Abstract
Eukaryotic microbial pathogens are major contributors to illness and death globally. Although much of their impact can be controlled by drug therapy as with prokaryotic microorganisms, the emergence of drug resistance has threatened these treatment efforts. Here, we discuss the challenges posed by eukaryotic microbial pathogens and how these are similar to, or differ from, the challenges of prokaryotic antibiotic resistance. The therapies used for several major eukaryotic microorganisms are then detailed, and the mechanisms that they have evolved to overcome these therapies are described. The rapid emergence of resistance and the restricted pipeline of new drug therapies pose considerable risks to global health and are particularly acute in the developing world. Nonetheless, we detail how the integration of new technology, biological understanding, epidemiology and evolutionary analysis can help sustain existing therapies, anticipate the emergence of resistance or optimize the deployment of new therapies.
The identification and use of antibiotics are some of the great medical achievements of the twentieth century, saving count-less lives by controlling the risk of infection from contagion, after injury, surgery or in immunosuppressed individuals. However, in only 80 years since the introduction of penicillin, resistance to a broad range of antibiotic drugs has become widespread, with the compounded risk from multidrug-resistant bacterial infections severely limiting treatment options. This has created justified concern and global attention, not only in the medical community but also at government level, in the media and the general public1.
While predominantly applied to control prokaryotic microbial infections, the threat of disease from eukaryotic microorganisms has also been contained by therapeutic drugs — preventing or controlling disease caused by eukaryotic parasites and fungi in both a human and animal health setting. These represent some of the most important disease-causing agents (Table 1), particularly in the tropics, where the distribution of a pathogen is frequently linked to the distribution of the arthropods that act as disease vectors. Such vector-borne parasites include malaria (Plasmodium spp.) and kineto-plastid parasites (Trypanosoma cruzi, which causes Chagas disease; Trypanosoma brucei gambiense and T. b. rhodesiense, which cause human African trypanosomiasis (HAT); and 17 Leishmania spp., which cause a variety of cutaneous and visceral diseases). Other clinically important protozoan parasite species not considered here are transmitted either orally (Toxoplasma, Giardia and Entamoeba) or venereally (Trichomonas). Distinct from the many obligate eukaryotic unicellular parasites, opportunistic fungal pathogens are global in distribution and include Candida, Aspergillus spp., Cryptococcus and Pneumocystis spp.
Table 1. Diseases caused by eukaryotic microorganisms, their vectors and frontline treatment options.
| Disease | Pathogen group | Vector | Pathogen | Frontline treatments* |
|---|---|---|---|---|
| Malaria | Apicomplexan | Anopheline mosquitoes | P. falciparum | Uncomplicated P. falciparum malaria: ACTs; artemether plus lumefantrine; artesunate plus amodiaquine; artesunate plus mefloquine; dihydroartemisinin plus piperaquine; artesunate plus sulfadoxine plus pyrimethamine. Severe malaria: parenteral (or rectal, children <6 years) artesunate followed by oral ACT (i.m. artemether or i.m. quinine if artesunate unavailable). |
|
P. vivax, P. ovale, P. malariae or P.knowlesi |
Blood stage infections: chloroquine (except in areas of chloroquine resistance); ACTs (except pregnant women and infants <6 months). Radical cure of liver (hypnozoite) infection: primaquine (close medical supervision with G6PD-deficient patients). | |||
| African trypanosomiasis | Kinetoplastid | Tsetse flies |
T. b. gambiense (chronic form) |
Haemolymphatic stage (no CNS involvement): pentamidine (i.m.). CNS stage: nifurtimox (oral) and eflornithine (i.v.) combination therapy (NECT) (melarsoprol if NECT unavailable). |
|
T. b. rhodesiense (acute form) |
Haemolymphatic stage (no CNS involvement): suramin (i.v.). CNS stage: melarsoprol (i.v.). |
|||
| American trypanosomiasis | Kinetoplastid | Triatomine bugs | T. cruzi | Benznidazole; nifurtimox. |
| Leishmaniasis | Kinetoplastid | Phlebotomine sandflies | Visceral disease: Leishmania donovani, L. infantum | Visceral disease: Amphotericin B (as liposomal or deoxycholate complex, i.v.); miltefosine (oral, contraindicated in pregnancy); paromomycin (i.m.); sodium stibogluconate (SSG) or meglumine antimonate, parenteral (except India and Nepal); SSG plus paromomycin (East Africa). |
| Mucocutaneous disease: L. braziliensis, L. panamensis | Mucocutaneous: SSG (systemic). | |||
| Cutaneous disease: For example, L. major, L. tropica, L. mexicana, L. amazonensis | Cutaneous: SSG (intralesional); paromomycin (ointment); miltefosine. | |||
| Invasive candidiasis | Fungal | Opportunistic |
C. albicans, C. glabrata, C. parapsilosis |
Echinocandins; fluconazole; liposomal amphotericin B. |
| Aspergillosis | Fungal | Opportunistic | A. fumigatus | Voriconazole (amphotericin B formulations; caspofungin; micafungin; posaconazole; itraconazole). |
| Pneumocystis pneumonia | Fungal | Opportunistic | Pneumocystis carinii | Sulfamethoxazole–trimethoprim (clindamycin–primaquine). |
| Cryptococcal meningitis | Fungal | Opportunistic | Cryptococcus neoformans | Amphotericin B plus flucytosine; amphotericin B plus fluconazole. |
Several of the parasitic pathogens are arthropod-transmitted, and in these cases the responsible vector is shown. Fungal pathogens are predominantly opportunistic. CNS, central nervous system; i.m., intramuscular; i.v. intravenous. *Second-line treatment options are given in parentheses. Data from the World Health Organization58,125–127 and other sources 57,128,129.
The control of these eukaryotic pathogens has often involved therapies whose development predates the use of penicillin and which, in some cases, have unacceptable toxicity profiles2. Nonetheless, as with the rise of antimicrobial resistance in bacteria, resistance has emerged or is emerging against therapies targeting these eukaryotic microorganisms, with potentially devastating consequences for exposed populations. This has received far less attention than antibacterial resistance, despite some commonality in its underlying causes. Here, we detail how the control of eukaryotic microorganisms poses both similar and distinct challenges to that of bacterial pathogens, the drugs used to combat these pathogens, and the resistance mechanisms they are evolving. Finally, we discuss how the latest methodological approaches can anticipate the emergence of drug resistance and support new therapeutic approaches, either through the development of new drugs, the maintenance of existing therapies or through the use of alternative approaches to limit the spread of drug resistance.
Common challenges for the control of microbial pathogens
The challenges of controlling eukaryotic microbial pathogens share many similarities with bacterial infections. Both replicate more rapidly than their hosts, such that resistance can be selected in a relatively short timescale in a treated host population. This is exacerbated by inappropriate treatment profiles, leading to sub-curative exposure in the context of infection3. Problems of suboptimal dosing are particularly acute when applied to tropical parasites. For example, in the case of antimalarial therapies, up to 35% of drugs may be of low quality, or have packaging and labelling that is poor or may be falsified4. The lower than optimal concentrations of the active agent found in these low-quality antimalarial drugs can rapidly select for resistance in exposed populations, as can under-dosing resulting from self-prescription. For zoonoses, parasite selection in livestock populations treated with antimicrobials in a context in which there is poor supply chain management, fraudulent provision or cost barriers to optimal dosing can also lead to the emergence of resistance. For African trypanosomes, this represents a considerable threat, since up to 50 million doses of trypanocides are used in sub-Saharan livestock annually, mainly as a prophylactic, and trypanocides represent 45% of animal health costs. Mirroring the situation with antibiotic use in veterinary contexts for bacterial infections, in which environmental contamination has generated substantial regulatory concern5, the agricultural use of fungicides might also contribute to the selection of azole-resistant Aspergillus fumigatus6.
