Abstract
Rationale: Patterns of longitudinal lung function growth and decline in childhood asthma have been shown to be important in determining risk for future respiratory ailments including chronic airway obstruction and chronic obstructive pulmonary disease.
Objectives: To determine the genetic underpinnings of lung function patterns in subjects with childhood asthma.
Methods: We performed a genome-wide association study of 581 non-Hispanic white individuals with asthma that were previously classified by patterns of lung function growth and decline (normal growth, normal growth with early decline, reduced growth, and reduced growth with early decline). The strongest association was also measured in two additional cohorts: a small asthma cohort and a large chronic obstructive pulmonary disease metaanalysis cohort. Interaction between the genomic region encompassing the most strongly associated single-nucleotide polymorphism and nearby genes was assessed by two chromosome conformation capture assays.
Measurements and Main Results: An intergenic single-nucleotide polymorphism (rs4445257) on chromosome 8 was strongly associated with the normal growth with early decline pattern compared with all other pattern groups (P = 6.7 × 10−9; odds ratio, 2.8; 95% confidence interval, 2.0–4.0); replication analysis suggested this variant had opposite effects in normal growth with early decline and reduced growth with early decline pattern groups. Chromosome conformation capture experiments indicated a chromatin interaction between rs4445257 and the promoter of the distal CSMD3 gene.
Conclusions: Early decline in lung function after normal growth is associated with a genetic polymorphism that may also protect against early decline in reduced growth groups.
Clinical trial registered with www.clinicaltrials.gov (NCT00000575).
Keywords: CSMD3, asthma, chronic obstructive pulmonary disease, longitudinal lung function patterns, genome-wide association studies
At a Glance Commentary
Scientific Knowledge on the Subject
In people with asthma, patterns of lung function growth and decline from early childhood through early adulthood, including reduced growth and early decline, may be indicative of future chronic airway obstruction, including development of chronic obstructive pulmonary disease in later life. These patterns may also lead to asthma–chronic obstructive pulmonary disease overlap syndrome because they identify young subjects with asthma meeting respiratory criteria for chronic obstructive pulmonary disease. A genetic investigation into these patterns is warranted.
What This Study Adds to the Field
We demonstrate evidence of a genetic effect on abnormal longitudinal lung function, integrating genetic, genomic, and chromatin interaction data. Using replication populations, we present evidence that the minor allele of rs4445257 on chromosome 8 is a risk factor for early decline following normal growth of lung function, but protective of early decline following reduced growth.
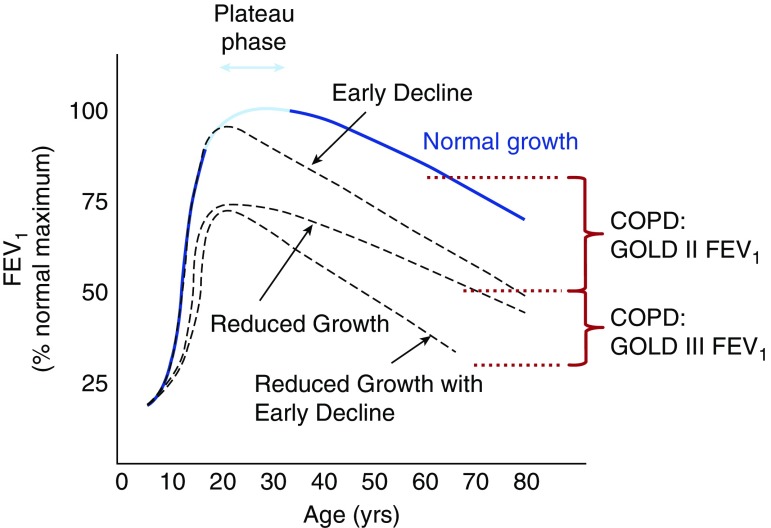
The natural history of lung function, measured by FEV1, in normal individuals is characterized by swift increase in adolescence, a leveling-off or plateauing in early adulthood, and a gradual decline into middle and old age (Figure 1) (1). In individuals with lung disease, including asthma, divergences from the canonical pattern can manifest as reduced growth, early decline, rapid decline, or a combination of these (2). A pattern of FEV1 growth and decline characterized by early decline has been associated with smoking and respiratory symptoms (3, 4), and reduced level of lung function has been associated with increased incidence of later-life chronic obstructive pulmonary disease (COPD) (5).
Figure 1.
Longitudinal lung function trajectories. Possible lung function trajectories over a person’s lifetime are shown; the lung function is plotted for each age as the percentage of the maximum FEV1 obtained for a normal individual (maximum usually attained in the 18- to 30-yr range). Normal lung function growth and decline (Normal growth) is characterized by a steep increase in adolescence, a plateau in early adulthood, and a gradual decline into old age. Abnormal trajectories (Reduced Growth, Early Decline, and Reduced Growth with Early Decline) are also shown. The red dotted line and red brace indicate levels of FEV1 that meet spirometric criteria for chronic obstructive pulmonary disease (COPD) Global Initiative for Chronic Obstructive Lung Disease (GOLD) stages 2 and 3.
Apart from the longitudinal patterns of lung growth and decline, reduced FEV1 is of clinical importance, because this is associated with later chronic airway obstruction (6–8) and with increased mortality (9). Reduced lung function is frequently associated with asthma incidence, including asthma recurrence and recurrent wheeze (10). Airway function is of particular interest in young people with asthma, a population at risk for chronic airway obstruction (8, 11–14).
A low level of lung function tends to remain stable during aging (15). Low FEV1, relative to a person’s age, sex, height, and race/ethnicity, tends to persist and track with growth from infancy or early childhood and into adulthood (16). Among genetic variants associated with asthma and COPD, some have subsequently been associated with rates of lung function decline among adults (17), whereas other single-nucleotide polymorphisms (SNPs) associated with reduced maximal lung function in the general population were not associated with decline of FEV1 in a large metaanalysis (18). Parallel genome-wide association studies (GWASs) of lung function in adults with asthma and in normal adults found little overlap among their strongest associations (18, 19).
We have previously investigated the determinants of FEV1 pattern and natural history of participants of the CAMP (Childhood Asthma Management Program) cohort, who were recruited at ages 5–12 years and followed for up to 18 years (2). We report here a GWAS of the impact of SNPs on longitudinal lung function growth patterns. Association results were extended to the Dutch Asthma Genetics cohort of young-adult subjects with asthma. Further association of candidate genetic variants in a COPD GWAS metaanalysis cohort provided generalization of these effects to a later-life low-FEV1 sample that realized the lung function endpoint projected for those with childhood asthma experiencing premature lung function decline. Some of the results of this study have been previously reported in the form of an abstract (20).
Methods
Cohort Methods
CAMP was a randomized, placebo-controlled trial of inhaled antiinflammatory treatments for mild to moderate childhood asthma followed by three phases of observational follow-up; the trial and all follow-up phases included at least annual spirometry (21, 22). A total of 1,041 participants enrolled in the trial between 1993 and 1995 at age 5–12 years; follow-up continued to 2012 when participants were age 22–30.
