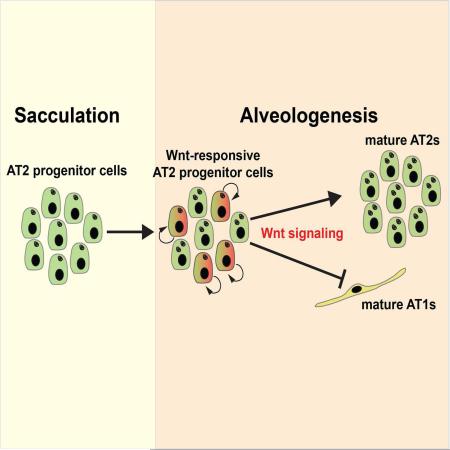
Abstract
Alveologenesis is the culmination of lung development and involves the correct temporal and spatial signals to generate the delicate gas exchange interface required for respiration. Using a novel Wnt signaling reporter system, we demonstrate the emergence of a Wnt-responsive alveolar epithelial cell sublineage that arises during alveologenesis called the axin2+ alveolar type 2 cell or AT2Axin2. The number of AT2Axin2 cells increases substantially during late lung development, correlating with a wave of Wnt signaling during alveologenesis. Transcriptome analysis, in vivo clonal analysis, and ex vivo lung organoid assays reveal that AT2sAxin2 promote enhanced AT2 cell growth during generation of the alveolus. Activating Wnt signaling results in expansion of AT2s whereas inhibition of Wnt signaling inhibits AT2 cell development and shunts alveolar epithelial development towards the alveolar type 1 cell lineage. These findings reveal a wave of Wnt-dependent AT2 expansion required for lung alveologenesis and maturation.
Graphical abstract
INTRODUCTION
Generation of the alveolus requires intricate interactions between multiple cell lineages to create the complex structure responsible for gas exchange in mammals (Morrisey and Hogan, 2010). Epithelial, mesenchymal, and endothelial cell lineages combine to expand the saccular structure at the distal tips of the branched airways starting around embryonic day 16.5 (E16.5) in mice (Whitsett and Weaver, 2015). Soon thereafter, this rudimentary structure remodels and promotes epithelial and mesenchymal cell communication, which helps integrate the developing vascular network. Remodeling of the alveolus continues postnatally concomitant with specification and maturation of alveolar type 1 (AT1) and type 2 (AT2) epithelial cells until lung maturity is reached at postnatal day 30 (PN30) in mice and into adolescence in humans (Branchfield et al., 2016; Herring et al., 2014; Mund et al., 2008). Despite the extensive knowledge of earlier stages of lung development including branching morphogenesis, little is known about the cell lineage specific interactions and molecular pathways governing the normal generation of the lung alveolus (Branchfield et al., 2016; El Agha et al., 2014; Yun et al., 2016). Since disruption of this process can be deleterious and result in neonatal diseases such as bronchopulmonary dysplasia (BPD) (Bourbon et al., 2005), a better understanding of the cellular growth and differentiation that occurs during this crucial stage of lung development is required.
Wnt signaling is a critical pathway important for self-renewal and specification of stem cells in multiple organs (Clevers et al., 2014). Components of the Wnt pathway are expressed in specific patterns during early lung development, and previous work has demonstrated essential roles for Wnt signaling in lung endoderm specification and early development (Cohen et al., 2009; De Langhe et al., 2008; Goss et al., 2009; Konigshoff and Eickelberg, 2010; Li et al., 2005; Li et al., 2002; Mammoto et al., 2012; Maretto et al., 2003; Miller et al., 2012; Okubo and Hogan, 2004; Rajagopal et al., 2008; Shu et al., 2005; Shu et al., 2002; van Amerongen et al., 2012). However, what role if any Wnt signaling plays in later stages of lung epithelial differentiation and maturation is unclear. Using a novel Wnt signaling reporter mouse line (Axin2CreERT2-TdTom), we reveal a previously unknown wave of Wnt signaling during alveologenesis. The Axin2CreERT2-TdTom reporter demarcates a sublineage of AT2s called AT2sAxin2, which emerges at the onset of alveologenesis. AT2sAxin2 promote lung organoid formation in ex vivo assays and have greater clonal growth potential in vivo during alveologenesis. Importantly, activation of Wnt signaling in the overall AT2 population elicits a similar self-renewal response, promoting enhanced organoid formation, increased proliferation, and increased clonal expansion during alveologenesis. Conversely, inhibition of Wnt signaling in the overall AT2 lineage inhibits organoid formation and AT2 self-renewal and shunts their differentiation towards the AT1 lineage. These data demonstrate a critical role for Wnt signaling during lung alveologenesis through expansion of the AT2 population via proliferation and balancing the ratio of AT2-AT1 cells.
RESULTS
The Axin2CreERT2-TdTom mouse line reveals dynamic Wnt-responsiveness during lung development
The Wnt signaling pathway is critical for lung endoderm specification and patterning of the branching lung and mesenchyme (Cohen et al., 2009; Goss et al., 2009; Harris-Johnson et al., 2009; Kadzik et al., 2014; Li et al., 2002; Miller et al., 2012; Mucenski et al., 2003; Rajagopal et al., 2008; Volckaert and De Langhe, 2015). However, the role for Wnt signaling during lung sacculation and alveologenesis is poorly understood. We have generated a novel Wnt signaling reporter mouse line to identify, purify, and characterize Wnt responsive lineages during lung development. The Axin2CreERT2-TdTom allele has an expression cassette consisting of a tamoxifen-inducible Cre recombinase linked to a TdTomato fluorescent protein by a 2A self-cleaving peptide inserted into the start codon of the mouse Axin2 gene (Figure S1A). Using this reporter line, we show that Axin2+ cells, marked by TdTomato expression, become restricted to the distal epithelium and the surrounding mesenchyme at E13.5 (Figure 1A and 1B). To track Axin2+ cells earlier in lung development, we crossed the Axin2CreERT2-TdTom mouse with the R26REYFP line and initiated a lineage trace at E10.5. Examination of Axin2CreERT2-TdTom:R26REYFP lungs at E14.5 shows that Axin2+ cells give rise to both proximal and distal lung epithelium and mesenchyme, corroborating previous data demonstrating the presence and importance of Wnt signaling in early lung development and the multipotency of the early lung endoderm (Figure 1C) (Al Alam et al., 2011; Goss et al., 2009; Mucenski et al., 2003; Shu et al., 2005).
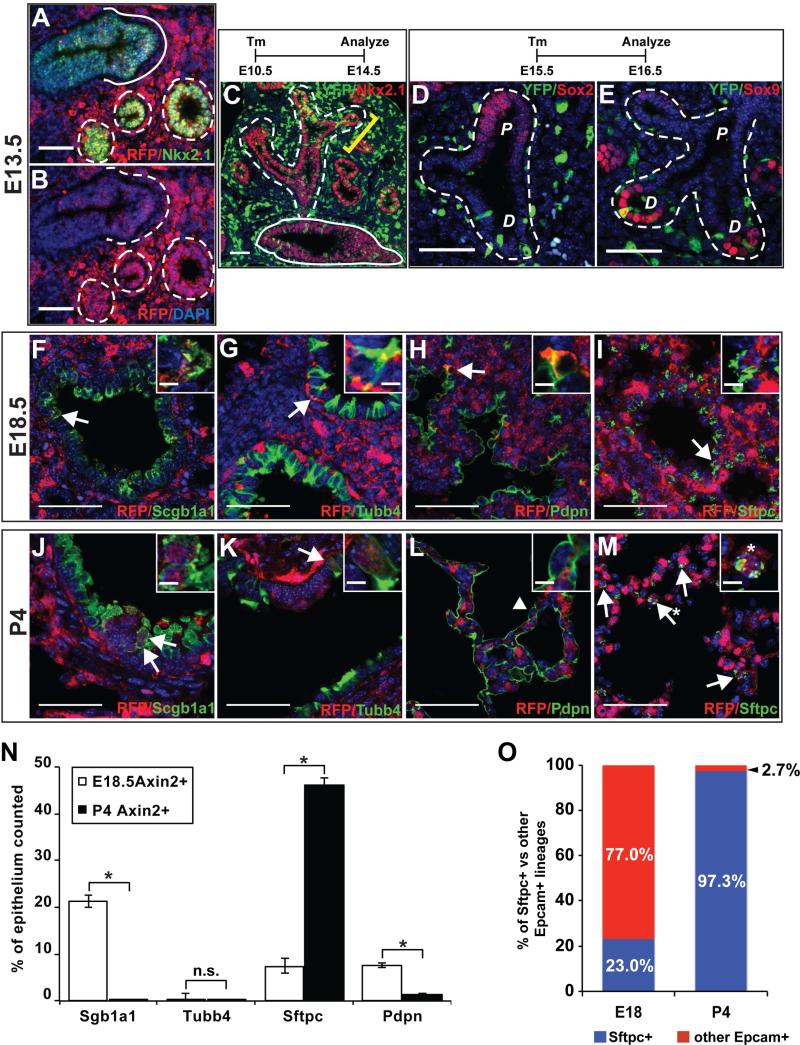
Figure 1. A novel reporter mouse defines Wnt-responsiveness in the developing lung.
