Abstract
Rationale: Recent studies suggest that baseline tuberculous sputum comprises a mixture of routinely culturable and differentially culturable tubercle bacteria (DCTB). The latter seems to be drug tolerant and dependent on resuscitation-promoting factors (Rpfs).
Objectives: To further explore this, we assessed sputum from patients with tuberculosis for DCTB and studied the impact of exogenous culture filtrate (CF) supplementation ex vivo.
Methods: Sputum samples from adults with tuberculosis and HIV-1 and adults with no HIV-1 were used for most probable number (MPN) assays supplemented with CF and Rpf-deficient CF, to detect CF-dependent and Rpf-independent DCTB, respectively.
Measurements and Main Results: In 110 individuals, 19.1% harbored CF-dependent DCTB and no Rpf-independent DCTB. Furthermore, 11.8% yielded Rpf-independent DCTB with no CF-dependent DCTB. In addition, 53.6% displayed both CF-dependent and Rpf-independent DCTB, 1.8% carried CF-independent DCTB, and 13.6% had no DCTB. Sputum from individuals without HIV-1 yielded higher CF-supplemented MPN counts compared with counterparts with HIV-1. Furthermore, individuals with HIV-1 with CD4 counts greater than 200 cells/mm3 displayed higher CF-supplemented MPN counts compared with participants with HIV-1 with CD4 counts less than 200 cells/mm3. CF supplementation allowed for detection of mycobacteria in 34 patients with no culturable bacteria on solid media. Additionally, the use of CF enhanced detection of sputum smear–negative individuals.
Conclusions: These observations demonstrate a novel Rpf-independent DCTB population in sputum and reveal that reduced host immunity is associated with lower prevalence of CF-responsive bacteria. Quantification of DCTB in standard TB diagnosis would be beneficial because these organisms provide a putative biomarker to monitor treatment response and risk of disease recurrence.
Keywords: tuberculosis, resuscitation-promoting factors, HIV, culturability, limiting dilution assay
At a Glance Commentary
Scientific Knowledge on the Subject
Sputum from patients with tuberculosis (TB) harbors drug-tolerant, differentially culturable tubercle bacteria (DCTB) that are unable to grow on solid media but can be recovered in liquid media supplemented with resuscitation-promoting factors, a group of bacterial growth stimulatory enzymes secreted by Mycobacterium tuberculosis.
What This Study Adds to the Field
In addition to culture filtrate (CF)-dependent DCTB, sputum from patients with TB harbors a significant proportion of resuscitation-promoting factor–independent DCTB. Enhanced recovery of DCTB through supplementation of sputum cultures with CF improved bacterial detection in sputum smear–negative patients. Sputum from individuals with TB and HIV-1 with CD4 counts greater than 200 cells/mm3 displayed higher levels of CF-responsive organisms than sputum from individuals with CD4 counts less than 200 cells/mm3. This study represents the most comprehensive analysis of DCTB to date and reports the presence of phenotypically distinct bacterial subpopulations in individuals with TB. These findings have important implications for diagnosis of TB, particularly in individuals with paucibacillary disease. Moreover, the quantitation of differentially culturable organisms now provides a novel biomarker to assess treatment response and risk of disease recurrence. Our data provide preliminary microbiologic evidence to validate the long-standing hypothesis that the host immune response to TB infection drives bacteria into phenotypically distinct, drug-tolerant states.
Tuberculosis (TB) remains a significant source of human suffering with approximately 1.5 million deaths and 9 million new infections annually (1). Attempts to eradicate this disease have been hampered by complex clinical presentation, delayed diagnosis, high rates of coincident HIV-1 infection, and rapid emergence of composite forms of drug resistance (1). Moreover, the 6-month TB treatment period is challenging for control programs of many countries with high incidences of the disease (2). In light of this, global research efforts are directed toward creating new treatment-shortening interventions and next-generation diagnostics, which collectively should result in more effective management of TB (3). Regrettably, these efforts have been plagued by a poor understanding of mycobacterial physiology during TB disease in humans and the lack of predictive biomarkers of treatment response and disease recurrence (4). In addition, surprisingly little is known about bacterial growth, or lack thereof, during latent TB infection, despite the fact that one-third of the world’s population is predicted to carry this form of TB (5–7).
Bacterial growth is a complex process modulated by various enzymes and the role of resuscitation-promoting factors (Rpfs), a group of secreted enzymes implicated in enhancing bacterial culturability, has been of particular interest (8–10). Mycobacterium tuberculosis possesses five rpf-like genes, designated rpfA–E, which are collectively dispensable for growth in vitro but are required for resuscitation from a nonculturable state and pathogenesis in a mouse model of TB infection (11). It has been demonstrated that a large proportion of sputum-derived bacteria required Rpf-supplementation for growth (12). These organisms, which we term differentially culturable tubercle bacteria (DCTB), were quantified in the sputum of patients with drug-susceptible active TB disease before the initiation of treatment (12). It is predicted that DCTB are unable to grow on solid media, but can be recovered in liquid media and a recent study also demonstrated that they display drug tolerance in vitro (12, 13). This is corroborated by studies that identified persisting/nonculturable bacterial populations in sputum, the recovery of which can be enhanced by supplementation of growth media with culture filtrate (CF) from axenic cultures of M. tuberculosis or recombinant Rpfs (13–15). The presence of DCTB has also been confirmed in the murine model of TB infection (16, 17).
Considering the importance of these observations and their consequences for the management and ultimate eradication of TB disease, we undertook a comprehensive validation of these preliminary findings in a large cross-sectional observational group of patients. We address key knowledge gaps highlighted by prior studies: (1) Do all patients with TB harbor DCTB? (2) Is the detection of DCTB dependent on the presence of Rpfs in CF? (3) Is there variance in the distribution of DCTB between individuals infected with HIV-1 and those uninfected with HIV-1? (4) How does the presence of these organisms correlate with existing TB diagnostics?