Prebronchodilator FEV1 values obtained on subjects without asthma in National Health and Nutrition Examination Survey (NHANES) III (23) adjusted for age, race/ethnicity, sex, and height were used to categorize CAMP participants into four patterns of lung function growth and decline: (1) normal growth with a normal plateau or maximum not yet reached (NG), (2) normal growth with early decline (NG/ED), (3) reduced growth with a normal plateau or maximum not yet reached (RG), and (4) reduced growth with early decline (RG/ED). NG was defined as a FEV1 growth curve predominantly above the 25th percentile of NHANES III, whereas RG was defined as a FEV1 growth curve below the 25th percentile. The presence of ED was indicated by a decrease from maximal level earlier than expected per NHANES III normal FEV1 growth curves. For further details, see McGeachie and coworkers (2).
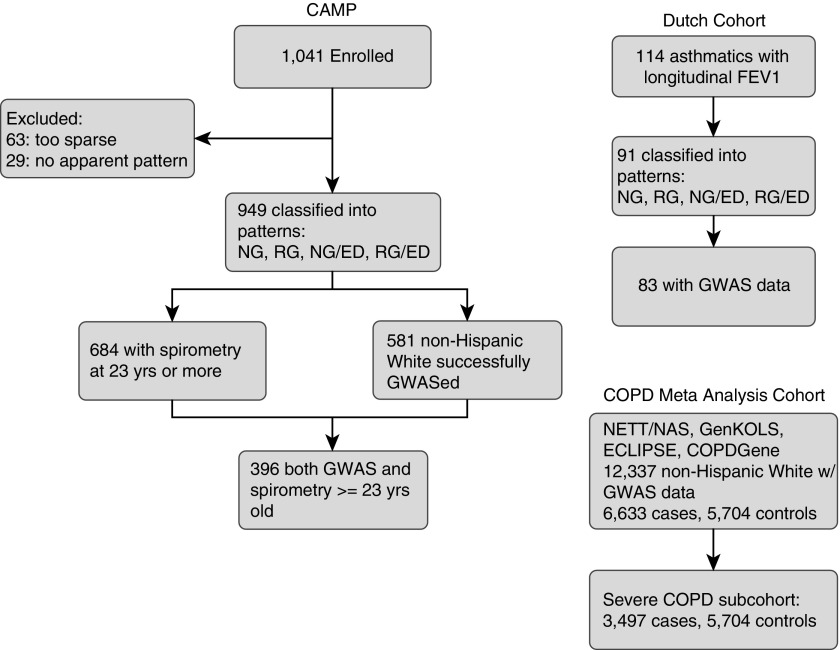
Of 1,041 CAMP participants, 63 were omitted for FEV1 measures too sparse to classify and 29 were omitted for FEV1 growth trajectories not matching any pattern. The 949 remaining participants were classified into one of the four patterns. Of these 949, a total of 684 (72%) had at least one FEV1 measurement at age 23 years or older; these 684 subjects were considered to have high-confidence pattern assignments (Figure 2).
Figure 2.
Diagram of included populations. CAMP (Childhood Asthma Management Program) is the primary discovery population; the Dutch cohort and chronic obstructive pulmonary disease metaanalysis cohort were used for generalization of the association to related lung-function cohorts. COPD = chronic obstructive pulmonary disease; ECLIPSE = Evaluation of COPD Longitudinally to Identify Predictive Surrogate End-points; ED = early decline; GenKOLS = Genetics of Chronic Obstructive Lung Disease study; GWAS = genome-wide association study; NAS = Normative Aging Study; NETT = National Emphysema Treatment Trial; NG = normal growth pattern (without early decline); NG/ED = normal growth with early decline pattern; RG = reduced growth pattern (without early decline); RG/ED = reduced growth with early decline pattern.
CAMP GWAS
To preserve as many participants as possible for genetic association tests, we performed a GWAS on 581 CAMP white non-Hispanic patients who were successfully genotyped using either the Illumina 550 or Illumina 610 microarrays (Illumina Inc., San Diego, CA) and had an initial pattern assignment but not necessarily a high-confidence classification. Genotypes were imputed using MaCH (24) to over 8 million SNPs by using phased genotype data from the 1,000 Genomes v3 project at the Channing Division of Network Medicine. Genotype data were filtered for quality by limiting investigation to SNPs with a minor allele frequency of at least 0.05 and probability of Hardy-Weinberg equilibrium of at least 0.001. A logistic regression test was used to assess the additive association of each SNP to longitudinal lung function growth pattern, using each group in turn and holding the other three groups as reference (NG vs. NG/ED + RG + RG/ED, NG/ED vs. NG + RG + RG/ED, RG vs. NG + NG/ED + RG/ED, and RG/ED vs. NG + NG/ED + RG). This resulted in four separate GWASs being executed each with 6,116,380 SNPs and 581 total participants in the four FEV1 category groups, with sex and age at enrollment included as covariates. Examination of ancestry-based principal components and a quantile-quantile plot with and without adjustment showed no evidence of population stratification (see Figure E1 in the online supplement) (genome inflation factor, 0.994; lambda, 1.016); principal components were not adjusted in the final model. For comparison purposes, we also reran the GWAS using the intersection of the 581 genotyped CAMP subjects with the 684 high-confidence pattern-assigned CAMP subjects (n = 396), following the previously mentioned methodology (Figure 2). SNP associations were calculated using PLINK 1.07 (25).
Dutch Asthma Genetics Cohort
A selection of 114 subjects with asthma taken from a cohort of 281 Dutch Asthma Genetics participants was used for replication based on longitudinal lung function data availability (14). This cohort comprised subjects with longitudinal prebronchodilator FEV1 ranging from ages 13 to 44 years. Subjects were chosen that had at least one FEV1 value before the age of 25 years and at least two measurements before the age of 29 (median [range], 18 [6–40] measurements before the age of 29). Longitudinal prebronchodilator FEV1 per subject were smoothed using LOESS splines (SPSS, version 22; IBM, Armonk, NY). A group of four asthma researchers (M.J.M., J.M.V., D.S.P., and S.T.W.) classified each subject as either ED, no ED, or unclassifiable. Classifications were initially performed separately and where discrepant compared with arrived at consensus classifications. This resulted in 91 total subjects with asthma classified as either ED or no ED. These subjects were then classified as RG or NG by comparison with NHANES III, as previously mentioned. Genome-wide genotyping was previously available on 83 of the 91 selected subjects with asthma, from the Illumina 317 and the Illumina 370 Duo chips (Illumina); the rs4445257 SNP passed quality control metrics. Complete details have been described previously (26). Association of rs4445257 to ED was performed using a logistic regression test (SPSS, version 22; IBM).
COPD Metaanalysis GWAS
COPD GWAS results from a combined analysis of subjects from the National Emphysema Treatment Trial/Normative Aging Study, Norway GenKOLS, Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints, and COPDGene studies were used to replicate the association observed in CAMP with early longitudinal lung function decline. Greater detail on this metaanalysis has been reported previously (27). Briefly, genotyping was performed on a total of 12,337 non-Hispanic white (n = 9,767) and African American (n = 2,570) subjects using Illumina platforms (HumanHap 550,Quad 610, or OmniExpress). These data were then imputed to 1,000 Genomes Phase 1 v3 reference samples using MaCH (24) and minimac (28). GWAS was performed separately in each cohort for both moderate to severe COPD (Global Initiative for Chronic Obstructive Lung Disease grade 2 and above) and severe COPD (Global Initiative for Chronic Obstructive Lung Disease grade 3 and above) adjusting for age, pack-years of smoking exposure, and principal components of genetic ancestry using PLINK 1.07 (25), and results were combined via fixed-effects metaanalysis using METAL (version 2010-08-01) (29).