(A and B) The Axin2CreERT2-TdTom allele produces TdTomato expression in Wnt-responsive (Axin2+) cells located in the mesenchyme and proximal and distal Nkx2.1+ epithelium (A) during branching morphogenesis with (B) showing the same image with RFP and DAPI staining only. Dashed white lines outline distal and solid lines outline proximal airways, respectively. (C) Induction of recombination at E10.5 in Axin2CreERT2-TdTom:R26REYFP mice followed by a four-day chase demonstrates that Axin2+ cells give rise to both proximal and distal Nkx2.1+ lung epithelium, with a larger number of cells labeled in the distal region (yellow bracket). (D and E) From E15.5-E16.5, lineage labeled Axin2+ cells become restricted to distal Sox2− (D), Sox9+ (E) lung epithelium. (F-I) At E17.5-E18, Axin2+ lung epithelial cells are sparsely located throughout distal bronchiolar and alveolar epithelium. Bronchiolar Axin2+ cells are predominantly Scgb1a1 secretory cells (F) with very rare Tubb4+ ciliated cells (G). In the alveolar region, there is a small number of Pdpn+ AT1s (H) and Sftpc+ AT2s (I). At P4, Axin2+ secretory (J) and ciliated (K) bronchiolar cells and AT1s (L) become very rare. However, there is a dramatic increase in Axin2+ AT2 (AT2Axin2) alveolar cells (M). (N) Quantification of Axin2+ secretory (Scgb1a1+) and ciliated (Tubb4+) bronchiolar epithelium, AT2s (Sftpc+), and AT1s (Pdpn+) reveals significant changes in lineage specific Axin2+ Wnt responsive lung epithelium. (O) At E18, a minority of the Axin2+ Wnt responsive epithelium is Sftpc+. However, by P4 more than 97% of the total Axin2+ epithelium is a sublineage of the total Sftpc+ AT2 lineage i.e. AT2Axin2s. Arrows in F-M indicate examples of a positive cell and insets are higher magnification of Axin2+ cells. D=distal lung endoderm/epithelium, P=proximal lung endoderm/epithelium. Quantification of cell numbers represented as mean ± SEM. Two-tailed student's t-test: *P < 0.05, n.s = non-significant; n=3 for each group. Scale bars = 50 μm.
As lung development progresses, the branching lung epithelium is patterned into distinct proximal and distal compartments(Morrisey and Hogan, 2010; Swarr and Morrisey, 2015). The emergence of these two compartments coincides with diminished Wnt-responsiveness with the few responsive cells confined to the Sox9+/Sox2− distal lung endoderm (Figure 1D and 1E).
By E17.5, lung sacculation is underway and the rudimentary alveolus expands to prepare the distal lung for the alveologenesis stage, where the saccules remodel to refine and expand the surface area required for high efficiency gas exchange in the postnatal period (Prodhan and Kinane, 2002). Axin2+ cells are found in the in lung mesenchyme from E17.5-E18.0 to postnatal day 4 (P4) (Figure S1B-E), with only a small increase in the number of Pdgfrβ+ mesenchymal cells during this phase (Figure S1J). In contrast, distinct changes occurred in the cellular composition of Wnt responsiveness in the maturing lung epithelium during this time frame. At E17.5-E18.0, a small number of Axin2+ proximal and distal epithelial cells exist (Figure 1F-I and 1N). As development progresses through the saccular stage and into the alveologenesis stage at P4, the number of Axin2+ secretory epithelial cells and Axin2+ AT1 cells decreased, and the Axin2+ multi-ciliated cells remained rare (Figure 1J-L). The remaining Axin2+ secretory epithelial cells were confined to the region surrounding Pgp9.5+ neuroendocrine bodies (Figure S1K and L). In contrast, a dramatic increase in the number of Axin2+ AT2 (AT2sAxin2) cells was observed (Figure 1I, M, N). This change represents an approximate six-fold increase in AT2sAxin2 during this late stage of sacculation into early alveologenesis (Figure 1N). By P4, greater than 97% of all Axin2+ epithelial cells are Sftpc+ AT2s (Figure 1O). These data reveal that Wnt responsive cells become restricted to the distal epithelium during sacculation and subsequently to a subpopulation of AT2s during alveologenesis.
We observed both a bright and dim TdTomato expressing population of cells in the lung at P4. To verify how faithful TdTomato expression correlated with Axin2 expression, we performed FACS and qPCR analysis of TdTomato expressing cells in the lung. Importantly, we recently showed that our Axin2CreERT2-TdTom mouse line faithfully recapitulates Axin2 expression and Wnt responsiveness in the intestinal crypts (Li et al., 2016). We observed both a dim and bright population of TdTomato expressing cells in the lung and Axin2 expression correlated with the levels of TdTomato expression in these two populations (Figure S2A and D). Furthermore, analysis of FACS-isolated cells using endogenous TdTomato expression and antibodies specific for epithelium (EpCAM) and Pdgfra+ (CD140a) mesenchymal lineages demonstrates that the epithelial cells are TdTomato dim and most of the mesenchymal cells are TdTomato (Figure S2B, C, E, F).
A wave of Wnt signaling appears in AT2s prior to alveologenesis
Our results suggest a wave of Wnt signaling occurs in the AT2 lineage coinciding with alveologenesis and maturation of the lung (Figure 2A). Careful examination for Wnt responsiveness between E17.5-E18.0 and P30 reveal that most AT2sAxin2 arise postnatally (Figure 2B-2G). The AT2Axin2 cells are scattered randomly throughout the alveolus with very occasional clustering of cells (Figure 2I). AT2Axin2 cells increase throughout the postnatal period and persist through P30 (Figure 2H and I). To determine whether the AT2sAxin2 are fixed sublineage fate or a dynamic state during alveologenesis, we performed a 3 week tamoxifen chase at P4 in Axin2CreERT2-TdTom:R26REYFP mice. Using FACS-based analysis, we noted that while many AT2Axin2 cells were both TdTomato and YFP double positive, there was a significant proportion of cells were YFP single positive and TdTomato single positive, suggesting that some AT2Axin2 cells lose their Wnt-responsiveness and some non-Axin2+ cells gain Wnt-responsiveness over time (Figure S2G and H). Confirmatory immunohistochemical staining corroborated these findings (Figure S2I and J).
Figure 2. A wave of Wnt signaling occurs in AT2s during alveologenesis.
(A) Graphic model illustrates the dynamic changes in Axin2+ Wnt-responsive lung epithelium during lung development. Late lung development is marked by restriction of Axin2+ epithelial cells to the distal alveolus and a significant increase in AT2Axin2 cells during alveologenesis. While there are few AT2Axin2 cells at E17.5-E18.0 (B and C), their numbers increase significantly by P1 (D and E) and P4 (F and G). AT2Axin2 cells continue to be observed at P30 (H and I). Dotted lines surround Sftpc+ AT2 and AT2Axin2 cells in insets C, E, G, I. (J-L) Lineage traced AT2Axin2 cells from E18.5 to P10 (J) and P4 to P10 (K) show a marked increase in AT2Axin2 cell expansion between these time points. Arrows indicate positive cells, which are quantified in (L). (M) Total AT2s isolated by FACS following a 24-hour tamoxifen induction. (N) Expression of Axin2 mRNA at P4 compared to E18.5 in AT2s showing a two-fold increase in expression. (O and P) Axin2+ lung epithelial cells are isolated using negative selection for CD31 and CD45 (upper FACS plot) and positive selection for TdTomato and EpCAM (lower FACS plot). (P) By flow cytometry, isolated Axin2+ lung epithelial cells are composed predominantly of AT2sAxin2 as measured by intracellular Sftpc and extracellular Pdpn staining. Quantification of cell numbers, qPCR data, and cell type percentage represented as mean ± SEM. Two-tailed student's t-test: P < 0.05; n=3-4 for each group. Scale bars: B, D, F, H, J, K = 50 μm; C, E, G, I = 10 μm; J, K = 5 μm.