Methods
Study Design
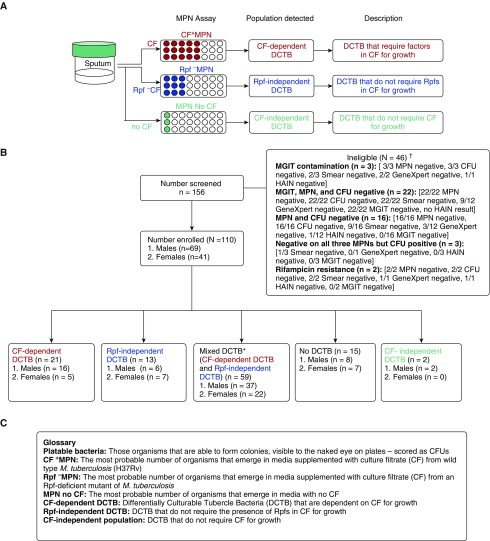
Ethics approval for this study was obtained from the Human Research Ethics Committee of the University of the Witwatersrand (clearance number: M110532). Potential study participants were approached at primary care clinics in Soweto, South Africa. Drug-sensitive patients with TB with a positive test result from either auramine-stained smear or by GeneXpert, obtained from the public sector National Health Laboratory Service (Johannesburg, South Africa), were eligible for recruitment. Patients willing to participate were asked to attend the study clinic where informed consent was obtained and a spot sputum was collected for processing in this study. Further details on the patient cohort, sputum processing, and most probable number (MPN) assays can be found in the online supplement. Briefly, the MPN assay was performed using CF supplementation (see Figure E1 in the online supplement). CF was isolated either from wild-type M. tuberculosis (referred to as CF) or from a quintuple rpf gene-knockout (referred to as Rpf− CF), which allowed for the detection of CF-dependent or Rpf-independent DCTB, respectively. As a control, MPN assays with no CF supplementation were also performed, which allowed for detection of CF-independent DCTB (Figure 1).
Figure 1.
Participant disposition flow chart. (A) Most probable number (MPN) assays were set up with culture filtrate (CF) and resuscitation-promoting factor (Rpf)− CF supplementation of growth media. CF was isolated from wild type Mycobacterium tuberculosis and Rpf− CF from a quintuple rpf gene-knockout mutant, allowing for the detection of Rpf-dependent or Rpf-independent differentially culturable tubercle bacteria (DCTB), respectively. To control for the CF effect, MPN assays with no CF supplementation were also performed, which allowed for detection of CF-independent DCTB. (B) A total of 156 patients were analyzed in this study. These included individuals that had strong clinical indication for TB disease either through a positive smear or positive GeneXpert result. Of these, 46 patients were excluded because of lack of culture-confirmed M. tuberculosis infection detected by MGIT, MPN, and/or CFU. Patients were also deemed ineligible if they were on antibiotic treatment or infected with drug-resistant strains. Of the 110 eligible patients, 21 and 13 had CF-dependent and Rpf-independent DCTB, respectively, whereas 59 patients had mixed DCTB populations. Sputum from 15 patients did not harbor DCTB, and two patients carried CF-independent DCTB. (C) A glossary of terms used in the figure. *Mixtures of CF-dependent DCTB and Rpf-independent DCTB populations. †In some cases, the data for laboratory diagnosis (GeneXpert, MGIT, HAIN, and smear status) were not available. HAIN = Hain Lifesciences Line Probe Assay Genotype MTBDR plus; MGIT = mycobacteria growth indicator tube.
Refinement of the MPN Assay
The MPN assay is based on a Poisson distribution of growth in a limiting dilution series (see Figure E1). Accurate quantification requires consistent dilution of organisms across a serial dilution series and in this context, a significant confounder was the possibility of bacterial clumping. To address this, dispersal of sputum-derived organisms was tested by vortexing in the presence of 2-mm glass beads for 10 seconds at maximum speed. Sputum samples from 19 patients were split equally and one sample was vortexed with beads and the other without beads. In many cases, bead treatment of the sputum sample allowed for a marginally higher MPN count and did not notably affect the viability of bacteria (see Figure E2). Moreover, in samples where the full quantum of organism was low, bead treatment allowed for the detection of low levels of DCTB (patients 54044 and 54060) (see Figure E2). Additional quality checks are detailed in the online supplement.
Data Analysis
Patients were stratified into HIV-1–infected and HIV-1–uninfected groupings based on two standard HIV rapid tests. Their bacillary load measures (CF+/Rpf−/MPN no CF, colony forming units [CFUs], mycobacteria growth indicator tube (MGIT) time to positivity [TTP] in days, smear status, and GeneXpert cycle threshold) were compared between groups, and medians with interquartile ranges were determined using the Kruskal-Wallis test. Frequencies and associated percentages were determined by lung pathology variables on chest radiograph and compared between the two groups using the chi-square test of proportions. Similarly, patient immunology and conventional TB diagnosis variables were compared using the chi-square test of proportions. For a further sensitivity analysis, we divided the individuals infected with HIV-1 into groups with CD4 counts less than/greater than 200 cells/mm3 and conducted a comparative analysis as described previously. All statistical analyses were two-sided and performed at a 95% level of significance using SAS Enterprise Guide version 5.1 (Statistical Analysis Software Institute, Cary, NC). Further detail is provided in the online supplement.