Genomic Validation
Our most strongly associated GWAS SNP was imputed in the CAMP cohort. We subsequently genotyped rs4445257 in CAMP using a TaqMan assay for allelic discrimination (Applied Biosystems, Foster City, CA), available as assay C__11868548_10. TaqMan analysis was performed using the QuantStudio 12K Flex Software version 1.2.2 (Thermo Fisher Scientific, Waltham, MA) according to manufacturer's specifications.
Candidate genes from our GWAS analysis were assessed for differential expression in a sample of 366 human fetal lung tissues (postconception age 54–137 d). Details of our fetal tissue sample collection and RNA isolation have been described previously (30). Expression was measured using the Affymetrix GeneChip Human Gene 1.0 ST array (Affymetrix, Santa Clara, CA). The complete microarray data are available through the Gene Expression Omnibus of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/geo/, accession number GSE68896). Differential expression across postconception age during human lung development was assessed by performing linear regression adjusted for sex and intrauterine smoke exposure status.
Hi-C Analysis of Non–Small Cell Lung Cancer Cells NCIH460
Genome-wide chromosome conformation capture (Hi-C) was performed as described next using two biologic replicates of NCIH460 cells (31, 32). Hi-C libraries were sequenced on an Hi-Seq2000 Illumina platform (paired end, 50 bases each). Reads were mapped to the human genome (hg19) using a previously published iterative mapping procedure, and polymerase chain reaction (PCR) duplicates were removed (33). Chromatin interaction data were then binned in 40-kb bins, and the data were corrected for intrinsic biases, such as mappability and restriction site density, as described previously (33). Hi-C interaction maps for biologic replicates were highly concordant (Pearson correlation >0.9) and were pooled. Hi-C data were produced as part of the ENCODE project and will be made publicly available according to ENCODE data release standards. The data are also available in Gene Expression Omnibus (accession number pending).
The combined Hi-C dataset was used to determine the positions of Topologically Associating Domain (TAD) boundaries: contiguous regions where loci display relatively high interactions, separated by boundaries that prevent interactions across them. Hi-C data for chromosome 8 from position 111 to 118 Mb (hg19 | chr8:111,000,000–118,000,000) were extracted from the genome-wide dataset to identify TAD boundaries in the relevant genomic region. The “insulation” score of each 40-kb bin was calculated by computing the average interaction signal within a square starting at the interaction matrix diagonal and extending 400 kb in each direction (10 × 10 bins), as described in more detail (34, 35). This captured the number of interactions across a given bin. Relative insulation scores were obtained by dividing each insulation score by the average of all insulation scores. Local minima representing TAD boundaries were then identified by scanning the entire region with a sliding window of size 800 kb and marking the midpoint of the 40-kb bin having the lowest insulation index value as a TAD boundary. Visual comparison of the insulation profile with the Hi-C interaction map confirmed the correct identification of TAD positions.
Circularized Chromosome Conformation Capture
A circularized chromosome conformation capture (4C) assay was used to assess the interaction frequency between rs4445257 and the promoters of neighboring genes. 4C templates were generated based on a published protocol with slight modifications (36, 37). Human bronchial epithelial cells (Beas-2B cell line) were cross-linked with 2% formaldehyde for 10 minutes at room temperature. After cross-linking, cell nuclei were isolated and DNA was digested with a primary restriction enzyme recognizing a 4-bp restriction site (MboI; NEB, cat. no. R0147). This was followed by proximity ligation. A secondary restriction enzyme digestion was performed with a 4-bp restriction enzyme (NlaIII; NEB, cat. no. R0125) recognizing a different sequence than the primary enzyme, followed again by proximity ligation. Approximately 100-ng template was used for the subsequent PCR reaction, with conditions as follows: 95°C denaturing for 3 minutes and 30 cycles of 95°C for 30 seconds, annealing at 56°C for 30 seconds, and extension at 72°C for 1 minute followed by extension at 72°C for 5 minutes. Primers used in 4C-PCR are listed in Table E1.
Results
We performed a discovery GWAS of each of four longitudinal lung function patterns (one group compared with the other three) in 581 non-Hispanic white CAMP subjects with both complete genotype and phenotype data (Figure 2). Baseline characteristics of this group are shown in Table 1. In the 581, we had 164 (28.2%) classified as NG, 116 (20.0%) as ED, 155 (26.7%) as RG, and 117 (20.1%) as RG with ED; additionally 29 (4.99%) had sparse data or an undeterminable pattern. The group of 581 used in GWAS was not significantly different from the remainder of the CAMP cohort (see Table E2), apart from racial homogeneity, and a lower proportion of sparse/undetermined patterns. We have previously conducted a misclassification analysis of pattern assignment in this cohort, finding that two qualified pulmonologists had high agreement on pattern determination (kappa, 0.92 and 0.89, for two replicates) (2). Additionally, this reference includes more details of the baseline and longitudinal differences between pattern groups.
Table 1.
Descriptive Characteristics of the CAMP Cohort by Lung Function Pattern
| Normal Growth (n = 164; 28.2%) | Normal Growth, Early Decline (n = 116; 20.0%) | Reduced Growth (n = 155; 26.7%) | Reduced Growth, Early Decline (n = 117; 20.1%) | P Value | |
|---|---|---|---|---|---|
| Sex, male, n (%) | 89 (54.27) | 71 (61.21) | 106 (68.39) | 70 (59.83) | 0.081 |
| Race, white, n (%) | 164 (100.00) | 116 (100.00) | 155 (100.00) | 117 (100.00) | — |
| Age, yr | 8.44 (±2.20) | 9.21 (± 2.02) | 8.56 (±2.10) | 9.44 (±2.01) | <0.001 |
| Age at diagnosis, yr | 3.30 (±2.49) | 3.40 (± 2.48) | 2.66 (±2.32) | 2.99 (±2.34) | 0.041 |
| Height, randomization, cm | 130.42 (±14.00) | 136.18 (±13.31) | 131.03 (±13.21) | 136.33 (±13.40) | <0.001 |
| Height, end of trial, cm | 152.64 (±13.10) | 158.07 (±14.05) | 153.64 (±13.99) | 158.30 (±12.91) | <0.001 |
| Weight, randomization, kg | 31.11 (±10.96) | 35.88 (±11.30) | 30.31 (±10.34) | 33.78 (±10.62) | <0.001 |
| Weight, end of trial, kg | 49.98 (±15.28) | 56.41 (±17.19) | 48.19 (±16.03) | 53.61 (±16.30) | <0.001 |
| BMI, randomization, kg/m2 | 17.80 (±2.94) | 19.01 (±3.37) | 17.19 (±2.95) | 17.78 (±2.97) | <0.001 |
| BMI, end of trial, kg/m2 | 20.93 (±3.87) | 22.23 (±4.56) | 19.89 (±4.05) | 21.04 (±4.57) | <0.001 |
| Treatment group, steroids, n (%) | 43 (26.22) | 36 (31.03) | 45 (29.03) | 37 (31.62) | 0.75 |
| ED/Hosp through trial, n | 0.71 (±1.51) | 0.47 (±1.04) | 0.97 (±1.89) | 0.61 (±1.40) | 0.053 |
| Serum IgE, log10 IU/ml | 2.59 (±0.67) | 2.48 (±0.73) | 2.59 (±0.65) | 2.73 (±0.71) | 0.052 |
| Eosinophils, log10 count | 2.43 (±0.61) | 2.45 (±0.65) | 2.50 (±0.50) | 2.62 (±0.41) | 0.028 |
| PC20, log mg/ml | 0.24 (±1.22) | 0.36 (±1.14) | −0.03 (±1.06) | −0.23 (±1.18) | <0.001 |
| Pre-BD FEV1, L | 1.70 (±0.48) | 1.87 (±0.48) | 1.50 (±0.42) | 1.61 (±0.41) | <0.001 |
| Post-BD FEV1, L | 1.84 (±0.51) | 2.01 (±0.50) | 1.66 (±0.44) | 1.81 (±0.45) | <0.001 |
| Pre-BD FEV1% predicted | 101.93 (±12.15) | 100.11 (±11.84) | 86.88 (±12.21) | 85.65 (±12.71) | <0.001 |
| Post-BD FEV1% predicted | 110.25 (±12.05) | 107.96 (±11.05) | 97.00 (±10.74) | 96.05 (±10.67) | <0.001 |
| Bronchodilator response, % | 0.09 (±0.07) | 0.08 (±0.09) | 0.12 (±0.11) | 0.13 (±0.11) | <0.001 |
Definition of abbreviations: BD = bronchodilator; BMI = body mass index; CAMP = Childhood Asthma Management Program; ED/Hosp = number of emergency department or hospitalizations required for asthma during trial period; PC20 = methacholine concentration required for a 20% reduction in airway volume.
