Summary
Members of the genus Bacteroides, mainly Bacteroides fragilis, can cause severe disease in man, especially after intestinal perforation in the course of abdominal surgery. Treatment is based on a small number of antibiotics, including metronidazole which has proved to be highly reliable throughout the last 40 to 50 years. Nevertheless, metronidazole resistance does occur in Bacteroides and has been mainly attributed to Nim proteins, a class of proteins with suggested nitroreductase function. Despite the potentially high importance of Nim proteins for human health, information on the expression of nim genes in Bacteroides fragilis is still lacking. It was the aim of this study to demonstrate expression of nim genes in B. fragilis at the protein level and, further, to correlate the level of Nim levels with the level of metronidazole resistance.
By application of two-dimensional gel electrophoresis, Nim proteins could be readily identified in nim-positive strains but their levels were not elevated to a relevant extent after induction of resistance to high doses of metronidazole. Thus, the presented data do not provide evidence for Nim proteins acting as nitroreductases using metronidazole as a substrate because no correlation of Nim levels and level of resistance could be observed. Further, no evidence was found that Nim proteins protect B. fragilis from metronidazole by sequestering activated metronidazole.
Introduction
The intestinal bacteria of the genus Bacteroides account for as much as approximately 30% of the human faecal isolates (Kuwahara et al., 2004) and utilise carbohydrate sources that are inaccessible to the human host. In general, Bacteroides spp. are beneficial to the host by producing volatile fatty acids that can be absorbed through the large intestine (Wexler, 2007). However, Bacteroides spp., i.e. predominantly B. fragilis, can also cause severe disease, especially after abdominal surgery or injury of the gastrointestinal tract (Aldridge & Sanders, 2002). Main symptoms include abscess formation (abdomen, liver, brain, lungs) and/or bacteraemia (Wexler, 2007). Treatment schemes mainly rely on ampicillin/sulbactam, clindamycin, carbapenems and the 5-nitroimidazole drug metronidazole (Wexler, 2007). Presently, carbapenems and metronidazole are the most reliable treatment options because resistance rates have remained very low (Hedberg & Nord, 2003; Sóki et al., 2013). Nevertheless, metronidazole-resistant Bacteroides isolates have been repeatedly isolated and seem to be even relatively common in some parts of the world, e.g. in the UK (Brazier et al., 1999). In most cases, metronidazole resistance is associated with the presence of nim genes which were first described as transmissible metronidazole resistance determinants (Breuil et al., 1989). They can either be chromosomal or plasmid-borne and are often, but not always, associated with insertion elements (Sóki et al., 2006). To date, 10 isoforms of nim have been described, i.e. nimA - J (Gal & Brazier, 2004; Husain et al., 2013), but there is currently no evidence that they act differently. In fact, the mode of action of Nim proteins has not been fully elucidated as yet but they are commonly believed to act as nitroreductases which reduce the nitromidazole drug’s nitro group to a hardly reactive amino group (Carlier et al., 1997). In contrast with this notion, however, only a fraction of nim-positive strains are metronidazole-resistant (Gal & Brazier, 2004; Löfmark et al., 2005) and the introduction of nimE and nimJ into B. fragilis 638R only had a very modest effect on metronidazole susceptibility (Husain et al., 2013). Possibly, this discrepancy can be reconciled by the observation that induction of high-level metronidazole-resistance is far easier in nim-positive Bacteroides isolates than in nim-negative isolates (Löfmark et al., 2005), suggesting elevated Nim expression in challenged isolates. Evidence for this, however, is presently lacking as no Nim expression studies have been conducted to date.
The present study aimed at filling this gap and, thereby, constitutes a first attempt to correlate Nim levels as such to metronidazole resistance. To this end, several B. fragilis isolates carrying a nim gene were studied by two-dimensional gel electrophoresis (2DE) in order to quantify abundancies of Nim. Further, it was assessed whether nim gene levels are being up-regulated in response to exposure to ever increasing concentrations of metronidazole. The data presented here might prove instrumental for a further assessment of the role of nim genes in the development of metronidazole resistance in Bacteroides spp.
Methods
Chemicals and growth media components
Metronidazole, tinidazole, haemin, dithiothreitol (DTT), CHAPS, urea, thiourea, acetone, trichloroacetic acid (TCA) and bisacrylamide were obtained from Sigma-Aldrich (St. Louis, USA). Brain-Heart-Infusion (BHI) broth and vitamin K1 were purchased from Carl Roth (Karlsruhe, Germany). Wilkins-Chalgren (WC) anaerobe agar was purchased from Oxoid (Basingstoke, England). Acrylamide, IPG-strips for isoelectric focussing, ampholytes, and iodoacetamide (IAA) were purchased from Bio-Rad (Hercules, USA). Yeast extract and Anaerocult A for anaerobic culture were obtained from Merck (Darmstadt, Germany). Coomassie Blue Brilliant was purchased from Serva (Heidelberg, Germany).