Using lineage tracing with the Axin2CreERT2-TdTom:R26REYFP mice, we found that Axin2+ endothelial and mesenchymal cell lineages remained mostly unchanged between E17.5-E18.0 through P10 (Figure S3A-H). However, we did observe a significant increase in alveolar epithelial progeny with little change in the bronchiolar epithelium when lineage traced from P4 to P10 (Figure S3I-O). Closer analysis of alveolar epithelial lineages shows that this increase is due to the marked expansion of lineage traced Axin2+ AT2s and not AT1s from P4 to P10 (Figure S3P).
Gene expression analysis on AT2s FACS isolated from SftpcCreERT2:R26REYFP E18.5 and P4 lungs revealed a 2-fold increase in Axin2 mRNA during this stage of lung development (Figure 2M and 2N). To verify that the increase in Axin2 expression correlates with an increased number of AT2Axin2 cells, we used FACS to isolate Axin2+ epithelial cells from Axin2CreERT2-TdTom lungs at P4 (Figure 2O) and subjected them to extracellular staining for the AT1 marker Pdpn and intracellular staining for the AT2 marker Sftpc (Figure S4A-C). When compared to E18.5 Axin2+ epithelial cells, there was a 2-fold increase in the number of AT2Axin2 cells at P4 with a concomitant reduction in the number of Axin2+ AT1s (Figure 2P). Of note, the efficiency of intracellular staining for Sftpc is around 50% (Figure S4D-G). Thus, a wave of Wnt signaling occurs during late sacculation into early alveologenesis that activates this pathway in a sublineage of AT2s called AT2Axin2 cells which are important for alveolar growth and maturation.
The AT2Axin2 sublineage promotes alveologenesis by promoting growth of the alveolar epithelium in vivo
AT2s undergo a significant increase in cell number during alveologenesis (Yang et al., 2016). To examine the proliferative potential of the AT2Axin2 population, we performed in vivo clonal analysis using the R26RBr2.1 reporter and 5-ethynyl-2-deoxyuridine (EdU) incorporation in the total AT2 population versus the AT2Axin2 sublineage. SftpcCreERT2:R26RBr2.1 and Axin2CreERT2-TdTom:R26RBr2.1 pups were administered tamoxifen at P4 followed by a chase to P30 (Fig. 3A and B). The average clone size differed between the AT2 and AT2Axin2 sublineage with the AT2Axin2-derived clones being larger (1.49 cells/clone for total AT2 vs. 1.65 cells/clone for AT2Axin2, P < 0.02) (Figure 3C). Importantly, the number of multi-cellular clones generated by the AT2Axin2 sublineage was significantly greater than the overall AT2 population. While the AT2Axin2 sublineage contained predominantly two and three cell clones, the majority of clones from the total AT2 population were comprised of one and two cells (Figure 3D). These data suggest that AT2sAxin2 have an increased ability for growth and expansion during lung alveologenesis.
Figure 3. AT2sAxin2 have increased clonal growth potential.
(A and B) Clonal expansion is enhanced in AT2sAxin2 during alveologenesis. Following lineage tracing with the R26RBr2.1 reporter, AT2sAxin2 can expand clonally to a greater extent than the AT2 lineage in general. Arrows indicate single cell clones, dashed lined circles indicate two cell clones, and solid lined circles indicate three cell clones. Note the three cell clone in the AT2Axin2 lineage trace versus the two cell clone in the Sftpc+ lineage trace. (C) Quantitatively, the average AT2Axin2 clone size is larger than AT2s in general. (D) The distribution of clone size exhibits a shift towards more two and three cell clones in AT2sAxin2 compared to the total AT2 population. (E-H) Assessment of proliferation by EdU incorporation at P4 shows that the majority of AT2sAxin2 are actively proliferating compared to a minority of Axin2- AT2s. Arrows represent EdU+ AT2sAxin2, and arrowheads indicate EdU+ Axin2− AT2s. (I) Quantification of average clone size and percent of EdU+ AT2s are represented as mean ± SEM, and size distribution is represented % of total clones. Two-tailed student's t-test: *P < 0.05 and Fisher's Exact Test: **P < 0.007, n=5 (58 clones) for AT2sAxin2, n=4 (191 clones) for total AT2s; Two-tailed student's t-test: ***P < 0.05, n=3 with 10 independent fields of view per mouse. Scale bars: A and B = 50 μm, E-H = 5 μm.
Next, we directly assessed the proliferative potential of the AT2Axin2 population at P4 and compared it to the overall AT2 lineage using EdU labeling (Salic and Mitchison, 2008). Following a short chase of 2 hours, lungs were harvested and immunostained to examine EdU incorporation, Sftpc, and RFP expression (Figure 3E-3H). Approximately 3% of the total AT2 lineage are proliferating at P4. Of this proliferating AT2/Sftpc+ population, 70% are AT2Axin2 cells while approximately 30% are the Sftpc+ non-AT2Axin2 population (Figure 3I). These results suggest that AT2Axin2 cells preferentially drive alveolar growth and expansion.
The AT2Axin2 sublineage is highly enriched in genes important for lung alveologenesis
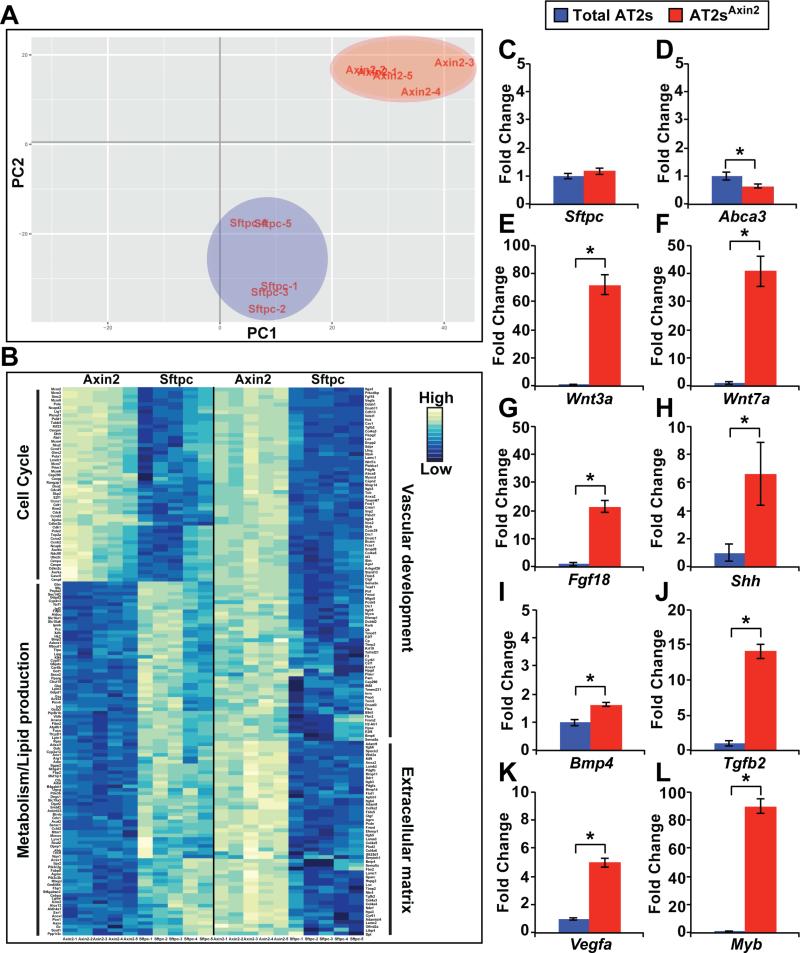
To define the gene expression characteristics behind the enhanced proliferation in the AT2Axin2 sublineage we performed transcriptome analysis on SftpcCreERT2:R26REYFP and Axin2CreERT2-TdTom P4 AT2 cells obtained by FACS. Principal component analysis using two variables illustrates significant gene expression differences between the total AT2 populations and the AT2Axin2 sublineage (Figure 4A). There were 4,101 differentially regulated genes between total AT2 and AT2Axin2 cells (see Supplemental Table 1 and GEO accession number GSE82154). Functional enrichment analysis of molecular functions and biological processes indicates that the AT2Axin2 sublineage is highly enriched in multiple biological processes associated with alveologenesis. These include the cell cycle, vascular development, and extracellular matrix production and interactions (Figure 4B). Interestingly, genes involved in metabolism/lipid metabolism, a function associated with mature AT2s, are down-regulated in AT2Axin2 cells. Despite this finding, AT2Axin2 cells express known AT2 marker genes such as Sftpc and Abca3 at levels equal or similar to the total AT2 population (Figure 4C and 4D). AT2Axin2 cells do not express or do not express different levels of early distal endoderm genes including Sox9, Id2, or Foxp2, indicating that they do not represent an immature developmental progenitor (see GEO accession number GSE82154). Importantly, the transcriptome data demonstrated that the AT2Axin2 sublineage expresses growth factor signaling components known to play important roles in lung development and stem cell biology including Fgf18, Shh, Bmp4, Tgfb2, Vegf, and Myb (Figure 4E-4L). In addition, enrichment of Wnt ligand, receptors, and co-receptors were observed in AT2Axin2 cells compared to the AT2 total population (Supplemental Table 2). These data indicate that the AT2Axin2 sublineage is distinct from the AT2 population in general and suggest that they act as signaling hubs for alveolar growth and differentiation.
