Abstract
In the last few years, protein and chemical microarrays have emerged as two important tools in the field of proteomics. Specific proteins, antibodies, small molecule compounds, peptides, and carbohydrates can now be immobilized on solid surfaces to form high-density microarrays. Depending on their chemical nature, immobilization of these molecules on solid support is accomplished by in situ synthesis, nonspecific adsorption, specific binding, nonspecific chemical ligation, or chemoselective ligation. These arrays of molecules can then be probed with complex analytes such as serum, total cell extracts, and whole blood. Interactions between the analytes and the immobilized array of molecules are evaluated with a number of different detection systems. In this paper, various components, methods, and applications of the protein and chemical microarray systems are reviewed.
INTRODUCTION
Chemical array is a form of the combinatorial library method that was first described by Geysen et al in 1984 [1]. Peptides were synthesized on polyethylene pins in a 96-well footprint and used for B-cell epitope mapping. Enzyme-linked immunosorbent assays were used for such analysis. In 1991, Foder et al of Affymax, Inc (Palto Alto, Calif) [2] reported the use of photolithography in conjunction with light-directed peptide synthesis to generate 1024 peptide on a 1.6-cm2 glass surface and the use of fluorescent microscopy to analyze interactions between the peptides and fluorescent-labeled antibodies. In 1992, Frank described the synthesis of a peptide array as spots on paper [3]. These three techniques all use parallel synthesis methods to generate arrays of peptides that are spatially separable and addressable. The identity of each peptide spot is known prior to any biological assay. In 1991, we reported the use of the highly efficient “split-mix” synthesis method to generate “one-bead one-compound” (OBOC) combinatorial peptide library (millions of peptides), in which each 80-μm bead displayed only one peptide entity [4]. This spatially separable but nonaddressable peptide microarray or library was screened with an enzyme-linked colorimetric assay, and individual color beads were then physically isolated for microsequencing. These early studies paved the road for the microarray field. In 1994, scientists from Affymetrix (Santa Clara, Calif), a spin-off company from Affymax, reported the use of photolithography/photochemistry approaches to synthesize the first DNA chip [5]. Microarrays of oligonucleotides were synthesized in situ on glass surface, and fluorescent-labeled octanucleotide probes were used to identify the complementary oligonucleotides on this 256 array. In 1995, Brown et al introduced a different form of a DNA microarray chip by using high-speed robot to spot array of different cDNAs on the glass surface. Fluorescent-labeled cDNA derived from mRNA of whole cell lysate was then used to probe the DNA microarray, allowing determination of expression levels of thousands of genes simultaneously [6, 7]. Several automatic arrayers have since become commercially available. The successful application of DNA microarrays to gene expression analysis [8, 9, 10], genetic diagnosis [11], and drug target identification [12], and the rapid development of the proteomics field have propelled many to think about the use of protein or chemical microarray as an efficient tool to evaluate the function of complex protein mixtures. In the last few years, several groups independently developed different chemical and protein microarray methods and applied them to study various biological and chemical problems [13, 14]. In this review, we will give an overview in the field and highlight recent developments.
TYPES OF MICROARRAY
Microarray, sometimes referred to as a chip or arrayed library, can be classified into two general types: biochips (biomicroarray) and chemical microarray. Biochips are usually generated from biochemical or biological components, such as, protein (including enzymes and antibodies), DNA, cell [15], and tissue [16]. Chemical microarrays consist of arrays of organic compounds including small organic molecules, peptides, and sugars. Based on how chemical microarrays are constructed, they can also be categorized as in situ synthesis array and spotting array. The chemistry of the in situ synthesis approach is more limited, particularly when photochemical reaction is a required synthetic step. As a result, only oligomeric molecules such as oligonucleotides or peptides are used in the in situ synthesis array. A spotting array refers to an array of compounds that are presynthesized and directly transferred and immobilized on a solid surface. This approach is more versatile and can be applied to generate a microarray of almost any molecules.
IN SITU SYNTHESIS MICROARRAY APPROACH
Two typical in situ synthesis approaches are SPOT-synthesis [3] and light-directed parallel synthesis [2]. The SPOT-synthesis method involves parallel peptide synthesis on membrane or paper. In this method, a small volume of solutions containing Fmoc-amino acids plus coupling reagents is dispensed onto the designated spot on the membrane. After the coupling reaction is complete, the whole membrane is washed and the N-terminal protecting group is deprotected prior to the next coupling cycle. Limited by possibility of contamination by reagents from adjacent spots, the distance between each spot cannot be too small. This results in a rather low-density array (eg, 25 spots/cm2). Production of peptide arrays with this technology has been reviewed recently [17]. Automated SPOT-synthesizers, such as the Auto-Spot Robot ASP222 (Intavis Inc, Germany), are now commercially available.
The light-directed in situ synthesis was initially developed for peptide synthesis [2] but has now been widely used for the synthesis of DNA microarrays. The commercially available Affymetrix chips are prepared by this approach. Figure 1 depicts the deprotection chemistries of light-directed parallel synthesis. The original method uses amino acids with a photolabile protecting group (eg nitroveratryloxycarbonyl, NVOC) as building blocks, and photolithographic method with appropriate masks is used to spatially deprotect the N-terminal protecting group during peptide synthesis. This approach was later adapted to oligonucleotide synthesis [5, 6]. In 1996, McGall et al reported a new photolithographic masking method that involves the use of a polymeric photoresistant film to construct a pattern onto the glass surface [18]. The patterned photoresistant film is used to mask selected regions of the substrate from exposure to standard chemical reagents during synthesis. The main advantage of this approach is that the resolution is superior, and therefore the density of the microarray can increase significantly.
Figure 1.
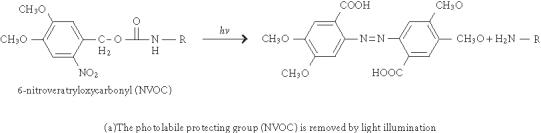
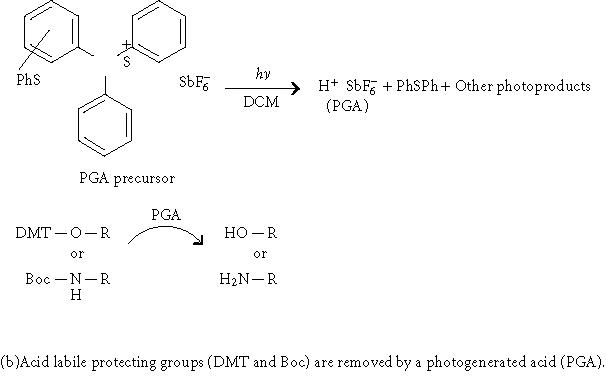
Deprotection chemistry in two approaches of light-directed parallel synthesis.