Bacterial strains and culture
The B. fragilis strains 638R, either without plasmid or with either pIP417 (nimA) or pIP421 (nimD), BF-8 (nimB, chromosomal), and 388/1 (nimE on plasmid pBF388C) were used for this study. The nimA gene on pIP417 is associated with IS1186 and whereas nimD on pIP421 is associated with IS1169. The nimB gene in BF-8 had been found to be associated with an IS1186-like insertion element (Haggoud et al., 1994) and nimE on pBF388C is associated with an IS-like element (ISBf6). Strains BF-8 (Sebald et al., 1990; Sóki et al., 2004; Sóki et al., 2006) and 388/1 (Sóki et al., 2004; 2006) had been found to be resistant to 8 mg/L and 16 mg/L metronidazole, respectively.
Cultures were routinely grown in anaerobic jars on Wilkins-Chalgren (WC) plates at 37°C. When batch cultures were needed for 2DE, bacteria were grown in 25 mL tissue culture flasks (Falcon, Becton Dickinson, Franklin Lakes, USA) in BHI broth supplemented with 0.5% yeast extract, 5 mg/L haemin, 1 mg/L vitamin K (BHI-S) as described in Jousimies-Somer et al., 2002. Bacterial culture was done in anaerobic jars (Merck, Darmstadt, Germany) using the Anaerocult A system.
Induction of high-level metronidazole resistance in B. fragilis and determination of minimal inhibitory concetrations (MIC)
High level resistance was induced by growing strains on WC agar plates with ever increasing concentrations of metronidazole until a maximal concentration of 256 mg/L was reached. The concentration of metronidazole in the plates was increased in doubling steps, i.e. 1, 2, 4, 8, 16, 32, 64, 128, and finally 256 mg/L. The minimal inhibitory concentration (MIC) was defined as the lowest metronidazole concentration at which no more B. fragilis growth on WC plates could be observed.
Exposure of B. fragilis 638R batch cultures to metronidazole
After 16 h, stationary phase cultures (20 mL) were divided equally into two 25 mL culture flasks and 10 mL of fresh BHI-S were added to each of the two flasks. Either 50 µM metronidazole or 50 µM tinidazole were added to one of the cultures, followed by 2 h incubation in an anaerobic jar at 37°C. Afterwards, samples were processed as described below.
Sample preparation for two-dimensional gel electrophoresis (2DE)
B. fragilis cultures (20 mL) were harvested at 3,000 × g for 10 min at 4°C in a Sigma 4K15 centrifuge. The pellet was re-suspended and washed in 1 × phosphate-buffered saline (PBS) and re-pelleted again at 3,000 × g for 10 min at 4°C. Afterwards bacteria were re-suspended in a small volume (400 µL) of ultrapure water and proteins were precipitated by adding 12.5% trichloroacetic acid (TCA) in acetone to the sample until a final concentration of 10% TCA was reached. Proteins were precipitated for at least 1 h at -20°C. The precipitates were pelleted in a Sigma 1-15PK cryo-centrifuge (20,000 × g, 20 min, 4°C) and pellets were washed in 90% acetone (20,000 × g, 20 min, 4°C). Afterwards pellets were dried and re-suspended in 2DE sample buffer (7 M urea, 2 M thiourea, 4% CHAPS, 1% DTT, 1% ampholytes (pH 3-10) for at least 3 h. Insoluble matter was removed by centrifugation at 20,000 × g for 20 min at 20°C. The protein concentration in the supernatant was determined by Bradford assay (Bradford, 1978) and 400 µg of protein were used for 2DE.
Two-dimensional gel electrophoresis (2DE)
Isoelectric focussing (IEF) was performed in 17 cm IPG strips with pH 3-10 non-linear (Bio-Rad) using a Protean IEF cell (Bio-Rad), using the following program: 12 h rehydration at 50 V, 1 h 150 V (rapid slope), 1 h 300 V (rapid slope), 1 h 2000 V (linear slope), 2 h 5000 V (linear slope), 7 h 8000 V (rapid slope). After IEF, gel strips were equilibrated in a two-step procedure in 6 M urea, 30% glycerol, 2% SDS, and 50 mM Tris pH 8.8 containing either 1% DTT (first step) or 4% IAA (second step). Strips were run vertically in a Protean II xi cell (Bio-Rad) at 22 mA over-night (4°C). After gel electrophoresis, gels were stained with Coomassie Brilliant Blue R-250 and scanned in an Epson V750 Pro scanner. Image analysis was performed using Melanie 4 software (GeneBio).