A further similarity between bacterial and eukaryotic microbial pathogens is the phenomenon of persister populations7. Persisters are a subpopulation of pathogen that survives after exposure to a chemotherapeutic agent (or vaccine). They can then re-establish patent infection while remaining drug sensitive (reviewed in ref. 8). The state of persistence is not heritable and resistance is not due to genetic alterations directly linked to rendering a drug ineffective. Rather, persistence is a physiological state in which the pathogen actively responds to drug assault. Persistence ensures incidental survival but does not future-proof a pathogen as genetically heritable resistance would. However, the combination of a persister population and suboptimal drug dosage could provide a reservoir from which resistance can emerge and may even provide a population predisposed to evolve resistance more readily. An example of this relating to parasite dormancy is the resistance of Plasmodium falciparum to artemisinin (and other antimalarials such as mefloquine and atovaquone), which was first characterized by degrees of persistence followed by the emergence of genomic changes now causally associated with resistance (see later). Similarly, fungal infections (for instance with Candida albicans) that are associated with biofilms are a good example of persister populations analogous to those found in bacterial communities9–11. The duration of persistence can range from days (P. falciparum) to life-long (for example, C. albicans). Mechanisms of persistence vary — persistence may emerge spontaneously, possibly through stochastic changes in gene expression that prepare a population of pathogens for survival in varying environmental conditions (‘bet hedging’). This is best described in bacteria12 but is a phenomenon recently characterized in P. falciparum13. Furthermore, environmental signals may induce persistence, such as the nutrient starvation typically encountered by C. albicans in biofilms9,14.
Distinct challenges for the control of microbial pathogens
Although bacterial and eukaryotic microorganisms share common features with respect to their responses to drug exposure, there are also differences, with some challenges specific to the control of eukaryotic pathogens. First, eukaryotic microorganisms are more similar to their hosts than prokaryotic pathogens in terms of their biochemistry and metabolism, genetic composition, cell architecture and biology. Consequently, drugs targeting eukaryotic microorganisms must focus on differences from the eukaryotic norm, or particular specialisms of each pathogen group. This can restrict the cross-specificity of drugs to a point where there are distinctions in sensitivity between different apicomplexans (malaria, toxoplasma) or between the evolutionarily divergent trypanosomes, T. brucei spp. and T. cruzi. Comprising a different evolutionary kingdom, fungi have many differences from other eukaryotic microbial pathogens, again necessitating drugs to be developed for, and targeted to, a particular pathogen. This increases the challenges for drug development and inevitably constricts the new drug pipeline.
Second, many eukaryotic microbial pathogens have evolved a parasitic lifestyle distinct from the opportunistic infections characteristic of most bacterial pathogens (but also fungi). The evolution of parasitism is often accompanied by the development of sophisticated immune evasion mechanisms, which increases the impact of the persister phenotypes described earlier. Specifically, bacteriostatic drugs can operate to clear infection in concert with the immune system15. However, drugs that generate cytostatic rather than cytocidal responses to infection with an immunosuppressive parasite can lead to recrudescence upon the removal of drug exposure. This, in turn, can predispose the population to selection for drug resistance. Similarly, adaptation to an intracellular lifestyle or particular body niche can protect parasites from drug exposure, a feature shared with some bacterial pathogens that have evolved to survive in cells rather than systemically (Legionella, Mycobacteria).
A third challenge relates to clinical diagnosis and screening for drug resistance in eukaryotic microbial pathogens16. In bacterial infections, screening for sensitivity to antibiotics is straightforward and routine. By contrast, eukaryotic parasites can require highly specialized growth media and considerable growth periods to determine their susceptibility or otherwise to potential drug therapies. Also, unlike bacterial susceptibility testing, in which a minimum inhibitory concentration (MIC) is determined, most parasitologists report half-maximum effective concentration (EC50) values without providing the Hill slope of the growth inhibition curve or calculating the 90% effective concentration (EC90) value. It is perfectly possible to obtain a resistant line with an identical EC50 to the susceptible isolate, yet that is still resistant owing to a shallower Hill slope. As a consequence, clinical diagnosis and the selection of appropriate clinical management can be slow or practically impossible in the context of all but the most specialized laboratories.
A fourth distinction from common bacterial infections is the economic challenge of treating diseases of the developing world. Diseases such as malaria, trypanosomiasis, leishmaniasis and cryptococcosis are common in the poorest parts of the world, where the economic capacity to develop or deliver treatments is very limited, often restricted to philanthropic and charitable donations or the concerted actions of intergovernmental agencies. This makes the threat of drug resistance even more acute since there is no financial incentive to develop new drugs to replace those to which resistance emerges. Nonetheless, some pharmaceutical companies are increasingly engaged in public–private partnerships, providing access to chemical compound collections and other resources helpful for discovering and developing new drugs for neglected tropical diseases. Excellent examples of this collaborative spirit include the Medicines for Malaria Venture (http://www.mmv.org/), the Drugs for Neglected Diseases initiative (http://www.dndi.org/) and the Tres Cantos Open Lab Foundation (http://www.openlabfoundation.org/).
One route to limit the impact of drug resistance has been the exploitation of combination therapies for parasitic infections. This approach has proved useful for cancer therapy as well as for the treatment of tuberculosis, leprosy and viral infections such as HIV. Results have also been encouraging for parasitic infections; for example, through artemisinin combination therapy (ACT)17,18 to limit the emergence and spread of artemisinin-resistant malaria, and for trypanosomes, for which nifurtimox–eflornithine combination therapy19 is proving more robust than eflornithine-based therapy alone. However, combination therapies for parasitic diseases require the availability of more than one effective drug or drug class, which is not always the case. Moreover, combination therapies have often been embraced only when resistance has already been detected to one of the frontline monotherapies, allowing multidrug-resistant parasites to be selected. In a situation like this, the use of drug combinations with different pharmacokinetics in plasma, as with artemisinin and piperaquine, can limit the emergence of resistance20. However, the cost of drugs for many parasites seen in the developing world can generate geographical discrepancies in the use of mono and combination therapies. In this scenario, the efficacy of combination therapies can be threatened by ingression of resistant parasites selected under monotherapy.
The final challenge for eukaryotic microorganisms that differs from many prokaryotic and viral pathogens has been the failure to formulate and use effective vaccines to prevent infection21. Malaria research has focused intensively on vaccine development without transformative success, whereas for African trypanosomes the immune evasion mechanism used by the parasite (antigenic variation) effectively renders vaccine approaches impossible. It has also proved challenging to produce safe, effective vaccines for other kinetoplastids, despite the widespread early use of ‘leishmanization’ for the cutaneous form of leishmaniasis, which has the risk of virulence in some individuals and immunosuppression22. Fungal pathogens have their greatest impact in immunocompromised individuals, rendering vaccines potentially less useful. At present there are no licenced fungal vaccines; nonetheless, there are promising developments for adhesion-like substance 3 (Als3)- and secreted aspartic protease 2 (Sap2)-based vaccines, although concerns have been raised over their univalency and the potential for C. albicans to circumvent their efficacy23.
Drugs and resistance mechanisms
Microorganisms have evolved numerous strategies to counteract cellular toxicity induced by diverse chemical stresses (xenobiotics, metals, reactive oxygen and reactive nitrogen species, and so on). Many of these generic defences have been co-opted for drug resistance. Figure 1 summarizes the major therapeutic agents used to target malaria, kinetoplastid parasites and fungi, highlighting the dates of introduction and the appearance of resistance for each. The principal methods of resistance (Fig. 2) involve either reduction of the free drug level at the target site of action, alterations in the drug target, reducing its drug-binding affinity, or overexpression of the target, restoring its essential function. In the case of inhibition of a metabolic pathway, the essential end product can be produced either by induction of an alternative pathway or by upregulation of a salvage pathway to obtain an essential metabolite from the host. Downstream consequences of target inhibition include damage to DNA, proteins and lipids such that upregulation of repair pathways can also contribute to resistance. Unlike bacteria, acquisition of resistance genes by lateral gene transfer on plasmids has not been observed for protozoan parasites or fungal pathogens. In Table 2 we summarize the drugs used to treat eukaryotic microbial pathogens, their mode of action and mechanisms of resistance where known. Here, we highlight specific examples in which drug resistance or the threat of resistance challenges current control efforts.
