Abstract
The isolation, structure determination and biological activities of a new linear pentapeptide caldoramide (5) from the marine cyanobacterium Caldora penicillata from Florida are described. Caldoramide (5) has structural similarities to the belamide A (4), dolastatin 10 (1) and dolastatin 15 (2). We profiled caldoramide against parental HCT116 colorectal cancer cells and isogenic cells lacking oncogenic KRAS or hypoxia-inducible factors 1α (HIF-1α) and 2α (HIF-2α). Caldoramide (5) showed differential cytotoxicity for cells containing both oncogenic KRAS and HIF over the corresponding knockout cells. LCMS dereplication indicated the presence of caldoramide (5) in a subset of C. penicillata samples.
Graphical Abstract

Organisms from the largely unexplored marine environment have proven to be a rich source of secondary metabolites that are potential therapeutics. Of these, the benthic marine cyanobacteria have been reported to be a source of compounds rich in diversity and biological activity.1–3 Cyanobacteria inhabit diverse environments which could explain the chemical diversity of the secondary metabolites that have been isolated from these marine organisms. Among the compounds that showed interesting biological activities were the small peptides and depsipeptides collectively named dolastatins, that were originally isolated from the sea hare Dolabella auricularia.4 The most potent among the series of dolastatins, dolastatin 10 (1), has been subsequently isolated from the cyanobacterium Symploca sp. (now known as Caldora penicillata) in a yield several magnitudes higher than from the sea hare.5,6 Specifically, dolastatins 10 (1) and 15 (2) have displayed extraordinary cytotoxicity to cancer cells.7,8 Their interesting biological activities have triggered synthetic efforts resulting in several drug leads that advanced to clinical trials.9 Inspired by the structure of dolastatin 10, a related cytotoxin served as the payload for the antibody-drug conjugate brentuximab vedotin (Adcetris), approved for the treatment of relapsed Hodgkin lymphoma and anaplastic large cell lymphoma.10 Largazole (3), a histone deacetylase (HDAC) inhibitor, has also been isolated from the cyanobacterium Symploca (now Caldora penicillata). Furthermore, a closely related structure to the dolastatins, belamide A (4), was reported as the major metabolite of the Panamanian cyanobacterium Symploca sp. and showed moderate cytotoxicity to the HCT116 colorectal cancer cell line.11

Here we report caldoramide (5), a new, closely related structural analogue of belamide A and dolastatin 15 (2), from the cyanobacterium Caldora penicillata. The taxonomy of this species, previously called Symploca sp., has recently been clarified.12 We have also detected via our dereplication method,13 that various geographically dispersed collections of C. penicillata consistently yielded known compounds such as dolastatin 10 (1) and/or the related homologue symplostatin 1 (6); these metabolites can serve as chemotaxonomic markers for this taxon.12,13 In addition to caldoramide (5), largazole14,15 and dolastatin 10 (1) 15 were isolated from this sample. Cytotoxicity profiling of caldoramide using three isogenic colorectal cancer cell lines16,17 revealed preferential targeting of oncogenic KRAS and hypoxia-inducible factor (HIF) pathways.
The wet sample of Caldora penicillata, a recollection from the Florida Keys, was stored frozen and freeze-dried prior to solvent extraction. The freeze-dried material was extracted first with EtOAc–MeOH (1:1) and then with EtOH–H2O (1:1) to give the non-polar and polar extracts, respectively. The two extracts were further partitioned to give EtOAc-soluble, n-BuOH-soluble and H2O-soluble fractions. The EtOAc-soluble fraction was subjected for further purification via repeated chromatography to give a new compound caldoramide (5), as well as the known cytotoxic compounds largazole (3) and dolastatin 10 (1).