In-gel tryptic digestion and mass-spectrometric analysis of isolated proteins
Tryptic digestion of Coomassie stained protein spots was performed by reversed phase LC ESI-ion Trap MS/MS on a Bruker amaZon ETD speed ion trap (Bruker Daltonics, Bremen, Germany) coupled to a Dionex Ultimate 3000 UHPLC system (Dionex, part of ThermoFisher, Germany) as described previously (Kolarich et al., 2012) with minor modifications as pointed out in detail in Supplementary Figure 1.
Data analysis was performed using ProteinScape 3 (Bruker Daltonics) and MASCOT 2.3 (MatrixScience, UK) using the following search parameters: Cysteine as carbamidomethyl was set as fixed modification, Deamidation (Asn/Gln) and oxidation (Met) were set as variable modifications. Up to 2 missed cleavages were allowed. Peptide tolerance (both MS and MS/MS) was set at ± 0.2 Da. The data was searched against the NCBI protein database.
qPCR analysis of the effect of metronidazole on plasmid copy number
Copy numbers of the nimA gene in B. fragilis 638R/pIP417 was determined by the ΔΔCT method using SYBR Green detection, the housekeeping glyceraldehyde-phosphate dehydrogenase (gap) gene as reference and the pIP417 repA and nimA genes as target genes. Copies of the nimA gene in clones of 638R/pIP417 adapted to 2 µg/mL and 256 µg/mL metronidazole, respectively (adaptation to metronidazole having been carried out as described above), were measured using unchallenged B. fragilis 638R/pIP417 as the control. Samples were prepared by the boiling method (Sóki et al., 2013) and the primers used were as follows: gap: gapd1BF 5’-AGCCATTGTAGCAGCTTTTT-3’ and gapd3BF 5’-GAAAACATCATCCCGTCT-3’, repA417: repA417-1 5’-TGAGCAACCAGAAAACTC-3’ and repA417-2 5’-TTTTTGCAGCATCCACAA-3’ and nimA: nimART1 5’-GTTCCTGCCGAGTTTACAAC-3’ and nimART2 5’-GATGGTCGAATCCCTTGTCT-3’. The PCR reactions included 5 μL SYBR Select mastermix (LifeTechnologies), 0.7 μM primers, and 1 μL of template preparations in 10 μL of final volumes in triplicate in 48-well PCR plates. Amplifications (95 °C 10 min; 95 °C 15 s, 57 °C 15 s, 72 °C 30 s, 35x and a melting curve from 72 °C to 95 °C) and analysis was made in the StepOne RT-PCR instrument (LifeTechnologies) and by the accompanying software (StepOne Software 2.1, LifeTechnologies).
Growth curve determinations of B. fragilis 638R with and without pIP417 (nimA)
The growth of the B. fragilis 638R and the same strain with the pIP417 plasmid was observed by densitometry at 600 nm using a UV/VIS spectrophotometer (BioMate, Thermo Electron Co.). Briefly, stationary cultures of the strains in BHI-S broth were diluted 40-fold in parallel in 3 mL aliquots into three glass test tubes of the same media and subsequently incubated anaerobically (BugBox, Ruskinn Technology Ltd) at 37 °C. After 5 h, 10 h, 15 h, 20 h, 25 h, and 30 h, aliquots in separate tubes were taken out from anaerobiosis, appropriately diluted to obtain readings in the range of 0.2-0.6 OD600 values, and their optical densities recorded.