Randomization and end of trial time points are approximately 48 months apart. All measures were taken at baseline unless otherwise indicated. P values were computed by two-sided analysis of variance test. Data are mean (±SD) unless otherwise indicated. Treatment group: randomized CAMP trial arm assignment to budesonide (inhaled corticosteroid) versus either nedocramil or placebo. BD response was computed as (post-BD FEV1 − pre-BD FEV1)/pre-BD FEV1.
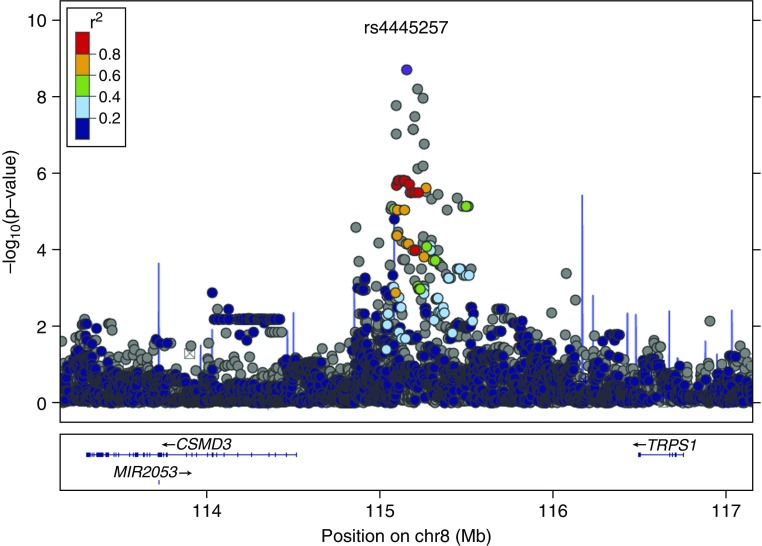
For GWAS statistical significance, we chose a threshold of P less than 1.25 × 10−8, representing the standard GWAS cutoff of P less than 5 × 10−8 (38), adjusted for four separate GWASs. We found evidence of a strong genetic association for the NG/ED pattern on the long arm of chromosome 8 (see Figure E2), represented by the SNP rs4445257 (P = 6.7 × 10−9) (Figure 3, Table 2). Each minor allele (G) conferred an odds ratio (OR) of 2.83 (95% confidence interval [CI], 1.98–4.04) of membership in the NG/ED pattern compared with the reference allele (T). Parallel analyses of the other three categories did not result in any genetic signals associated with FEV1 pattern at the genome-wide significance level (P = 1.25 × 10−8). The SNP rs4445257 lies between the Cub and Sushi multiple domain 3 (CSMD3) and trichorhinophalangeal syndrome I (TRPS1) genes, and has moderate prevalence in populations of European ancestry (minor allele G, frequency 28%; reference allele T). This SNP was imputed from CAMP genotypes with very high confidence (MaCH imputation r2 value of 0.99). Subsequent direct genotyping of this SNP in CAMP confirmed the imputed association, although attenuated (P = 7.9 × 10−6; OR, 2.22 [95% CI, 1.57–3.16]), with 26.5 discordant alleles and 10 missing genotypes. A sensitivity analysis using only subjects with the smaller high-confidence phenotypes (n = 396) resulted in less significant associations overall, but rs4445257 nonetheless remained the strongest signal (P = 2.2 × 10−6) (see Figure E3).
Figure 3.
Locus plot of single-nucleotide polymorphism associations with normal growth and early decline on chromosome 8. The peak at rs4445257 is approximately 633 kbp upstream from CSMD3 and approximately 1.4 Mbp downstream from TRPS1. Color indicates linkage disequilibrium from rs4445257 (r2).
Table 2.
Association Tests of SNP rs4445257
| P Value | Odds Ratio (Minor Allele G) | |
|---|---|---|
| CAMP | ||
| Base model | ||
| NG/ED vs. (NG + RG + RG/ED) | 6.7 × 10−9 | 2.83 (1.98–4.04) |
| Interaction analysis (multiple regression) | ||
| (NG/ED + RG/ED) vs. (NG + RG) | ||
| rs4445257 | 5.6 × 10−6 | 11.5 (4.0–33.2) |
| RG/ED vs. NG/ED | 2.8 × 10−4 | 6.4 (2.3–17.4) |
| rs4445257 × (RG/ED vs. NG/ED) | 2.7 × 10−4 | 0.27 (0.13–0.55) |
| Likelihood ratio of interaction vs. base model | 8.3 × 10−4 | |
| Stratified analysis | ||
| NG/ED vs. NG | 1.4 × 10−6 | 3.1 (1.9–5.0) |
| RG/ED vs. RG | 0.51 | 0.83 (0.48–1.4) |
| Dutch Asthma Cohort | ||
| (NG/ED + RG/ED) vs. (NG + RG) | 0.051 | 0.38 |
| COPD metaanalysis | ||
| COPD case vs. control | 0.0487 | 0.89 (0.80–0.98) |
| Severe COPD case vs. control | 0.016 | 0.91 (0.84–0.99) |
Definition of abbreviations: CAMP = Childhood Asthma Management Program; COPD = chronic obstructive pulmonary disease; NG = normal growth pattern (without early decline); NG/ED = normal growth with early decline pattern; RG = reduced growth pattern (without early decline); RG/ED = reduced growth with early decline pattern; SNP = single-nucleotide polymorphism.
All tests are logistic regression. P values less than 0.05 are statistically significant.