Caldoramide (5) was obtained as an optically active white powder. HRESI/TOFMS supported the molecular formula of C37H60N5O6. Following the interpretation of DQF COSY, edited HSQC and HMBC experiments (Table 1), the 1H and 13C NMR signals were assignable to one OMe group, N,N-dimethylvaline (C-35 to C-42), valine (C-29 to N-34), N-Me-valine (C-22 to C-28), N-Me-isoleucine (C-14 to C-21), a putative phenylalanine (C-4 to C-12) and a conjugated enol ether (C-1 to C-3). The remarkable chemical shifts recorded for a methine carbon (C-2, δC 94.8; δH 4.80) adjacent to a non-protonated carbon (C-3, δC 178.1) suggested oxygenation at the latter position. Further, HMBC correlations between H-2 (δH 4.80) and C-1 (δC 169.1) and C-4 (δC 59.7); H-4 (δH 4.83) and C-2 (δC 94.8); H2-6 (δH 3.30, 3.08) and C-3 (δC 178.1) were observed and these data established a substituted pyrrolinone ring (C-1 to C-5) similar to that reported for belamide A (4).11 The HMBC correlation connected the OMe (δH 3.72) to the C-3 (δC 178.1) position in the pyrrolinone ring. Additional HMBC correlations from the H2-6 methylene protons (δH 3.30, 3.08) to C-3 (δC 178.1), C-7 (δC 134.7) and C-8/12 (δC 129.6) connected the benzyl moiety to the C-4 position of the pyrrolinone ring. These data supported a 3-methoxy-4-benzylpyrrolinone moiety present at the C-terminus, and therefore, it was apparent that the N,N-dimethylvaline residue formed the N-terminus. Sequencing of the remaining three amino acid residues was accomplished by analyzing the HMBC and NOESY data (Table 1). NOESY correlations of H-34 (N-H, δH 6.92) to H-36 (δH 2.44) and H3-41/42 (N,N-dimethyl, δH 2.23) of the N,N-dimethyl-valine, and to H-30 (δH 4.81) and H-31(δH 1.96) of the valine moiety indicated the connection between these two amino acids. HMBC correlations of NH-34 to C-30 (δC 53.6) and C-35 (δC 171.8) and H-36 to C-35 confirmed the connection between the N,N-dimethyl-valine and valine moieties. HMBC correlations between H3-28 (N-Me, δH 3.10) and C-23 (δC 58.3), C-29 (δC 172.8) and NOESY correlations shown between H3-28 and H-23 (δH 5.27) and H3-28 and H-30 (δH 4.81) of valine linked the valine and N-Me-valine residues. The NOESY correlations observed between H3-21 (δH 3.21) and H-15 (δH 6.45), H-16 (δH 2.09), and H-23 (δH 5.27) and HMBC correlations between H3-21 and C-15 (δC 58.7) and C-22 (δC 171.2) connected the N-Me-valine to the N-Me-isoleucine, which in turn is connected to the remaining C-terminal unit to satisfy the molecular formula. These data established the planar structure of caldoramide (5) as N,N-diMe-Val-Val-N-Me-Val-N-Me-Ile-3-O-Me-4-benzylpyrrolinone.
Table 1.
NMR Spectroscopic Data (600 MHz, CDCl3) for Caldoramide (5)
| unit | C/H No | δC mult. | δH (J in Hz) | COSYa | HMBCb | NOESYc |
|---|---|---|---|---|---|---|
| Bn-O-Me-pyrrolinone | 1 | 169.1, C | ||||
| 2 | 94.8, CH | 4.80, s | 1, 4 | 13 | ||