Figure 4. The AT2Axin2 sublineage is highly enriched in genes important for alveologenesis.
(A) Principal component analysis illustrates unique segregation of the AT2Axin2 sublineage from the total AT2 population. (B) Microarray analysis comparing the total AT2 population versus the AT2Axin2 sublineage demonstrates enrichment of up-regulated genes for cell cycle, vascular development, and extracellular matrix interactions and production and down-regulation of genes associated with metabolism and lipid metabolism in the AT2Axin2 sublineage. (C-L) qPCR analysis on genes important in alveolar growth and differentiation confirms microarray findings on selected genes. Quantification of qPCR data is represented as mean ± SEM. Two-tailed student's t-test: *P < 0.05, n=4 for each group.
Increased Wnt signaling activity enhances ex vivo lung organoid formation from alveologenesis stage AT2s
To further characterize the ability of the AT2Axin2 sublineage to promote alveolar growth during the alveologenesis stage of lung development, we took advantage of an ex vivo lung organoid model and adapted it for use in early postnatal lung alveolar epithelium (Barkauskas et al., 2013). FACS was used to isolate AT2s from SftpcCreERT2:R26REYFP and AT2Axin2 cells from Axin2CreERT2-TdTom P4 pups. Five thousand isolated Sftpc+ or Axin2+ alveolar epithelial cells were co-cultured with 50,000 adult lung fibroblasts to form lung alveolar organoids (Figure 5A and 5B). Quantification of colony forming efficiency (CFE) demonstrated an increase in CFE for the AT2Axin2 sublineage compared to the total Sftpc+ AT2 population (Figure 5C). Both total AT2s and AT2sAxin2 have the ability for self-renewal (Figure 5C), and both are capable of multi-lineage differentiation as noted by expression of Sftpc and Pdpn for AT2 and AT1 cells, respectively (Figure 5D-5G).
Figure 5. AT2sAxin2 exhibit enhanced ex vivo lung organoid formation.
(A-C) AT2sAxin2 exhibit an increase in colony forming efficiency for lung alveolar organoid formation compared to total AT2s at passage 0 (P0) and passage 1 (P1). By passage 2 and 3 (P2-P3), these differences are no longer significant. (D and F) Hematoxylin and eosin (H&E) staining showing organoid structure in total AT2s and AT2sAxin2. (E and G) Immunohistochemical staining for the AT2 and AT1 markers, Sftpc and Pdpn, respectively, demonstrate that total AT2s and AT2sAxin2 are composed of both differentiated AT2s and AT1s. (H-L) Wnt3a treatment increases CFE while IWP2 treatment inhibits CFE in AT2 derived organoids. Treatment with both IWP2 and Wnt3a shows that inhibition can be rescued, in part, with exogenous Wnt ligand. Quantification of CFE is represented as average CFE ± SEM. Two-tailed student's t-test: *P < 0.05, n > 3 for each group in 4 independent experiments for A-I, n > 2 for each group in 3 independent experiments for J-N.
Wnt signaling and Wnt-responsiveness have been shown to be important for intestinal organoid growth and Wnt agonists are used for standard organoid culture (Farin et al., 2016; Farin et al., 2012; Yin et al., 2014). Therefore, we asked whether activating Wnt signaling in the overall AT2 cell lineage would result in enhanced organoid formation. Treatment of the AT2 lineage with Wnt3a resulted in a significant increase in CFE (Figure 5H-L). Conversely, treating with the Wnt ligand secretion inhibitor IWP-2 (Chen et al., 2009), resulted in inhibition of lung organoid formation, which was partially rescued with addition of Wnt3a to the culture media (Figure 5K and L). Importantly, AT2Axin2 cells loose TdTomato expression after they are placed in culture (Figure S5A-C), likely due to disruption of the alveolar niche. Treatment with recombinant Wnt3a can maintain TdTomato expression (Figure S5D), suggesting that a persistent Wnt signal is required for continued organoid formation.
Wnt activation promotes clonal expansion of AT2 cells during alveologenesis in vivo
Our ex vivo lung organoid assays suggest that AT2Axin2 cells promote alveolar growth during alveologenesis. To determine whether Wnt signaling is important for alveolar epithelial growth in vivo, we generated SftpcCreERT2:Ctnnb1fl(Ex3)/+):R26REYFP to activate Wnt/β-catenin signaling using the activated β-catenin allele (Ctnnb1fl(Ex3)/+)) (Harada et al., 1999). Of note, activating Wnt/β-catenin with the Axin2CreERT2-TdTom allele results in rapid neonatal lethality due to non-pulmonary related defects, precluding their use in these studies. Wnt/β-catenin signaling was activated by tamoxifen treatment at P4 and lungs were examined at P30. SftpcCreERT2:Ctnnb1fl(Ex3)/+):R26REYFP lungs exhibited increased cellularity with an increase in AT2 cell number (Figure 6A-6C and Figure S6A and S6B). Proliferation was increased by more than three-fold in SftpcCreERT2:Ctnnb1fl(Ex3)/+):R26REYFP AT2s compared to control mice (Figure 6D-6F).
Figure 6. Wnt activation increases AT2 proliferation and expansion during alveologenesis in vivo.
(A-C) Activation of Wnt signaling increases total alveolar epithelial cell and AT2 cell numbers when lineage tracing from P4 to P30 compared to SftpcCreERT2:Ctnnb1+/+ control mice. Both total Nkx2.1+ and Sftpc+ cells are increased in the alveolar region. (D-F) Proliferation as measured by Ki67 staining is increased in SftpcCreERT2:Ctnnb1fl(Ex3)/+ mutants compared to controls. Arrows indicate Ki67+ Sftpc lineage marked cells. (G-J) Wnt activation in AT2s results in clonal expansion of AT2s as noted by increased cell number per clone and increased clone size. Note the larger size of the CFP clone in H as compared to the YFP and RFP clones in G. (K-M) AT1 cell differentiation is unaffected by activation of Wnt signaling in AT2s as noted by co-staining for the YFP lineage mark and Aqp5. Quantification of cell number counts, %Ki67+, average clone size, and % of YFP+ cells is represented as mean ± SEM. Two-tailed student's t-test: *P < 0.05, n > 4 for each group; Two-tailed student's t-test: **P < 0.05 and Fisher's exact test: ***P <0.05, n=142 clones for SftpcCreERT2:Ctnnb1+/+ and n=258 clones for SftpcCreERT2:Ctnnb1fl(Ex3)/+ mutants.
To determine whether activation of Wnt/β-catenin signaling promoted clonal expansion of AT2 cells during alveologenesis, we utilized SftpcCreERT2:Ctnnb1(+/+):R26RBr2.1 and SftpcCreERT2:Ctnnb1fl(Ex3)/+):R26RBr2.1 treated with tamoxifen at P4 to track the expansion of AT2 cells. Wnt activation in AT2s resulted in a significant increase in clone size compared to control mice after four weeks (Figure 6G-I). Whereas the clone size distribution ranged from one to three clones with predominantly one cell clones in control mice, SftpcCreERT2:Ctnnb1fl(Ex3)/+):R26RBr2.1 mutant mice developed clones ranging from one cell to six cells with the majority of clones containing two or more cells (Figure 6J). These data indicate that AT2s with enhanced Wnt activity exhibit increased clonal expansion.