Instead of using photolithographic masks, Singh-Gasson et al [19], reported a maskless light-directed array synthesizer, which utilizes a digital micromirror device consisting of a 600 × 800 array of micromirrors to form virtual masks. With 1 : 1 imaging, the synthesizer can produce 480,000 pixels of synthetic oligomers in a 10 × 14 mm area. A similar maskless microarray setup was also reported by Gao et al to synthesize oligonucleotide [20] and peptide [21] microarrays. However, this method is unique in that it uses light to generate acid in situ to deprotect the acid labile protecting groups of the growing chain (4,4'-dimethoxytrityl (DMT) for nucleotides or Boc for amino acids, Figure 1) instead of using photolabile protecting groups. As a result, commercially available standard building blocks for both peptide and oligonucleotide synthesis can be used with this method.
SPOTTING MICROARRAY APPROACH
As previously indicated, in situ synthesis methods are mainly used to generate arrays of oligomeric compounds such as oligonucleotide and peptides. In contrast, the spotting microarray approach, in conjunction with a proper immobilization method, can generate many copies of the same chip more efficiently since the compounds need to be synthesized only once. Although macromolecules such as proteins, DNA, and larger peptides can easily be adsorbed onto solid surface through noncovalent interactions, immobilization of small organic molecules, short peptides, or simple sugars will require covalent attachment, preferably through chemoselective site-specific ligation.
IMMOBILIZATION VIA PHYSICAL ADSORPTION
The first and simplest type of immobilization is through surface adsorption. This approach is particularly useful for proteins, and it has been used in standard ELISA, dot blot, and Western blot for many years. The commonly used solid supports are hydrophobic plastics such as polystyrene. Protein microarrays have also been generated on an aminosilane cationic surface [22], nitrocellulose membrane [23], and Hybond ECL membrane [24, 25]. Through adsorption, Wang et al have prepared carbohydrate arrays on nitrocellulose bonded slides [26]. They discovered that the efficiency of adsorption of dextran to nitrocellulose increases with size. The 2000-kd dextran bound significantly stronger than the 20-kd dextran. Wang et al [27] have used HydroGel-coated slides for arraying 43 monoclonal antibodies (mAb) against cytokines and chemokines, and used a fluorescence-based multiplexed immunoassays to quantitate the level of these proteins. We spotted peptide-protein and peptide-agarose conjugates on polystyrene slide as a way to display peptides (Xu and Lam et al, unpublished data, May 2003). Fang et al [28] have developed a membrane protein microarray by spotting cell membrane preparations containing G-protein-coupled receptors on γ-aminopropylsilane-derivatized glass or gold-coated glass surface.
IMMOBILIZATION VIA SPECIFIC SURFACE INTERACTION
In addition to immobilization via nonspecific physical adsorption, molecules can be tagged and immobilized through specific noncovalent interactions between the tag and the already immobilized capturing molecule for the tag. A typical example is the biotin-streptavidin system for immobilizing biotinylated proteins onto streptavidin-coated surfaces [29]. Likewise, a small molecule can also be biotinylated and printed onto a surface that has been precoated with a monolayer of streptavidin or neutravidin. Neutravidin is sometimes preferred as it has less nonspecific interaction with other proteins. We were able to successfully print a microarray of biotinylated synthetic peptides onto a neutravidin-coated polystyrene microscope slide using a Wittech 03 arrayer (Wittech, Inc, Taiwan) [30]. Lesaicherre et al have recently reported [31] the derivatization of glass slides with avidin for immobilization of biotinylated peptides. The poly-His-Ni2+ system has also been reported for protein microarray [32]. In this method, proteins containing a poly-histidine tag were printed onto Ni2+-chelating surfaces. Another approach is to use anti-GST antibody or glutathione-coated slides to capture a series of GST-fusion proteins. Boronic acid groups are known to form very stable complex with some moieties such as cis or coaxial 1,2-diol. Immobilized phenylboronic acids have been used in chromatography [33] and protein immobilization [34]. Similar strategies can, in principle, be applied to protein or chemical microarrays as well.
IMMOBILIZATION VIA COVALENT ATTACHMENT
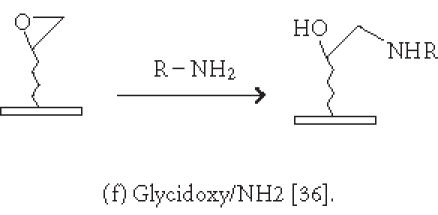
Although nonspecific physical adsorption has been used successfully for generating a microarray of macromolecules, this approach is less useful for the preparation of small molecule or small peptide microarrays. These small molecules can be conjugated to a tag which in turn binds to the immobilized capturing agent (see above). Alternatively, they can be immobilized via covalent attachment to a functional group on the solid surface. Figures 2 and 3 summarize some of the common chemistries used in generating microarrays by covalent attachment. Chemical modification of the solid surface is necessary to create functional groups for covalent immobilization and to achieve homogeneous immobilization. Commercially available aldehyde-derivatized glass slides that have been used for DNA immobilization can also be used for protein microarrays [35]. The aldehyde groups on the glass surface react with primary amines on the protein to form Schiff's base linkages. BSA is used to block the remaining unreacted aldehyde groups or other nonspecific binding sites. Zhu et al [32, 36] have described the use of a 3-glycidoxypropyltrimethoxysilane (GPTS) to activate polydimethylsiloxane (PDMS) on the slide surface prior to protein immobilization. Lin et al have reported [37] the printing of protein microarrays on an aminopropyltrimethoxysilane surface activated with bis-sulfosuccinimidyl suberate. Furthermore, Benters et al [38] have demonstrated the use of succinimidyl ester- or isocyanate-functionalized dendrimer on a solid surface for nucleic acid and protein microarrays.
Figure 2.