We then performed a generalization analysis of our strongest signal in two additional respiratory disease cohorts that display related but distinct lung-function phenotypes. We first assessed rs4445257 for association with NG/ED in a cohort of 83 Dutch subjects with asthma. Of Dutch subjects successfully classified into the four lung function patterns, 16 had NG, four had NG/ED, 38 had RG, and 25 RG/ED. Because few subjects had the NG/ED phenotype, we combined NG/ED and RG/ED into one category and observed a nearly significant protective effect for ED of the G minor allele of rs4445257, when compared with the two no-decline groups (P = 0.051; OR, 0.38 per minor allele). This observed effect was in the opposite direction from our discovery asthma cohort. To further elucidate the role of rs4445257, we tested association with COPD case status (P = 0.0487) in a large metaanalysis of COPD cohorts (6,633 cases and 5,704 control subjects) (27). The association with COPD was stronger when restricted to patients with severe COPD phenotypes (3,497 cases and 5,704 control subjects; P = 0.016). Importantly, however, the minor allele was protective of COPD (OR, 0.89 [95% CI, 0.80–0.98] and OR, 0.91 [95% CI, 0.84–0.99] in severe COPD), again in the opposite direction of effect from our discovery cohort (Table 2).
Because the Dutch Cohort was composed of mostly RG/ED rather than NG/ED, we conducted an interaction analysis of rs4445257 with RG in CAMP, as follows. We used joint ED (combining NG/ED and RG/ED) as a phenotype, and using RG as a covariate, using a regression composed as follows: ED = RG + SNP + RG × SNP (Table 2). We observed significant effects of rs4445257 on ED, where minor alleles predisposed to ED (P = 5.6 × 10−6; OR, 11.5 [95% CI, 4.0–33.2]); of RG on ED (P = 2.8 × 10−4; OR, 6.4 [95% CI, 2.3–17.4]), where RG predisposed to ED; and also a significant interaction effect of rs4445257 and RG, where minor alleles of the SNP in the presence of RG were protective of ED (2.7 × 10−4; OR, 0.27 [95% CI, 0.13–0.55]). This interaction model provided a better fit than the model with just rs4445257 (P = 8.3 × 10−4, likelihood ratio test). In stratified analysis within the NG group, rs4445257 was significantly associated with ED (P = 1.4 × 10−6; OR, 3.1 [95% CI, 1.9–5.0]); but within the RG group, rs4445257 was nonsignificant (P = 0.51; OR, 0.83 [95% CI, 0.48–1.4]). This evidence shows that rs4445257 may interact with RG in its effect on ED; it may be a risk factor for ED in cases of NG, but did not have a significant protective effect in cases of RG.
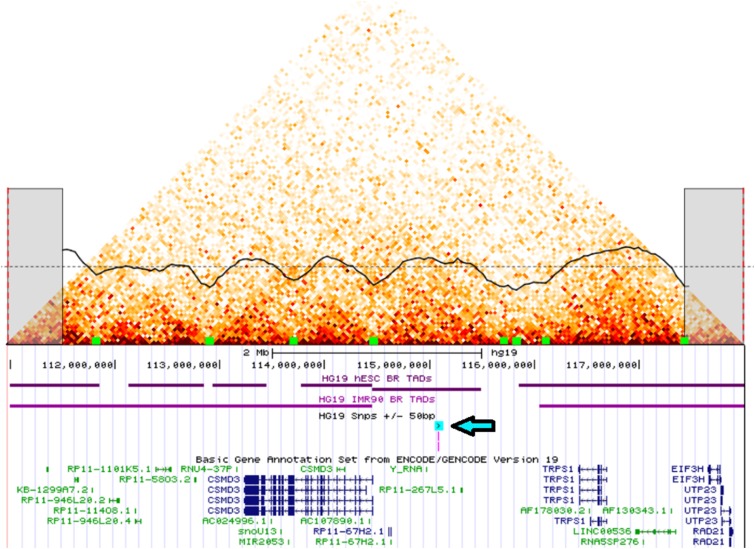
To identify possible biologic mechanisms of rs4445257, we used chromatin experiments to localize its interactions to nearby genes. The SNP rs4445257 is located 633 kb from the nearest gene (CSMD3) and over 1.4 Mb from the nearest gene in the other direction (TRPS1), making direct assignment of a relevant target gene difficult. We applied the Hi-C methodology in the small-cell lung cancer cell line NCIH460 to identify interaction domains (TADs) and their boundaries (39). Figure 4 shows the Hi-C interaction map of an 8-Mb region surrounding rs4445257. rs4445257 was located in a large TAD (bp 114,440,001–115,800,001, chromosome 8, GRCh37.p13), with the CSMD3 promoter located in the 40-kb bin that contained the TAD boundary. TRPS1 was located in a different TAD separated from rs4445257 by a cluster of TAD boundaries, making it a less likely target gene. To confirm the interaction of rs4445257 with CSMD3, we performed a 4C assay followed by PCR to assess the possible chromatin interaction between rs4445257 and the promoter of CSMD3 in Beas-2b cells, a bronchial epithelial cell line. Consistent with the Hi-C data, we observed PCR products amplified from primer pairs targeting rs4445257 and the promoter of CSMD3, in contrast to pairs targeting rs4445257 and the TRPS1 promoter (see Figure E4). This indicates that chromatin arrangement brings rs4445257 in contact with the CSMD3 promoter, enabling a possible molecular interaction between the two loci. Together, these physical interaction results strongly implicated CSMD3 as the relevant target gene of rs4445257.
Figure 4.
Hi-C interaction data (40 kb bins) in NCI-H460 lung epithelium cells on chromosome 8 from 110 to 118 Mbp. Clear domains of increased chromatin interaction are observed as triangles that correspond to topologically associating domains (TADs), separated by boundaries. The dark line through the heatmap represents the TAD insulation score calculated from the Hi-C data. Dips in the plot represent domain boundaries that are also indicated as green blocks at the bottom of the heatmap. Single-nucleotide polymorphism (SNP) rs4445257 is shown with a blue highlighted arrow in the genome snapshot under the heatmap and is located in a large domain. The promoter of CSMD3 is at the boundary of this domain, indicating possible interaction with the SNP; the promoter of TRPS1 is located in a different domain.
The public GTEx project database indicated significant evidence that rs4445257 is an eQTL for CSMD3 in nerve tissue (P = 0.009) (40). In a sample of bronchial epithelial cells from 45 subjects with asthma from the ABRIDGE cohort (41), a neighboring SNP, rs73706006, in high LD with rs4445257 (r2 ≥ 0.99) showed significant association with the expression of CSMD3 (P = 0.0004) (see Figure E5). In both GTEx and ABRIDGE, minor alleles were associated with a decrease in expression.
The fact that at least reduced lung function trajectories seem to be set early in life (16) suggests a possible developmental role for CSMD3. Therefore, we investigated the role of the CSMD3 gene in human lung development using genome-wide gene expression profiles available from 366 human fetal lung tissue samples (30). CSMD3 demonstrated lower expression as postconception age of the sample was higher (P = 0.007), indicating a potential role in lung programming that manifests later in life, or a gene with multiple separate effects in the lung.
To determine if previous SNP and gene associations with asthma and/or lung function–related phenotypes were also associated with longitudinal lung function patterns, we performed a lookup of known SNPs in our GWAS results (see Tables E4–E7). Of 484 identified SNPs, 436 were assayed or imputed in our CAMP cohort. For each of these, we report the association strength with NG (see Table E4), NG/ED (see Table E5), RG (see Table E6), and RG/ED (see Table E7). None of these SNPs reached statistical significance after multiple testing correction for 436 tests.