Results
The participant disposition flow chart is given in Figure 1. We numerically defined DCTB as the population that is detected when bacillary load, as quantified by the MPN assay, exceeded the number of platable organisms (CFU). Our experimental design consisted of three distinct MPN assays for each sputum sample, including (1) CF+ MPN, (2) Rpf− MPN, and (3) MPN no CF, which allowed for the relative quantification of CF-dependent, Rpf-independent, and CF-independent DCTB in each patient (Figure 1).
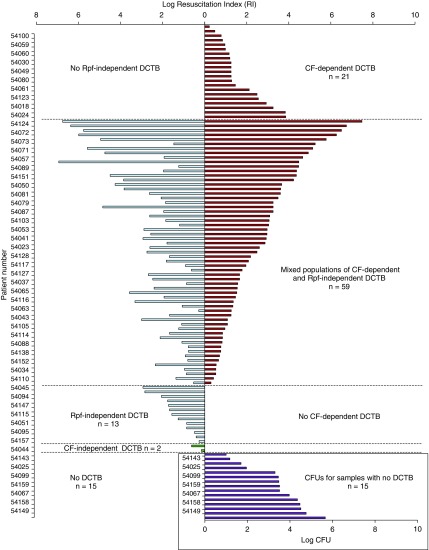
We recruited 156 patients of whom 110 were culture positive for TB. The median age was 37.0 years, 63% were men, and 56% were HIV-infected (Table 1). From those who had a prior GeneXpert test, 84% were positive, 74% were smear positive, and 93% were positive on MGIT culture, with median TTP of 13.5 days (Table 1). From these sputum samples, 21 of 110 (19.1%) yielded CF-dependent DCTB, with no other detectable DCTB (Figure 2; see Table E1). Conversely, 13 of 110 (11.8%) displayed Rpf-independent DCTB, with no detectable CF-dependent DCTB (Figure 2; see Table E1). In these cases, it is possible that the addition of CF resulted in suppression of growth, as measured by the increase in growth stimulation observed when the same sputum sample was supplemented with Rpf− CF. This inhibitory effect was also observed, albeit to a lesser extent, in sputum samples displaying mixed DCTB populations, where some sputa displayed higher levels of Rpf-independent DCTB when compared with CF-dependent DCTB (see Table E1). Most sputum samples in our study, 59 (53.6%), had both populations of CF-dependent and Rpf-independent DCTB (Figure 2; see Table E1). Of the remaining samples, 15 (13.6%) had no DCTB, where the quantum of organisms detected by the MPN assay was equivalent or lower than the CFU. A further two patients (1.8%) had CF-independent DCTB where the number of organisms in the MPN no CF was higher than either the CF or Rpf− CF supplemented MPN assays (Figure 2, inset; see Table E1). We also noted significant variation in the quantum of various DCTB populations between sputum samples, ranging from 0.2 to 7.5 log, and in some cases, this is higher than previously observed (12).
Table 1.
Demographics, Microbiology, and Diagnostic Data for Patients with TB Categorized by HIV-1 Infection Status and CD4 T-Cell Counts
| Variable | HIV Status |
CD4 Classification |
|||||
|---|---|---|---|---|---|---|---|
| Overall (n = 110) | HIV Negative (n = 49) | HIV Positive (n = 61) | P Value*† | CD4 <200 (n = 34) | CD4 >200 (n = 25) | P Value*† | |
| Demographics | |||||||
| Male, n (%) | 69.0 (62.7) | 35.0 (71.4) | 34.0 (55.7) | 0.0907 | 20.0 (58.8) | 13.0 (52) | 0.6019 |
| Age, yr, median (IQR) | 37.0 (30.0–44.0) | 32.0 (25.0–40.0) | 41.0 (34.0–44.0) | 0.0001 | 40.0 (33.0–44.0) | 42.0 (34.0–51.0) | 0.1380 |
| BMI‡ | |||||||
| Underweight, n (%) | 45.0 (40.9) | 23.0 (46.9) | 22.0 (36.1) | 0.25 | 10.0 (29.4) | 11.0 (44.0) | 0.2475 |
| Normal, n (%) | 58.0 (52.7) | 22.0 (46.9) | 35.0 (57.4) | 0.19 | 22.0 (67.6) | 11.0 (44.0) | 0.1134 |
| Overweight, n (%) | 7.0 (6.4) | 3.0 (6.1) | 4.0 (6.6) | 0.93 | 1.0 (2.9) | 3.0 (12.0) | 0.1714 |
| Median (IQR), kg/m2 | 19.2 (17.2–22.0) | 18.8 (16.8–20.6) | 20.1 (17.6–22.4) | 0.1293 | 20.2 (18.3–22.5) | 19.2 (17.5–22.2) | 0.6773 |
| Lung pathology | |||||||
| Cavitation§ | |||||||
| Cavity vs. no cavity, n (%) | 33.0 (33.0) vs. 67.0 (67.0) | 10.0 (23.8) vs. 32.0 (76.2) | 23.0 (39.7) vs. 35.0 (60.4) | 0.1061 | 12.0 (43.0) vs. 16.0 (57.0) | 9.0 (45.0) vs. 11.0 (55.0) | 0.4733 |
| Extent of disease, n (%)|| | |||||||