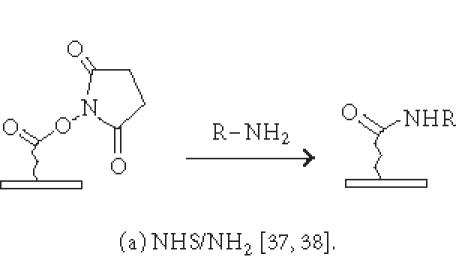
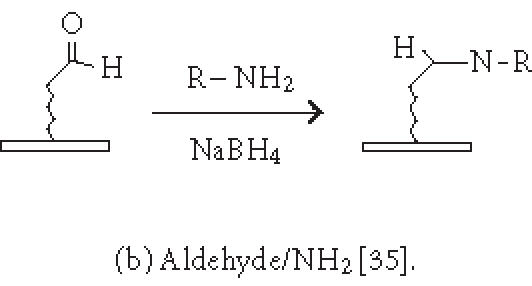
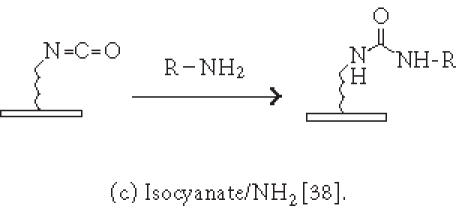
Chemistries of covalent immobilization (nonselective ligation).
Figure 3.
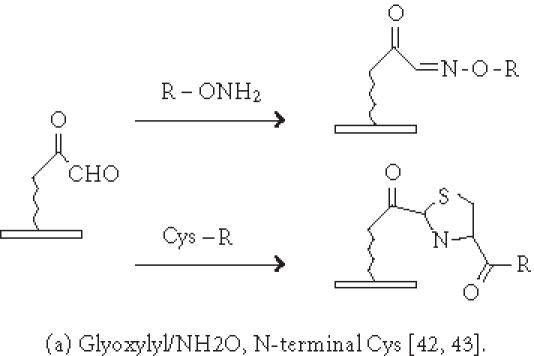
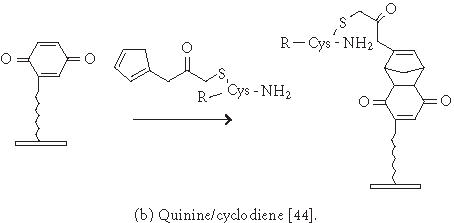
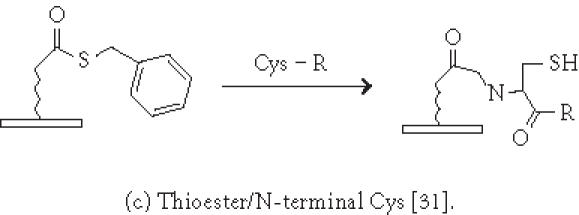
Chemistries of covalent immobilization (chemoselective ligation).
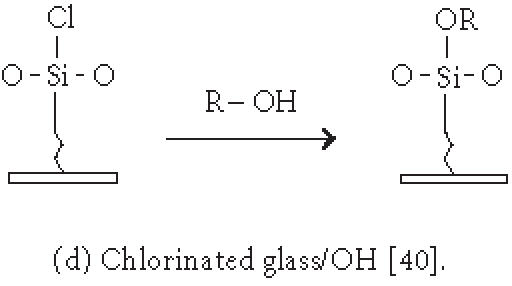
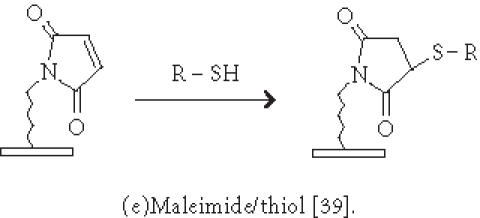
Immobilization of small molecules or short peptides often requires covalent linkage of the compounds onto the solid support. Michael addition has been used by Schreiber's group to ligate thiol-containing compounds to maleimide-derivatized glass slides to form a microarray of small molecules [39]. They have also described [40] the covalent attachment of alcohol-containing small molecule compounds onto chlorinated glass slides. In covalent immobilization of molecules, the site of ligation is often important, especially for molecules containing multiple biologically important functional groups. For example, if a peptide requires its free amino and sulfhydryl group to be biologically active, ligation with standard aldehyde, N-hydroxyl succimide, isocyanate, or maleimide chemistries will not work. Carlson and Beal [41] have observed that the attachment point of immobilized peptide-acridine conjugates significantly affects RNA binding. In our group, we have applied the highly selective chemoselective ligation reaction to covalently attach peptides or small molecules onto the glyoxylyl-derivatized glass slide via oxime bond or thiazolidine ring formation [42]. With this chemoselective ligation method, peptides or small molecules are immobilized on the solid support at a predetermined site (aminoxyl or 1,2 aminothiol functional groups), and other α-amino groups, ε-amino groups, or isolated thiol groups present in the molecules will not react. This eliminates the risk of inactivating the compounds during immobilization. We have recently reported a new strategy to significantly improve the loading of glyoxylyl groups onto glass surfaces by using acrylic acid as a starting material [43]. Further improvement of loading can be accomplished by adding hydrophilic linkers and bifurcating amino acids such as lysine between the glyoxylyl group and the glass surface (Xu and Lam, May 2002). Lesaicherre et al [31] have recently reported the derivatization of glass slides with thioester for chemoselective ligation of peptides with an N-terminal cysteine (1,2 aminothiol group) to form an amide bond. Houseman et al [44] have described a peptide chip prepared by the Diels-Alder-mediated immobilization of peptides on the quinone-functionalized surfaces. In this method, a cyclodiene moiety is incorporated into peptides for the purpose of immobilization. As an alternative to immobilizing peptide-protein conjugates onto plastic slides via physical adsorption, we have used a chemically derivatized protein scaffold (eg, glyoxylyl-functionalized BSA) to first coat the polystyrene slide via nonspecific adsorption. Peptides or small molecules are then printed onto the functionalized protein-coated slides. After incubation, these compounds are immobilized onto the coating surface via a site-specific ligation reaction. In some applications, immobilization of the ligand onto the glass surface via a long hydrophilic linker or a protein may be beneficial for biological activity. For example, we have found that a long linker is needed for phosphorylation of a peptide substrate by p60c-src protein tyrosine kinase [42].
CD, MICROFLUIDICS, MULTIPLEX-BEADS, AND FIBER-OPTIC MICROARRAYS
Kido et al [45] have developed a compact disc-based microarray system for immunoassays, which uses a piezoelectric inkjet applicator to generate a high-density antigen microarray (75-μm spot) on a polycarbonate disc. Competitive inhibition immunoassays with fluorescent antibodies are performed on the disc, and the final read-out is accomplished with a commercially available fluorescence scanner.
Microfluidics is another format of microarray that is based on microchannel systems. Shi et al [46] have developed a radial capillary array electrophoresis microplate system, which consists of 96 separate microfabricated separation channels that connect a central common anode reservoir to 96 injectors at the perimeter of the 10-cm diameter wafer. A laser-excited rotary confocal scanner with four-color detection channels is used for detection. A high-quality restriction map of the 96 samples can be obtained in less than 120 seconds with this device.