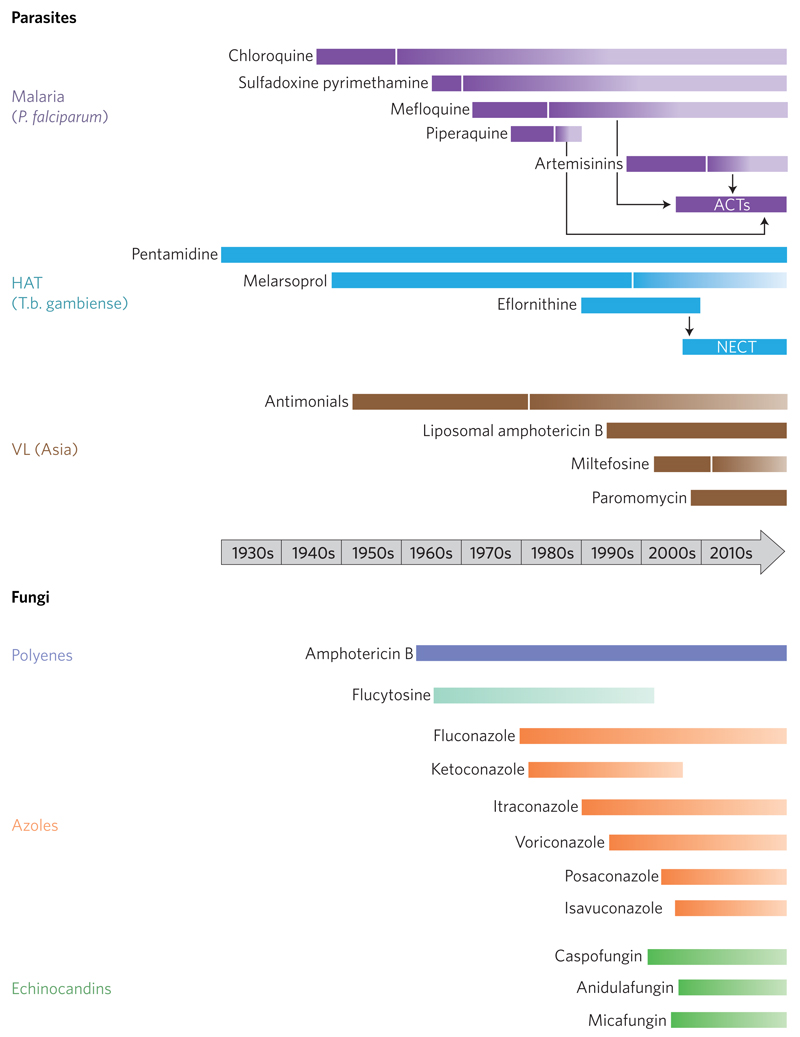
Figure 1. Timelines for emergence of drug resistance in parasitic diseases and fungi.
The solid bars represents the time from first widespread clinical use to the first year drug resistance was suspected or confirmed. The shading gradient indicates that certain drugs are still in use for particular indications or in specific geographical locations. NECT, nifurtimox–eflornithine combination therapy; VL visceral leishmaniasis. For fungal pathogens, insensitive or resistant strains have been identified shortly after the introduction of all of the major classes of antifungal agents. In the case of amphotericin B, there remains very little resistance — and differences in sensitivity mainly reflect the relative inherent sensitivity of different species to this agent.
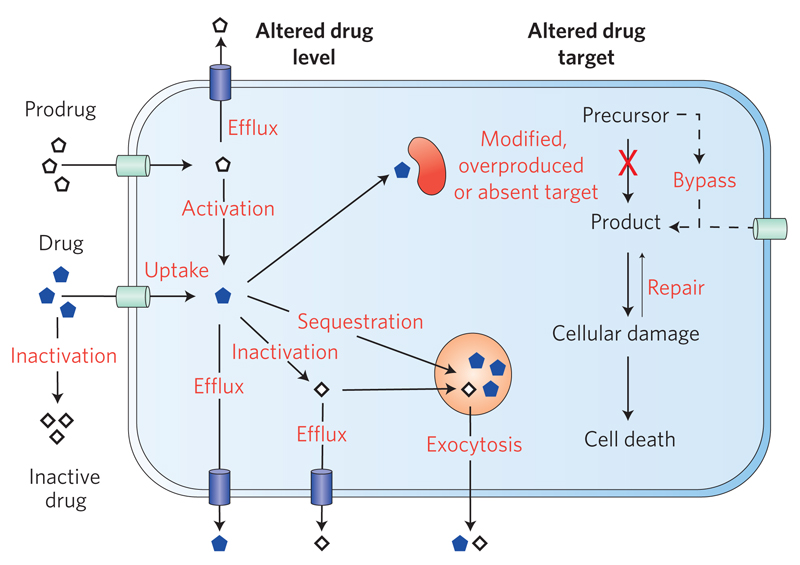
Figure 2. Molecular mechanisms of drug resistance.
Eukaryotic microbial pathogens can exhibit drug resistance through reducing the overall intracellular concentration of the drug (less uptake, more efflux), by inactivating or failing to activate the drug, or by sequestering the drug away from its target. Resistance can also be mediated by reducing affinity of the drug for the target by mutation or by reducing the drug effect by overexpression of the target. Salvage and bypass pathways can also lower the overall impact of the drug action, as can the activation of pathways to repair any damage caused.
Table 2. Modes of action and mechanisms of drug resistance in eukaryotic microorganisms.
| Pathogen | Drug and date of resistance reported | Drug class | Mode of action | Resistance mechanism |
|---|---|---|---|---|
| Plasmodium | Chloroquine: 1957 (Southeast Asia); 1960 (South America); mid-1980s (Africa) | 4-Aminoquinoline | Chloroquine interferes with the detoxification of haem into chemically inert haemozoin, resulting in accumulation of toxic chloroquine ferric haem complex and subsequent parasite lysis130. | K76T (ref. 27) mutation in a digestive-vacuole-sited, ATP-dependent, 10-transmembrane-domain transporter PfCRT27; a range of more than 30 different mutations might interact epistatically131–133. These stimulate the active efflux of chloroquine by mutant PfCRT or the passive efflux of diprotonated chloroquine133. Other genes contributing to resistance include: the P. falciparum multidrug resistance transporter 1 (PfMDR1) homologue; multipass transmembrane transporter CG2; and PfNHE1 and a sodium hydrogen antiporter also associated with quinine resistance134. The specific genetic background of the parasite and the range of mutations in genes other than PfCRT are also key to the manifestation of chloroquine resistance135. An independent mutation in PfCRT (C350R) can reverse chloroquine resistance and also increase susceptibility of the parasite to other antimalarials (mefloquine, quinine and lumefantrine, but not piperiquine136). The mutation N326D confers increased resistance to the antimalarial amodiaquine40. |
| Mefloquine: 1982 (Thailand) | Quinoline-4-methanol | Blockade of haemozoin formation and binding to phospholipids. | PfMDR1 is associated with mefloquine resistance137 but may also modulate chloroquine resistance through compensatory mutations that counteract PfCRT mutations that compromise parasite fitness138. | |
| Artesunate, dihydroartemisinin and artemether: 2008 (Southeast Asia)29 | Sesquiterpene lactone endoperoxides | Form a carbon-centred free radical or reactive electrophilic intermediate that alkylates a number of malaria proteins139 after activation by haem or free iron. | Dormancy resulting in an extended ring stage phase of development in the erythrocyte promotes resistance30–32. Multiple independent mutations in a gene encoding KELCH13 confer resistance33–37. This results in its enhanced association with PI(3)K, which is subsequently underubiquitinated and accumulates along with its lipid product, PI(3)P. The specific genetic background of the parasite and the range of mutations in genes other than kelch13 may also be key to the manifestation of resistance to artemisinin33. |
|
| Sulfadoxine/pyrimethamine: 1967 (Thailand); 1980s (Africa) | Antifols | Sulfadoxine: inhibition of dihydropteroate synthase (DHPS). Pyrimethamine: inhibition of dihydrofolate reductase (DHFR). Synergistic effect on thymidylate synthesis. |
Decreased affinity of both drugs for their respective targets. Resistance to sulfadoxine involves DHPS point mutations. DHPS variant A437G confers moderate resistance, with the additional mutations S436F and A613S conferring high-level resistance140. Pyrimethamine clinical resistance involves DHFR point mutations at S108N in Africa and Southeast Asia. Additional mutations that confer high-level resistance are N51I and C59R (ref. 141). Increased GTP cyclohydrolase (copy number variations) enhances folate biosynthesis, compensating for loss of fitness141. | |
| Proguanil | High-level resistance to cycloguanil (a metabolite of proguanil) involves DHFR mutation of Ser108 to threonine. The triple mutations (C59R, S108N and I164L) confer cross-resistance to both pyrimethamine and cycloguanil142. | |||
| Atovaquone (in combination with proguanil for prophylaxis or treatment) | Effective resistance to atovaquone involves one of a range of mutations in cytochrome b, most commonly Y268S. Other mutations associated with such resistance include I258M, Y268C, M133I and V259L143. | |||
| African trypanosomes | Suramin | Naphthylamine trisulfonic acid | Mode of action unknown. | Laboratory-generated resistance mediated through the silencing of invariant surface glycoprotein (ISG75), the AP1 adaptin complex, lysosomal proteases and major lysosomal transmembrane protein, as well as spermidine and N-acetylglucosamine biosynthesis108. |
| Pentamidine: clinical resistance is not significant | Diamidine | Mode of action unknown. | Resistance is associated with loss of uptake on the P2 adenine/adenosine transporter144, (AT1)145. Cross-resistance between melaminophenyl arsenicals and diamidines is mediated by AQP2 (ref. 146). A chimaeric AQP2/AQP3 gene is associated with cross-resistance to melarsoprol and pentamidine in laboratory-generated49,146,147 and clinical isolates148,149 |
|
| Melarsoprol: treatment failures have been reported in the Democratic Republic of Congo, Uganda, Angola and Sudan2 | Trivalent melaminophenyl arsenical | Forms a cyclic complex with trypanothione known as MelT (ref. 46). Inhibits trypanothione reductase and probably other targets. | Resistance is associated with loss of uptake on the P2 adenine/adenosine transporter144,145. A non-functional mutant has been identified in melarsoprol-resistant field isolates150. See also AQP in pentamidine section. |
|
| Eflornithine (difluoromethylornithine) | Fluorinated amino acid | Mechanism-based inhibitor of ornithine decarboxylase, required for biosynthesis of polyamines and trypanothione. | Laboratory-generated resistance is due to loss of a non-essential amino acid transporter151,152. There is no detected resistance in T. b. gambiense, but there is inherent resistance in some clinical isolates of T. b. rhodesiense2. | |
| Nifurtimox: poor efficacy as monotherapy, used in combination therapy with eflornithine (NECT) | Nitrofuran | Prodrug activated by an oxygen-insensitive mitochondrial nitroreductase (NTR)153 to form highly reactive drug metabolites154 that kill trypanosomes via unknown mechanisms155. | A genome-scale RNA interference screen identified NTR and a number of other genes possibly associated with NTR function108. NTR is also the key resistance determinant in laboratory-generated lines156,157, showing cross-resistance to fexinidazole, an oral nitroimidazole currently undergoing phase II/III clinical trials for HAT. | |