Results and Discussion
NimA, NimB, NimD, and NimE are clearly expressed and can be visualized by 2DE
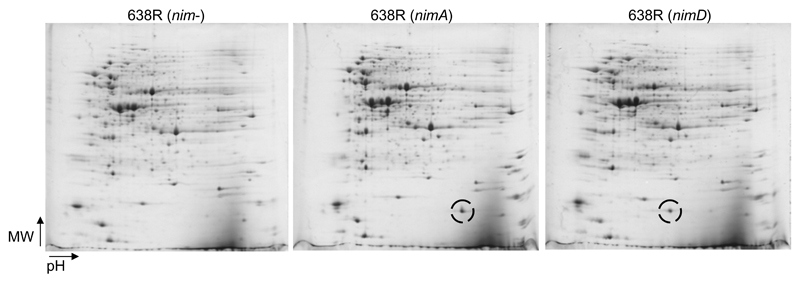
In order to demonstrate that nim genes are in fact expressed at the protein level, 2DE was performed with cultures of several B. fragilis strains which harbour a nim gene (Table 1). As a first step, strain B. fragilis 638R was analysed because direct comparison was possible between clones without plasmid (nim-negative) and clones bearing either pIP417 (nimA) or pIP421 (nimD). 2D-gels of 638R clones, carrying either nim gene, were searched for additional protein spots as compared to the nim-negative clone. Indeed, additional proteins in the expected molecular weight range of NimA and NimD (18 – 20 kDa) were readily found (Fig. 1). The proteins were isolated from the gels, analysed by mass spectrometry, and identified as NimA and NimD, respectively (Supplementary Figure 1). NimA levels, as determined densitometrically with Melanie 2DE-imaging software in 2D-gels from four different gel runs at four different time points (four 2D-gels in total), ranged from 0.7% to 1.5% of the total protein visualised (1.1 ± 0.3%). Thus, the method enables reproducible measurements of protein levels but does not allow reliable conclusions on differences in abundance below 100%. NimD levels were found to account for 0.58% of the total protein visualized. NimB and NimE were also identified in 2D-gels from strains BF8 and 388/1, respectively (Table 2). Interestingly, levels of NimB (0.29% of total protein) and NimE (0.14% of total protein) were clearly lower than levels of NimA and NimD in 638R (Table 2). In case of nimB this might not only be attributable to a lower rate of transcription and/or translation per se but also to a lower copy number due to its chromosomal location. In contrast, nimA, nimD, and nimE are all episomal (Table 1).
Table 1.
The B. fragilis strains used in this study. The minimal inhibitory concentration (MIC) was defined as the lowest metronidazole concentration at which no more B. fragilis growth on WC plates could be observed.
| B. fragilis Strain/Clone | nim gene | Accession Numbers of Nim proteins | Location of nim gene | MIC of metronidazole observed in this study | reference |
|---|---|---|---|---|---|
| 638R | none | - | < 1 mg/L | Sebald et al. 1990 | |
| 638R (nimA) | nimA | gi|435265 | pIP417 | 4 mg/L | Stubbs et al. 2000 |
| 638R (nimD) | nimD | gi|440387 | pIP421 | 4 mg/L | Stubbs et al. 2000 |
| 388/1 | nimE | gi|435261 | pBF388C | 32 mg/L | Sóki et al., 2004 |
| BF8 | nimB | gi|70559779 | chromosome | 2 mg/L | Sebald et al., 1990 |
Figure 1.
Representative images of 2D-gels (pH range 3-10 non-linear, 12.5% PAA) from strain 638R, either without nim gene (left image), with plasmid pIP417 (nimA, IS1186), nimA-positive, (middle), or with plasmid pIP421 (nimD, IS1169), nimD-positive, (right). The Nim proteins (encircled) are easily discernible as prominent spots in the lower molecular mass range of the gel. The theoretical molecular weights of NimA and NimD amount to 20.2 kDa and 18.5 kDa, respectively. Directions of increasing molecular mass (MW) and increasing pH are indicated by arrows.
Table 2.
Levels of NimA, NimB and NimE before and after induction of high level metronidazole resistance (256 mg/L). Percentages refer to the relative abundance of the Nim proteins as compared to total protein visualized by 2DE. The strain carrying the respective nim gene is indicated in brackets.
| Nim A (Bf 638R) | Nim B (BF8) | Nim E (388/1) | |
|---|---|---|---|
| Original level (%) | 0.7 | 0.29 | 0.14 |
| Induced level (%) | 0.87 | 0.4 | 0.18 |
Levels of NimA, NimB, and NimE are not further increased upon adaptation to high concentrations of metronidazole
After visualisation and measurement of NimA, B, D, and E levels in B. fragilis, it was assessed whether adaptation to higher concentrations of metronidazole leads to increased levels of Nim proteins. Since Nim proteins have been suggested to act as nitroreductases which reduce metronidazole and other nitroimidazole drugs to nonreactive aminoimidazoles (Carlier et al., 1997), their abundance is expected to be positively correlated to the level of metronidazole resistance. In order to test this, high level metronidazole resistance (256 mg/L) was induced in 638R (nimA), BF8 (nimB) and 388/1 (nimE). Interestingly, high level metronidazole resistance was most easily induced in 388/1, although this strain expresses the lowest level of Nim (Table 2). 638R (nimA) and 638R (nimD) already initially grew well on plates with 1 µg/mL metronidazole and somewhat slower on plates with 2 µg/mL metronidazole, whereas 638R (nim-negative) did not grow at 1 µg/mL metronidazole, indicating a protective effect of NimA and NimD. High level metronidazole resistance in 638R (nim-negative) could not be induced and attempts to generate clones which were resistant to metronidazole concentrations higher than 16 mg/L remained unsuccessful. This corroborates previous results (Löfmark et al., 2005) which indicated that nim-positive strains can be adapted to high metronidazole concentrations much more easily than nim-negative strains.