Han et al have described [47] the development of quantum-dot-tagged microbeads for multiplexed optical coding of biomolecules. This group immobilizes different peptides, proteins, or nucleic acids on various samples of microbeads into which different-size hydrophobic quantum dots are embedded. These quantum-dot-tagged microbeads are used in suspension in conjunction with flow cytometry to track the identity of the immobilized molecules when mixed with the analytes. These investigators have used DNA hybridization studies to demonstrate that the coding and target signals can be read at a single bead level simultaneously. Similar technology using traditional fluorophores to encode microbeads has been commercialized by Luminex (Austin, Tex).
Walt has pioneered the fiber-optic microarray biosensor technology, which is commercially known as Illumina BeadArray [48]. It involves the patterning of an array of wells at the tip of an optical imaging fiber bundle. The depth of each well can be controlled by etching time and the concentration of hydrofluoric acid used. The fiber-optic tip is then dipped into a suspension of a library of OBOC expressing microbeads (3 micron diameter). The chemicals on the microbeads can be synthesized by “split-mix” synthesis or through parallel synthesis. Alternatively, proteins can be adsorbed on the beads in parallel. Through surface tension, each of the microwells is filled with just one microbead. This random microbead array is decoded [49] for a specific application, prior to analysis of unknown samples. This technology has been applied for the detection of a variety of analytes ranging from DNA to organic vapors [50, 51, 52]. One major advantage of this method is that the packing density of the array is extremely high with center-to-center distance of each bead at 5 μm. However, the necessary decoding step is a major drawback.
PRINTING METHODS AND INSTRUMENTATION
Photolithography, mechanical microspotting, and inkjet application are the three general methods for microarray printing. The mask and maskless photolithography methods [3, 19, 20, 21] that involve in situ light-directed synthesis of oligomers can generate high-quality, high-density microarrays. The mechanical spotting (or surface contact) technique uses a pin to transfer liquid samples to the slides by direct surface contact. The pin-ring technique (eg, GSM 417 arrayer, Affymetrix) and the stamp microcontact technique (eg, SpotBot Personal Microarrayer, TeleChem International, Inc, Sunnyvale, Calif) allow for fairly reproducibly sized spots. The inkjet method employs an electric current to dispense the liquid sample onto the solid support. This noncontact piezoelectric technique accurately and rapidly dispenses monodisperse droplets. Moerman et al [53] have reported the use of electrospraying in a stable cone-jet mode to generate a highly reproducible spot of biological material 130–350 μm in diameter, and as small as 50 pL. More recently, Avseenko et al [54] have used the electrospray method to deposit dry protein onto a dextran-grafted surface, followed by incubation in a 100% humidity chamber. This method was able to generate a 0.6 × 0.6 mm2 array with each spot 30–40 μm in diameter and consisting of 28 different protein antigens and allergens. Ringeisen et al [55] have utilized a laser transfer technique, which allows accurate deposition of picoliter volumes of active proteins onto standard microarray substrates.
MICROARRAY APPLICATION
Table 1 summarizes the various aspects of the microarray technology: attachment chemistries, immobilized molecules, analytes, and detection methods. In most cases, the analytes that have been used to probe the microarray are complex mixtures of molecules: for example, mRNA or cDNA preparations, total cell extracts or intact cells, complex protein mixtures, body fluids such as serum or whole blood, fermentation broths, environmental samples, or microorganisms. In some cases, a pure protein, such as a protein kinase or a protease, is used to probe the peptide microarray to determine the substrate profile of these enzymes. Protein microarrays are frequently used to study different protein functions such as protein-protein interactions. Zhu et al [32] have identified many new calmodulin- and phospholipid-interacting proteins through assaying a microarray of 5800 yeast proteins using known proteins and phospholipids as probes. Similar technique was also used to evaluate protein substrate profile of 119 yeast protein kinases [36]. Sreekumar et al [56] have applied protein microarrays to discover novel radiation-regulated proteins. Huang et al [25] have used antibody arrays to quantitate a large number of different cytokines from sera and culture media. Knezevic et al [57] have reported antibody microarrays to analyze protein expression in cancer tissue of the oral cavity. Ziauddin and Sabatini have reported the use of plasmid DNA microarray to transfect cells in situ to form a new cell microarray with newly expressed proteins encoded by the plasmid DNA [15]. The use of protein microarrays for serum marker detection and discovery using prostate cancer as a model disease has recently been reviewed [58]. Mezzasoma et al [59] have reported the use of microbial antigen arrays to detect serum antibodies against the ToRCH antigens in a panel of characterized human sera. Antibody microarrays have been used for proteomic profiling of the cancer microenvironment [57]. Paweletz et al [60] have reported the development of the so-called reverse-phase protein microarrays to quantitate proteins derived from microdissected tissues. They have immobilized serial dilution of total cell lysates from microdissected tissues on nitrocellulose-coated glass slides and have used different enzyme-linked antibodies to probe specific proteins or phosphoproteins.
Table 1.
Summary of microarray methods and detection techniques.
| Attachment chemistries |
| -In situ synthesis via covalent bond: spot synthesis [3, 17]; light-directed parallel synthesis [2, 19, 20, 21]. |
| -Nonspecific adsorption: polystyrene or polymer-coated surface [23, 24, 25, 26]; glass surface, and cationic surface amino groups [22]. |
| -Nonspecific covalent attachment via activated surfaces: aldehyde [35]; succinimidyl ester [37, 38]; isocyanate [38]; glycidoxy [36]; chlorine [40]. |
| -Chemoselective ligation via activated surfaces: glyoxylyl [42, 43]; quinone [44]; thioester [31]; maleimide [39]. |
| Arrayed molecules: mode of immobilization |
| -Direct link with solid support: DNAs [6, 7]; target proteins [32, 59]; antibodies [6, 19, 25, 20, 57]; alcohol-containing small molecules [40]; carbohydrates [26]; thiolated small molecules [39]. |
| -Indirect link with solid support via immobilized capturing molecules: anti-tag Ab/tagged-target proteins, streptavidin/biotin-target proteins, streptavidin/biotin-carbohydrates, streptavidin/biotin-peptides [31, 65, 42]; streptavidin/biotin-organic molecules [42]; glutathione/GST-fusion proteins, Ni/His tag proteins [32]; gold/thiolated DNAs, peptides, and proteins. |
| Analytes |
| mRNAs, cDNAs, total cell extract [57]; protein mixture, body fluid, serum [25, 58, 59]; environmental sample [58]; pure enzyme, microorganisms, and intact cells [42, 65]. |
| Detection methods |
| Fluorescence, fluorescent-quenching, chemiluminescence, luminescence, FRET, color-dye, enzyme-linked, radiolabel: Phosphoryl imager or autoradiogram [42]; scintillation proximity [71, 72]; protease activity, protein kinase activity [31, 42, 44, 66, 71]; plasmon resonance spectroscopy [70]; whole cell [42, 61]; and cell function [42]. |
Park and Clark [61] have described a sol-gel-encapsulated enzyme array to screen biocatalytic activity or enzyme inhibition. Rakow and Suslick [62] have developed a colorimetric sensor array for detection of volatile chemicals at a concentration below two parts per million. A chemical microarray technique has also been applied to monitor chemical reactions by determining the enantiomeric excess of thousands of samples [63].