| South American trypanosomes | Benznidazole, nifurtimox: natural resistance in some T. cruzi isolates | Nitroheterocyclics | Benznidazole is activated by mitochondrial NTR153,158 to form electrophilic drug metabolites159,160. | Drug efflux via an ABCG-like transporter161. The NAD(P)H flavin oxidoreductase (old yellow enzyme) is downregulated in resistant lines162,163. However, this enzyme does not reduce benznidazole and only reduces nifurtimox under anaerobic conditions164. |
| Visceral leishmaniasis | Sodium stibogluconate, meglumine antimonite: 1990s widespread resistance in India and Nepal, not widespread in sub-Saharan Africa or Brazil | Pentavalent antimonials | SbV is reduced to SbIII to attack intracellular amastigotes. Likely to bind multiple targets including trypanothione reductase165,166, tryparedoxin peroxidase167 and CCHC zinc finger proteins166. | Selection for resistance to trivalent arsenic results in cross-resistance to trivalent antimony in vitro61 and in vivo62. Resistance is multifactorial through several mechanisms: (1) Decreased reduction of SbV to SbIII. (2) SbIII is taken up via an aquaglyceroporin73 and modulation of expression of AQP1 affects SbIII susceptibility71–73. (3) Elevated intracellular trypanothione levels168 or increased biosynthetic potential65,66,78,79. (4) Increased levels of tryparedoxin peroxidase confer resistance to SbIII (ref. 169) and are found in clinical resistant isolates167. (5) MRPA (also known as PgpA or ABCC3), a member of the ATP-binding cassette (ABC) transporters, is amplified in some resistant lines170–172 and sequesters SbIII in an intracellular vacuolar compartment close to the flagellar pocket80. (6) Chaperones and stress-related proteins are upregulated67,68, potentially reducing or repairing cellular damage induced by antimonials173. |
| Paromomycin | Aminoglycoside | Inhibition of protein synthesis. | Added to World Health Organization essential medicines list in 2007. No notable clinical resistance. Laboratory-derived resistant lines show decreased drug uptake and increased expression of ribosomal proteins174. | |
| Miltefosine: 2012 (Indian subcontinent) | Alkylphosphocholine | Miltefosine markedly perturbs lipid metabolism175–177, but the targets and precise mechanism of action are not fully understood178. | Resistance involves either: loss-of-function mutations or underexpression of an aminophospholipid translocase (LdMT)179–181 or its regulatory subunit LdRos3 (ref. 182); or drug efflux by ABC transporters183,184. Laboratory-generated resistant lines show alterations in lipid metabolism and gene expression85,185, but WGS in another study identified mutations only in the miltefosine transporter, pyridoxal kinase and an α-adaptin-like protein176. |
|
| Amphotericin B (deoxycholate or liposomal formulation): no notable clinical resistance reported | Polyene macrolide antibiotics | See below. | Not applicable, as no notable clinical resistance has been reported. | |
| Fungi | Amphotericin B, amphotericin deoxycholate | Polyene macrolide antibiotics | Binds ergosterol more avidly than human cholesterol, disrupting the semipermeable membrane and causing leakage of essential metabolites and the collapse of electrochemical gradients. Binding of low-density lipoprotein receptors and amphotericin-mediated oxidative damage may also contribute. | Laboratory mutants with lower ergosterol content are less sensitive to amphotericin B, but are rare clinically. Aspergillus terreus is intrinsically less amphotericin sensitive but resistant strains have a normal ergosterol content, suggesting that membrane permeability may not be the only mechanism of amphotericin action186. Binding to ergosterol might contribute to its mode of action187. |
| Fluconazole, itraconazole, voriconazole, posaconazole, ravuconazole, isavuconazole | Azoles | Bind haem groups and inhibit the P450-mediated 14α-demethylation (Erg11p or Cyp51p) of lanosterol in the ergosterol biosynthetic pathway. Leads to impaired membrane permeability, membrane protein action and cell wall synthesis188. | Resistance involves the overexpression of drug efflux pumps and point mutations in the target ERG11/CYP51A gene product, along with promoter mutations in these genes189–191. Changes in the levels of three fungal main efflux pumps, Cdr1, Cdr2 and Mdr1, and mutations in the genes encoding the Tac1, Upc2, Pdr1 and Mrr1 transcription factors required for efflux pump upregulation, represent major causes of decreased drug sensistivity192,193. This type of azole resistance can be acerbated by isochromosome formation and aneuploidy, which can increase the copy number of key resistance genes such as ERG11 and TAC1 (refs 194–196). Interference with the RNA-polymerase-II-interacting mediator complex can re-sensitize Pdr1-dependent regulation of drug efflux pumps197. Chaperone Hsp90 can mitigate against stress induced damage198 and also contribute to multidrug resistance with echinocandins. TR34/L98H and the more recently identified TR46/Y121F/T289A alleles that confer clinical azole resistance are likely to have arisen from environmentally generated mutations. |
|
| Caspofungin, micafungin, anidulafungin, CD101 (formerly biofungin) | Echinocandins | Cyclic hexapeptides with an antifungal bioactive lipid side chain that binds the fungal-specific β-1,3-glucan synthase Fks cell membrane proteins, disrupting cell wall integrity. | Resistance through point mutations in two major hotspots in the β-1,3 glucan synthase genes FKS1, and, in C. glabrata, FKS2 (refs 86, 101, 199); these reduce drug binding57,181,182. Upregulation of cell wall chitin can protect cell wall damage184–186. Hsp90 chaperone can mitigate against stress-induced damage170. |
|
| Flucytosine (5-fluorocytosine) | Fluoropyrimdines | Converted to 5-fluorouracil by cytosine deaminase, which becomes incorporated into RNA, resulting in inhibition of DNA synthesis. | Resistance results from mutations in the genes encoding cytosine permease transporter, cytosine deaminase, which converts 5-fluorocytosine to 5-fluorouracil, or the uracil phosphoribosyl transferase required to convert 5-fluorocytosine into a substrate for nucleic acid synthesis200. Their impact is lessened by the use of 5-fluorocytosine in combination therapy. |
Malaria
The most successful antimalarial in history to date has been chloroquine, a 4-aminoquinoline derivative of quinine (itself the world’s first mass-distributed antimalarial), first synthesized in 1934 (ref. 24). Chloroquine was cheap and remained effective for decades. However, due to massive overuse and suboptimal compliance, resistance to chloroquine emerged in Southeast Asia in 1957 and in South America in 1960, and — by the mid-1980s — it was barely possible to use even in Africa25. Although disputed by some26, the leading candidate for resistance to chloroquine is the P. falciparum chloroquine resistance transporter PfCRT (ref. 27). However, despite reports that PfCRT functions as a chloride channel, a proton pump, an activator of Na+/H+ exchangers or a cation channel, the physiological function of PfCRT remains unclear28. Nonetheless, PfCRT is central to much antimalarial resistance, the precise profile of which is modulated by associated mutations in other genes.