Discussion
Finding genetic associations to abnormal longitudinal lung function patterns may help to identify patients at risk for chronic airway obstruction and the genetic effects that influence airway disease. The SNP rs4445257 was associated with early lung function decline in two asthma cohorts, and also associated with COPD. In a model including RG and an interaction term, there was a positive effect of interaction, suggesting that this SNP may operate differently in cohorts with reduced growth before reaching lung function plateau in early adulthood, and with normal growth with normal lung function plateau. Although false-positive associations may explain our observed results, we hypothesize that rs4445257 minor alleles are protective of early lung function decline in the presence of reduced lung function growth, while being a risk factor for early decline in the presence of normal lung function growth. Thus, this SNP may interact with an unknown genomic or environmental factor that correlates with maximally attained FEV1 (i.e., RG vs. NG), which would also account for the allele’s observed interaction effect with RG on ED in the CAMP cohort (42).
The interaction of two genomic loci (our rs4445257 locus and another, unknown locus associated with RG) is perhaps likely in light of the polygenic and pleiotropic genomic architecture of complex diseases and traits (43). Although longitudinal data would be required to make a definitive assessment, it is possible the COPD cohort is one where the RG phenotype dominates, similar to the cohorts discussed by Lange and coworkers (5). The complex genetic relationship uncovered here parallels the complex phenotypic development that characterizes mild and moderate persistent asthma from childhood into adulthood. That other SNPs previously associated with asthma and lung function–related phenotypes were not associated with longitudinal lung function patterns may indicate that the conditions have separate genetic etiologies, and that longitudinal phenotypes are different from cross-sectional ones.
The SNP rs4445257 was part of a major locus of high association with the NG/ED category (Figure 3), a locus between CSMD3 and TRPS1 and functionally linked to CSMD3 through Hi-C and 4C experiments. Together, these results suggested potential regulation of the expression of CSMD3 by a narrow region containing rs4445257. This region contains several SNPs in high LD with rs4445257, including rs73706006 4 kbp away (Figure 3). More research is required to determine if other variants in this region effect CSMD3 expression.
The CSMD3 gene has not previously been associated with conditions related to lung function or to respiratory diseases, such as asthma or COPD. CSMD3 encodes a transmembrane protein that is primarily expressed in the brain, nervous tissue, and testis (UniGene [44], GTEx [40]), although CSMD3 expression has also been observed in lung tissue (Illumina Body Map [45], Human Protein Atlas [46]). Although its biologic function is unclear (47), CSMD3 encodes a protein structurally similar to that of CSMD1, which itself has been implicated in asthma–COPD overlap syndrome (48). In lung carcinomas, mutations in CSMD3 were found (49) and suppression of CSMD3 in airway epithelial cells resulted in increased proliferation of those cells in culture (49).
Gene-expression pathway analysis of brain tissue has implicated CSMD3 in GABAergic neuronal fate and neuronal development (47). γ-Aminobutyric acid signaling is primarily responsible for maintenance of muscle tone (50), including airway smooth muscle tone (51, 52). There is growing evidence that airway smooth muscle tone is a predisposing factor for airway hyperresponsiveness (53, 54), and may contribute to reduced prebronchodilator lung volume in people with asthma (55). It is possible that rs4445257 influences CSMD3 expression to affect airway epithelial proliferation and related epithelial integrity and/or airway smooth muscle tone, either of which predisposes for asthma, asthma exacerbations, and lower lung function; however, further investigation is needed to address such possibilities.
The present study has several limitations. Our best interpretation of our results leads to an unusual mixed effect of rs4445257 on early decline of FEV1, where it seems to predispose to ED in the presence of NG but be protective for early decline in the presence of RG. Larger sample sizes would be helpful in evaluating this effect further, as would additional cohorts with detailed longitudinal lung function measured throughout adolescence and young adulthood, although samples meeting these criteria are difficult to obtain. The COPD generalization cohort, although large, does not contain longitudinal pulmonary function assessments over extended time periods, and as such we cannot accurately assess lung function patterns in that group. Therefore we are unable to determine if this population is predominantly a cohort of RG, as would be consistent with our hypothesis, or of ED, or of rapid decline, and this or a combination of these patterns resulted in COPD. Finally, our chromatin studies have demonstrated that the action of rs4445257 on CSMD3’s promoter is likely, and some evidence of association to CSMD3 expression was found in GTEx data, but further work is required to verify this and determine what exact variant is the functional variant.
Early decline of FEV1 after normal growth is potentially associated with a genetic polymorphism (rs4445257) that may be protective of early decline in reduced growth groups. This SNP physically interacts with the CSMD3 promoter in vitro, and is associated with expression of the CSMD3 gene.
Footnotes
Supported through a grant from the Parker B. Francis Foundation (M.J.M.); P01-HL083069 and U01-HL065899 (S.T.W.); U01-HL105569 (R.A.W.); U01HL075408 (K.P.Y., A.L.S., M.L.V.N., R.A.W., and J.T.); National Institutes of Health (NIH) grants R01HL127200 and R21HL120794 (X.Z.); R01-HL113264 (M.H.C.); K01-HL127265 (D.C.C.-C.); K08-HL102265 and R01-HL124233 (P.J.C.); R01-HL075478, R01-HL089856, and P01-HL105339 (E.K.S.); U01-HL091075 (R.C.S.); K25 HL091124 (A.T.K.); and U01-HL065899 and R01-NR013391 (K.G.T.). Work in the Dekker laboratory is supported by the National Human Genome Research Institute (grants HG003143 and HG007010), the NIH common fund (DK 107980), and the Human Frontier Science Project Organization. The Childhood Asthma Management Program trial and CAMP Continuation Study were supported by contracts NO1-HR-16044, 16045, 16046, 16047, 16048, 16049, 16050, 16051, and 16052 with the NHLBI and General Clinical Research Center grants M01RR00051, M01RR0099718-24, M01RR02719-14, and RR00036 from the National Center for Research Resources. The CAMP Continuation Study/Phases 2 and 3 were supported by grants U01HL075232, U01HL075407, U01HL075408, U01HL075409, U01HL075415, U01HL075416, U01HL075417, U01HL075419, U01HL075420, and U01HL075408 from the NHLBI. The National Jewish Health site was also supported in part by Colorado CTSA grant UL1RR025780 from NCRR/NIH and UL1TR000154 from National Center for Advancing Translational Sciences/NIH. The Dutch Asthma Genetics study was supported by grants from the Netherlands Lung Foundation (AF 95.09, AF 98.48, AF 3.2.02.51, and AF 3.2.07.015) and a grant from the University Medical Center Groningen.