All the assay methods developed for on-bead screening of OBOC combinatorial libraries [4, 64] are applicable to chemical microarrays. In our laboratory, we use the OBOC combinatorial library method (100,000 to a few million different compounds per library) to identify ligands or substrates that are of biological interest. The positive hits are then resynthesized and spotted in a microarray format, in multiple replicate sets for subsequent probing with a number of different analytes and under different conditions. In principal, this stepwise approach will enable us to focus our attention on a finite number of ligands that can subsequently be characterized and developed into a diagnostic chip. For example, we have used whole cell binding assay to screen random peptide libraries for leukemia cell binding ligands. The positive ligands are then resynthesized, immobilized on plastic or glass slides, and these peptide microarrays can be used to probe whole blood derived from patients with leukemia [65]. Our ultimate goal is to develop microarrays of cancer targeting peptides that can be probed, allowing physicians to rapidly identify the therapeutic peptide cocktail effective for a specific patient. Similarly, we have used an on-bead functional assay to screen OBOC combinatorial libraries for protein kinase substrates [66]. Unique peptide substrates for a number of protein kinases have been identified using this approach [67, 68, 69, 70]. Immobilization of these peptides with a long hydrophilic linker to form a peptide microarray, in principle, will enable the development of protein kinase substrate chips [42]. These chips can be used to profile the protein kinase activities of whole cell extracts derived from biopsy samples of cancer patients. Other groups have also reported the use of peptide array to profile protein kinase activities [31, 44, 71].
METHODS OF DETECTION
Standard immunodetection techniques, such as enzyme-linked colorimetric, fluorescent, FRET, chemiluminescence, or luminescence methods, are useful to analyze chemical microarrays. As mentioned above, arrayed peptides or protein substrates can be phosphorylated by protein kinases in the presence of [γ 33P]ATP, followed by detection with autoradiography or by a phosphor imager [42, 66]. Cell adhesion assays can be performed on a microarray and detected by microscopy using cell staining [42, 65]. One very useful detection method that does not need reporter systems or tags is surface plasmon resonance (SPR) spectroscopy [72]. SPR applies unlabeled probes (eg, antibodies, proteins, and drug candidates) to the surface of a gold-coated glass chip where testing molecules are immobilized. The chip is scanned from below by a light beam. The beam is reflected back by the gold layer, and the angle of reflection varies according to the mass of the molecules attached. Compounds captured by the immobilized array can therefore be located and quantitated by measuring this angle on each array spot or every pixel corresponding to the entire surface of the slide. Furthermore, this technology may also provide a measure of the affinity as well as the “on” and “off” rate of binding between the capturing agents and the captured molecules. Morozov et al [73] have described the use of a charge-coupled device to quantitatively detect isotope-labeled ligands bound to a protein microarray. Although it has not been applied in microarray detection, in principle, a homogenous assay such as scintillation proximity assay [74, 75] can also be used. In this case the analyte has to be radiolabeled (eg, by 35S), and the surface of the slide coated with a layer of scintillant.
PERSPECTIVES AND CHALLENGES
Microarray technologies enable the evaluation of thousands to tens of thousands of molecular interactions simultaneously in a high-throughput manner. Microarrays have made significant impact on biology, medicine, drug discovery, and many other related fields and are considered indispensable in genomic and proteomic research pursuits. In the field of drug discovery, microarray techniques can be utilized to identify drug targets that are unique to a disease. Chemical microarray, a form of combinatorial libraries, can also be used for lead identification, as well as optimization of these leads. The impact of microarray on medicine in the future will be significant. Just to mention a few, genetic diseases will be routinely diagnosed and confirmed by gene chips; hundreds to thousands of blood tests will be performed simultaneously on a chip using a few drops of blood from the patient; nucleic acids or proteins derived from a cancer specimen will be analyzed on a chip and a correct diagnosis will be made immediately and, based on the analysis, target-specific anticancer drugs will be prescribed by the physician. In this era of bioterrorism, the development of a chip capable of detecting a multitude of biological or chemical agents in the environment will be of great interest to the law enforcement agencies.
Challenges in this field include the development of novel material or surfaces with minimal nonspecific binding of biological molecule and yet allow for specific ligation of the testing molecule on the surface. Better, more site-specific ligation chemistries for immobilization of synthetic ligands or proteins to the solid support must also be developed. Although it has been reported that compounds derived from individual beads of the OBOC library can be recovered and immobilized on glass surface to form multiple replicates of chemical microarray [76, 77], consistent recovery of enough material from every bead and efficient ligation of the minute amount of material to the glass surface in a site-specific manner remains a big challenge.
Homogenous assays are popular in drug screen but wide application of these detection approaches to microarrays needs to be developed. In principle, the use of plasmon resonance spectroscopy to analyze the entire microarray for real-time association and dissociation among every spot is feasible, but successful development and commercialization of such an instrument remains to be developed. Mass spectrometry, in principle, can be used to (a) identify the molecular masses of all the captured molecules that bind to each of the microarray spot without the requirement of labeling or amplification [78] and (b) determine the amino acid sequence of some of the peptide fragments obtained from the captured proteins. However, to achieve these goals, a more sensitive mass spectrometer with special labeling techniques and sampling devices will be needed. Microarray technologies have already proven to be invaluable in the field of genomics and proteomics and solutions to these challenges will undoubtedly facilitate the development of clinically useful diagnostic chips in the foreseeable future.
Acknowledgments
ACKNOWLEDGMENT
We would like to thank Amanda Enstrom for editorial assistance. This work was supported by Grants NIH CA78868, NIH CA78909, and NIH CA86364.