Artemisinin and its derivatives are fast-acting but short-lived antimalarials that have been globally successful. In particular, ACTs (for example, artemether–lumefantrine, artesunate–amodiaquine and dihydroartemisinin–piperaquine) were recommended by the World Health Organization in 2001 to ensure high cure rates of falciparum malaria and to reduce the spread of drug resistance to other frontline drugs. However, clinical resistance was confirmed in 2008 (ref. 29), characterized by a failure to clear parasites rapidly in patients around the Thai–Cambodian border30,31. Resistant parasites were characterized by transcriptomics32, large-scale whole-genome sequencing (WGS) of clinical isolates33,34 and classical generation of resistant mutants by in vitro culture followed by WGS (ref. 35). This pinpointed multiple independent mutations in a gene encoding a Kelch propeller protein (KELCH13), which was then causally linked to resistance by reverse genetics36,37. Large-scale genomic epidemiological evidence suggests that artemisinin resistance is not as straightforward as the simple acquisition of mutations in kelch13. Indeed, nonsynonymous mutations in ferredoxin, apicoplast ribosomal protein S10, multidrug resistance protein 2 and PfCRT also showed strong associations with artemisinin resistance29. These mutations appear to act as markers of a genetic landscape upon which artemisinin-resistance-conferring kelch13 mutations are more likely to occur. These landscape mutations also correlate with the current geographical limits of artemisinin resistance29. This concept is further supported by additional genomic epidemiological evidence that demonstrates that many of the 20 or so mutations in kelch13 that have been implicated in the Southeast Asian manifestation of artemisinin resistance are also found in African P. falciparum isolates. However, these mutations are present at no greater frequency in the African strains than other P. falciparum genes, indicating a lack of selective pressure in that continent and that these strains lack the enabling genetic background observed in Southeast Asia38.
Kelch propeller domain proteins are subcellular organizers of multiprotein complexes, and indeed artemisinin-resistance-associated mutation of KELCH13 results in its reduced association with phosphatidylinositol-3-OH kinase (PI(3)K)39. Experimental overexpression of PI(3)K results in enhanced artemisinin resistance, and phosphatidylinositol 3-phosphate (PI(3)P) levels are predictive of resistance to artemisinin39. In addition, upregulation of the chaperonin complexes PROSC and TRiC, involved in the unfolded protein stress response in other eukaryotes, may contribute to artemisinin resistance32. Worryingly, resistance to some of the various ACT regimens (involving lumefantrine, amodiaquine and PfCRT) is becoming evident40–43. However, the framework for the rapid evaluation of genome evolution in the face of drugs is in place and will hopefully swiftly indicate any further potential mechanisms.
Human African trypanosomiasis
The vast majority of reported cases of HAT are caused by T. b. gambiense, with less than 2% caused by T. b. rhodesiense44. Treatment involves either pentamidine or suramin for stage 1 infection (before central nervous system involvement), whereas melarsoprol, eflornithine or nifurtimox/eflornithine combination therapy are used once the parasite crosses the blood–brain barrier2, with the combination therapy reducing the duration of treatment regimens. Given the limited chemotherapeutic options for the treatment of HAT (Table 2), drug resistance could seriously compromise efforts to eliminate this epidemic disease as a public health problem44. Fortunately, resistance emergence for pentamidine has not been marked, despite continuous use of pentamidine since the 1940s, including a mass chemoprophylactic campaign in the 1950s in the then Belgian Congo. However, cross-resistance to pentamidine and melarsoprol, used for stage 2 of infection, is frequently observed. Melarsoprol is a trivalent melaminophenyl arsenical that has a propensity to react covalently with vicinal dithiols, including the parasite-specific dithiol, trypanothione45, to form a cyclic complex known as MelT (ref. 46). Melarsoprol has a high incidence of severe (lethal) toxicities and high rates of treatment failures have been reported in the Democratic Republic of Congo, Uganda, Angola and Sudan2. Although therapeutic failure does not necessarily equate with drug resistance, it appears that the high relapse rate in northwest Uganda is associated with reduced susceptibility to melarsoprol47,48. The recent report that the aquaglyceroporin AQP2 appears to function as a transporter for large drugs such as pentamidine and melarsoprol was surprising, given that aquaglyceroporins are channels facilitating the passive transport of water and small neutral molecules across cell membranes. Nonetheless, there is strong evidence that AQP2 is synonymous with the high-affinity pentamidine transporter (HAPT1)49, with a recent report indicating that pentamidine binds and inhibits the transporter and is then internalized via endocytosis50.
Chagas disease
For Trypanosoma cruzi, an intracellular parasite with a wide tissue tropism, infection has three phases: an acute phase associated with high parasitaemia; an asymptomatic (indeterminate) phase lasting anywhere between 10 to 30 years, during which parasitaemia is controlled by the immune response; and a chronic phase in about 30–40% of patients characterized by either cardiac disease or digestive disease (mega-oesophagus and megacolon). For treatment, benznidazole and nifurtimox have considerable activity in the acute phase51 and benznidazole also eliminates parasitaemia in the indeterminate and chronic phases of the disease52,53. However, a large multi-centre, randomized trial of benznidazole for chronic Chagas cardiomyopathy failed to significantly reduce cardiac clinical deterioration at up to 5 years follow-up53. Whether this is due to differences in drug susceptibility, pharmacokinetic/pharmacodynamic issues or the pathophysiology of the disease is not known. The results of two recent clinical trials with azole ergosterol inhibitors, posaconazole and E1224 (a pro-drug of ravuconazole) have been equally disappointing52,54,55.
Visceral leishmaniasis
Treatment of visceral leishmaniasis, cutaneous and mucocutaneous leishmaniasis is limited to four main drugs: pentavalent antimonial complexes (sodium stibogluconate and meglumine antimonate); amphotericin B (as deoxycholate or liposomal formulations); the aminoglycoside paromomycin; and the alkylphosphocholine miltefosine56,57. Treatment varies according to geographical location, the immune status and other co-morbidities of the patient, and the disease classification58.
Of these treatments, widespread resistance to antimonial drugs is specific to South Asia and is not seen in sub-Saharan Africa or Brazil. Indeed, antimonial drugs are not recommended in India or Nepal owing to treatment failures starting in the 1990s and now reported to be as high as 60% in some regions59. This has been attributed to inappropriate treatment in an unregulated private health system or to the use of substandard antimonial drugs. However, South Asia is the only region where arsenic exposure and widespread antimonial resistance co-exist. Thus, environmental pollution and exposure of patients to arsenic in food and drinking water was proposed as an alternative hypothesis for the high rates of resistance60. Arsenic and antimony are both metalloids and selection of leishmania parasites for resistance to trivalent arsenic results in cross-resistance to trivalent antimony in vitro61, but its physiological relevance is uncertain. Chronic exposure of infected mice to arsenic in drinking water at environmentally relevant levels showed that it is possible to generate resistance to pentavalent antimony in vivo62. A retrospective clinico-epidemiological study identified a trend towards increased treatment failure in arsenic-exposed patients, but failed to reach statistical significance63.