| Limited | 33.0 (35.5) | 8.0 (20.0) | 25.0 (47.2) | 0.007 | 12.0 (40.0) | 12.0 (57.1) | 0.2274 |
| Moderate | 28.0 (30.1) | 19.0 (47.5) | 9.0 (17.0) | 0.002 | 7.0 (23.3) | 1.0 (4.8) | 0.0727 |
| Extensive | 32.0 (34.4) | 13.0 (32.5) | 19.0 (35.8) | 0.74 | 11.0 (36.7) | 8.0 (38.1) | 0.9173 |
| Patient immunology | |||||||
| CD4 count (only HIV-infected), cells/mm3, median (IQR)¶ | N/A | N/A | 139.0 (83.0–331.0) | N/A | 89.0 (74.0–119.0) | 350.0 (263.0–414.0) | <0.0001 |
| HAART treatment, n (%)** | N/A | N/A | 12.0 (19.7) | N/A | 4.0 (11.8) | 6.0 (24.0) | 0.2158 |
| Conventional TB diagnosis, n (%) | |||||||
| Smear grade positive†† | 81.0 (73.6) | 41.0 (83.7) | 40.0 (65.6) | 0.0322 | 23.0 (67.6) | 16.0 (64) | 0.7700 |
| Scanty/+ | 16.0 (14.5) | 9.0 (18.4) | 7/61 (11.5) | 0.308 | 5.0 (14.7) | 1.0 (4.0) | 0.1788 |
| ++ | 12.0 (10.9) | 4.0 (8.2) | 8/61 (13.1) | 0.408 | 4.0 (11.8) | 4.0 (16.0) | 0.6387 |
| +++ | 53.0 (48.2) | 28.0 (57.1) | 25/61 (41.0) | 0.092 | 14.0 (41.2) | 11.0 (44.0) | 0.8283 |
| GeneXpert result‡‡ | |||||||
| High, n (%) | 14.0 (15.1) | 9.0 (23.1) | 5.0 (9.3) | 0.066 | 3.0 (9.4) | 2.0 (9.5) | 0.9855 |
| Medium, n (%) | 24.0 (25.8) | 16.0 (41.0) | 8.0 (14.8) | 0.004 | 4.0 (12.5) | 4.0 (19) | 0.5149 |
| Low, n (%) | 40.0 (43.0) | 9.0 (23.1) | 31.0 (57.4) | 0.001 | 21.0 (65.6) | 10.0 (47.6) | 0.1932 |
| None, n (%) | 15.0 (16.1) | 5.0 (12.8) | 10.0 (18.5) | 0.46 | 4.0 (12.5) | 5.0 (23.8) | 0.2835 |
| Median (IQR) GeneXpert cycle threshold | 21.3 (15.8–26.3) | 18.3 (15.3–22.4) | 23.6 (16.1–27.7) | 0.0278 | 24.3 (17.9–28.1) | 23.0 (12.8–25.8) | 0.3525 |
| MGIT time to positivity, d, median (IQR) | 13.5 (10–20)§§ | 12.0 (9–18) | 15.0 (11–20) | 0.0958 | 15.5 (13–22) | 13.0 (8.5–20) | 0.3277 |
| MPN | |||||||
| CF+ MPN, log median (IQR) | 2.9 (1.7–4.7) | 3.5 (1.9–5.2) | 2.7 (1.3–3.9) | 0.0725|||| | 1.9 (1.3–3.3) | 3.6 (2.3–6.2) | 0.0236¶¶ |
| Rpf− MPN, log median (IQR) | 2.6 (1.3–4.5) | 2.7 (0.9–5.2) | 2.6 (1.3–4.3) | 0.5555|||| | 2.2 (1.3–3.7) | 2.7 (1.9–4.5) | 0.2483¶¶ |
| MPN no CF, log median (IQR) | 0.0 (0.0–0.9) | 0.0 (0.0–0.9) | 0.0 (0.0–0.9) | 0.9259|||| | 0.0 (0.0–0.9) | 0.0 (0.0–1.7) | 0.4000¶¶ |
| MPN time to positivity, d, median (IQR) | 21.0 (14.0–21.0) | 21.0 (14.0–21.0) | 21.0 (14.0–21.0) | 0.7110|||| | 21.0 (14.0–28.0) | 14.0 (14.0–21.0) | 0.3071¶¶ |
| CFU, median (IQR) | 1.7 (0.0–3.5) | 2.2 (0.8–3.6) | 1.6 (1.0–2.8) | 0.0867|||| | 1.1 (1.0–2.4) | 1.7 (1.0–4.0) | 0.0936¶¶ |
| Strain typing, n (%) | |||||||
| Beijing | 20.0 (18.2) | 8.0 (16.3) | 12.0 (19.7) | 0.6511 | 6.0 (17.6) | 5.0 (20.0) | 0.8186 |
| Non-Beijing | 61.0 (55.5) | 26.0 (53.1) | 35.0 (57.4) | 0.6508 | 20.0 (58.8) | 15.0 (60.0) | 0.9276 |
| Mixed | 21.0 (19.1) | 13.0 (26.5) | 8.0 (13.1) | 0.0764 | 3.0 (8.8) | 4.0 (16.0) | 0.3996 |
| Not determined | 8.0 (7.3) | 2.0 (4.1) | 6.0 (9.8) | 0.2481 | 5.0 (14.7) | 1.0 (4.0) | 0.1788 |
Definition of abbreviations: BMI = body mass index; CF = culture filtrate; HAART = highly active antiretroviral therapy; IQR = interquartile range; MGIT = mycobacteria growth indicator tube; MPN = most probable number; N/A = not applicable; Rpf = resuscitation-promoting factors; TB = tuberculosis.
P value compares HIV-positive versus HIV-negative and HIV-positive individuals with CD4 count below 200 and above 200; significant at P less than 0.05 (95% confidence interval), shown in bold.
All the proportion comparisons by HIV status and CD4 classification were conducted by chi-square test; MPN comparison was conducted by analysis of covariance adjusting for sex and BMI.
One participant did not have data for weight and height. Underweight: BMI <18.5 kg/m2; normal: BMI 18.5 to <25.0 kg/m2; overweight: BMI 25.0 to <30.0 kg/m2.