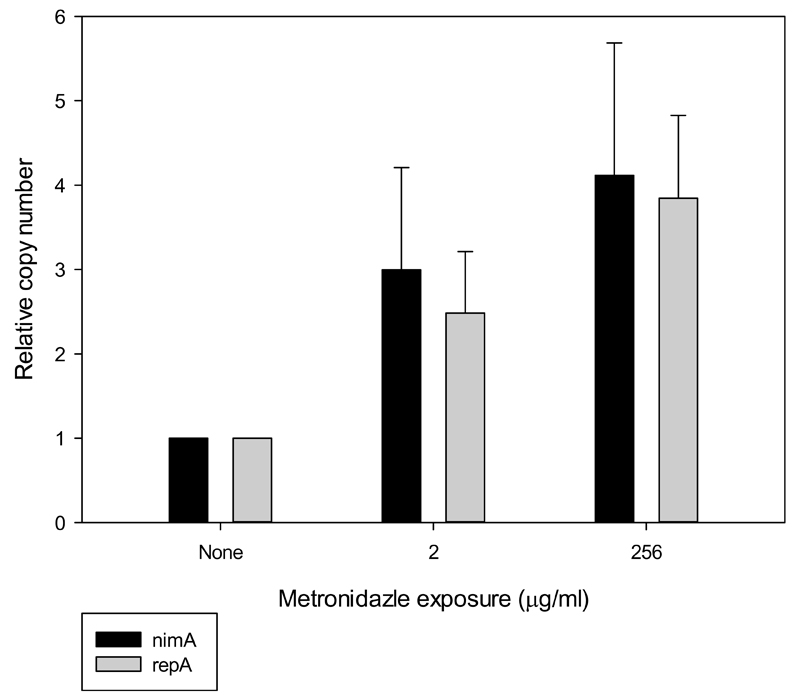
When Nim levels were measured by 2DE in the highly resistant clones, however, no relevant up-regulation as compared to the respective parent clones was found (Table 2). In fact, the observed levels of Nim B and E were slightly higher as compared to the unchallenged clones (Table 2), but this is not statistically significant when taking into consideration the observed variance in abundance of NimA in unchallenged 638R (nimA) (1.1 ± 0.3% of total protein visualised). Despite the lack of correlation between Nim levels and the level of metronidazole resistance, copy numbers of plasmid pIP417 (nimA) were three to four-fold higher when 638R (nimA) was grown on plates containing metronidazole than when grown without metronidazole (Fig. 2). Interestingly, copy numbers of the nimA gene were not found to be significantly increased in the 638R (nimA) clone adapted to 256 mg/L metronidazole when compared to 638R (nimA) routinely grown with 2 mg/L metronidazole (Fig. 2). Therefore, the reason for the elevated plasmid numbers in 638R (nimA) in presence of metronidazole is unclear. The notion of a selection of clones with higher copy numbers in response to increasing metronidazole concentrations is neither supported by the measured NimA levels (Table 2) nor by the insignificant increase of plasmid numbers in the clone adapted to 256 mg/L metronidazole as compared to the clone adapted to 2 mg/L. However, as metronidazole is mutagenic (Sisson et al., 2000), higher copy numbers of the nimA gene might, at least in the first phase of the development of resistance, increase chances to retain a copy of the gene without deleterious mutations. It is important to note, however, that these speculations only apply for B. fragilis strains having a plasmid-borne nim gene, and not for strains which have a chromosomal nim gene, e.g. BF8 (Table 1). Still, the selection of clones with a higher number of chromosomal copies of nim genes can presently not be ruled out because copy numbers of nimB in BF8 were not determined in this study.
Figure 2.
Relative copy number determination of the nimA plasmid (pIP417) depending on metronidazole exposure/adaptation. Error bars indicate SEM with 95% confidence.
These considerations notwithstanding, no evidence was found for induction of Nim levels upon exposure to increasing doses of metronidazole, nor for a selection of clones expressing higher levels of Nim with increasing level of resistance. In fact, given the comparably low abundance of Nim proteins in the strains BF8 (nimB-positive) and 388/1 (nimE-positive), no dependence of the protective effect of Nim proteins on copy number in the cell could be found at all. It is important to note, however, that NimB and NimE could function differently from NimA and NimD and that the different genetic background of BF8 and 388/1 could allow lower amounts of Nim to be equally as effective as the higher amounts of NimA and NimD in 638R.