Recent data in adult mice suggest that AT1 and AT2 cells can exhibit bidirectional differentiation (Barkauskas et al., 2013; Jain et al., 2015). Therefore, we examined the relative ratios of AT2s and AT1s in Ctnnb1fl(Ex3)/+ mutant mice and compared them with their control mice counterparts. We performed lineage tracing from P4 to P30 in mice with the R26RBr2.1 reporter that can outline the extended structure of AT1s and AT2s derived from Sftpc+ cells with CFP, YFP, GFP, or RFP. In addition we utilized the R26REYFP reporter with subsequent antibody staining for Aqp5 and Sftpc. We found no significant differences in the percent of lineage traced AT2s versus AT1s in the two groups examined (Figure 6K-6M and S6C), suggesting that activation of Wnt signaling did not affect AT2-AT1 differentiation at this stage of development.
Inhibition of Wnt signaling impedes AT2 cell expansion and shunts differentiation towards the AT1 lineage during lung alveologenesis
To examine the impact of loss of Wnt/β-catenin signaling on AT2 proliferation and differentiation during lung alveologenesis, we generated SftpcCreERT2:Ctnnb1fl/fl:R26REYFP to conditionally delete β-catenin in AT2s at P4 and analyze alveolar development through P30. Loss of β-catenin in AT2s at this stage of lung development did not appear to grossly alter lung morphology (Figure S7A and S7B). However, immunostaining revealed a statistically significant decrease in AT2s (Figure S7C-E). Likewise, there was a notable 2.5-fold decrease in AT2 proliferation as measured by Ki67 staining (Figure 7A-7C).
Figure 7. Loss of Wnt signaling in AT2s inhibits proliferation and promotes differentiation into AT1s.
(A-C) Proliferation is reduced in SftpcCreERT2:Ctnnb1fl/fl mutant AT2s compared to controls following deletion of β-catenin from P4 to P30. (D-F) Loss of Wnt signaling in SftpcCreERT2:Ctnnb1fl/fl:R26RBr2.1 mutants from P4 to P30 leads to increased differentiation into AT1s during alveologenesis as noted by the increase in clones exhibiting the spread and flattened morphology of AT1 cells. (G-I) These data were confirmed using the R26RmTmG reporter to outline the cell surface of the AT1 cells along with Aqp5 co-immunostaining. (J) Illustration depicting the role of re-emergent Wnt signaling in balancing epithelial growth and differentiation during lung alveologenesis. Quantification of % Ki67+, average % of clone, and % of YFP+ cells is represented by mean ± SEM. Two-tailed student's t-test: *P < 0.05, n > 4 for each group; Two-tailed student's t-test: **P < 0.05, n=133 clones for controls and n=203 clones for SftpcCreERT2:Ctnnb1fl/fl mutants.
Interestingly, loss of β-catenin expression in SftpcCreERT2:Ctnnb1fl/fl:R26RBr2.1 mutants, resulted in a marked increase in the number of AT1 cells in the clones derived from Sftpc+ AT2s (Figure 7D-7F). These findings suggested that Wnt signaling normally restricts the differentiation of the AT2 lineage into the AT1 lineage during lung alveologenesis. To further corroborate whether Wnt signaling is essential for the restriction of AT2-AT1 differentiation, we generated SftpcCreERT2:Ctnnb1fl/fl:R26RmTmG mutants and controls to outline the full cell surface of lineage traced cells (Muzumdar et al., 2007). Following induction of recombination at P4, SftpcCreERT2:Ctnnb1fl/fl:R26RmTmG mutant mice produced an approximately five-fold increase in lineage traced AT1 cells (Figure 7G-7I). These studies reveal dynamic Wnt-responsiveness in late lung development that promotes AT2 growth and inhibits differentiation of AT1 cells during alveologenesis (Figure 7J).
DISCUSSION
Generation of the lung alveolus is the definitive step in the development of a functional respiratory unit for gas exchange. Pathway-mediated differentiation and maintenance of alveolar epithelial cell populations during alveologenesis have remained a poorly understood aspect of lung development. While a role for Wnt signaling in promoting early lung distal endoderm progenitor fate has been described (Shu et al., 2005; Volckaert et al., 2013), the pathways mediating later saccular and alveolar epithelial development are less understood. Our data indicate that Wnt signaling, an important regulator of early lung development, is reactivated during alveologenesis to provide a wave of AT2 cell growth for the final important stages of lung growth and development.
Wnt signaling plays critical roles during early lung endoderm specification, branching morphogenesis, and lung mesenchymal development (Cohen et al., 2009; De Langhe et al., 2008; Goss et al., 2009; Kadzik et al., 2014; Konigshoff and Eickelberg, 2010; Li et al., 2005; Li et al., 2002; Mammoto et al., 2012; Maretto et al., 2003; Miller et al., 2012; Okubo and Hogan, 2004; Rajagopal et al., 2008; Shu et al., 2005; Shu et al., 2002; van Amerongen et al., 2012). Our data now show that as lung development progresses, Wnt-responsiveness in lung epithelial cells wanes until early alveologenesis, when there is a re-emergence of Wnt-responsive alveolar epithelial cells called AT2sAxin2. While previous studies have examined some aspects of the structural remodeling that occurs during the alveolar stage of lung development (Amy et al., 1977; Bourbon et al., 2005; Branchfield et al., 2016; Massaro and Massaro, 2007), little is known about how the alveolus expands during early postnatal lung growth and in particular the molecular pathways that regulate this process. As the lung grows into its final mature size in the adult, the AT2 and AT1 lineages expand to accommodate this growth. However, recent studies suggest that AT1 and AT2 cell expansion is not balanced during alveologenesis. While AT1s stop proliferating during the first few days after birth, AT2s continue to proliferate and expand throughout alveologenesis (Yang et al., 2016). This difference may be due to the ability of the AT1 cell to dramatically remodel and spread over a great distance in the alveolus, allowing it to grow through cellular remodeling rather than proliferation (Wang et al., 2016; Weibel, 2015; Yang et al., 2016). Given the role that the AT2 lineage plays in synthesis of pulmonary surfactant and as an important monitor of the innate immune response, the ability to expand and grow during alveologenesis likely allows the lung in general to respond to challenges during this critical early postnatal period of life (Mulugeta et al., 2015). Our data shows that the re-emergence of Wnt signaling promotes growth of the postnatal alveolar epithelium and balances the epithelial composition of the alveolus through control of AT2-AT1 differentiation.
Disruption in alveolar growth underlies many important pediatric lung diseases including bronchopulmonary dysplasia (Bourbon et al., 2005). Moreover, premature infants that are born prior to the completion of alveologenesis often have long-term consequences including impaired lung function and pulmonary hypertension (Islam et al., 2015; Stenmark and Abman, 2005). While clinicians have recognized the importance of alveologenesis in lung function for many decades, the basic understanding of the cellular interactions and molecular pathways involved in promoting this late developmental process has remained an enigma. Cellular proliferation is generally thought to be low during this developmental period but our data suggests that AT2 cell expansion, primarily through expansion of the AT2Axin2 sublineage, plays a critical role in postnatal alveolar growth. The wave of Wnt signaling that occurs during early postnatal lung alveologenesis identified in the present study is critical for the final stages of lung development by promoting a burst of AT2 expansion to meet the needs of the adult organ. Interestingly, while loss of β-catenin-dependent Wnt signaling in AT2s results in the transition of some of the AT2 population into AT1s, further increases in β-catenin-dependent Wnt signaling did not result in inhibition of AT2 to AT1 differentiation. This suggests that there is a threshold of Wnt signaling required to maintain the AT2 fate, and an additional increase in Wnt signaling only promotes AT2 cell proliferation. As such, studies pharmacologically modulating Wnt signaling following experimental injury could provide important insight into whether this pathway could be used therapeutically to treat neonatal lung disease.