Nineteen participants did not have cavitation results in the HIV stratification category, and 10 patients did not have cavitation data in the CD4 classification category.
Seventeen participants did not have results for extent of disease in the HIV status category. Eight participants with CD4 count records did not have results for extent of disease. Limited lesions involve a total lung area less than one-quarter the area of the entire thoracic cavity. Moderate lesions are bigger than limited lesions and even with bilateral involvement; these have a total lung area of less than one-half the area of the entire thoracic cavity. Extensive lesions involve a total lung area equal to or more than half the area of the entire thoracic cavity. Radiographs are viewed either anteroposterior or posteroanterior.
Two participants had missing CD4 counts.
Two participants were on HAART with missing CD4 counts in the HIV status category, and two participants had missing HAART results in the CD4 classification category.
Includes scanty, +, ++, and +++.
Seventeen patients did not have a GeneXpert result; one patient had an error reading on the GeneXpert in the HIV status category; six participants had no GeneXpert result; and one patient had an error reading for GeneXpert in the CD4 classification category.
Ninety-eight patients had a positive MGIT result out of 106 patients, the remaining four had contamination/missing information in the HIV status category, and 56 patients had a positive MGIT result out of 59 patients. The remaining three had contamination/missing information in the CD4 classification category.
P values presented are unadjusted. When adjusted for BMI and sex, P values are 0.1504, 0.3992, 0.5564, 0.8756, and 0.2965 for log median CF+ MPN, log median Rpf− MPN, log median MPN no CF, median MPN time to positivity, and median CFU, respectively.
P values presented are unadjusted. When adjusted for BMI and sex, P values are 0.0176, 0.2067, 0.8721, 0.6079, and 0.0942 for log median CF+ MPN, log median Rpf− MPN, log median MPN no CF, median MPN time to positivity, and median CFU, respectively.
Figure 2.
Distribution of differentially culturable tubercle bacteria (DCTB) in a cross-sectional group of patients with tuberculosis. Shown on the y-axis are individual patients with their relative proportions of DCTB (given as the quantum of resuscitatable bacteria, reported as the resuscitation index [RI = MPN/CFU]) on the x-axis. Culture filtrate (CF)-dependent DCTB, calculated as the log(CF+ MPN/CFU), is reflected in red. If the calculation of CF-dependent or Rpf-independent DCTB yielded a negative value, this was adjusted to 0 because only the growth stimulatory effects of CF/Rpf− CF were considered. Rpf-independent DCTB, calculated as the log(Rpf− MPN/CFU), is reflected in blue. CF-independent DCTB, calculated as the log(MPN No CF/CFU), is shown in green. In cases where the CFU was zero, a value of 1 was used to reflect the absence of culturable bacteria, which indicates that the entire population detected in the MPN assay constituted DCTB. The combined colored bars reflect patients with both CF-dependent and independent DCTB populations and absence of bars indicated no detectable DCTB. Inset depicts log CFU counts in samples with no detectable DCTB population. MPN = most probable number; Rpf = resuscitation-promoting factors.
Thirty-four patients in our study displayed no culturable bacteria on solid media for CFU determination and eight samples did not grow in routine liquid culture (MGIT). However, sputum from all of these patients displayed growth of M. tuberculosis in the MPN assays, albeit with variable dependency on Rpfs (see Table E1) demonstrating the benefit of CF-supplementation in the diagnosis of TB. Correlation between detectable CFUs and HIV-1 infection status revealed that samples that displayed no CFUs were largely individuals infected with HIV-1 (23 of 34 [68%]) but these sputum samples yielded positive growth in MPN assays. In our group of patients, a greater proportion of individuals infected with HIV-1 were male (n = 34; 55.7%) and older than their counterparts not infected with HIV-1 (41 vs. 32 yr; P = 0.0001). Additionally, a larger proportion of patients infected with HIV-1 had limited lung pathology, negative smears, and a GeneXpert diagnosis with a high median cycle threshold (Table 1).
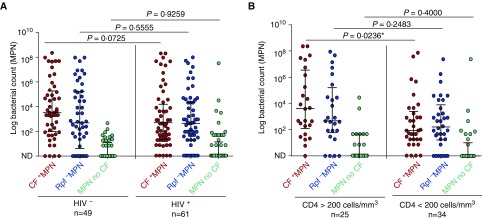
We hypothesized that individuals with compromised immunity retain altered proportions of MPN-responsive organisms when compared with individuals not infected with HIV-1. Sputum samples from patients infected with HIV-1 displayed lower levels of CF+ MPN counts when compared with sputum from patients not infected with HIV-1 (2.7 vs. 3.5; not significant with a 95% confidence interval; P = 0.0725, unadjusted) (Figure 3A and Table 1). This observation suggested that compromised host immunity is associated with a reduction in DCTB and culturable bacteria. When adjusted for sex and body mass index, this difference remains statistically insignificant (P = 0.1504). No significant differences were found in the proportion of Rpf− MPN bacterial subpopulations between participants infected and not infected with HIV-1 (Table 1).
Figure 3.
Measures of bacterial load stratified by HIV-1 infection status or CD4 T-cell counts. (A) Scatterplot depicting bacterial load distributions in HIV infected/uninfected individuals. (B) Scatterplot depicting bacterial load distributions in individuals with high versus low CD4 T-cell counts. Error bars represent medians and interquartile ranges. To determine statistical significance, the Mann–Whitney U test was used with a 95% confidence interval. CF+ MPN (red), Rpf− MPN (blue), MPN no CF (green). Two participants had missing CD4 counts. In all categories the CF+ MPN/Rpf− MPN yielded higher bacterial counts when compared with the MPN no CF (P < 0.0001). *Significant with a 95% confidence interval. CF = culture filtrate; MPN = most probable number; ND = no bacterial growth detected; Rpf = resuscitation-promoting factors.