The protective effect of nim genes and an increased propensity of nim-positive strains to adapt to high metronidazole concentrations (Löfmark et al., 2005) remains undisputed and is supported by our results presented here. And although our data do not provide any support for the notion of Nim proteins reducing metronidazole, it can presently not be ruled out that Nim proteins do indeed contribute to metronidazole resistance but require an inducible host factor, either an enzyme or a cofactor, for activity. In this case it would not be the induction of Nim expression that is critical for resistance to be demonstrated but induction of the hypothetical host factor. The expression level of this host factor could vary from strain to strain, explaining why protective levels of Nim are so differently high in different strains (Table 2). Currently, however, there is no indication that such a host factor exists. It is also interesting to note that all our attempts to measure nitroreductase activity of recombinantly expressed NimA in vitro have so far been unsuccessful (data not shown).
NimA is not bound by metronidazole and does not affect the growth rate of B. fragilis 638R
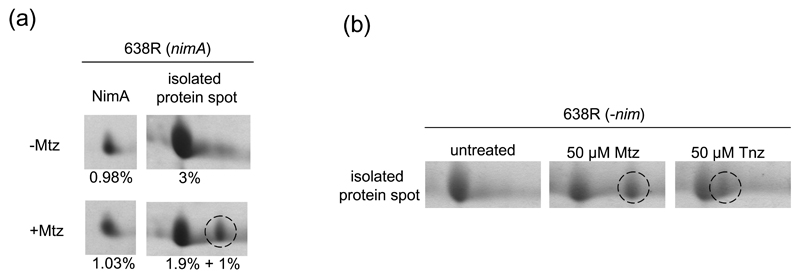
Since no correlation of Nim levels and the observed level of metronidazole resistance was observed, it was hypothesised that Nim proteins might protect sensitive target molecules in B. fragilis from low doses of metronidazole by acting as bait and sequestering activated metronidazole. This assumption was mainly based on the observed high levels of Nim (Table 2) and on the propensity of activated metronidazole and other nitroimidazoles to form covalent adducts with defined target proteins (Leitsch et al., 2007; 2009; 2012). As adduct formation with metronidazole can be visualised by 2DE due to a shift of pI of the bound protein to a more the alkaline pH (Leitsch et al., 2007; 2009; 2012; Williams et al., 2012), it was speculated that covalent adduct formation of Nim proteins and metronidazole would be discernible on 2D-gels. Alternatively, metronidazole treatment could also lead to the degradation of target proteins as observed with elongation factor 1-γ in Giardia lamblia (Leitsch et al., 2012). However, when the levels and the integrity of NimA in untreated 638R (nimA) and 638R (nimA) exposed to 50 µM metronidazole for 2 h were compared, neither a shift nor any degradation of the protein could be observed (Fig. 3a), suggesting that NimA is not a target of metronidazole and, therefore, cannot sequester activated metronidazole. Importantly, adduct formation of metronidazole with several other B. fragilis proteins was observed. Of these, one prominent protein spot (2 – 3% of all protein visualized) was isolated and analysed by mass spectrometry. The isolated spot contained four proteins of similar size and pI (Supplementary Figure 1): a putative oxidoreductase, a putative D-3-phosphoglycerate dehydrogenase, glutaminase, and a putative purine nucleoside phosphorylase. It was not determined which component of the spot was bound by metronidazole but it is interesting to note that purine nucleoside phosphorylase in the protist parasite Entamoeba histolytica had been identified earlier as a target of metronidazole (Leitsch et al., 2007). Formation of adducts was confirmed in 638R (nim-negative) with metronidazole and tinidazole (Fig. 3b). Tinidazole forms covalent adducts with the same proteins as metronidazole but leads to narrower shifts in pI of bound proteins, presumably due to the different charge of its side chain (Leitsch et al., 2007; 2009; 2012; Williams et al., 2012). Due to this easily discernible difference with regard to the width of pI shift, tinidazole has proven instrumental in distinguishing adduct formation from unspecific changes in proteins upon nitroimidazole treatment. The extent of adduct formation observed in 638R (nim-negative) compared well with that in 638R (nimA), further arguing against an immediate protective effect of NimA (Figs. 3a and 3b).
Figure 3.
2DE analysis of metronidazole treated 638R.
(a), integrity and relative abundance of NimA and the putative reductase (E1WVQ6_BACF6) were checked on 2D-gels after 2 h of incubation of equally divided 638R (nimA) stationary phase cultures either without drug or with 50 µM of metronidazole. The broken circle indicates the location of the metronidazole adduct of the isolated protein spot. Percentages refer to the relative abundance of the measured proteins as compared to total protein visualized.
(b), validation of metronidazole adduct formation in 638R (nim-negative). Broken circles indicate adducts of the metronidazole adduct of the isolated protein spot with either metronidazole or tinidazole.