| 3 | 178.1, C | |||||
| 4 | 59.7, CH | 4.83, dd (4.8, 3.4) | 6 | 3, 7 | 6 | |
| N-5 | ||||||
| 6 | 35.6, CH2 | 3.30, dd (13.7, 4.8) | 4 | 3, 4, 7, 8/12 | 4, 8/12 | |
| 3.08, dd (13.7, 3.4) | 4 | 3, 4, 7, 8/12 | 4, 8/12 | |||
| 7 | 134.7, C | |||||
| 8/12 | 129.6, CH | 7.00, dd (7.5, 1.3) | 9/11 | 6, 10 | 4, 11 | |
| 9/11 | 128.3, CH | 7.20, m | 8/12, 10 | 7 | ||
| 10 | 127.0, CH | 7.14, m | 9/11 | 8/12 | ||
| 13 | 58.4, CH3 | 3.72, s | 3 | 2, 8/12 | ||
| N-Me-Ile | 14 | 172.2, C | ||||
| 15 | 58.7, CH | 6.45, d (10.9) | 16 | 14, 17, 19, 21 | 17, 19 | |
| 16 | 34.3, CH | 2.09, m | 15, 17, 19 | 15, 17, 18 | 17, 18, 19 | |
| 17 | 25.1, CH2 | 1.26, m | 16, 18 | 15, 16, 18, 19 | 15, 16, 18 | |
| 1.10, m | 16, 18 | 15, 16, 18, 19 | 15, 16, 18 | |||
| 18 | 10.6, CH3 | 0.84, t (7.5) | 17 | 16, 17 | 16, 17 | |
| 19 | 14.5, CH3 | 0.89, d (6.8) | 16 | 15, 16, 17 | 15, 16 | |
| N-20 | ||||||
| 21 | 31.8, CH3 | 3.21, s | 15, 22 | 15, 16, 23 | ||
| N-Me-Val | 22 | 171.2, C | ||||
| 23 | 58.3, CH | 5.27, d (10.3) | 24 | 22, 24, 25, 28 | 21, 25, 26, 28 | |
| 24 | 27.2, CH | 2.38, m | 23, 25, 26 | 22, 23, 25 | 23, 25, 26, 28 | |
| 25 | 19.6, CH3 | 0.89, d (6.9) | 24 | 23, 24, 26 | 23, 24, 28 | |
| 26 | 18.3, CH3 | 0.75, d (6.9) | 24 | 23, 24, 25 | 23, 24, 28, | |
| N-27 | ||||||
| 28 | 30.5, CH3 | 3.10, s | 23, 29 | 23, 24, 30, 31 | ||
| Val | 29 | 172.8, C | ||||
| 30 | 53.6, CH | 4.81, dd (8.9, 3.4) | 31, 34 | 29, 31, 32, 33 | 28, 31, 32, 34 | |
| 31 | 31.0, CH | 1.96, m | 30, 32, 33 | 29, 30, 32, 33 | 30, 32, 33, 34 | |
| 32 | 17.8, CH3 | 0.93, d (6.9) | 31 | 30, 31, 33 | 30, 31, 34 | |
| 33 | 19.9, CH3 | 0.98, d (6.9) | 31 | 30, 31, 32 | 30, 31 | |
| NH-34 | 6.92, d (8.9) | 30 | 30, 35 | 30, 31, 36, 41/42 | ||
| N,N-diMe-Val | 35 | 171.8, C | ||||
| 36 | 76.6, CH | 2.44, d (6.2) | 37 | 35, 38, 39, 41 | 34, 37, 38, 41/42 | |
| 37 | 27.7, CH | 2.08, m | 36, 38, 39 | 35, 36, 38, 39 | 36, 38, 39, 41/42 | |
| 38 | 20.2, CH3 | 0.99, d (6.9) | 37 | 36, 37, 39 | 36, 37, 41/42 | |
| 39 | 17.7, CH3 | 0.92, d (6.9) | 37 | 36, 37, 38 | 36, 37, 41/42 | |
| 40-N | ||||||
| 41 | 43.0, CH3 | 2.23, s | 36 | 34, 36, 38, 39 | ||
| 42 | 43.0, CH3 | 2.23, s | 36 | 34, 36, 38, 39 |
COSY correlations are from proton(s) stated to the indicated proton(s).
HMBC correlations, optimized for 2/3JCH = 8 Hz, are from proton(s) stated to the indicated carbon.
NOESY correlations are from proton(s) stated to the indicated proton(s).
The absolute configuration was determined by enantioselective HPLC analysis. Determination of the configuration at C-4 required ozonolysis of 5 prior to hydrolysis with 6 N HCl. The ozonolysis18 prior to hydrolysis yielded free phenylalanine in the hydrolyzate.11 The retention times of the components of the hydrolyzate were compared to the amino acid standards and indicated the L-configuration for all amino acids in caldoramide (5).