In individuals infected with HIV-1, when stratified into CD4 counts less than/greater than 200 cells/mm3, there were no differences in demographics, chest radiograph presentation, and GeneXpert cycle threshold (Table1). However, a significant difference in CF+ MPN counts was noted where individuals with CD4 counts greater than 200 cells/mm3 had a higher median CF+ MPN compared with those with CD4 counts less than 200 cells/mm3 (3.6 vs. 1.9; P = 0.0236, unadjusted; P = 0.0176, adjusted for sex and body mass index) (Figure 3B and Table 1).
Next we investigated the relationship between DCTB and other diagnostic methods used to detect mycobacterial load, such as TTP in MGIT and GeneXpert cycle threshold. We stratified our patient group by smear status and found a strong correlation between the CF+ MPN and TTP or GeneXpert cycle threshold in both smear-positive and smear-negative patients (Table 2). This effect is lost in smear-negative patients in the absence of Rpfs (Rpf− MPN) (Table 2). Rpfs also enhanced bacterial detection by the MPN assay, because the absence of Rpfs results in a greater proportion of samples with no growth in the MPN assay (24 of 110 with no growth in Rpf− MPN vs. 9 of 110 with no growth in CF+ MPN) (see Table E1).
Table 2.
Diagnostic Benefit of CF Supplementation versus No Rpf Supplementation in Smear-Negative and Smear-Positive Individuals
| Variables | Smear-Positive Correlation* | Smear-Negative Correlation* |
|---|---|---|
| CF+ MPN | ||
| MGIT TTP† vs. log CF+ MPN | −0.54‡ (P < 0.0001) | −0.54‡ (P = 0.0049) |
| GeneXpert cycle threshold vs. log CF+ MPN | −0.62‡ (P < 0.0001) | −0.74‡ (P = 0.0004) |
| Rpf− MPN | ||
| MGIT TTP† vs. log Rpf− MPN | −0.53‡ (P < 0.0001) | −0.13§ (P = 0.53) |
| GeneXpert cycle threshold vs. log Rpf− MPN | −0.61‡ (P < 0.0001) | −0.01§ (P = 0.95) |
Definition of abbreviations: CF = culture filtrate; MGIT = mycobacteria growth indicator tube; MPN = most probable number; Rpf = resuscitation-promoting factors; TTP = time to positivity.
Pearson coefficient.
TTP in days.
Significant negative linear relationship between variable and log CF+ MPN/Rpf− MPN.
A weak nonsignificant negative linear relationship between variable and log Rpf− MPN in smear-negative patients.
The growth stimulatory effect observed with CF supplementation of sputum cultures in previous studies has been ascribed to the activity of Rpfs (12). A limitation in some of these studies was that dependency on Rpfs was not assessed by using a control CF without Rpfs. Our data thus far illustrated that in some cases, Rpfs contribute to increasing bacterial growth in sputum-derived bacteria. However, we now report the detection of an Rpf-independent DCTB population in TB-diseased patients. To further test the contributions of putative growth stimulatory enzymes, the CF and Rpf− CF was subjected to heat treatment to inactivate secreted enzymes. Heat treatment resulted in a reduction, but not abrogation, of CF+ MPNs in most cases, with complete loss of growth stimulatory activity in 9 of 35 (26.0%) samples (see Figure E3). In the Rpf− MPN, heat treatment also led to a loss of growth in 6 of 35 (17%) samples (see Figure E3). The residual growth stimulation detected in most samples with heat-treated CF or Rpf− CF suggests that growth stimulation in MPN assays with these supplements is most likely the result of interplay between heat-labile and heat-resistant factors.
Finally, we assessed strain diversity in our study population to gain a better understanding of the distribution of circulating strains within the study community and to assess if any strains preferentially responded to Rpf supplementation. From the 110 samples, spoligotyping results were obtained for 102 cultures. In 81 of these, a single strain was detected, which we classified either into Beijing or non-Beijing. There was no significant difference in the distribution of these strains between patients with HIV-1 versus patients without HIV-1 or in patients with HIV-1 with CD4 counts less than/greater than 200 cells/mm3 (Table 1). However, specimens from patients with Rpf-independent DCTB (and no other detectable DCTB population) displayed no Beijing strains, whereas these strains were found in patients with only CF-dependent DCTB or mixed DCTB populations (see Figure E4). These data suggest that patients infected with Beijing strains harbor organisms that are responsive to Rpfs. In the remaining 21 patients, we detected mixed strain infections (Table 1).
Discussion
The ability to detect and quantify DCTB in patients with either active TB or latent TB infection is critically important for assessing the risk for reactivation of disease and monitoring responses to TB treatment. In this study, we determined the proportion of bacterial populations with differential culturability in patients with TB with and without HIV-1. The lack of culturability after periods of stress has commonly been used as a surrogate measure for metabolic quiescence and drug tolerance (18) and we hypothesize that our measure of DCTB in sputum-derived bacteria provides a useful readout of the proportion of nonreplicating organisms in the host. The presence of DCTB in the sputum of patients with active TB disease is documented (12).
