Sputum rules how we diagnose and manage tuberculosis (TB). Not all patients with TB produce sputum, and those who do furnish a self-produced biopsy of one or more cavities, lesions in which Mycobacterium tuberculosis (Mtb) faces different environmental conditions than in other sites in the body (1). Yet it is sputum that diagnosticians smear, stain, experimentally infect with fluorescent phage, plate on agar, culture in liquid in mycobacterial growth indicator tubes in BACTEC devices, and dispense to cassettes in GeneXpert machines in a combined effort to make the diagnosis, quantify the bacterial burden, and test sensitivity to available drugs (2, 3). The therapist uses the mycobacterial count in sputum to follow the impact of combination therapy and to adjust its length. Clinical trialists subject sputum to the early bactericidal activity test as a gateway through which any new TB drug candidate must pass if it is to go on to larger, longer, and far more expensive tests of efficacy in combination with other drugs (4).
TB diagnosticians, clinical microbiologists, therapists, and clinical trialists already cope with enormous burdens. Now imagine telling them that the tubercle bacilli that they detect and enumerate in sputum from most patients are just a fraction—often a miniscule fraction—of the viable Mtb present; that the ones they count are killed faster by standard TB drugs than the ones to which their tests are blind; and that many sputum specimens that they consider culture negative after several months of treatment are teeming with viable Mtb. The physicians and scientists would probably be intrigued, alarmed—and skeptical.
Such a reaction greeted a landmark article published in the Journal by Mukamolova and colleagues in 2010 (5). Those authors reported that 80 to 99.9% of viable Mtb in the sputa of 20 of 25 treatment-naive patients with TB were only revealed by limiting dilution in fresh Mtb culture filtrate (CF) compared with counting cfu on solid media. Treatment of five patients (out of eight) for 7 to 11 days reduced the number of Mtb detected in their sputa by a standard cfu assay far more than it reduced the differentially culturable tubercle bacilli (DCTB), that is, those revealed by limiting dilution. In four patients monitored for 14 to 115 days of treatment, cfu dropped below the limit of detection, but the limiting dilution assay detected 1.3 to 6 log10 Mtb (5).
The 2010 findings had enormous potential significance but also raised questions. Mtb is notoriously clumpy. If clumps come apart during serial dilution, the estimation of the starting number by the statistical method called “most probable number” can lead to gross exaggeration. Mukamolova and colleagues reported that the detection of far greater numbers of Mtb by limiting dilution than by cfu required the inclusion in the dilution medium of at least one of the five resuscitation-promoting factors (Rpfs)—a name given to a family of mutually homologous cell wall–cleaving enzymes encoded by Mtb (5). Yet, there was very little evidence for detection of DCTB with pure Rpf. Other authors, using in vitro systems to generate DCTB, did not find evidence for dependence on exogenous Rpfs and nominated other candidates as the active factors, including peptides, phospholipids, fatty acids, and cyclic adenosine monophosphate (6–8). The importance of the question attracted a growing number of researchers but produced no reports of independent confirmation.
Enter a second landmark study, published in this issue of the Journal, by Chengalroyen and colleagues (pp. 1532–1540) (9). A meticulous analysis of samples from 110 patients with TB confirmed the observations of Mukamolova and colleagues (5) that DCTB predominate over cfu in sputum from 86% of the subjects. However, the findings of Chengalroyen and colleagues challenged the role of the Rpfs and revealed even greater complexity in the population structure of Mtb. Samples from more than half the patients contained DCTB both when the limiting dilution was performed using CF from wild-type Mtb, which might contain Rpfs, or from Mtb in which Chengalroyen and colleagues had deleted all five rpf genes, which could not contain Rpfs (9). Moreover, another 11.8% of patients provided samples in which DCTB could only be detected if the rpf genes were deleted from the Mtb furnishing the CF. In contrast, in 19.1% of the samples, the CF had to come from wild-type Mtb for DCTB to be detected. Could it get more complicated? Yes: in 1.8% of samples, DCTB were detected only if CF was omitted altogether, and in 13.6% of samples, DCTB were not detected. One way of interpreting these findings—but not the only way—is that sputum may deliver a biopsy of sites where Mtb can predominate in any one of five states, as operationally defined by its detectability as cfu or in various combinations of dependency on factors from the two types of CF, as summarized in Table 1.