EXPERIMENTAL PROCEDURES
Animals
Description and genotyping information regarding Ctnnb1fl/fl, Ctnnb1fl(Ex3)/+, R26REYFP, R26RBr2.1, and R26RmTmG mouse lines has been previously described (Brault et al., 2001; Harada et al., 1999; Madisen et al., 2010; Muzumdar et al., 2007; Snippert et al., 2010). The SftpcCreERT2 mouse line was a generous gift from Hal Chapman at UCSF, and its construction has been previously described (Chapman et al., 2011). The Axin2CreERT2-TdTomato mouse was constructed by inserting an expression cassette, consisting of a creERT2 cDNA linked to a 2A self-cleaving peptide sequence linked to a TdTomato cDNA, into the start codon of the Axin2 locus. Full details on the construction of this reporter line are available upon request. All animal studies were performed under guidance of the University of Pennsylvania Institutional Animal Care and Use Committee.
Tamoxifen induction of cell lineage tracing
Tamoxifen (Sigma) was dissolved in 100% ethanol and diluted with corn oil (Sigma) to produce a 10% ethanol:tamoxifen:corn oil mixture at 20 mg/mL. Mice were injected intraperitoneally (IP) with the indicated doses to induce recombination based on the observed efficiency of each cre and reporter line. For lineage tracing in SftpcCreERT2 mice, they were IP injected with 100 μg/gm of mouse for the R26REYFP and R26RmTmG alleles. For clonal analysis using the R26RBr2.1 allele, 5-10 μg/gm was used. Induction of recombination in Axin2CreERT2-TdTomato P4 pups was accomplished using 100 μg/gm of mouse for the R26REYFP and R26RmTmG alleles and 200 μg/gm for the R26RBr2.1 allele.
Histology
Lungs were inflated with 2% paraformaldehyde under constant pressure of 30 cm water and allowed to fix overnight. Tissue was embedded in paraffin and sectioned. Hematoxylin and eosin staining was performed to examine tissue morphology. Immunohistochemistry was used to detect protein expression using the following antibodies on paraffin sections: GFP (chicken, Aves, 1:500), GFP (goat, Abcam, 1:100), RFP (rabbit, Rockland, 1:250), Nkx2.1 (rabbit, Santa Cruz, 1:50), Sox2 (rabbit, Seven Hills, 1:500), Sox9 (rabbit, Santa Cruz, 1:100), Pecam (rat, HistoBioTec, 1:20), SM22α (goat, Abcam, 1:100), Pdgfrα (rabbit, Cell Signaling, 1:50), Pdgfrα (goat, R&D Systems, 1:50), Pdgfrβ (rabbit, Cell Signaling, 1:100), Pdgfrβ (goat, R&D Systems, 1:400), Scgb1a1 (goat, Santa Cruz, 1:20), Tubb4 (mouse, BioGenex, 1:20), Sftpc (rabbit, Millipore, 1:250), Sftpc (goat, Santa Cruz, 1:50), Pdpn (mouse, Hybridoma Bank, 1:50), Aqp5 (rabbit, Abcam, 1:100), and Ki67 (rabbit, Abcam, 1:50).
Quantification of alveolar epithelial cell number
Immunostaining using the indicated cell lineage specific markers was used to identify lineage traced alveolar epithelial cell types. Confocal microscopy using a Leica TCS SP8 confocal scope was employed to capture images. For each mouse, confocal Z-stack images were taken in at least 10 random alveolar areas. All images were subject to ImageJ software analysis. Cell counts were performed manually using the Cell Counter plug-in for ImageJ. At least 10 different regions of each lung or 1000 cells were counted for each mouse.
Quantification of proliferation
Proliferation was assessed using either the Click-iT EdU Alexa Fluor 647 Imaging Kit (ThermoFisher Scientific) or Ki67 immunostaining. For proliferation analysis using EdU, P4 pups were administered 50 mg/kg EdU via IP injection. Two hours post injection, the lungs were harvested for tissue processing as described previously. Following double immunostaining for RFP and Sftpc, lung sections were processed with the Click-iT EdU reaction. Z-stack optical sections were attained via confocal microscopy and 10 independent alveolar fields of view were assessed for RFP, Sftpc, and EdU expression. Cell counts were performed using ImageJ software.
Quantification of clonal analysis
Clonal analysis was performed after induction of recombination at P4 followed by a four-week chase. Lungs were harvested as described and fixed overnight in 2% paraformaldehyde. The next day, the lungs were washed with PBS for 2 hours. The lungs were embedded in OCT compound (Fisher HealthCare) and frozen on dry ice and ethanol. 12 μm lung sections were cut and subjected to immunostaining for Sftpc to identify all AT2s followed by counterstaining with DAPI (4′,6-diamidino-2-phenylindole). At least 10 Z-stack sections were analyzed from random fields. Each single-colored clone was analyzed for number of cells in each clone. Single-colored cells within 50 μm of one another were considered in the same clone. Average number of cells per clone was calculated in addition to the categorical range of distribution for clone size.
Lung alveolar epithelial cell isolation
Lungs from Axin2CreERT2-TdTomato pups were harvested at P4 and processed into single cell suspensions using a dispase (Collaborative Biosciences)/collagenase (Life Technologies)/DNase solution. Axin2+ AT2s were sorted from the single suspension using either a MoFlo Astrios EQ (Beckman Coulter) or FACSJazz (BD Biosciences) flow cytometer with antibody staining for CD31-PECy7 (eBioscience), CD45-PECy7 (eBioscience), and EpCAM-APC (eBioscience). Following negative selection for CD31 and CD45, Axin2 AT2s were positively selected for TdTomato and EpCAM and sorted into a modified MTEC-Plus media (SABM (Lonza) instead of DMEM/F12), called MTECSAGM. The total AT2 population (Sftpc+ AT2s) was isolated from lungs of P4 SftpcCreERT2:R26REYFP pups following induction of recombination on P3. Cells were sorted from single cell suspensions after enzyme digestion and isolated based on EYFP fluorescence. Lung fibroblasts for organoid assays were isolated from adult wild type mice. Briefly, lungs were harvested, digested, and processed into a single cell suspension with subsequent plating on plastic tissue culture dishes. Cells were maintained in DMEM/F12 (Life Technologies) supplemented with 10% FBS and 1X Anti-Anti- (Life Technologies). Following three passages, the highly enriched mesenchymal population was analyzed to be greater than 99% pure mesenchyme as determined by flow cytometry assessment using antibodies for epithelial (EpCAM-APC), endothelial (CD31-PECy7), hematopoietic (CD45-PECy7), and mesenchymal cells (CD140a:Pdgfrα (eBioscience) and CD140b:Pdgfrβ (eBioscience) (Peng et al., 2015)
Ex vivo lung alveolar organoid assay
For lung alveolar organoid formation, 5,000 Axin2+ or Sftpc+ AT2s were mixed with 50,000 primary lung fibroblasts and centrifuged into cell pellets in microcentrifuge tubes. Pellets were resuspended in a 1:1 mixture of MTEC-SAGM:Matrigel (45 μl of modified MTEC-SAGM followed by the addition of 45 μl of growth factor-reduced, phenol-free Matrigel (Corning)). The mixture was then aliquoted into 24 well cell culture insert (Falcon) and allowed to solidify at 37°C. MTEC-SAGM was then placed into each well of the 24-well plate. The use of ligands and chemical antagonists were added to both the insert and well culture media at initial formation of organoids. Wnt3a was used at a concentration of 200 ng/uL. IWP-2 was used at 5 μM. DMSO was used for vehicle treatment. Media with or without treatments were replaced every other day until day 14 where organoids were assayed for total number by manual counts. For self-renewal assays, after 14 days organoids were incubated with dispase (Stem Cell Technology) for 30 minutes then incubated in 0.25% Trypsin-EDTA for 30 minutes. Single cell suspensions were obtained, and the cells were re-sorted to regenerate organoids.
Flow cytometry for expression of Sftpc and Pdpn in Axin2+ cells
Cells obtained by FACS from the Axin2CreERT2-TdTomato lungs were subjected to further analysis for expression of Sftpc and Pdpn. Cells were first stained with the extracellular Pdpn antibody (Pdpn-eFluor660, eBioscience, 1:100) followed by fixation and permeabilization using a standard intracellular staining kit (eBioscience). Cells were stained with anti-Sftpc primary antibody (goat, Santa Cruz, 1:50) or its goat IgG isotype control both followed by Alexa Fluor 488 secondary antibody. Cells were assessed using flow cytometer (Accuri C6) and results were expressed at percentage of Axin2+ EpCAM+ cells.