References
- Geysen H M, Meloen R H, Barteling S J. Use of peptide synthesis to probe viral antigens for epitopes to a resolution of a single amino acid. Proc Natl Acad Sci USA. 1984;81(13):3998–4002. doi: 10.1073/pnas.81.13.3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor S P, Read J L, Pirrung M C, Stryer L, Lu A T, Solas D. Light-directed, spatially addressable parallel chemical synthesis. Science. 1991;251(4995):767–773. doi: 10.1126/science.1990438. [DOI] [PubMed] [Google Scholar]
- Frank R. Spot-synthesis: An easy technique for the positionally addressable, parallel chemical synthesis on a membrane support. Tetrahedron. 1992;48(42):9217–9232. [Google Scholar]
- Lam K S, Salmon S E, Hersh E M, Hruby V J, Kazmierski W M, Knapp R J. A new type of synthetic peptide library for identifying ligand-binding activity. Nature. 1991;354(6348):82–84. doi: 10.1038/354082a0. [DOI] [PubMed] [Google Scholar]
- Pease A C, Solas D, Sullivan E J, Cronin M T, Holmes C P, Fodor S P.A. Light-generated oligonucleotide arrays for rapid DNA sequence analysis. Proc Natl Acad Sci USA. 1994;91(11):5022–5026. doi: 10.1073/pnas.91.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena M, Shalon D, Davis R W, Brown P O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270(5235):467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Schena M, Shalon D, Heller R, Chai A, Brown P O, Davis R W. Parallel human genome analysis: microarray-based expression monitoring of 1000 genes. Proc Natl Acad Sci USA. 1996;93(20):10614–10619. doi: 10.1073/pnas.93.20.10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart D J, Winzeler E A. Genomics, gene expression and DNA arrays. Nature. 2000;405(6788):827–836. doi: 10.1038/35015701. [DOI] [PubMed] [Google Scholar]
- Kurella M, Hsiao L L, Yoshida T, et al. DNA microarray analysis of complex biologic processes. J Am Soc Nephrol. 2001;12(5):1072–1078. doi: 10.1681/ASN.V1251072. [DOI] [PubMed] [Google Scholar]
- Cuzin M. DNA chips: a new tool for genetic analysis and diagnostics. Transfus Clin Biol. 2001;8(3):291–296. doi: 10.1016/s1246-7820(01)00141-0. [DOI] [PubMed] [Google Scholar]
- Maughan N J, Lewis F A, Smith V. An introduction to arrays. J Pathol. 2001;195(1):3–6. doi: 10.1002/path.924. [DOI] [PubMed] [Google Scholar]
- Marton M J, DeRisi J L, Bennett H A, et al. Drug target validation and identification of secondary drug target effects using DNA microarrays. Nat Med. 1998;4(11):1293–1301. doi: 10.1038/3282. [DOI] [PubMed] [Google Scholar]
- Lam K S, Renil M. From combinatorial chemistry to chemical microarray. Curr Opin Chem Biol. 2002;6(3):358–353. doi: 10.1016/s1367-5931(02)00326-5. [DOI] [PubMed] [Google Scholar]
- Khandurina J, Guttman A. Microchip-based high-throughput screening analysis of combinatorial libraries. Curr Opin Chem Biol. 2002;6:359–366. doi: 10.1016/s1367-5931(02)00323-x. [DOI] [PubMed] [Google Scholar]
- Ziauddin J, Sabatini D M. Microarrays of cells expressing defined cDNAs. Nature. 2001;411(6833):107–110. doi: 10.1038/35075114. [DOI] [PubMed] [Google Scholar]
- Moch H, Kononen T, Kallioniemi O P, Sauter G. Tissue microarrays: what will they bring to molecular and anatomic pathology? Adv Anat Pathol. 2001;8(1):14–20. doi: 10.1097/00125480-200101000-00002. [DOI] [PubMed] [Google Scholar]
- Reineke U, Volkmer-Engert R, Schneider-Mergener J. Applications of peptide arrays prepared by the SPOT-technology. Curr Opin Biotechnol. 2001;12(1):59–64. doi: 10.1016/s0958-1669(00)00178-6. [DOI] [PubMed] [Google Scholar]
- McGall G, Labadie J, Brock P, Wallraff G, Nguyen T, Hinsberg W. Light-directed synthesis of high-density oligonucleotide arrays using semiconductor photoresists. Proc Natl Acad Sci USA. 1996;93(24):13555–13560. doi: 10.1073/pnas.93.24.13555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh-Gasson S, Green R D, Yue Y, et al. Maskless fabrication of light-directed oligonucleotide microarrays using a digital micromirror array. Nat Biotechnol. 1999;17(10):974–978. doi: 10.1038/13664. [DOI] [PubMed] [Google Scholar]
- LeProust E, Pellois J P, Yu P, et al. Digital light-directed synthesis. A microarray platform that permits rapid reaction optimization on a combinatorial basis. J Comb Chem. 2000;2(4):349–354. doi: 10.1021/cc000009x. [DOI] [PubMed] [Google Scholar]
- Pellois J P, Wang W, Gao X. Peptide synthesis based on t-Boc chemistry and solution photogenerated acids. J Comb Chem. 2000;2(4):355–360. doi: 10.1021/cc0000139. [DOI] [PubMed] [Google Scholar]
- Martin B D, Gaber B P, Patterson C H, Turner D C. Direct protein microarray fabrication using a hydrogel stamper. Langmuir. 1998;14:3971–3975. [Google Scholar]
- Ge H. UPA, a universal protein array system for quantitative detection of protein-protein, protein-DNA, protein-RNA and protein-ligand interactions. Nucleic Acids Res. 2000;28(2):e3. doi: 10.1093/nar/28.2.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R-P. Detection of multiple proteins in an antibody-based protein microarray system. J Immunol Methods. 2001;255(1-2):1–13. doi: 10.1016/s0022-1759(01)00394-5. [DOI] [PubMed] [Google Scholar]
- Huang R-P, Huang R, Fan Y, Lin Y. Simultaneous detection of multiple cytokines from conditioned media and patient's sera by an antibody-based protein array system. Anal Biochem. 2001;294(1):55–62. doi: 10.1006/abio.2001.5156. [DOI] [PubMed] [Google Scholar]
- Wang D, Liu S, Trummer B J, Deng C, Wang A. Carbohydrate microarrays for the recognition of cross-reactive molecular markers of microbes and host cells. Nat Biotechnol. 2002;20(3):275–281. doi: 10.1038/nbt0302-275. [DOI] [PubMed] [Google Scholar]