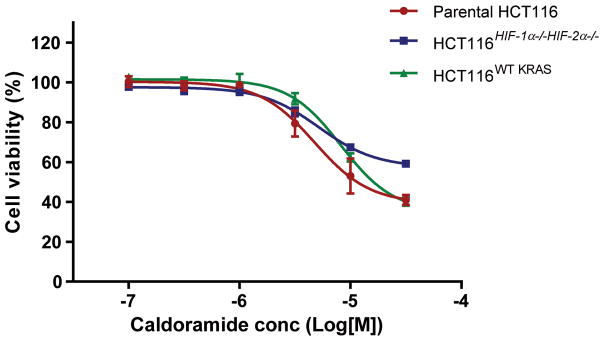
Differential cytotoxicity profiling of isogenic knockouts of cancer genes can be used to determine target pathway specificity. Oncogenic KRAS and activated HIF pathways have been linked to angiogenesis, cell growth, survival and metastasis.16,17,19 To test if caldoramide’s mechanism of action involves these pathways, we used the parental HCT116 colorectal cancer cell line that contains one oncogenic KRAS allele and one wildtype KRAS allele, HCT116 cells without the oncogenic KRAS allele (HCT 116WT KRAS), and HCT116 cells depleted of HIF-1α and HIF-2α (HCT116HIF-1α−/−HIF-2α−/−).16 In a recently reported gene expression profiling of isogenic HIF and KRAS knockout cell lines, we showed that there is a significant overlap between global gene expression affected by oncogenic KRAS with genes affected by both HIF-1α and HIF-2α.17 This signature overlap indicates that HIF transcriptional activity is regulated by oncogenic KRAS. Caldoramide (5) showed preferential antiproliferative activity against cells containing oncogenic KRAS or HIF transcription factors, viz. parental HCT 116 over HCT116HIF-1α−/−HIF-2α−/− and over HCT116WT KRAS (Figure 1). The potency of caldoramide in HCT116HIF-1α−/−HIF-2α−/− was slightly decreased (IC50 shifts from 3.9 μM to 5.2 μM) and maximum efficacy reduced to 67% compared with parental HCT116 cells. The IC50 shift in HCT116WT KRAS was more pronounced (3.9 μM to 8.6 μM), while efficacy was unaffected.
Figure 1.
Effects of caldoramide (5) on cell viability of isogenic HCT116 cell lines.
The cytotoxicity profiles of both dolastatin 10 (1) and largazole (3) have been previously determined using the same cell lines. Knockout cells were much less susceptible to dolastatin 10 with >50-fold shifts in IC50 values compared with parental cells with concomitant reduced efficacy (Table 2).17 The HDAC inhibitor largazole, also showed differential cytotoxicity for parental HCT116 over HCT116HIF-1α−/−HIF-2α−/− but this difference in cytotoxicity is only seen with HCT116HIF-1α−/−HIF-2α−/− and not in the cells lacking oncogenic KRAS (Table 2).17 Caldoramide (5), although structurally related, is less potent than dolastatin 1017 across the three cell lines investigated. Thus, all three compounds have varying activity profiles against the HCT116 isogenic cell lines that were tested, suggesting that the dose-response analyses of these cell lines might be used as a platform to group antiproliferative compounds by mechanism.
Table 2.
Comparison of Cytotoxicity Profiles of the Three Co-produced Metabolites Against Isogenic HCT116 Cells
| Compound | IC50
|
Efficacy Relative to Parental HCT116, %
|
||||
|---|---|---|---|---|---|---|
| Parental | HIF-1α−/−2α −/− | Oncogenic KRAS ko (HCT116WT KRAS) | Parental | HIF-1α−/−2α −/− | Oncogenic KRAS ko (HCT116WT KRAS) | |
| Caldoramide (5) | 3.9 μM | 5.2 μM | 8.6 μM | 100 | 67 | 100 |
| Dolastatin 10 (1)a | 0.48 nM | 65 nM | 24 nM | 100 | 74 | 83 |
| Largazole (3)a | 6.41 nM | 18 nM | 3.7 nM | 100 | 70 | 67 |
Values taken from reference 17.