Resistance to antimonials is multifactorial, and most of the mechanisms shown in Fig. 2 have been implicated in Leishmania. Studies on experimental and clinical resistant isolates strongly support the hypothesis that trypanothione has a pivotal role in antimonial resistance. However, none of the following mechanisms are universal in resistant isolates. Decreased biological reduction of SbV to SbIII has been reported in resistant leishmania amastigotes64 and two candidate ‘antimony reductases’ have been identified, although genetic65,66 and proteomic studies67,68 have not identified any changes in either TDR1 (ref. 69) or ArsC (ref. 70). The mechanism of uptake of SbV is not known, but modulation of expression of AQP1 affects SbIII susceptibility71–73. AQP1 copy number and expression levels correlate with susceptibility to SbIII in some, but not all, clinical isolates74,75. However, interpretation of this observation is complicated by the fact that AQP1 is located on chromosome 31, which is frequently trisomic or tetrasomic76 in these mosaic aneuploid parasites77. Upregulation of trypanothione and ancillary biosynthetic pathways has also been observed in genomic65,66 and metabolomic78,79 studies. MRPA is responsible for ATP-dependent efflux of SbIII as a thiol conjugate into membrane vesicles80 and a homodimeric ABC half-transporter (ABCI4) is one possible candidate for efflux across the plasma membrane81.
Miltefosine, the only oral treatment for visceral leishmaniasis, was first approved for use in India in 2002. However, a decade on there is an increasing rate of clinical relapse82,83, which threatens to undermine the Kala-Azar Elimination Program in the Indian subcontinent. Stable resistance is readily generated in the laboratory with no cross-resistance to other anti-leishmanial drugs84,85.
Fungi
Several classes of antifungals are used clinically (Tables 1 and 2) — each with very different drug resistance profiles. The oldest antifungals are the polyene macrolide antibiotics, exemplified by amphotericin B, which remains a frontline choice for a broad-spectrum agent for fungal infections of unknown aetiology. Amphotericin deoxycholate is toxic to the kidneys, a side effect that is substantially ameliorated in lipid carrier formulations such as AmBisome, which also has potent anti-Leishmania activity. As with other eukaryotic pathogens, resistance to antifungal drugs has become an increasingly important clinical problem86,87. A few recognized cases exist of inherent resistance of certain fungi to specific antifungals, but mostly resistance is due to induced changes and mutations.
The imidazoles and more modern triazoles (collectively known as the ‘azoles’) constitute the main class of antifungals used in the treatment of infections. Various modifications of the triazole ring have generated a series of antifungals including fluconazole (used mainly in the treatment of Candida infections), and itraconazole, voriconazole, posaconazole, ravuconazole and the recently licenced isavuconazole, which have improved activity against Aspergillus and filamentous fungal species. These compounds have important differences in antifungal potencies, spectrum of activities, bioavailability, drug interactions and potential toxicity. For example, some patients treated with voriconazole suffer from photosensitivity and an elevated risk of skin carcinoma88. Other sterol inhibitors include the allylamines squalene epoxidase inhibitors and phenylmorpholine Erg24 D14 reductase and Erg2 D8-D7 isomerase inhibitors that are used topically against dermatophytic infections for which clinical resistance is low.
Although some fungi such as Candida krusei are inherently azole resistant, multiply triazole-resistant strains are now emerging89,90, as well as strains with cross-resistance to azoles and echinocandins, suggesting the emergence of concerning multidrug-resistant phenotypes in medically important fungi91. A threat from multi-azole-resistant strains of A. fumigatus may have arisen under the selective pressure of agricultural azole fungicides and subsequent transmission of azole-resistant strains to the clinic by spore dispersal92–95. The prevalence of these alleles is increasing in Europe and now in other parts of the world90,96,97. In Candida, mutants harbouring azole resistance have a fitness deficit98; however, multidrug-resistant strains of Aspergillus do not seem to have markedly decreased fitness, implying that they may become stably represented in the environment.
The most recently developed major class of antifungal is the echinocandin antibiotics, of which caspofungin, micafungin and anidulafungin are used clinically. These have similar pharmaco-kinetic properties, although a new echinocandin (CD101; formerly biofungin) is in clinical trials and has improved stability in vivo and requires less frequent intravenous dosing. Echinocandins are fungicidal against Candida species and fungistatic or fungicidal against Aspergillus, causing hyphal or bud tip lysis, but they are not efficacious against Pneumocystis jiroveci and some other species.
Hsp90-mediated changes in drug tolerance have also been implicated in determining echinocandin sensitivity99. Recently, multidrug azole/echinocandin resistance has been identified in fungi and this is particularly frequent in strains of C. glabrata, which is common in patients with haematological malignancies and solid tumours100,101. These multidrug-resistant strains of C. glabrata can then only be treated with intravenous amphotericin, and since this agent has poor penetration into urine such infections are essentially untreatable.
Outstanding challenges and future prospects
We began by highlighting the similarities and differences between the emergence of drug resistance in prokaryotic and eukaryotic microorganisms. The control of emerging drug resistance in eukaryotic microbial pathogens also has similarities and distinctions from prokaryotic drug resistance, and the challenge for the future is to ensure best practice is employed for both groups. One effective mechanism to control the spread of drug resistance in bacterial pathogens is the application of appropriate antibiotic stewardship, applying the right drug at the right dose, at the right time, for the right duration. This approach operates effectively where there is well-regulated healthcare, effective and rapid screening, a selection of available drugs as contingency, and the necessary education and engagement between the patient and healthcare provider. Moreover, bacterial drug resistance is a global phenomenon in which resistance selected through poor stewardship in one geographical area may be contained by stringent practices in other areas, or combatted by an investment in new pharmaceutical development in wealthy countries. These containment measures are inevitably less effective where primary care is limited or too expensive, education is lacking or where the diseases involved do not have a direct impact in the developed world. As a consequence, the limitation of many eukaryotic pathogens to the poorer parts of the world makes a coordinated response to resistance emergence more difficult to achieve.
The drivers of the emergence of resistance are also more difficult to mitigate for many eukaryotic pathogens. As highlighted earlier, drug provenance and effective delivery is a considerable challenge in the developing world. Delivery is a particular challenge for prospective mass drug administration programmes, in which delivery to a population on a broad or local scale, if incomplete, can counteract its intention to contain the spread of existing resistance in target regions. A further complication in low- and middle-income countries is the effects of co-infection or malnutrition in populations treated with drugs targeting a particular pathogen (discussed previously102). Notably, the pharmacokinetic behaviour of drugs in malnourished individuals may be variable and unpredictable, leading to inadvertent under-dosing, which can drive resistance. When combined with immunosuppression induced by many parasites, or the hospital-induced immunosuppression of patients that become susceptible to fungal infection, drug concentrations that would clear infections in the context of a robust immune system may fall short in its absence. The ecological balance between distinct pathogens in patients with co-infections can also lead to unanticipated consequences, in which the removal of one pathogen can create a niche exploited by a distinct pathogen, or in which the normal interactions between pathogens with each other, and with the immune system, is perturbed with drug pressure. The resistance mechanisms selected in drug-treated populations can also alter pathogen phenotypes with the risk of enhanced virulence.
Although the factors that drive drug resistance are well known, it remains essential to identify when drug resistance arises and to respond rapidly and effectively. As with health care, surveillance is a key challenge for diseases in the developing world, where populations may be inaccessible, reluctant to engage or where treatment failure can have multiple causes beyond the emergence of drug resistance. Moreover, resistance can show considerable variation among populations or in different geographical settings. Here, accurate and rapid detection is critical to understand resistance epidemiology and thereby the best treatment to deliver, but this can be difficult to achieve. Despite this, developments in field polymerase chain reaction (PCR) assays and next-generation sequencing permit the sensitive identification and tracking of emergent resistance, allowing earlier control responses than could be previously achieved. Hence, an integration of improved therapeutic delivery and treatment monitoring are critical control points to reduce the emergence of resistance, in tandem with the discovery of the relevant resistance mechanisms and the search for new drug therapies. These combined approaches span the individual scientific researcher to clinicians, to health agencies, governments and populations, all of which must be well integrated and alert, with effective and rapid communication between different levels to allow appropriate responses to be put into action if needed.