Author Contributions: Substantial analysis, M.J.M., K.P.Y., X.Z., F.G., A.L.S., M.L.V.N., S.S., A.T.K., M.H.C., D.C.C.-C., P.J.C., G.J., A.S., Y.Z., B.R.L., J.D., J.M.V., I.H., Q.L., J.T., R.C.S., and S.T.W. Writing, M.J.M., K.P.Y., A.L.S., M.L.V.N., J.T., R.C.S., and S.T.W. Planning, M.J.M., K.P.Y., A.L.S., M.L.V.N., R.A.W., S.J.S., J.D., J.S., R.A.C., R.S.Z., N.F.A., P.V.W., H.W.K., H.G., D.S.P., B.A.R., A.L.F., K.G.T., E.K.S., J.T., R.C.S., and S.T.W. Funding, R.A.W., S.J.S., J.D., J.S., R.A.C., R.S.Z., N.F.A., P.V.W., H.W.K., H.G., G.H.K., D.S.P., B.A.R., A.L.F., K.G.T., E.K.S., J.T., R.C.S., and S.T.W. Data acquisition, X.Z., F.G., A.L.S., M.L.V.N., R.A.W., S.J.S., S.S., A.T.K., J.D., J.S., R.A.C., R.S.Z., N.F.A., P.V.W., H.W.K., H.G., J.M.V., G.H.K., D.S.P., B.A.R., I.H., Q.L., A.L.F., K.G.T., E.K.S., J.T., R.C.S., and S.T.W. All authors participated in the intellectual revision and drafting of the manuscript, and approved the final version.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.201602-0250OC on July 1, 2016
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Speizer FE, Tager IB. Epidemiology of chronic mucus hypersecretion and obstructive airways disease. Epidemiol Rev. 1979;1:124–142. doi: 10.1093/oxfordjournals.epirev.a036206. [DOI] [PubMed] [Google Scholar]
- 2.McGeachie MJ, Yates KP, Zhou X, Guo F, Sternberg AL, Van Natta ML, Wise RA, Szefler SJ, Sharma S, Kho AT, et al. CAMP Research Group. Patterns of growth and decline in lung function in persistent childhood asthma. N Engl J Med. 2016;374:1842–1852. doi: 10.1056/NEJMoa1513737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tager IB, Segal MR, Speizer FE, Weiss ST. The natural history of forced expiratory volumes. Effect of cigarette smoking and respiratory symptoms. Am Rev Respir Dis. 1988;138:837–849. doi: 10.1164/ajrccm/138.4.837. [DOI] [PubMed] [Google Scholar]
- 4.Wang X, Mensinga TT, Schouten JP, Rijcken B, Weiss ST. Determinants of maximally attained level of pulmonary function. Am J Respir Crit Care Med. 2004;169:941–949. doi: 10.1164/rccm.2201011. [DOI] [PubMed] [Google Scholar]
- 5.Lange P, Celli B, Agustí A, Boje Jensen G, Divo M, Faner R, Guerra S, Marott JL, Martinez FD, Martinez-Camblor P, et al. Lung-function trajectories leading to chronic obstructive pulmonary disease. N Engl J Med. 2015;373:111–122. doi: 10.1056/NEJMoa1411532. [DOI] [PubMed] [Google Scholar]
- 6.Weiss S, Speizer F.Epidemiology and natural historyIn: Weiss EB, Stein M, editors. Bronchial asthma mechanisms and therapeutics, 3rd ed. Boston: Little, Brown; 1993. p. 15
- 7.Fletcher C. The natural history of chronic bronchitis and emphysema: an eight-year study of early chronic obstructive lung disease in working men in London. New York: Oxford University Press; 1976. [Google Scholar]
- 8.Vonk JM, Jongepier H, Panhuysen CI, Schouten JP, Bleecker ER, Postma DS. Risk factors associated with the presence of irreversible airflow limitation and reduced transfer coefficient in patients with asthma after 26 years of follow up. Thorax. 2003;58:322–327. doi: 10.1136/thorax.58.4.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Young RP, Hopkins R, Eaton TE. Forced expiratory volume in one second: not just a lung function test but a marker of premature death from all causes. Eur Respir J. 2007;30:616–622. doi: 10.1183/09031936.00021707. [DOI] [PubMed] [Google Scholar]
- 10.Sears MR, Greene JM, Willan AR, Wiecek EM, Taylor DR, Flannery EM, Cowan JO, Herbison GP, Silva PA, Poulton R. A longitudinal, population-based, cohort study of childhood asthma followed to adulthood. N Engl J Med. 2003;349:1414–1422. doi: 10.1056/NEJMoa022363. [DOI] [PubMed] [Google Scholar]
- 11.Ulrik CS. Outcome of asthma: longitudinal changes in lung function. Eur Respir J. 1999;13:904–918. doi: 10.1034/j.1399-3003.1999.13d35.x. [DOI] [PubMed] [Google Scholar]
- 12.Jamrozik E, Knuiman MW, James A, Divitini M, Musk AW. Risk factors for adult-onset asthma: a 14-year longitudinal study. Respirology. 2009;14:814–821. doi: 10.1111/j.1440-1843.2009.01562.x. [DOI] [PubMed] [Google Scholar]
- 13.Svanes C, Sunyer J, Plana E, Dharmage S, Heinrich J, Jarvis D, de Marco R, Norbäck D, Raherison C, Villani S, et al. Early life origins of chronic obstructive pulmonary disease. Thorax. 2010;65:14–20. doi: 10.1136/thx.2008.112136. [DOI] [PubMed] [Google Scholar]
- 14.Dijkstra A, Vonk JM, Jongepier H, Koppelman GH, Schouten JP, ten Hacken NH, Timens W, Postma DS. Lung function decline in asthma: association with inhaled corticosteroids, smoking and sex. Thorax. 2006;61:105–110. doi: 10.1136/thx.2004.039271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grol MH, Gerritsen J, Vonk JM, Schouten JP, Koëter GH, Rijcken B, Postma DS. Risk factors for growth and decline of lung function in asthmatic individuals up to age 42 years. A 30-year follow-up study. Am J Respir Crit Care Med. 1999;160:1830–1837. doi: 10.1164/ajrccm.160.6.9812100. [DOI] [PubMed] [Google Scholar]
- 16.Stern DA, Morgan WJ, Wcenter AL, Guerra S, Martinez FD. Poor airway function in early infancy and lung function by age 22 years: a non-selective longitudinal cohort study. Lancet. 2007;370:758–764. doi: 10.1016/S0140-6736(07)61379-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poon AH, Houseman EA, Ryan L, Sparrow D, Vokonas PS, Litonjua AA. Variants of asthma and chronic obstructive pulmonary disease genes and lung function decline in aging. J Gerontol A Biol Sci Med Sci. 2014;69:907–913. doi: 10.1093/gerona/glt179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imboden M, Bouzigon E, Curjuric I, Ramasamy A, Kumar A, Hancock DB, Wilk JB, Vonk JM, Thun GA, Siroux V, et al. Genome-wide association study of lung function decline in adults with and without asthma. J Allergy Clin Immunol. 2012;129:1218–1228. doi: 10.1016/j.jaci.2012.01.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Postma DS, Meyers DA, Jongepier H, Howard TD, Koppelman GH, Bleecker ER. Genomewide screen for pulmonary function in 200 families ascertained for asthma. Am J Respir Crit Care Med. 2005;172:446–452. doi: 10.1164/rccm.200407-864OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGeachie MJ, Yates KP, Cho MH, Croteau-Chonka DC, Castaldi PJ, Silverman EK, Zhou X, Wise RA, Tonascia J, Sternberg AL, et al. Phenotypic and genetic risk factors for reduced growth and early decline in FEV1 into early adulthood in childhood asthmatics [abstract] Am J Respir Crit Care Med. 2014;189:A1004. [Google Scholar]
- 21.Childhood Asthma Management Program Research Group. The Childhood Asthma Management Program (CAMP): design, rationale, and methods. Control Clin Trials. 1999;20:91–120. [PubMed] [Google Scholar]
- 22.The Childhood Asthma Management Program Research Group. Long-term effects of budesonide or nedocromil in children with asthma. N Engl J Med. 2000;343:1054–1063. doi: 10.1056/NEJM200010123431501. [DOI] [PubMed] [Google Scholar]