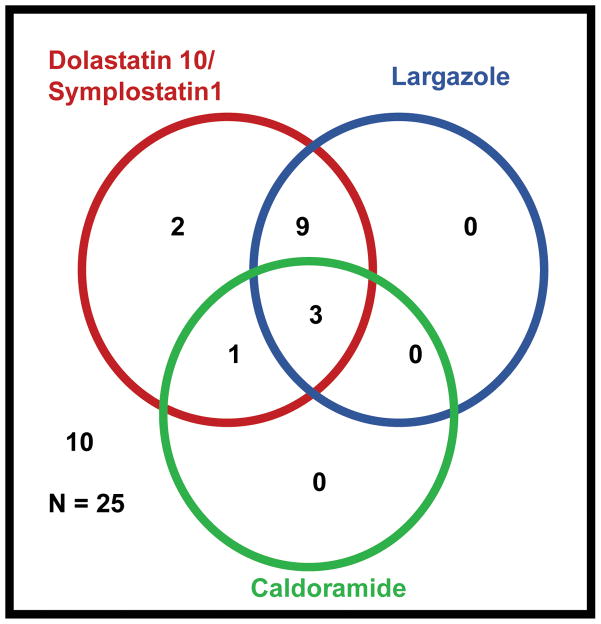
Extracts, both non-polar and polar, for the different collections of cyanobacterial samples were eluted with MeOH through C18 Solid Phase Extraction columns to remove interference from the polar compounds for HPLC-MS analysis. Detection of known compounds has provided a guide into prioritization of extracts at the early stage for further purification. The sensitive, reliable, and fast HPLC tandem mass spectrometry fragmentation (HPLC-MS/MS), was used to screen several extracts that were collected from various sites for the presence of known compounds. We monitored the parent ion and the most stable fragment ion to detect the presence of each compound. Using this technique, we have simultaneously scanned for the presence of dolastatin 10 (1), symplostatin 1 (6), largazole (3), and caldoramide (5) in an array of extracts and partitions from various collections of cyanobacterial samples (Figure 2). Most samples contained the metabolites dolastatin 10 (1) and/or the structurally related symplostatin 1 (6), as well as largazole (3). Furthermore, we found that in the extracts tested, caldoramide (5), did not elute alone, but has been consistently co-produced with dolastatin 10 (1) or symplostatin 1 (6). This characteristic has been shown by largazole (3), which was reported to be co-produced with the same metabolites.13 Lastly, from the extracts tested, caldoramide (5) was detected in four samples, all of which belong to Caldora penicillata. This suggests that the presence of caldoramide (5) is genus or species specific.
Figure 2.
Distribution of the three structural classes of compounds isolated from marine cyanobacteria from Florida.
We have described the isolation of a modified peptide, caldoramide (5) from the marine cyanobacterium Caldora penicillata. Our tandem MS/MS method has revealed that this compound, like largazole, is co-produced with dolastatin 10 (1) and/or its homologue symplostatin 1 (6). While only moderately cytotoxic, similarly as described for belamide A (4), we have shown that caldoramide (5) is able to inhibit the growth of cells that have both oncogenic KRAS and HIF-1α and HIF-2α transcription factors (parental HCT 116) to a greater extent than cells lacking those genes (HCT116HIF-1α−/−HIF-2α−/− and HCT116WT KRAS). A related yet not identical selectivity profile has been determined for dolastatin 10 (1) and largazole (3).17 Consequently, all three coproduced compounds derived from the same extract appear to act as indirect HIF inhibitors and exert different profiles, suggesting complementary mechanisms.
Experimental Section
General Experimental Procedures
The optical rotations were recorded on a Rudolph Research Analytical Autopol III automatic polarimeter. UV spectrophotometric data was acquired on a Shimadzu PharmaSpec UV–visible spectrophotometer. NMR data were collected on a JEOL ECA-600 spectrometer operating at 600.17 MHz for 1H and 150.9 MHz for 13C. The edited-HSQC experiment was optimized for JCH = 140 Hz and the HMBC spectrum was optimized for 2/3JCH = 8 Hz. 1H NMR chemical shifts (referenced to residual CHCl3 observed at δH 7.25) were assigned using a combination of data from 2D DQF COSY and multiplicity-edited HSQC experiments. Similarly, 13C NMR chemical shifts (referenced to CDCl3 observed at δC 77.0) were assigned on the basis of multiplicity-edited HSQC experiments. The HRMS data were obtained using an Agilent 6210 LC-TOF mass spectrometer equipped with an APCI/ESI multimode ion source detector at the Mass Spectrometer Facility at the University of California, Riverside, California. Silica gel 60 (EMD Chemicals, Inc. 230–400 mesh) was used for column chromatography. All solvents used were of HPLC grade (Fisher Scientific).
