleic Acid Chemistry. 2012;Chapter 8(Unit8.8) doi: 10.1002/0471142700.nc0808s51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paul A, Nanjunda R, Kumar A, Laughlin S, Nhili R, Depauw S, Deuser SS, Chai Y, Chaudhary AS, David-Cordonnier M-H, Boykin DW, Wilson WD. Bioorg. Med. Chem. Lett. 2015;25:4927–4932. doi: 10.1016/j.bmcl.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harika NK, Paul A, Stroeva E, Chai Y, Boykin DW, Germann MW, Wilson WD. Nucleic Acids Res. 2016;44:4519–4527. doi: 10.1093/nar/gkw353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y, Kumar A, Depauw S, Nhili R, David-Cordonnier M-H, Lee MP, Ismail MA, Farahat AA, Say M, Chackal-Catoen S, Batista-Parra A, Neidle S, Boykin DW, Wilson WD. J. Am. Chem. Soc. 2011;133:10171–10183. doi: 10.1021/ja202006u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vázquez O, Sánchez MI, Martínez-Costas J, Vázquez ME, Mascareñas JL. Org. Lett. 2010;12:216–219. doi: 10.1021/ol902501j. [DOI] [PubMed] [Google Scholar]
- 34.Sánchez MI, Vázquez O, Martínez-Costas J, Vázquez ME, Mascareñas JL. Chem. Sci. 2012;3:2383–2387. [Google Scholar]
- 35.Kielkopf CL, White S, Szewczyk JW, Turner JM, Baird EE, Dervan PB, Rees DC. Science. 1998;282:111–115. doi: 10.1126/science.282.5386.111. [DOI] [PubMed] [Google Scholar]
- 36.Saha A, Hashiya F, Kizaki S, Asamitsu S, Hashiya K, Bando T, Sugiyama H. Chem. Commun. 2015;51:14485–14488. doi: 10.1039/c5cc05104e. [DOI] [PubMed] [Google Scholar]
- 37.Dervan PB, Edelson BS. Curr. Opin. Struct. Biol. 2003;13:284–299. doi: 10.1016/s0959-440x(03)00081-2. [DOI] [PubMed] [Google Scholar]
- 38.Koeller KJ, Harris GD, Aston K, He G, Castaneda CH, Thornton MA, Edwards TG, Wang S, Nanjunda R, Wilson WD, Fisher C, Bashkin JK. Med. Chem. (Los Angeles) 2014;4:338–344. doi: 10.4172/2161-0444.1000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buchmueller KL, Staples AM, Howard CM, Horick SM, Uthe PB, Le NM, Cox KK, Nguyen B, Pacheco KAO, Wilson WD, Lee M. J. Am. Chem. Soc. 2005;127:742–750. doi: 10.1021/ja044359p. [DOI] [PubMed] [Google Scholar]
- 40.Paul A, Chai Y, Boykin DW, Wilson WD. Biochemistry. 2015;54:577–587. doi: 10.1021/bi500989r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chenoweth DM, Poposki JA, Marques MA, Dervan PB. Bioorg. Med. Chem. 2007;15:759–770. doi: 10.1016/j.bmc.2006.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chenoweth DM, Viger A, Dervan PB. J. Am. Chem. Soc. 2007;129:2216–2217. doi: 10.1021/ja0682576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brucoli F, Guzman JD, Maitra A, James CH, Fox KR, Bhakta S. Bioorg. Med. Chem. 2015;23:3705–3711. doi: 10.1016/j.bmc.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 44.Chavda S, Liu Y, Babu B, Davis R, Sielaff A, Ruprich J, Westrate L, Tronrud C, Ferguson A, Franks A, Tzou S, Adkins C, Rice T, Mackay H, Kluza J, Tahir SA, Lin S, Kiakos K, Bruce CD, Wilson WD, Hartley JA, Lee M. Biochemistry. 2011;50:3127–3136. doi: 10.1021/bi102028a. [DOI] [PubMed] [Google Scholar]
- 45.Clark TT. Wiley Interdiscip. Rev.: Comput. Mol. Sci. 2012;3:13–20. [Google Scholar]
- 46.Murray JS, Lane P, Politzer P. Int. J. Quantum Chem. 2008;108:2770–2781. [Google Scholar]
- 47.Tilly D, Chevallier F, Mongin F. Synthesis. 2015;48:184–199. [Google Scholar]
- 48.Beno BR, Yeung K-S, Bartberger MD, Pennington LD, Meanwell NA. J. Med. Chem. 2015;58:4383–4438. doi: 10.1021/jm501853m. [DOI] [PubMed] [Google Scholar]
- 49.Farahat AA, Boykin DW. J. Heterocycl. Chem. 2013;50:585–589. doi: 10.1002/jhet.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laughlin S, Wang S, Kumar A, Farahat AA, Boykin DW, Wilson WD. Chem. Eur. J. 2015;21:5528–5539. doi: 10.1002/chem.201406322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Farahat AA, Kumar A, Say M, Barghash AE-DM, Goda FE, Eisa HM, Wenzler T, Brun R, Liu Y, Mickelson L, Wilson WD, Boykin DW. Bioorg. Med. Chem. 2010;18:557–566. doi: 10.1016/j.bmc.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 52.Shi X, Chaires JB. Nucleic Acids Res. 2006;34:e14. doi: 10.1093/nar/gnj012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nanjunda R, Munde M, Liu Y, Wilson W. In: Methods for Studying Nucleic Acid/Drug Interactions. Wanunu Y, editor. CRC Press; 2011. pp. 91–119. [Google Scholar]
- 54.Neubauer H, Gaiko N, Berger S, Schaffer J, Eggeling C, Tuma J, Verdier L, Seidel CAM, Griesinger C, Volkmer A. J. Am. Chem. Soc. 2007;129:12746–12755. doi: 10.1021/ja0722574. [DOI] [PubMed] [Google Scholar]
- 55.Wang M, Mao Z, Kang TS, Wong CY, Mergny JL, Leung, leung C-H, Ma D-L. Chem. Sci. 2016;7:2516–2523. doi: 10.1039/c6sc00001k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eriksson M, Nordén B. Drug-Nucleic Acid Interactions vol. Vol. 340. Elsevier; 2001. pp. 68–98. [DOI] [PubMed] [Google Scholar]
- 57.Fornander LH, Wu L, Billeter M, Lincoln P, Nordén B. J. Phys. Chem. B. 2013;117:5820–5830. doi: 10.1021/jp400418w. [DOI] [PubMed] [Google Scholar]
- 58.Laughlin S, Wilson WD. Int. J. Mol. Sci. 2015;16:24506–24531. doi: 10.3390/ijms161024506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trott O, Olson AJ. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.