- Wang C C, Huang R-P, Sommer M, et al. Array-based multiplexed screening and quantitation of human cytokines and chemokines. J Proteome Res. 2002;1(4):337–343. doi: 10.1021/pr0255203. [DOI] [PubMed] [Google Scholar]
- Fang Y, Frutos A G, Lahiri J. Membrane protein microarrays. J Am Chem Soc. 2002;124(11):2394–2395. doi: 10.1021/ja017346+. [DOI] [PubMed] [Google Scholar]
- Ruiz-Taylor L A, Martin T L, Zaugg F G, et al. Monolayers of derivatized poly(L-lysine)-grafted poly(ethylene glycol) on metal oxides as a class of biomolecular interfaces. Proc Natl Acad Sci USA. 2001;98(3):852–857. doi: 10.1073/pnas.98.3.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aina O H, Sroka T C, Chen M L, Lam K S. Therapeutic cancer targeting peptides. Biopolymers. 2002;66(3):184–199. doi: 10.1002/bip.10257. [DOI] [PubMed] [Google Scholar]
- Lesaicherre M L, Uttamchandani M, Chen G Y.J, Yao S Q. Developing site-specific immobilization strategies of peptides in a microarray. Bioorg Med Chem Lett. 2002;12(16):2079–2083. doi: 10.1016/s0960-894x(02)00379-7. [DOI] [PubMed] [Google Scholar]
- Zhu H, Bilgin M, Bangham R, et al. Global analysis of protein activities using proteome chips. Science. 2001;293(5537):2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- Singhal R P, DeSilva S S.M. Boronate affinity chromatography. Adv Chromatogr. 1992;31:293–335. [PubMed] [Google Scholar]
- Stolowitz M L, Ahlem C, Hughes K A, et al. Phenylboronic acid-salicylhydroxamic acid bioconjugates. 1. A novel boronic acid complex for protein immobilization. Bioconjug Chem. 2001;12(2):229–239. doi: 10.1021/bc0000942. [DOI] [PubMed] [Google Scholar]
- MacBeath G, Schreiber S L. Printing proteins as microarrays for high-throughput function determination. Science. 2000;289(5485):1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- Zhu H, Klemic J F, Chang S, et al. Analysis of yeast protein kinases using protein chips. Nat Genet. 2000;26(3):283–289. doi: 10.1038/81576. [DOI] [PubMed] [Google Scholar]
- Lin S C, Tseng F G, Huang H M, Huang C Y, Chieng C C. Microsized 2D protein arrays immobilized by micro-stamps and micro-wells for disease diagnosis and drug screening. Fresenius J Anal Chem. 2001;371(2):202–208. doi: 10.1007/s002160100991. [DOI] [PubMed] [Google Scholar]
- Benters R, Niemeyer C M, Wohrle D. Dendrimer-activated solid supports for nucleic acid and protein microarrays. Chembiochem. 2001;2(9):686–694. doi: 10.1002/1439-7633(20010903)2:9<686::AID-CBIC686>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- MacBeath G, Koehler A N, Schreiber S L. Printing small molecules as microarrays and detecting protein-ligand interactions en masse. J Am Chem Soc. 1999;121:7967–7968. [Google Scholar]
- Hergenrother P J, Depew K M, Schreiber S L. Small-molecule microarrays: Covalent attachment and screening of alcohol containing small molecules on glass slides. J Am Chem Soc. 2000;122:7849–7850. [Google Scholar]
- Carlson C B, Beal P A. Point of attachment and sequence of immobilized peptide-acridine conjugates control affinity for nucleic acids. J Am Chem Soc. 2002;124(29):8510–8511. doi: 10.1021/ja026029f. [DOI] [PubMed] [Google Scholar]
- Falsey J R, Renil M, Park S, Li S, Lam K S. Peptide and small molecule microarray for high throughput cell adhesion and functional assays. Bioconjug Chem. 2001;12(3):346–353. doi: 10.1021/bc000141q. [DOI] [PubMed] [Google Scholar]
- Xu Q, Lam K S. An efficient approach to prepare glyoxylyl functionality on solid-support. Tetrahedron Letters. 2002;43(25):4435–4437. [Google Scholar]
- Houseman B T, Huh J H, Kron S J, Mrksich M. Peptide chips for the quantitative evaluation of protein kinase activity. Nat Biotechnol. 2002;20(3):270–274. doi: 10.1038/nbt0302-270. [DOI] [PubMed] [Google Scholar]
- Kido H, Maquieira A, Hammock B D. Disc-based immunoassay microarrays. Anal Chim Acta. 2000;411(1-2):1–11. [Google Scholar]
- Shi Y, Simpson P C, Scherer J R, et al. Radial capillary array electrophoresis microplate and scanner for high-performance nucleic acid analysis. Anal Chem. 1999;71(23):5354–5361. doi: 10.1021/ac990518p. [DOI] [PubMed] [Google Scholar]
- Han M, Gao X, Su J Z, Nie S. Quantum-dot-tagged microbeads for multiplexed optical coding of biomolecules. Nat Biotechnol. 2001;19(7):631–635. doi: 10.1038/90228. [DOI] [PubMed] [Google Scholar]
- Walt D R. Techview: molecular biology. Bead-based fiber-optic arrays. Science. 2000;287(5452):451–452. doi: 10.1126/science.287.5452.451. [DOI] [PubMed] [Google Scholar]
- Albert K J, Gill D S, Pearce T C, Walt D R. Automatic decoding of sensor types within randomly ordered, high-density optical sensor arrays. Anal Bioanal Chem. 2002;373(8):792–802. doi: 10.1007/s00216-002-1406-8. [DOI] [PubMed] [Google Scholar]
- Albert K J, Walt D R. High-speed fluorescence detection of explosives-like vapors. Anal Chem. 2000;72(9):1947–1955. doi: 10.1021/ac991397w. [DOI] [PubMed] [Google Scholar]
- Ferguson J A, Steemers F J, Walt D R. High-density fiber-optic DNA random microsphere array. Anal Chem. 2000;72(22):5618–5624. doi: 10.1021/ac0008284. [DOI] [PubMed] [Google Scholar]
- Stitzel S E, Cowen L J, Albert K J, Walt D R. Array-to-array transfer of an artificial nose classifier. Anal Chem. 2001;73(21):5266–5271. doi: 10.1021/ac010111w. [DOI] [PubMed] [Google Scholar]
- Moerman R, Frank J, Marijnissen J C.M, Schalkhammer T G.M, van Dedem G W.K. Miniaturized electrospraying as a technique for the production of microarrays of reproducible micrometer-sized protein spots. Anal Chem. 2001;73(10):2183–2189. doi: 10.1021/ac001041g. [DOI] [PubMed] [Google Scholar]