Collection, Extraction, and Isolation
The sample of cyanobacterial assemblage of Caldora penicillata for this study was collected from Big Pine Ledges (24.55302 N x 81.38028 W), a reef site near Big Pine Key, Florida, on March 30, 2015. This sample was a recollection of FK 12–18, collected at the same location and well characterized by both microscopic and molecular methods in our recent paper that described this species,12 and was consistent with the prior collection morphologically and chemically.12,13 A voucher specimen of the recollection (FK 15-1) is maintained at the Smithsonian Marine Station, Fort Pierce, FL. The freeze dried material (96.6 g) was first extracted with EtOAc–MeOH (1:1) followed by H2O–EtOH (1:1). The combined lipophilic and polar extract was partitioned between EtOAc and H2O and the aqueous portion subsequently partitioned between n-BuOH and H2O. Concentration of these extracts furnished 0.775 g (0.8%) of an EtOAc-soluble fraction and 2.275 g (2.3%) of a BuOH-soluble fraction. The EtOAc-soluble fraction (0.760 g) was purified on a column of SiO2 (25 g) using a step gradient system of hexanes–30%EtOAc, EtOAc, EtOAc–10% MeOH and MeOH to give four sub-fractions (1a–1d). The sub-fraction 1b (0.106 g), eluting with EtOAc was further chromatographed on column of C18 silica (7 g) using a step gradient system of MeOH–10% H2O, MeOH and EtOAc to give three sub-fractions. The first sub-fraction (0.068 g) eluting with MeOH–10% H2O was re-chromatographed on a Si-column (6 g) using a step gradient system of hexanes–30 % EtOAc, hexanes–60% EtOAc, EtOAc and MeOH to give another four sub-fractions. The last two sub-fractions were combined (0.053 g) and pushed through a SiO2 Sep-Pak with EtOAc followed by MeOH. The fraction 1 (0.019 g), eluting with EtOAc was further purified by reversed-phase HPLC (semi-prep 250 × 10 mm, 5 μm, RP-18, flow 3.0 mL/min) using MeOH-8.5% H2O to give a known compound largazole (3) 2.0 mg (tR = 8.8 min, yield, 0.002% dry wt) and caldoramide (5) 5.9 mg (tR = 9.4 min, yield, 0.006% dry wt). The sub-fraction 1d (0.194 g) eluting with MeOH was separated on column of C18 (12 g) using a step gradient system of MeOH-H2O followed by MeOH to give four sub-fractions. The sub-fraction 2 (6.5 mg) eluting with MeOH-10%H2O was further purified by reversed-phase HPLC (semi-prep 250 × 10 mm, 5 μm, RP-18, flow 3.0 mL/min) using MeOH-23% H2O to give the known compound dolastatin 10 (1) 1.3 mg (tR = 24 min, yield, 0.001% dry wt). The two known compounds largazole14 and dolastatin 1015 were identified by LCMS data and their structures were confirmed by comparing 1H NMR spectra with the 1H NMR spectra published in the literature.
Caldoramide (5): colorless, amorphous powder; [α]25D +11.1 (c 0.36, MeOH); UV (MeOH) λmax (log ε) 240 (4.19) nm; 1H and 13C NMR data, Table 1; HRESI/TOFMS m/z 670.4527 [M + H]+ (calcd for C37H60N5O6, 670.4538).