Transcriptome Analysis
Comparison of the transcriptomes of P4 Axin2CreERT2-TdTomato and SftpcCreERT2:R26REYFP pups was performed using microarray analysis. FACS-based isolation of AT2 or AT2Axin2 cells from the lungs of five pups for each genotype was performed as described above. Cells were collected in RNAProtect Cell Reagent (Qiagen) and subjected to RNA extraction using a combined Trizol (Life Technologies) and RNEasy MiniElute Cleanup Kit (Qiagen). Biotinylated cRNA probe libraries were constructed and then hybridized to Affymatrix Mouse Gene 2.0ST arrays. Microarray data were analyzed using the Oligo package available at the Bioconductor Web site (http://www.bioconductor.org). The raw data were background-corrected by the robust multichip average (RMA) method and then normalized by an invariant set method. Genes with 80% of samples with an expression signal above the negative control probes were considered detectable or present. Differential gene expression analysis was performed using the limma package available at the Bioconductor Web site. P-values were adjusted for multiple comparisons using a false discovery rate. Principal component analysis was performed in R using the prcomp function. Gene Ontology enrichment analysis was performed using the ToppFun tool available from the ToppGene suite website (https://toppgene.cchmc.org). The Gene Expression Omnibus accession number for the microarray data is GSE82154.
Statistical Analysis
Statistical analysis was performed on the data using Excel and R. Two-tailed student t-test was used for comparison of two experimental groups. Using the R, a Fishers’ exact test was performed to determine an association between clone size and genotype/treatment in all experiments. Statistical data was considered significant if P < 0.05.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful for the technical support of the Penn Cardiovascular Institute Histology Core for histology and immunohistochemical services. In addition, they would like to extend thanks to Andrea Stoudt for confocal microscopy technical support. Support for these studies come from grants from the NIH to E.E.M. (HL087825, HL100405, HL110942), D.B.F. (NIH T32 HL007915 and K12 HD043245), and T.P. (NIH K08 HL121146). In addition, D.B.F. is supported by a fellowship award from the Parker B. Francis Fellowship Program and a grant from the Pulmonary Hypertension Association's Matthew & Michael Wojciechowski Pediatric PH Research & Mentoring Grant.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
D.B.F, T.P. and E.E.M. designed experiments and participated in writing the manuscript. D.B.F., J.Z., M.S., T.L.V., S.Z., M.M.L completed experiments and data analysis. Bioinformatics analysis was performed by M.P.M.
REFERENCES
- Al Alam D, Green M, Tabatabai Irani R, Parsa S, Danopoulos S, Sala FG, Branch J, El Agha E, Tiozzo C, Voswinckel R, et al. Contrasting expression of canonical Wnt signaling reporters TOPGAL, BATGAL and Axin2(LacZ) during murine lung development and repair. PloS one. 2011;6:e23139. doi: 10.1371/journal.pone.0023139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amy RW, Bowes D, Burri PH, Haines J, Thurlbeck WM. Postnatal growth of the mouse lung. Journal of anatomy. 1977;124:131–151. [PMC free article] [PubMed] [Google Scholar]
- Barkauskas CE, Cronce MJ, Rackley CR, Bowie EJ, Keene DR, Stripp BR, Randell SH, Noble PW, Hogan BL. Type 2 alveolar cells are stem cells in adult lung. The Journal of clinical investigation. 2013;123:3025–3036. doi: 10.1172/JCI68782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourbon J, Boucherat O, Chailley-Heu B, Delacourt C. Control mechanisms of lung alveolar development and their disorders in bronchopulmonary dysplasia. Pediatric research. 2005;57:38R–46R. doi: 10.1203/01.PDR.0000159630.35883.BE. [DOI] [PubMed] [Google Scholar]
- Branchfield K, Li R, Lungova V, Verheyden JM, McCulley D, Sun X. A three-dimensional study of alveologenesis in mouse lung. Developmental biology. 2016;409:429–441. doi: 10.1016/j.ydbio.2015.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brault V, Moore R, Kutsch S, Ishibashi M, Rowitch DH, McMahon AP, Sommer L, Boussadia O, Kemler R. Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development. 2001;128:1253–1264. doi: 10.1242/dev.128.8.1253. [DOI] [PubMed] [Google Scholar]
- Chapman HA, Li X, Alexander JP, Brumwell A, Lorizio W, Tan K, Sonnenberg A, Wei Y, Vu TH. Integrin alpha6beta4 identifies an adult distal lung epithelial population with regenerative potential in mice. The Journal of clinical investigation. 2011;121:2855–2862. doi: 10.1172/JCI57673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Dodge ME, Tang W, Lu J, Ma Z, Fan CW, Wei S, Hao W, Kilgore J, Williams NS, et al. Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nature chemical biology. 2009;5:100–107. doi: 10.1038/nchembio.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H, Loh KM, Nusse R. Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science. 2014;346:1248012. doi: 10.1126/science.1248012. [DOI] [PubMed] [Google Scholar]
- Cohen ED, Ihida-Stansbury K, Lu MM, Panettieri RA, Jones PL, Morrisey EE. Wnt signaling regulates smooth muscle precursor development in the mouse lung via a tenascin C/PDGFR pathway. The Journal of clinical investigation. 2009;119:2538–2549. doi: 10.1172/JCI38079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Langhe SP, Carraro G, Tefft D, Li C, Xu X, Chai Y, Minoo P, Hajihosseini MK, Drouin J, Kaartinen V, et al. Formation and differentiation of multiple mesenchymal lineages during lung development is regulated by beta-catenin signaling. PloS one. 2008;3:e1516. doi: 10.1371/journal.pone.0001516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Agha E, Herold S, Al Alam D, Quantius J, MacKenzie B, Carraro G, Moiseenko A, Chao CM, Minoo P, Seeger W, et al. Fgf10-positive cells represent a progenitor cell population during lung development and postnatally. Development. 2014;141:296–306. doi: 10.1242/dev.099747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farin HF, Jordens I, Mosa MH, Basak O, Korving J, Tauriello DV, de Punder K, Angers S, Peters PJ, Maurice MM, et al. Visualization of a short-range Wnt gradient in the intestinal stem-cell niche. Nature. 2016;530:340–343. doi: 10.1038/nature16937. [DOI] [PubMed] [Google Scholar]
- Farin HF, Van Es JH, Clevers H. Redundant sources of Wnt regulate intestinal stem cells and promote formation of Paneth cells. Gastroenterology. 2012;143:1518–1529. e1517. doi: 10.1053/j.gastro.2012.08.031. [DOI] [PubMed] [Google Scholar]
- Goss AM, Tian Y, Tsukiyama T, Cohen ED, Zhou D, Lu MM, Yamaguchi TP, Morrisey EE. Wnt2/2b and beta-catenin signaling are necessary and sufficient to specify lung progenitors in the foregut. Developmental cell. 2009;17:290–298. doi: 10.1016/j.devcel.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada N, Tamai Y, Ishikawa T, Sauer B, Takaku K, Oshima M, Taketo MM. Intestinal polyposis in mice with a dominant stable mutation of the beta-catenin gene. The EMBO journal. 1999;18:5931–5942. doi: 10.1093/emboj/18.21.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris-Johnson KS, Domyan ET, Vezina CM, Sun X. beta-Catenin promotes respiratory progenitor identity in mouse foregut. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:16287–16292. doi: 10.1073/pnas.0902274106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herring MJ, Putney LF, Wyatt G, Finkbeiner WE, Hyde DM. Growth of alveoli during postnatal development in humans based on stereological estimation. American journal of physiology Lung cellular and molecular physiology. 2014;307:L338–344. doi: 10.1152/ajplung.00094.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam JY, Keller RL, Aschner JL, Hartert TV, Moore PE. Understanding the Short- and Long-Term Respiratory Outcomes of Prematurity and Bronchopulmonary Dysplasia. American journal of respiratory and critical care medicine. 2015;192:134–156. doi: 10.1164/rccm.201412-2142PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R, Barkauskas CE, Takeda N, Bowie EJ, Aghajanian H, Wang Q, Padmanabhan A, Manderfield LJ, Gupta M, Li D, et al. Plasticity of Hopx(+) type I alveolar cells to regenerate type II cells in the lung. Nature communications. 2015;6:6727. doi: 10.1038/ncomms7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadzik RS, Cohen ED, Morley MP, Stewart KM, Lu MM, Morrisey EE. Wnt ligand/Frizzled 2 receptor signaling regulates tube shape and branch-point formation in the lung through control of epithelial cell shape. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:12444–12449. doi: 10.1073/pnas.1406639111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konigshoff M, Eickelberg O. WNT signaling in lung disease: a failure or a regeneration signal? American journal of respiratory cell and molecular biology. 2010;42:21–31. doi: 10.1165/rcmb.2008-0485TR. [DOI] [PubMed] [Google Scholar]