Based on previous results, it was hypothesised that Nim proteins might have an indirect effect on metronidazole sensitivity, e.g. by impeding the metabolism of B. fragilis. In fact, metabolically generated reductive power is a prerequisite for the reduction of nitroimidazole drugs (Samuelson, 1999). It was argued that growth rate is a function of metabolic capacity and growth curves of 638R (nim-negative) and 638R (nimA) were determined in BHI-S broth and in unsupplemented BHI broth which lacks haemin and vitamin K. However, no influence of NimA on growth of 638R (nimA) as compared to 638R (nim-negative) could be found, doubling times (1.39 ± 0.02/h and 1.45 ± 0.04/h, respectively) and maximal culture densities (OD600= 1.52 ± 0.02 and 1.46 ± 0.03, respectively) of both clones being practically identical. This result argues against any influence of NimA on B. fragilis metabolism.
Conclusion
This study constitutes the first attempt to quantify Nim levels in B. fragilis. No correlation between Nim levels and the level of metronidazole resistance was found which contrasts with the established notion of Nim proteins inactivating or sequestering metronidazole directly. Further studies on the function of Nim proteins will be necessary to obtain a more reliable notion of their function.
Supplementary Material
Acknowledgements
This work was supported by the Austrian Science Fund (grant P22546) and the Max Planck Society (Daniel Kolarich).
References
- Aldridge KE, Sanders CV. Susceptibility trending of blood isolates of the Bacteroides fragilis group over a 12-year period to clindamycin, ampicillin-sulbactam, cefoxitin, imipenem, and metronidazole. Anaerobe. 2002;8:301–5. doi: 10.1016/S1075-9964(03)00028-3. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1978;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Brazier JS, Stubbs SLJ, Duerden BI. Metronidazole resistance among clinical isolates belonging to the Bacteroides fragilis group: time to be concerned? J Antimicrob Chemother. 1999;44:577–82. doi: 10.1093/jac/44.4.580. [DOI] [PubMed] [Google Scholar]
- Breuil J, Dublanchet A, Truffaut N, Sebald M. Transferable 5-nitroimidazole resistance in Bacteroides fragilis group. Plasmid. 1989;21:151–54. doi: 10.1016/0147-619x(89)90060-7. [DOI] [PubMed] [Google Scholar]
- Carlier JP, Sellier N, Rager MN, Reysset G. Metabolism of a 5-nitroimidazole in susceptible and resistant isogenic of Bacteroides fragilis. Antimicrob Agents Chemother. 1997;41:1495–9. doi: 10.1128/aac.41.7.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal M, Brazier JD. Metronidazole resistance in Bacteroides spp. carrying nim genes and the selection of slow-growing metronidazole-resistant mutants. J Antimicrob Chemother. 2004;54:109–16. doi: 10.1093/jac/dkh296. [DOI] [PubMed] [Google Scholar]
- Haggoud A, Reysset G, Azeddoug H, Sebald M. Nucleotide sequence analysis of two 5-nitroimidazole resistance determinants from Bacteroides strains and of a new insertion sequence upstream of the two genes. Antimicrob Agents Chemother. 1994;38:1047–51. doi: 10.1128/aac.38.5.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedberg M, Nord CE. Antimicrobial susceptibility of Bacteroides fragilis group isolates in Europe. Clin Microbiol Infect. 2003;9:475–88. doi: 10.1046/j.1469-0691.2003.00674.x. [DOI] [PubMed] [Google Scholar]
- Husain F, Veeranagouda Y, His J, Meggersee R, Abratt V, Wexler HM. Two multidrug-resistant clinical isolates of Bacteroides fragilis carry a novel metronidazole resistance nim gene (nimJ) Antimicrob Agents Chemother. 2013;57:3767–3774. doi: 10.1128/AAC.00386-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jousimies-Somer HR, Summanen P, Citron DM, Baron EJ, Wexler HM, Finegold SM. Wadsworth - KTL Anaerobic Bacteriology Manual. Sixth Edition. Belmont, California, USA: Star Publishing Company; 2002. [Google Scholar]
- Kolarich D, Jensen PH, Altmann F, Packer NH. Determination of site-specific glycan heterogeneity on glycoproteins. Nat Protoc. 2012;7:1285–98. doi: 10.1038/nprot.2012.062. [DOI] [PubMed] [Google Scholar]