Ozonolysis, Acid Hydrolysis and Enantioselective HPLC Analysis
Compound 5 (1.0 mg) was dissolved in 1.5 mL of CH2Cl2 and ozonized at −78 °C. The solvent was evaporated and the product was suspended in 6 N HCl (0.5 mL) and heated at 110 °C for 12 h in a sealed tube. The hydrolyzate was concentrated to dryness. The residue was reconstituted in 0.2 mL of H2O and analyzed by enantioselective HPLC, comparing the retention times with those of authentic standards [Phenomenex Chirex (D) Penicillamine, 4.6 x 250 mm, 5 μm]; solvent 2.0 mM CuSO4 and mixtures of 2.0 mM CuSO4–CH3CN (95:5 or 92:8); detection at 254 nm. Using 2.0 mM CuSO4 with a flow rate of 0.8 mL/min, the retention times (tR min) for authentic standards were N,N-diMe-L-Val (11.5) and N,N-diMe-D-Val (14.4), and with a flow rate of 1.0 mL/min, the retention times (tR min) for authentic standards were N-Me-L-Val (19.5), N-Me-D-Val (31.6). Using 2.0 mM CuSO4-CH3CN (95:5) with a flow rate of 1.0 mL/min, the retention times (tR min) for authentic standards were L-Val (14.4) and D-Val (19.4) and with a flow rate of 0.8 mL N-Me-L-allo-Ile (25.3), N-Me-D-allo-Ile (37.1) and N-Me-L-Ile (29.1). N-Me-D-Ile was not available for this analysis. N-Me-D-Ile and N-Me-D-allo-Ile are reported to have close retention times.18 No peak appeared close to the retention time of 37 min in the hydrolyzate. Using 2.0 mM CuSO4-CH3CN (92:8) with a flow rate of 1.0 mL/min, the retention times (tR min) for authentic standards were L-Phe (80.3) and D-Phe (84.5). The absolute configurations of the amino acid moieties in the hydrolyzate were confirmed by comparing the retention times in their respective chromatography traces as N,N-diMe-L-Val (11.5), N-Me-L-Val (19.5), L-Val (14.4), N-Me-L-Ile (29.1) and L-Phe (80.3).
HPLC-MS Profiling
Cyanobacteria samples that were previously fractionated using 100% MeOH through C18 Solid Phase Extraction column were reconstituted in MeOH to 1 mg/mL concentration. Samples for analysis were diluted to 10,000 ng/mL in MeCN from the methanolic stock solutions. For each sample, 10 μL was injected and analysis was done using the following conditions: 1) HPLC: 0.1% HCOOH in MeCN–0.1% HCOOH in H2O [20% –100% MeCN for 25 min followed by 100% MeCN for 5 min, flow rate 0.25 mL/min, on a Hypersil GOLD aQ C18 Polar Endcapped LC column; 2) MS and MS/MS: Thermo Fisher LCQ-Deca system with ESIMS in positive ion mode and CID 35%. Detection of largazole (3), symplostatin 1 (6), and dolastatin 10 (1) using MRM was previously reported.13
Cell Viability Assay
Human colorectal cancer cell lines HCT116, HCT116HIF-1α−/−HIF-2α−/− and HCT 116WT KRAS were cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% Fetal Bovine Serum and maintained in 5% CO2 at 37 °C. The cells were seeded in a 96-well plate (10,000 cells/well), and treated with different concentrations of 5 after 24 h. The cell viability was measured 48 h following treatment with MTT dye using manufacturer’s protocol (Promega).
Supplementary Material
Acknowledgments
This research was supported by the National Institutes of Health, NCI grant R01CA172310. Financial support from the Smithsonian Competitive Grants Program for Science supported collection and an internship for C.G. We thank the Harbor Branch Oceanographic Institute at Florida Atlantic University spectroscopy facility for 600 MHz NMR spectrometer time and optical rotation measurements. The high resolution mass spectrometric analysis was performed by the UCR mass spectrometer facility, Department of Chemistry, University of California at Riverside. We are grateful to E. Bartels and the staff at the Mote Tropical Research Laboratory for assistance with collections. E. Bartels also kindly provided the photograph for the TOC graphic. This is contribution number XXX from the Smithsonian Marine Station at Fort Pierce.