Fortunately, while drug resistance is emerging in many eukaryotic microbial pathogens, new tools and methodologies are being developed to (1) predict resistance mechanisms, (2) identify modes of drug action and potential escape pathways, and (3) understand pathogen biochemistry as a means of discovering possible new therapies. With respect to drug resistance, the advent of cost-effective and rapid genome resequencing allows signatures of selection to be identified103–106, while genome-wide RNA interference screens allow the mapping of resistance pathways107,108, and overexpression libraries109 can assist with drug target deconvolution through selective screens. These genetic tools are complemented by improvements in proteomics such that adaptations accompanying drug resistance can be pinpointed, providing information on resistance mechanisms, and potential diagnostic tools to detect the emergence of resistance110. Combined with the improved sensitivity and resolution of metabolomics analysis111, biochemical pathways can also be mapped in the context of drug exposure, allowing bypass mechanisms to be highlighted, if present. These each provide the essential early warning systems necessary to identify and combat the spread of drug resistance. Furthermore, certain combination therapies might offer novel transmission blocking strategies: very recently, resistance to the antimalarial atovaquone, a component (with proguanil) of the widely used and successful treatment marketed as Malarone, was further characterized. Resistance mutations that appear during the blood stage of infection localize to the mitochondrial protein cytochrome b, one of the few proteins encoded by the highly reduced Plasmodium mitochondrial genome. All atovaquone-resistance mutations examined generate a deficient mitochondrion and a parasite that, while viable in the blood, is incapable of development in the mosquito and thereby cannot be transmitted112. Thus, despite the fact that resistance to atovaquone might arise repeatedly, each incident is isolated. Drugs that target cytochrome b could form part of combination therapies that are self-limiting in terms of spread of drug resistance and may delay any transmission of resistance that arises to the drug it is partnered with.
Concluding remarks
Drug resistance in eukaryotic microorganisms is an increasing global problem that threatens the advances in health care made over the last 50 years. This mirrors the situation for bacterial and viral pathogens but is particularly acute given the abundance of eukaryotic pathogens in the poorest regions of the world. These countries have the least capacity to respond to the emergence of resistance through the development of new drugs, vaccines and diagnostics, while developed countries lack financial incentives to assist. Nonetheless, there are opportunities to respond to this threat owing to the distinct biology of many major eukaryotic pathogens, uncovered through basic research. Furthermore, many eukaryotic microorganisms are arthropod-borne diseases, such that targeting transmission can be a route to pathogen control not available for opportunistic pathogens. This can take the form of transmission-blocking vaccines or drugs targeting Plasmodium113, or the application of vector control measures such as insecticide-impregnated bed nets114, peri-domestic and indoor residual insecticide spraying114,115, tsetse traps116, or improved housing117. Sterile insect release is also a route to limiting the vector population and so restricting disease spread118,119. Eukaryotic microorganisms have also, like some bacterial pathogens, been found to show cooperative and social behaviours to optimize their establishment and transmission in their hosts or vectors120,121. These social responses can control parasite density or the development of transmission stages122–124, such that blocking or mimicking signals for communication or their transduction pathways provides new routes for limiting the impact of the pathogens using strategies that might be less susceptible to the emergence of resistance.
Whether or not new targets or approaches can be identified, there is a real need to optimize the delivery and deployment of drugs. Control of drug quality and distribution, and the supply of cost-effective drugs is crucial. Furthermore, the application of both epidemiological modelling and evolutionary theory to guide drug treatment policies is important in prolonging the lifespan of drugs and thereby maximizing the return on the considerable cost associated with developing and introducing a new drug. Targeted therapy as opposed to mass drug administration is key to limiting the emergence of resistance, or containing resistance when it is detected. This requires an integration of epidemiology, diagnosis, detection and supply chain control, as well as investment in a pipeline of new therapeutics ready to be deployed when resistance inevitably emerges. Only through slowing the emergence of resistance and accelerating new drug discovery will the control successes achieved against eukaryotic microbial pathogens be sustained.
Acknowledgements
This Review is a collaboration between the Wellcome Trust funded centres of Infectious Disease Research in Scotland (IDRIS). This comprises the Edinburgh University ‘Centre for Immunity, Infection and Evolution’ (Wellcome Trust grant reference 095831), Glasgow University ‘Wellcome Trust Centre for Molecular Parasitology’ (Wellcome Trust grant reference WT104111AIA), Dundee University Wellcome Trust strategic award in ‘Chemical Biology for Target Identification’ (Wellcome Trust grant reference 10502) and the Aberdeen University Wellcome Trust Strategic Award in ‘Medical Mycology and Fungal Immunology’ (Wellcome Trust grant reference 097377). Personal research support involved Wellcome Trust senior investigator awards to K.R.M. (Wellcome Trust grant reference 103740/Z/14/Z) and N.A.R.G. (Wellcome Trust grant reference 101873), and Wellcome Trust Principal Research fellowships to A.H.F. (Wellcome Trust grant reference 079838) and A.P.W. (Wellcome Trust grant reference 107046/Z/15/Z).
Footnotes
Author contributions
All authors contributed equally to the preparation of the manuscript.
Additional information
Reprints and permissions information is available online at www.nature.com/reprints. Correspondence should be addressed to K.R.M.
Competing interests
The authors declare no competing financial interests.
References
- 1.Woolhouse M, Farrar J. Policy: an intergovernmental panel on antimicrobial resistance. Nature. 2014;509:555–557. doi: 10.1038/509555a. [DOI] [PubMed] [Google Scholar]
- 2.Lutje V, Seixas J, Kennedy A. Chemotherapy for second-stage human African trypanosomiasis. Cochrane Database Syst Rev. 2013;6:CD006201. doi: 10.1002/14651858.CD006201.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fernandez FM, Green MD, Newton PN. Prevalence and detection of counterfeit pharmaceuticals: a mini review. Ind Eng Chem Res. 2008;47:585–590. [Google Scholar]
- 4.Nayyar GM, Breman JG, Newton PN, Herrington J. Poor-quality antimalarial drugs in southeast Asia and sub-Saharan Africa. Lancet Infect Dis. 2012;12:488–496. doi: 10.1016/S1473-3099(12)70064-6. [DOI] [PubMed] [Google Scholar]
- 5.Woolhouse M, Ward M, van BB, Farrar J. Antimicrobial resistance in humans, livestock and the wider environment. Philos Trans R Soc Lond B Biol Sci. 2015;370:20140083. doi: 10.1098/rstb.2014.0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verweij PE, Chowdhary A, Melchers WJ, Meis JF. Azole resistance in Aspergillus fumigatus: can we retain the clinical use of mold-active antifungal azoles? Clin Infect Dis. 2016;62:362–368. doi: 10.1093/cid/civ885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewis K. Persister cells, dormancy and infectious disease. Nature Rev Microbiol. 2007;5:48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- 8.Cohen NR, Lobritz MA, Collins JJ. Microbial persistence and the road to drug resistance. Cell Host Microbe. 2013;13:632–642. doi: 10.1016/j.chom.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.LaFleur MD, Kumamoto CA, Lewis K. Candida albicans biofilms produce antifungal-tolerant persister cells. Antimicrob Agents Chemother. 2006;50:3839–3846. doi: 10.1128/AAC.00684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kucharikova S, Tournu H, Lagrou K, Van DP, Bujdakova H. Detailed comparison of Candida albicans and Candida glabrata biofilms under different conditions and their susceptibility to caspofungin and anidulafungin. J Med Microbiol. 2011;60:1261–1269. doi: 10.1099/jmm.0.032037-0. [DOI] [PubMed] [Google Scholar]