- 23.Hankinson JL, Odencrantz JR, Fedan KB. Spirometric reference values from a sample of the general U.S. population. Am J Respir Crit Care Med. 1999;159:179–187. doi: 10.1164/ajrccm.159.1.9712108. [DOI] [PubMed] [Google Scholar]
- 24.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Portelli MA, Siedlinski M, Stewart CE, Postma DS, Nieuwenhuis MA, Vonk JM, Nurnberg P, Altmuller J, Moffatt MF, Wardlaw AJ, et al. Genome-wide protein QTL mapping identifies human plasma kallikrein as a post-translational regulator of serum uPAR levels. FASEB J. 2014;28:923–934. doi: 10.1096/fj.13-240879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cho MH, McDonald ML, Zhou X, Mattheisen M, Castaldi PJ, Hersh CP, Demeo DL, Sylvia JS, Ziniti J, Laird NM, et al. NETT Genetics, ICGN, ECLIPSE and COPDGene Investigators. Risk loci for chronic obstructive pulmonary disease: a genome-wide association study and meta-analysis. Lancet Respir Med. 2014;2:214–225. doi: 10.1016/S2213-2600(14)70002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genome-wide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharma S, Tantisira K, Carey V, Murphy AJ, Lasky-Su J, Celedón JC, Lazarus R, Klanderman B, Rogers A, Soto-Quirós M, et al. A role for Wnt signaling genes in the pathogenesis of impaired lung function in asthma. Am J Respir Crit Care Med. 2010;181:328–336. doi: 10.1164/rccm.200907-1009OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Belton JM, McCord RP, Gibcus JH, Naumova N, Zhan Y, Dekker J. Hi-C: a comprehensive technique to capture the conformation of genomes. Methods. 2012;58:268–276. doi: 10.1016/j.ymeth.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Imakaev M, Fudenberg G, McCord RP, Naumova N, Goloborodko A, Lajoie BR, Dekker J, Mirny LA. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat Methods. 2012;9:999–1003. doi: 10.1038/nmeth.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crane E, Bian Q, McCord RP, Lajoie BR, Wheeler BS, Ralston EJ, Uzawa S, Dekker J, Meyer BJ. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature. 2015;523:240–244. doi: 10.1038/nature14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith EM, Lajoie BR, Jain G, Dekker J. Invariant TAD boundaries constrain cell-type-specific looping interactions between promoters and distal elements around the CFTR locus. Am J Hum Genet. 2016;98:185–201. doi: 10.1016/j.ajhg.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van de Werken HJ, Landan G, Holwerda SJ, Hoichman M, Klous P, Chachik R, Splinter E, Valdes-Quezada C, Oz Y, Bouwman BA, et al. Robust 4C-seq data analysis to screen for regulatory DNA interactions. Nat Methods. 2012;9:969–972. doi: 10.1038/nmeth.2173. [DOI] [PubMed] [Google Scholar]
- 37.Simonis M, Kooren J, de Laat W. An evaluation of 3C-based methods to capture DNA interactions. Nat Methods. 2007;4:895–901. doi: 10.1038/nmeth1114. [DOI] [PubMed] [Google Scholar]
- 38.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Croteau-Chonka DC, Qiu W, Martinez FD, Strunk RC, Lemanske RF, Jr, Liu AH, Gilliland FD, Millstein J, Gauderman WJ, Ober C.et al.; Asthma BioRepository for Integrative Genomic Exploration (Asthma BRIDGE) Consortium. Gene expression profiling in blood provides reproducible molecular insights into asthma control Am J Respir Crit Care Med[online ahead of print] 5 Aug 2016; DOI: 10.1164/rccm.201601-0107OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin PI, Vance JM, Pericak-Vance MA, Martin ER. No gene is an island: the flip-flop phenomenon. Am J Hum Genet. 2007;80:531–538. doi: 10.1086/512133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee SH, Yang J, Goddard ME, Visscher PM, Wray NR. Estimation of pleiotropy between complex diseases using single-nucleotide polymorphism-derived genomic relationships and restricted maximum likelihood. Bioinformatics. 2012;28:2540–2542. doi: 10.1093/bioinformatics/bts474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pontius JU, Wagner L, Schuler G. UniGene: a unified view of the transcriptome. The NCBI Handbook. Bethesda, MD: National Center for Biotechnology Information; 2003. [Google Scholar]
- 45.Petryszak R, Burdett T, Fiorelli B, Fonseca NA, Gonzalez-Porta M, Hastings E, Huber W, Jupp S, Keays M, Kryvych N, et al. Expression Atlas update--a database of gene and transcript expression from microarray- and sequencing-based functional genomics experiments. Nucleic Acids Res. 2014;42:D926–D932. doi: 10.1093/nar/gkt1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 47.Pandey AK, Lu L, Wang X, Homayouni R, Williams RW. Functionally enigmatic genes: a case study of the brain ignorome. PLoS One. 2014;9:e88889. doi: 10.1371/journal.pone.0088889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hardin M, Cho M, McDonald ML, Beaty T, Ramsdell J, Bhatt S, van Beek EJ, Make BJ, Crapo JD, Silverman EK, et al. The clinical and genetic features of COPD-asthma overlap syndrome. Eur Respir J. 2014;44:341–350. doi: 10.1183/09031936.00216013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu P, Morrison C, Wang L, Xiong D, Vedell P, Cui P, Hua X, Ding F, Lu Y, James M, et al. Identification of somatic mutations in non-small cell lung carcinomas using whole-exome sequencing. Carcinogenesis. 2012;33:1270–1276. doi: 10.1093/carcin/bgs148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Watanabe M, Maemura K, Kanbara K, Tamayama T, Hayasaki H. GABA and GABA receptors in the central nervous system and other organs. Int Rev Cytol. 2002;213:1–47. doi: 10.1016/s0074-7696(02)13011-7. [DOI] [PubMed] [Google Scholar]
- 51.Gallos G, Townsend E, Yim P, Virag L, Zhang Y, Xu D, Bacchetta M, Emala CW. Airway epithelium is a predominant source of endogenous airway GABA and contributes to relaxation of airway smooth muscle tone. Am J Physiol Lung Cell Mol Physiol. 2013;304:L191–L197. doi: 10.1152/ajplung.00274.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mizuta K, Xu D, Pan Y, Comas G, Sonett JR, Zhang Y, Panettieri RA, Jr, Yang J, Emala CW., Sr GABAA receptors are expressed and facilitate relaxation in airway smooth muscle. Am J Physiol Lung Cell Mol Physiol. 2008;294:L1206–L1216. doi: 10.1152/ajplung.00287.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Price D, Fromer L, Kaplan A, van der Molen T, Román-Rodríguez M. Is there a rationale and role for long-acting anticholinergic bronchodilators in asthma? NPJ Prim Care Respir Med. 2014;24:14023. doi: 10.1038/npjpcrm.2014.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Canning BJ, Woo A, Mazzone SB. Neuronal modulation of airway and vascular tone and their influence on nonspecific airways responsiveness in asthma. J Allergy (Cairo) 2012;2012:108149. doi: 10.1155/2012/108149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kelly VJ, Brown NJ, Sands SA, Borg BM, King GG, Thompson BR. Effect of airway smooth muscle tone on airway distensibility measured by the forced oscillation technique in adults with asthma. J Appl Physiol (1985) 2012;112:1494–1503. doi: 10.1152/japplphysiol.01259.2011. [DOI] [PubMed] [Google Scholar]