Footnotes
Supporting Information. 1H, 13C, COSY, HMBC, 2D NOESY, HSQC NMR spectra of caldoramide (5) in CDCl3. The Supporting Information is available free of charge on the ACS Publications website at DOI:
References
- 1.Blunt JW, Copp BR, Keyzers RA, Munro MHG, Prinsep MR, Northcote PT, Prinsep MR. Nat Prod Rep. 2014;31:160–258. doi: 10.1039/c3np70117d. [DOI] [PubMed] [Google Scholar]
- 2.Tan LT. Phytochemistry. 2007;68:954–979. doi: 10.1016/j.phytochem.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 3.Van Wagoner R, Drummond AK, Wright JLC. Adv Appl Microbio. 2007;61:89–217. doi: 10.1016/S0065-2164(06)61004-6. [DOI] [PubMed] [Google Scholar]
- 4.Yamada K, Kigoshi H. Bull Chem Soc Jpn. 1997;70:1479–1489. [Google Scholar]
- 5.Pettit GR, Kamano Y, Herald CL, Fujii Y, Kizu H, Boyd M, Boettner FE, Doubek DL, Schmidt JM, Chapuis JC. Tetrahedron. 1993;49:9151–9170. [Google Scholar]
- 6.Luesch H, Moore RE, Paul VJ, Mooberry SL, Corbett TH. J Nat Prod. 2001;64:907–910. doi: 10.1021/np010049y. [DOI] [PubMed] [Google Scholar]
- 7.Pettit GR, Kamano Y, Herald CL, Tuinman A, Boettner FE, Kizu H, Schmidt JM, Baczynskyj L, Tomer KB, Bontems RJ. J Am Chem Soc. 1987;109:6883–6885. [Google Scholar]
- 8.Pettit GR, Kamano Y, Dufresne C, Cerny RL, Herald CL, Schmidt JM. J Org Chem. 1989;54:6005–6006. [Google Scholar]
- 9.Flahive E, Srirangam J. In: Anticancer Agents from Natural Products. Cragg GM, Kingston DGI, Newman DJ, editors. Chapter 11. CRC Press, Taylor and Francis Group; Boca Raton, Florida: 2005. pp. 191–213. [Google Scholar]
- 10.Senter PD, Sievers EL. Nat Biotechnol. 2012;30:631–637. doi: 10.1038/nbt.2289. [DOI] [PubMed] [Google Scholar]
- 11.Simmons TL, McPhail KL, Ortega-Barría E, Mooberry SL, Gerwick WH. Tetrahedron Lett. 2006;47:3387–3390. [Google Scholar]
- 12.Engene N, Tronholm A, Salvador-Reyes LA, Luesch H, Paul VJ. J Phycol. 2015;51:670–681. doi: 10.1111/jpy.12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salvador-Reyes LA, Engene N, Paul VJ, Luesch H. J Nat Prod. 2015;78:486–492. doi: 10.1021/np500931q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taori K, Paul VJ, Luesch H. J Am Chem Soc. 2008;130:1806–1807. doi: 10.1021/ja7110064. [DOI] [PubMed] [Google Scholar]
- 15.Salvador-Reyes LA, Luesch H. Nat Prod Rep. 2015;32:478–503. doi: 10.1039/c4np00104d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burkitt K, Chun SY, Dang DT, Dang LH. Mol Cancer Ther. 2009;8:1148–1156. doi: 10.1158/1535-7163.MCT-08-0944. [DOI] [PubMed] [Google Scholar]
- 17.Bousquet MS, Ma JJ, Ratnayake R, Havre PA, Yao J, Dang NH, Paul VJ, Carney TJ, Dang LH, Luesch H. ACS Chem Biol. 2016;11:1322–1331. doi: 10.1021/acschembio.5b00860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taori K, Liu Y, Paul VJ, Luesch H. ChemBioChem. 2009;10:1634–1639. doi: 10.1002/cbic.200900192. [DOI] [PubMed] [Google Scholar]
- 19.Talks KL, Turley H, Gatter KC, Maxwell PH, Pugh CW, Ratcliffe PJ, Harris AL. Am J Pathol. 2000;157:411–421. doi: 10.1016/s0002-9440(10)64554-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.