Consistent with this, we were able to robustly detect populations of CF-dependent DCTB in a large proportion of patients. It should be recognized that the use of CF does not necessitate the direct involvement of Rpfs in unmasking CF-dependent DCTB. This could be the result of other factors in the CF or the products of Rpf-mediated cleavage of the cell wall. Additionally, we report a second, distinct Rpf-independent DCTB population that emerges in MPN assays with Rpf− CF supplementation. These Rpf-independent DCTB are present in patients either as a single detectable population or as mixed populations with CF-dependent DCTB. A notable proportion of sputum samples from patients with HIV-1 was associated with the lack of culturable bacteria on standard solid media and in these cases, the MPN assay with CF supplementation may be a useful TB diagnostic test because it allows for identification of bacteria that do not grow on solid media. The CF+ MPN assay also provided significant benefit in detecting bacteria in sputum samples that were smear negative. However, it should also be noted that some samples in our study harbored bacilli with improved growth on solid agar. In these cases, bacillary load would be underestimated in liquid media, highlighting the possibility that a single diagnostic test may not be sufficient to capture all the phenotypic diversity present in sputum.
Rpfs are lytic transglycosylases with the ability to hydrolyze peptidoglycan in the bacterial cell wall and as such, play an important role in regulating degradation of the bridge between cells during division to promote daughter cell separation (10, 19, 20). In this regard, exogenous supplementation of bacterial cultures with Rpfs (or CF) may promote cell separation in dividing cells and thereby increase the number of singlet cells in the culture. This would manifest in the higher MPN score observed with CF supplementation and consequently lead to an overestimation of the quantum of DCTB present. Although we cannot unequivocally rule this out in patients with DCTB, the following two observations suggest that this cannot be the sole explanation for the occurrence of DCTB in our patient population: a notable proportion of patients in this study had Rpf-independent DCTB, where supplementation with Rpf− CF results in enhanced bacterial recovery, indicating that growth stimulation of DCTB is mediated through CF-dependent and Rpf-independent mechanisms; and analysis of cell sizes in a random sampling of patients confirms that most sputum-derived bacteria exist as singletons, with a small proportion of bacteria that are longer than 3.5 μM, which would represent dividing bacteria (see Figure E5).
Thus, hydrolysis of the septum by cell wall hydrolases cannot solely account for the occurrence of DCTB in MPN assays. To further address the mechanistic basis of CF-mediated growth stimulation, MPN assays were conducted with heat-treated filtrate to eliminate enzymatic activity. Heat-treated CF and Rpf− CF displayed reduced growth stimulation but in most cases, the stimulatory effect was not completely abolished. This suggests that growth stimulation of organisms in sputum is mediated by both enzymatic and nonenzymatic effects. In this regard, it has been demonstrated that cyclic AMP and fatty acids retain the capacity to modulate the growth of nonreplicating mycobacteria (21–23).
Host immunity has been predicted to play an important role in determining the environmental stresses that tubercle bacteria experience during infection and inflammation associated with chronic granulomatous TB disease. Immune responses, together with drug treatment, may drive the formation of, or select for, drug-tolerant populations in pulmonary lesions. We sought to test this hypothesis by studying the distribution of DCTB in patients with HIV-1 infection and found that individuals with a comparatively better immune competency (as assessed by the lack of HIV-1 infection or CD4 T cell counts >200 cells/mm3 in individuals with HIV-1) display higher CF+ MPN values. This suggests that a reduced host immune response affects the distribution of CF+ MPN-responsive organisms. However, these experiments are insufficient to unequivocally associate modulation of host immunity with changes in DCTB profiles and further work is required in this regard.
In conclusion, our data confirm the presence of DCTB in a group of South African patients with TB at baseline. Detection of both CF-dependent and -independent DCTB in the sputum of patients with TB points to a dynamic interplay between distinct subpopulations in the infected human lung. How these populations respond to therapy and the manner in which they change with resolution of disease now awaits further investigation.
Acknowledgments
Acknowledgment
The authors thank Caroline Tiemessen, William Mac Kenzie, Galina Mukamolova, Michael Barer, and CBTBR laboratory members for insightful comments and technical advice. The authors are grateful to the participants in this study.
Footnotes
Supported by the National Institutes of Health (U01 AI069453-07), National Research Foundation of South Africa, South African Medical Research Council, Centre for Aids Prevention Research in South Africa, and the Howard Hughes Medical Institute.
Author Contributions: B.D.K., R.H., and G.C. conceived the study. M.D.C. and G.M.B. executed experimental work. E.M.S. and R.W. assisted with spoligotyping. K.O. conducted statistical analysis. N.M. was responsible for patient recruitment. B.G.G. provided assistance with study design, experimental analysis, and study management. B.D.K., B.G.G., and M.D.C. compiled the manuscript. All authors approved the final version of the manuscript.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.201604-0769OC on July 7, 2016