- Li C, Hu L, Xiao J, Chen H, Li JT, Bellusci S, Delanghe S, Minoo P. Wnt5a regulates Shh and Fgf10 signaling during lung development. Developmental biology. 2005;287:86–97. doi: 10.1016/j.ydbio.2005.08.035. [DOI] [PubMed] [Google Scholar]
- Li C, Xiao J, Hormi K, Borok Z, Minoo P. Wnt5a participates in distal lung morphogenesis. Developmental biology. 2002;248:68–81. doi: 10.1006/dbio.2002.0729. [DOI] [PubMed] [Google Scholar]
- Li N, Yousefi M, Nakauka-Ddamba A, Tobias JW, Jensen ST, Morrisey EE, Lengner CJ. Heterogeneity in readouts of canonical wnt pathway activity within intestinal crypts. Developmental dynamics : an official publication of the American Association of Anatomists. 2016;245:822–833. doi: 10.1002/dvdy.24423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nature neuroscience. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammoto T, Chen J, Jiang E, Jiang A, Smith LE, Ingber DE, Mammoto A. LRP5 regulates development of lung microvessels and alveoli through the angiopoietin-Tie2 pathway. PloS one. 2012;7:e41596. doi: 10.1371/journal.pone.0041596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maretto S, Cordenonsi M, Dupont S, Braghetta P, Broccoli V, Hassan AB, Volpin D, Bressan GM, Piccolo S. Mapping Wnt/beta-catenin signaling during mouse development and in colorectal tumors. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:3299–3304. doi: 10.1073/pnas.0434590100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massaro D, Massaro GD. Developmental alveologenesis: longer, differential regulation and perhaps more danger. American journal of physiology Lung cellular and molecular physiology. 2007;293:L568–569. doi: 10.1152/ajplung.00258.2007. [DOI] [PubMed] [Google Scholar]
- Miller MF, Cohen ED, Baggs JE, Lu MM, Hogenesch JB, Morrisey EE. Wnt ligands signal in a cooperative manner to promote foregut organogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:15348–15353. doi: 10.1073/pnas.1201583109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrisey EE, Hogan BL. Preparing for the first breath: genetic and cellular mechanisms in lung development. Developmental cell. 2010;18:8–23. doi: 10.1016/j.devcel.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucenski ML, Wert SE, Nation JM, Loudy DE, Huelsken J, Birchmeier W, Morrisey EE, Whitsett JA. beta-Catenin is required for specification of proximal/distal cell fate during lung morphogenesis. The Journal of biological chemistry. 2003;278:40231–40238. doi: 10.1074/jbc.M305892200. [DOI] [PubMed] [Google Scholar]
- Mulugeta S, Nureki S, Beers MF. Lost after translation: insights from pulmonary surfactant for understanding the role of alveolar epithelial dysfunction and cellular quality control in fibrotic lung disease. American journal of physiology Lung cellular and molecular physiology. 2015;309:L507–525. doi: 10.1152/ajplung.00139.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mund SI, Stampanoni M, Schittny JC. Developmental alveolarization of the mouse lung. Developmental dynamics : an official publication of the American Association of Anatomists. 2008;237:2108–2116. doi: 10.1002/dvdy.21633. [DOI] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Okubo T, Hogan BL. Hyperactive Wnt signaling changes the developmental potential of embryonic lung endoderm. Journal of biology. 2004;3:11. doi: 10.1186/jbiol3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng T, Frank DB, Kadzik RS, Morley MP, Rathi KS, Wang T, Zhou S, Cheng L, Lu MM, Morrisey EE. Hedgehog actively maintains adult lung quiescence and regulates repair and regeneration. Nature. 2015;526:578–582. doi: 10.1038/nature14984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prodhan P, Kinane TB. Developmental paradigms in terminal lung development. BioEssays : news and reviews in molecular, cellular and developmental biology. 2002;24:1052–1059. doi: 10.1002/bies.10177. [DOI] [PubMed] [Google Scholar]
- Rajagopal J, Carroll TJ, Guseh JS, Bores SA, Blank LJ, Anderson WJ, Yu J, Zhou Q, McMahon AP, Melton DA. Wnt7b stimulates embryonic lung growth by coordinately increasing the replication of epithelium and mesenchyme. Development. 2008;135:1625–1634. doi: 10.1242/dev.015495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu W, Guttentag S, Wang Z, Andl T, Ballard P, Lu MM, Piccolo S, Birchmeier W, Whitsett JA, Millar SE, et al. Wnt/beta-catenin signaling acts upstream of N-myc, BMP4, and FGF signaling to regulate proximal-distal patterning in the lung. Developmental biology. 2005;283:226–239. doi: 10.1016/j.ydbio.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Shu W, Jiang YQ, Lu MM, Morrisey EE. Wnt7b regulates mesenchymal proliferation and vascular development in the lung. Development. 2002;129:4831–4842. doi: 10.1242/dev.129.20.4831. [DOI] [PubMed] [Google Scholar]
- Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- Stenmark KR, Abman SH. Lung vascular development: implications for the pathogenesis of bronchopulmonary dysplasia. Annual review of physiology. 2005;67:623–661. doi: 10.1146/annurev.physiol.67.040403.102229. [DOI] [PubMed] [Google Scholar]
- Swarr DT, Morrisey EE. Lung endoderm morphogenesis: gasping for form and function. Annual review of cell and developmental biology. 2015;31:553–573. doi: 10.1146/annurev-cellbio-100814-125249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Amerongen R, Bowman AN, Nusse R. Developmental stage and time dictate the fate of Wnt/beta-catenin-responsive stem cells in the mammary gland. Cell stem cell. 2012;11:387–400. doi: 10.1016/j.stem.2012.05.023. [DOI] [PubMed] [Google Scholar]
- Volckaert T, Campbell A, Dill E, Li C, Minoo P, De Langhe S. Localized Fgf10 expression is not required for lung branching morphogenesis but prevents differentiation of epithelial progenitors. Development. 2013;140:3731–3742. doi: 10.1242/dev.096560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volckaert T, De Langhe SP. Wnt and FGF mediated epithelialmesenchymal crosstalk during lung development. Developmental dynamics : an official publication of the American Association of Anatomists. 2015;244:342–366. doi: 10.1002/dvdy.24234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Frank DB, Morley MP, Zhou S, Wang X, Lu MM, Lazar MA, Morrisey EE. HDAC3-Dependent Epigenetic Pathway Controls Lung Alveolar Epithelial Cell Remodeling and Spreading via miR-17-92 and TGF-beta Signaling Regulation. Developmental cell. 2016;36:303–315. doi: 10.1016/j.devcel.2015.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weibel ER. On the tricks alveolar epithelial cells play to make a good lung. American journal of respiratory and critical care medicine. 2015;191:504–513. doi: 10.1164/rccm.201409-1663OE. [DOI] [PubMed] [Google Scholar]
- Whitsett JA, Weaver TE. Alveolar development and disease. American journal of respiratory cell and molecular biology. 2015;53:1–7. doi: 10.1165/rcmb.2015-0128PS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Hernandez BJ, Martinez Alanis D, Narvaez del Pilar O, Vila-Ellis L, Akiyama H, Evans SE, Ostrin EJ, Chen J. The development and plasticity of alveolar type 1 cells. Development. 2016;143:54–65. doi: 10.1242/dev.130005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Farin HF, van Es JH, Clevers H, Langer R, Karp JM. Niche-independent high-purity cultures of Lgr5+ intestinal stem cells and their progeny. Nature methods. 2014;11:106–112. doi: 10.1038/nmeth.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun EJ, Lorizio W, Seedorf G, Abman SH, Vu TH. VEGF and endothelium-derived retinoic acid regulate lung vascular and alveolar development. American journal of physiology Lung cellular and molecular physiology. 2016;310:L287–298. doi: 10.1152/ajplung.00229.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.