- Kuwahara T, Yamashita A, Hirakawa H, Nakayama H, Toh H, Okada N, Kuhara S, Hattori M, Hayashi T, Ohnishi Y. Genomic analysis of Bacteroides fragilis reveals extensive DNA inversions regulating cell surface adaption. Proc Natl Acac Sci USA. 2004;101:14919–24. doi: 10.1073/pnas.0404172101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiros HKS, Kozielski-Stuhrmann S, Kapp U, Terradot L, Leonard GA, McSweeney SM. Structural basis of 5-nitroimidazole antibiotic resistance. J Biol Chem. 2004;279:55840–9. doi: 10.1074/jbc.M408044200. [DOI] [PubMed] [Google Scholar]
- Leitsch D, Kolarich D, Wilson IBH, Altmann F, Duchêne M. Nitroimidazole action in Entamoeba histolytica: a central role for thioredoxin reductase. PLoS Biol. 2007;5:1820–1834. doi: 10.1371/journal.pbio.0050211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitsch D, Kolarich D, Binder M, Stadlmann J, Altmann F, Duchêne M. Trichomonas vaginalis: metronidazole and other nitroimidazole drugs are reduced by the flavin enzyme thioredoxin reductase and disrupt the cellular redox system. Implications for nitroimidazole toxicity and resistance. Mol Microbiol. 2009;72:518–536. doi: 10.1111/j.1365-2958.2009.06675.x. [DOI] [PubMed] [Google Scholar]
- Leitsch D, Schlosser S, Burgess A, Duchêne M. Nitroimidazole drugs vary in their mode of action in the human parasite Giardia lamblia. Int J Parasitol Drugs Drug Res. 2012;2:166–170. doi: 10.1016/j.ijpddr.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löfmark S, Fang H, Hedberg M, Edlund C. Inducible metronidazole resistance and nim genes in clinical Bacteroides fragilis group isolates. Antimicrob Agents Chemother. 2005;49:1253–6. doi: 10.1128/AAC.49.3.1253-1256.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelson J. Why metronidazole is active against both bacteria and parasites. Antimicrob Agents Chemother. 1999;43:1533–41. doi: 10.1128/aac.43.7.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebald M, Reysset G, Breuil J. What’s new in 5-nitroimidazole resistance in the Bacteroides fragilis group? In: Borriello SP, editor. Clinical and Molecular Aspects of Anaerobes. Petersfield. UK: Wrightson Biomedical Publishing Ltd; 1990. pp. 217–225. [Google Scholar]
- Sisson G, Jeong JY, Goodwin A, Bryden L, Rossler N, Lim-Morrison S, Raudonikiene A, Berg DE, Hoffman PS. Metronidazole activation is mutagenic and causes DNA fragmentation in Helicobacter pylori and in Escherichia coli containing a cloned H pylori rdxA+ (nitroreductase) gene. J Bacteriol. 2000;182:5091–5096. doi: 10.1128/jb.182.18.5091-5096.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sóki J, Eitel Z, Urbán E, Nagy E. ESCMID Study Group on Anaerobic Infections. Molecular analysis of the carbapenem and metronidazole resistance mechanisms of Bacteroides strains reported in a Europe-wide antibiotic resistance survey. Int J Antimicrob Agents. 2013;41:122–5. doi: 10.1016/j.ijantimicag.2012.10.001. [DOI] [PubMed] [Google Scholar]
- Sóki J, Gal M, Brazier JS, Rotimi VO, Urbán E, Nagy E, Duerden BI. Molecular investigation of genetic elements contributing to metronidazole resistance in Bacteroides strains. J Antimicrob Chemother. 2006;57:212–20. doi: 10.1093/jac/dki443. [DOI] [PubMed] [Google Scholar]
- Sóki J, Fodor E, Hecht DW, Edwards R, Rotimi VO, Kerekes I, Urbán E, Nagy E. Molecular characterization of imipenem-resistant, cfiA-positive Bacteroides fragilis isolates from the USA, Hungary and Kuwait. J Med Microbiol. 2004;53:413–9. doi: 10.1099/jmm.0.05452-0. [DOI] [PubMed] [Google Scholar]
- Stubbs SL, Brazier JS, Talbot PR, Duerden BI. PCR-restriction fragment length polymorphism analysis for identification of Bacteroides spp. and characterization of nitroimidazole resistance genes. J Clin Microbiol. 2000;38:3209–3213. doi: 10.1128/jcm.38.9.3209-3213.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wexler HM. Bacteroides: the good, the bad, and the nitty-gritty. Clin Microbiol Rev. 2007;20:593–621. doi: 10.1128/CMR.00008-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CF, Lloyd D, Kolarich D, Alagesan K, Duchêne M, Cable J, Williams D, Leitsch D. Disrupted intracellular redox balance of the diplomonad fish parasite Spironucleus vortens by 5-nitroimidazoles and garlic-derived compounds. Vet Parasitol. 2012;190:62–73. doi: 10.1016/j.vetpar.2012.05.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.