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Dheda K, Barry CE, 3rd, Maartens G. Tuberculosis. Lancet. 2015;387:1211–1226. doi: 10.1016/S0140-6736(15)00151-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zumla A, Kim P, Maeurer M, Schito M. Zero deaths from tuberculosis: progress, reality, and hope. Lancet Infect Dis. 2013;13:285–287. doi: 10.1016/S1473-3099(13)70039-2. [DOI] [PubMed] [Google Scholar]
- 3.Zumla A, Nahid P, Cole ST. Advances in the development of new tuberculosis drugs and treatment regimens. Nat Rev Drug Discov. 2013;12:388–404. doi: 10.1038/nrd4001. [DOI] [PubMed] [Google Scholar]
- 4.Wallis RS, Kim P, Cole S, Hanna D, Andrade BB, Maeurer M, Schito M, Zumla A. Tuberculosis biomarkers discovery: developments, needs, and challenges. Lancet Infect Dis. 2013;13:362–372. doi: 10.1016/S1473-3099(13)70034-3. [DOI] [PubMed] [Google Scholar]
- 5.Young M, Mukamolova GV, Kaprelyants AS.Mycobacterial dormancy and its relation to persistence Parish T.editor. Mycobacterium: molecular biology Norwich, UK: Horizon Scientific; 2005265–320. [Google Scholar]
- 6.Chao MC, Rubin EJ. Letting sleeping dos lie: does dormancy play a role in tuberculosis? Annu Rev Microbiol. 2010;64:293–311. doi: 10.1146/annurev.micro.112408.134043. [DOI] [PubMed] [Google Scholar]
- 7.Esmail H, Barry CE, III, Young DB, Wilkinson RJ. The ongoing challenge of latent tuberculosis. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130437. doi: 10.1098/rstb.2013.0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kana BD, Mizrahi V. Resuscitation-promoting factors as lytic enzymes for bacterial growth and signaling. FEMS Immunol Med Microbiol. 2010;58:39–50. doi: 10.1111/j.1574-695X.2009.00606.x. [DOI] [PubMed] [Google Scholar]
- 9.Kana BD, Mizrahi V. Resuscitation promoting factors in bacterial population dynamics during TB infection. Drug Discov Today Dis Mech. 2010;7:e13–e18. [Google Scholar]
- 10.Nikitushkin VD, Demina GR, Shleeva MO, Guryanova SV, Ruggiero A, Berisio R, Kaprelyants AS. A product of RpfB and RipA joint enzymatic action promotes the resuscitation of dormant mycobacteria. FEBS J. 2015;282:2500–2511. doi: 10.1111/febs.13292. [DOI] [PubMed] [Google Scholar]
- 11.Kana BD, Gordhan BG, Downing KJ, Sung N, Vostroktunova G, Machowski EE, Tsenova L, Young M, Kaprelyants A, Kaplan G, et al. The resuscitation-promoting factors of Mycobacterium tuberculosis are required for virulence and resuscitation from dormancy but are collectively dispensable for growth in vitro. Mol Microbiol. 2008;67:672–684. doi: 10.1111/j.1365-2958.2007.06078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mukamolova GV, Turapov O, Malkin J, Woltmann G, Barer MR. Resuscitation-promoting factors reveal an occult population of tubercle bacilli in sputum. Am J Respir Crit Care Med. 2010;181:174–180. doi: 10.1164/rccm.200905-0661OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turapov O, O’Connor BD, Sarybaeva AA, Williams C, Patel H, Kadyrov AS, Sarybaev AS, Woltmann G, Barer MR, Mukamolova GV. Phenotypically adapted Mycobacterium tuberculosis populations from sputum are tolerant to first line drugs. Antimicrob Agents Chemother. 2016;60:2476–2483. doi: 10.1128/AAC.01380-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kolwijck E, Friedrich SO, Karinja MN, van Ingen J, Warren RM, Diacon AH. Early stationary phase culture supernatant accelerates growth of sputum cultures collected after initiation of anti-tuberculosis treatment. Clin Microbiol Infect. 2014;20:O418–O420. doi: 10.1111/1469-0691.12441. [DOI] [PubMed] [Google Scholar]
- 15.Dhillon J, Fourie PB, Mitchison DA. Persister populations of Mycobacterium tuberculosis in sputum that grow in liquid but not on solid culture media. J Antimicrob Chemother. 2014;69:437–440. doi: 10.1093/jac/dkt357. [DOI] [PubMed] [Google Scholar]
- 16.Hu Y, Liu A, Ortega-Muro F, Alameda-Martin L, Mitchison D, Coates A. High-dose rifampicin kills persisters, shortens treatment duration, and reduces relapse rate in vitro and in vivo. Front Microbiol. 2015;6:641. doi: 10.3389/fmicb.2015.00641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turapov O, Glenn S, Kana B, Makarov V, Andrew PW, Mukamolova GV. The in vivo environment accelerates generation of resuscitation-promoting factor-dependent mycobacteria. Am J Respir Crit Care Med. 2014;190:1455–1457. doi: 10.1164/rccm.201407-1289LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barer MR, Harwood CR. Bacterial viability and culturability. Adv Microb Physiol. 1999;41:93–137. doi: 10.1016/s0065-2911(08)60166-6. [DOI] [PubMed] [Google Scholar]
- 19.Hett EC, Chao MC, Deng LL, Rubin EJ. A mycobacterial enzyme essential for cell division synergizes with resuscitation-promoting factor. PLoS Pathog. 2008;4:e1000001. doi: 10.1371/journal.ppat.1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hett EC, Chao MC, Steyn AJ, Fortune SM, Deng LL, Rubin EJ. A partner for the resuscitation-promoting factors of Mycobacterium tuberculosis. Mol Microbiol. 2007;66:658–668. doi: 10.1111/j.1365-2958.2007.05945.x. [DOI] [PubMed] [Google Scholar]
- 21.Shleeva M, Goncharenko A, Kudykina Y, Young D, Young M, Kaprelyants A. Cyclic AMP-dependent resuscitation of dormant Mycobacteria by exogenous free fatty acids. PLoS One. 2013;8:e82914. doi: 10.1371/journal.pone.0082914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Yang Y, Woods A, Cotter RJ, Sun Z. Resuscitation of dormant Mycobacterium tuberculosis by phospholipids or specific peptides. Biochem Biophys Res Commun. 2001;284:542–547. doi: 10.1006/bbrc.2001.4993. [DOI] [PubMed] [Google Scholar]
- 23.Nazarova EV, Shleeva MO, Morozova NS, Kudykina YK, Vostroknutova GN, Ruzhitsky AO, Selishcheva AA, Sorokoumova GM, Shvets VI, Kaprelyants AS. Role of lipid components in formation and reactivation of Mycobacterium smegmatis “nonculturable” cells. Biochemistry (Mosc) 2011;76:636–644. doi: 10.1134/S0006297911060034. [DOI] [PubMed] [Google Scholar]