- Avseenko N V, Morozova T Y, Ataullakhanov F I, Morozov V N. Immunoassay with multicomponent protein microarrays fabricated by electrospray deposition. Anal Chem. 2002;74(5):927–933. doi: 10.1021/ac010970k. [DOI] [PubMed] [Google Scholar]
- Ringeisen B R, Wu P K, Kim H, et al. Picoliter-scale protein microarrays by laser direct write. Biotechnol Prog. 2002;18(5):1126–1129. doi: 10.1021/bp015516g. [DOI] [PubMed] [Google Scholar]
- Sreekumar A, Nyati M K, Varambally S, et al. Profiling of cancer cells using protein microarrays: discovery of novel radiation-regulated proteins. Cancer Res. 2001;61(20):7585–7593. [PubMed] [Google Scholar]
- Knezevic V, Leethanakul C, Bichsel V E, et al. Proteomic profiling of the cancer microenvironment by antibody arrays. Proteomics. 2001;1(10):1271–1278. doi: 10.1002/1615-9861(200110)1:10<1271::AID-PROT1271>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Miller J C, Butler E B, Teh B S, Haab B B. The application of protein microarrays to serum diagnostics: prostate cancer as a test case. Dis Markers. 2001;17(4):225–234. doi: 10.1155/2001/272394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezzasoma L, Bacarese-Hamilton T, Di Cristina M, Rossi R, Bistoni F, Crisanti A. Antigen microarrays for serodiagnosis of infectious diseases. Clin Chem. 2002;48(1):121–130. [PubMed] [Google Scholar]
- Paweletz C P, Charboneau L, Bichsel V E, et al. Reverse phase protein microarrays which capture disease progression show activation of pro-survival pathways at the cancer invasion front. Oncogene. 2001;20(16):1981–1989. doi: 10.1038/sj.onc.1204265. [DOI] [PubMed] [Google Scholar]
- Park C B, Clark D S. Sol-gel encapsulated enzyme arrays for high-throughput screening of biocatalytic activity. Biotechnol Bioeng. 2002;78(2):229–235. doi: 10.1002/bit.10238. [DOI] [PubMed] [Google Scholar]
- Rakow N A, Suslick K S. A colorimetric sensor array for odour visualization. Nature. 2000;406(6797):710–713. doi: 10.1038/35021028. [DOI] [PubMed] [Google Scholar]
- Korbel G A, Lalic G, Shair M D. Reaction microarrays: a method for rapidly determining the enantiomeric excess of thousands of samples. J Am Chem Soc. 2001;123(2):361–362. doi: 10.1021/ja0034747. [DOI] [PubMed] [Google Scholar]
- Lam K S, Lebl M, Krchnak V. The “one-bead-one-compound” combinatorial library method. Chem Rev. 1997;97(2):411–448. doi: 10.1021/cr9600114. [DOI] [PubMed] [Google Scholar]
- Healey B G, Walt D R. Fast temporal response fiber-optic chemical sensors based on the photodeposition of micrometer-scale polymer arrays. Anal Chem. 1997;69(11):2213–2216. doi: 10.1021/ac961058s. [DOI] [PubMed] [Google Scholar]
- Wu J, Ma Q N, Lam K S. Identifying substrate motifs of protein kinases by a random library approach. Biochemistry. 1994;33(49):14825–14833. doi: 10.1021/bi00253a022. [DOI] [PubMed] [Google Scholar]
- Lam K S, Wu J, Lou Q. Identification and characterization of a novel synthetic peptide substrate specific for src-family protein tyrosine kinases. Int J Pept Protein Res. 1995;45(6):587–592. doi: 10.1111/j.1399-3011.1995.tb01323.x. [DOI] [PubMed] [Google Scholar]
- Lou Q, Leftwich M E, Lam K S. Identification of GIYWHHY as a novel peptide substrate for human p60c-src protein tyrosine kinase. Bioorg Med Chem. 1996;4(5):677–682. doi: 10.1016/0968-0896(96)00063-6. [DOI] [PubMed] [Google Scholar]
- Wu J J, Phan H, Lam K S. Comparison of the intrinsic kinase activity and substrate specificity of c-Abl and Bcr-Abl. Bioorg Med Chem Lett. 1998;8(17):2279–2284. doi: 10.1016/s0960-894x(98)00413-2. [DOI] [PubMed] [Google Scholar]
- Wu J J, Afar D E, Phan H, Witte O N, Lam K S. Recognition of multiple substrate motifs by the c-Abl protein tyrosine kinase. Comb Chem High Throughput Screen. 2002;5(1):83–91. doi: 10.2174/1386207023330516. [DOI] [PubMed] [Google Scholar]
- Lesaicherre M L, Uttamchandani M, Chen G Y, Yao S Q. Antibody-based fluorescence detection of kinase activity on a peptide array. Bioorg Med Chem Lett. 2002;12(16):2085–2088. doi: 10.1016/s0960-894x(02)00378-5. [DOI] [PubMed] [Google Scholar]
- Rich R L, Myszka D G. Advances in surface plasmon resonance biosensor analysis. Curr Opin Biotechnol. 2000;11(1):54–61. doi: 10.1016/s0958-1669(99)00054-3. [DOI] [PubMed] [Google Scholar]
- Morozov V N, Gavryushkin A V, Deev A A. Direct detection of isotopically labeled metabolites bound to a protein microarray using a charge-coupled device. Biochem Biophys Methods. 2002;51(1):57–67. doi: 10.1016/s0165-022x(01)00245-7. [DOI] [PubMed] [Google Scholar]
- Cook N D. Scintillation proximity assays: a versatile high throughput screening technology. Drug Discov Today. 1996;1(7):287–294. [Google Scholar]
- Picardo M, Hughes K T. Marcel Dekker; New York, NY: 1997. Scintillation proximity assays; pp. 307–316. High Throughput Screening . [Google Scholar]
- Blackwell H E, Pérez L, Stavenger R A, et al. A one-bead, one-stock solution approach to chemical genetics: part 1. Chem Biol. 2001;8(12):1167–1182. doi: 10.1016/s1074-5521(01)00085-0. [DOI] [PubMed] [Google Scholar]
- Clemons P A, Koehler A N, Wagner B K, et al. A one-bead, one-stock solution approach to chemical genetics: part 2. Chem Biol. 2001;8(12):1183–1195. doi: 10.1016/s1074-5521(01)00086-2. [DOI] [PubMed] [Google Scholar]
- Fung E T, Thulasiraman V, Weinberger S R, Dalmasso E A. Protein biochips for differential profiling. Curr Opin Biotechnol. 2001;12(1):65–69. doi: 10.1016/s0958-1669(00)00167-1. [DOI] [PubMed] [Google Scholar]