Table 1.
Operationally Distinct Subpopulations of Culturable Mycobacterium tuberculosis in Sputum from Treatment-Naive Subjects
| Subpopulation | Numerically Abundant when Assayed by: |
|||
|---|---|---|---|---|
| LD + WT CF | LD + KO CF | LD − CF | cfu | |
| 1 | +++ | +++ | − | − |
| 2 | ++ | − | − | − |
| 3 | − | ++ | − | − |
| 4 | − | − | + | − |
| 5 | − | − | − | ++ |
Definition of abbreviations: CF = culture filtrate; KO = knockout, deleted for five rpf genes; LD = limiting dilution; WT = wild type.
The number of plus signs represents the proportion of subjects with differentially culturable tubercle bacilli or cfu as indicated: +, <2%; ++, 2–50%; +++, >50%.
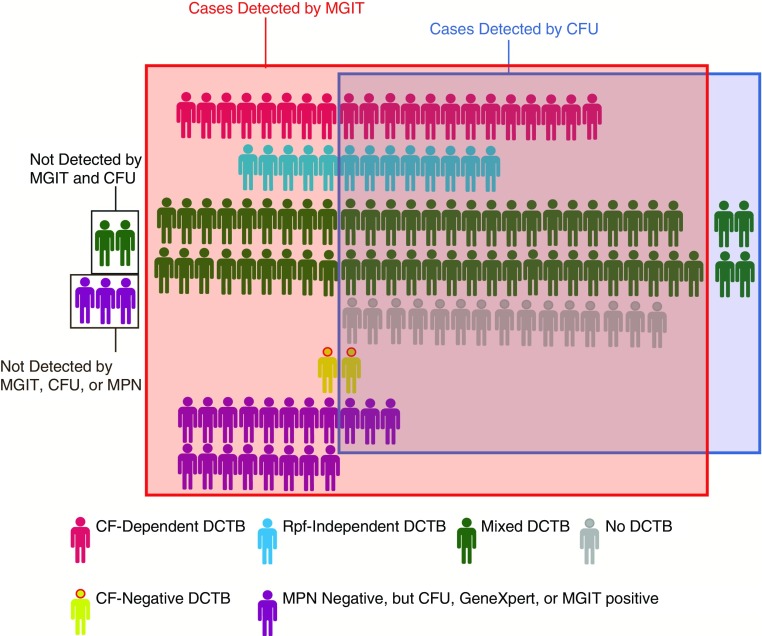
These important and fascinating findings raise a series of questions of their own, on which the study by Chengalroyen and colleagues (9) casts some light, but not yet enough. What drives Mtb into the differentially culturable state? The higher proportion of DCTB in subjects without HIV infection observed by Chengalroyen and colleagues offers a tantalizing but underpowered glimpse into the potential role of host immunity in altering Mtb subpopulations (9). Sputum is known to contain both extracellular and intracellular Mtb bacilli, not only in macrophages but also in neutrophils (10). The relative proportion of these populations varies from patient to patient, which might contribute to the variability of responses to CF with or without Rpfs present. What is the role of CF and of Rpfs? How many different assays on sputum (among those used in this study) is it necessary to apply to maximize diagnosis, to monitor conventional treatment, or to test a drug candidate (Figure 1)? For routine purposes, what combination of assays would best marry yield with feasibility?
Figure 1.
Intersubject variability in culture methods detecting Mycobacterium tuberculosis (Mtb) in sputum and implications for case detection. Each of the 110 subjects for whom results were analyzed is represented according to Mtb culture method positivity (9). Also represented are 22 subjects who were excluded from analysis but were described as having a positive mycobacterial growth indicator tube (MGIT), cfu, or GeneXpert test. The individual’s color classifies the predominant population identified, and they are grouped according to detection method positivity. Not all subjects’ sputum was tested by every method, and available information did not allow subgrouping by sputum smear positivity and GeneXpert positivity. In this patient population preselected by GeneXpert or smear positivity, no single culture method detected all cases. CF = culture filtrate; DCTB = differentially culturable tubercle bacilli; MPN = most probable number; Rpf = resuscitation-promoting factors.
Answers to these questions are needed before rolling out limiting dilution assays into routine practice. To establish the clinical relevance of the “resuscitatable” or DCTB populations, coordinated experimental schemes could be adopted in preclinical efficacy studies, early bactericidal activity trials, and later-stage studies of clinical development, transmission, and epidemiology in the context of both active disease and latent infection. An approach similar to that taken by Chengalroyen and colleagues would inform clinicians and drug discovery and development teams as to whether DCTB play a role in disease progression, therapy outcome, and transmission, and if so, what the most appropriate assays may be to take DCTB into account in diagnosis and treatment (9).
There are still further questions, the urgency of which is increased by the study of Chengalroyen and colleagues (9), but which that study was not designed to address: What molecular features distinguish Mtb in the five operationally defined categories listed in Table 1? What are the implications for actions of drugs? How well do these different states represent subpopulations of Mtb in infected sites other than the cavities that contribute sputum? Is there a way to generate DCTB in vitro to produce Mtb that resemble sputum DCTB well enough to serve for testing drug candidates for activity against DCTB?
Footnotes
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Lenaerts A, Barry CE, III, Dartois V. Heterogeneity in tuberculosis pathology, microenvironments and therapeutic responses. Immunol Rev. 2015;264:288–307. doi: 10.1111/imr.12252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wallis RS, Doherty TM, Onyebujoh P, Vahedi M, Laang H, Olesen O, Parida S, Zumla A. Biomarkers for tuberculosis disease activity, cure, and relapse. Lancet Infect Dis. 2009;9:162–172. doi: 10.1016/S1473-3099(09)70042-8. [DOI] [PubMed] [Google Scholar]
- 3.Shenai S, Amisano D, Ronacher K, Kriel M, Banada PP, Song T, Lee M, Joh JS, Winter J, Thayer R, et al. Exploring alternative biomaterials for diagnosis of pulmonary tuberculosis in HIV-negative patients by use of the GeneXpert MTB/RIF assay. J Clin Microbiol. 2013;51:4161–4166. doi: 10.1128/JCM.01743-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diacon AH, Donald PR. The early bactericidal activity of antituberculosis drugs. Expert Rev Anti Infect Ther. 2014;12:223–237. doi: 10.1586/14787210.2014.870884. [DOI] [PubMed] [Google Scholar]
- 5.Mukamolova GV, Turapov O, Malkin J, Woltmann G, Barer MR. Resuscitation-promoting factors reveal an occult population of tubercle bacilli in sputum. Am J Respir Crit Care Med. 2010;181:174–180. doi: 10.1164/rccm.200905-0661OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shleeva M, Goncharenko A, Kudykina Y, Young D, Young M, Kaprelyants A. Cyclic AMP-dependent resuscitation of dormant Mycobacteria by exogenous free fatty acids. Plos One. 2013;8:e82914. doi: 10.1371/journal.pone.0082914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y, Yang Y, Woods A, Cotter RJ, Sun Z. Resuscitation of dormant Mycobacterium tuberculosis by phospholipids or specific peptides. Biochem Biophys Res Commun. 2001;284:542–547. doi: 10.1006/bbrc.2001.4993. [DOI] [PubMed] [Google Scholar]
- 8.Manina G, McKinney JD. A single-cell perspective on non-growing but metabolically active (NGMA) bacteria. Curr Top Microbiol Immunol. 2013;374:135–161. doi: 10.1007/82_2013_333. [DOI] [PubMed] [Google Scholar]
- 9.Chengalroyen MD, Beukes GM, Gordhan BG, Streicher EM, Churchyard G, Hafner R, Warren R, Otwombe K, Martinson N, Kana BD. Detection and quantification of differentially culturable tubercle bacteria in sputum from patients with tuberculosis. Am J Respir Crit Care Med. 2016;194:1532–1540. doi: 10.1164/rccm.201604-0769OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eum SY, Kong JH, Hong MS, Lee YJ, Kim JH, Hwang SH, Cho SN, Via LE, Barry CE., III Neutrophils are the predominant infected phagocytic cells in the airways of patients with active pulmonary TB. Chest. 2010;137:122–128. doi: 10.1378/chest.09-0903. [DOI] [PMC free article] [PubMed] [Google Scholar]