- 11.LaFleur MD, Qi Q, Lewis K. Patients with long-term oral carriage harbor high-persister mutants of Candida albicans. Antimicrob Agents Chemother. 2010;54:39–44. doi: 10.1128/AAC.00860-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kussell E, Kishony R, Balaban NQ, Leibler S. Bacterial persistence: a model of survival in changing environments. Genetics. 2005;169:1807–1814. doi: 10.1534/genetics.104.035352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rovira-Graells N, et al. Transcriptional variation in the malaria parasite Plasmodium falciparum. Genome Res. 2012;22:925–938. doi: 10.1101/gr.129692.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K. Persister cells and tolerance to antimicrobials. FEMS Microbiol Lett. 2004;230:13–18. doi: 10.1016/S0378-1097(03)00856-5. [DOI] [PubMed] [Google Scholar]
- 15.Pankey GA, Sabath LD. Clinical relevance of bacteriostatic versus bactericidal mechanisms of action in the treatment of Gram-positive bacterial infections. Clin Infect Dis. 2004;38:864–870. doi: 10.1086/381972. [DOI] [PubMed] [Google Scholar]
- 16.Laufer MK, Djimde AA, Plowe CV. Monitoring and deterring drug-resistant malaria in the era of combination therapy. Am J Trop Med Hyg. 2007;77:160–169. [PubMed] [Google Scholar]
- 17.Peters W. The prevention of antimalarial drug resistance. Pharmacol Ther. 1990;47:499–508. doi: 10.1016/0163-7258(90)90067-c. [DOI] [PubMed] [Google Scholar]
- 18.White NJ, Olliaro PL. Strategies for the prevention of antimalarial drug resistance: rationale for combination chemotherapy for malaria. Parasitol Today. 1996;12:399–401. doi: 10.1016/0169-4758(96)10055-7. [DOI] [PubMed] [Google Scholar]
- 19.Priotto G, et al. Nifurtimox-eflornithine combination therapy for second-stage African Trypanosoma brucei gambiense trypanosomiasis: a multicentre, randomised, phase III, non-inferiority trial. Lancet. 2009;374:56–64. doi: 10.1016/S0140-6736(09)61117-X. [DOI] [PubMed] [Google Scholar]
- 20.Nosten F, White NJ. Artemisinin-based combination treatment of falciparum malaria. Am J Trop Med Hyg. 2007;77:181–192. [PubMed] [Google Scholar]
- 21.Hotez PJ, Bottazzi ME, Strych U. New vaccines for the world’s poorest people. Annu Rev Med. 2015;67:405–417. doi: 10.1146/annurev-med-051214-024241. [DOI] [PubMed] [Google Scholar]
- 22.Kumar R, Engwerda C. Vaccines to prevent leishmaniasis. Clin Transl Immunology. 2014;3:e13. doi: 10.1038/cti.2014.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cassone A. Development of vaccines for Candida albicans: fighting a skilled transformer. Nature Rev Microbiol. 2013;11:884–891. doi: 10.1038/nrmicro3156. [DOI] [PubMed] [Google Scholar]
- 24.Wellems TE, Plowe CV. Chloroquine-resistant malaria. J Infect Dis. 2001;184:770–776. doi: 10.1086/322858. [DOI] [PubMed] [Google Scholar]
- 25.Wongsrichanalai C, Pickard AL, Wernsdorfer WH, Meshnick SR. Epidemiology of drug-resistant malaria. Lancet Infect Dis. 2002;2:209–218. doi: 10.1016/s1473-3099(02)00239-6. [DOI] [PubMed] [Google Scholar]
- 26.Awasthi G, Das A. Genetics of chloroquine-resistant malaria: a haplotypic view. Mem Inst Oswaldo Cruz. 2013;108:947–961. doi: 10.1590/0074-0276130274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fidock DA, et al. Mutations in the P-falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell. 2000;6:861–871. doi: 10.1016/s1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pulcini S, et al. Mutations in the Plasmodium falciparum chloroquine resistance transporter, PfCRT, enlarge the parasite’s food vacuole and alter drug sensitivities. Sci Rep. 2015;5 doi: 10.1038/srep14552. 14552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noedl H, et al. Evidence of artemisinin-resistant malaria in western Cambodia. N Engl J Med. 2008;359:2619–2620. doi: 10.1056/NEJMc0805011. [DOI] [PubMed] [Google Scholar]
- 30.Amaratunga C, et al. Artemisinin-resistant Plasmodium falciparum in Pursat province, western Cambodia: a parasite clearance rate study. Lancet Infect Dis. 2012;12:851–858. doi: 10.1016/S1473-3099(12)70181-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dondorp AM, et al. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2009;361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mok S, et al. Drug resistance. Population transcriptomics of human malaria parasites reveals the mechanism of artemisinin resistance. Science. 2015;347:431–435. doi: 10.1126/science.1260403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miotto O, et al. Multiple populations of artemisinin-resistant Plasmodium falciparum in Cambodia. Nature Genet. 2013;45:648–655. doi: 10.1038/ng.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takala-Harrison S, et al. Genetic loci associated with delayed clearance of Plasmodium falciparum following artemisinin treatment in Southeast Asia. Proc Natl Acad Sci USA. 2013;110:240–245. doi: 10.1073/pnas.1211205110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ariey F, et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghorbal M, et al. Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system. Nature Biotechnol. 2014;32:819–821. doi: 10.1038/nbt.2925. [DOI] [PubMed] [Google Scholar]
- 37.Straimer J, et al. Drug resistance. K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science. 2015;347:428–431. doi: 10.1126/science.1260867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.MalariaGEN Plasmodium falciparum Community Project. Genomic epidemiology of artemisinin resistant malaria. Elife. 2016;5:e08714. doi: 10.7554/eLife.08714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mbengue A, et al. A molecular mechanism of artemisinin resistance in Plasmodium falciparum malaria. Nature. 2015;520:683–687. doi: 10.1038/nature14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gabryszewski SJ, Modchang C, Musset L, Chookajorn T, Fidock DA. Combinatorial genetic modeling of pfcrt-mediated drug resistance evolution in Plasmodium falciparum. Mol Biol Evol. 2016 doi: 10.1093/molbev/msw037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sisowath C, et al. In vivo selection of Plasmodium falciparum parasites carrying the chloroquine-susceptible pfcrt K76 allele after treatment with artemether-lumefantrine in Africa. J Infect Dis. 2009;199:750–757. doi: 10.1086/596738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Venkatesan M, et al. Polymorphisms in Plasmodium falciparum chloroquine resistance transporter and multidrug resistance 1 genes: parasite risk factors that affect treatment outcomes for P. falciparum malaria after artemether-lumefantrine and artesunate-amodiaquine. Am J Trop Med Hyg. 2014;91:833–843. doi: 10.4269/ajtmh.14-0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amaratunga C, et al. Dihydroartemisinin-piperaquine resistance in Plasmodium falciparum malaria in Cambodia: a multisite prospective cohort study. Lancet Infect Dis. 2016;16:357–365. doi: 10.1016/S1473-3099(15)00487-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Franco JR, Simarro PP, Diarra A, Ruiz-Postigo JA, Jannin JG. The journey towards elimination of gambiense human African trypanosomiasis: not far, nor easy. Parasitology. 2014;141:748–760. doi: 10.1017/S0031182013002102. [DOI] [PubMed] [Google Scholar]
- 45.Fairlamb AH, Blackburn P, Ulrich P, Chait BT, Cerami A. Trypanothione: a novel bis(glutathionyl)spermidine cofactor for glutathione reductase in trypanosomatids. Science. 1985;227:1485–1487. doi: 10.1126/science.3883489. [DOI] [PubMed] [Google Scholar]
- 46.Fairlamb AH, Henderson GB, Cerami A. Trypanothione is the primary target for arsenical drugs against African trypanosomes. Proc Natl Acad Sci USA. 1989;86:2607–2611. doi: 10.1073/pnas.86.8.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matovu E, et al. Melarsoprol refractory T.b. gambiense from Omugo, north-western Uganda. Trop Med Int Health. 2001;6:407–411. doi: 10.1046/j.1365-3156.2001.00712.x. [DOI] [PubMed] [Google Scholar]
- 48.Robays J, et al. High failure rates of melarsoprol for sleeping sickness, Democratic Republic of Congo. Emerg Infect Dis. 2008;14:966–967. doi: 10.3201/eid1406.71266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Munday JC, et al. Trypanosoma brucei aquaglyceroporin 2 is a high-affinity transporter for pentamidine and melaminophenyl arsenic drugs and the main genetic determinant of resistance to these drugs. J Antimicrob Chemother. 2014;69:651–663. doi: 10.1093/jac/dkt442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Song J, et al. Pentamidine is not a permeant but a nanomolar inhibitor of the Trypanosoma brucei aquaglyceroporin-2. PLoS Pathog. 2016;12:e1005436. doi: 10.1371/journal.ppat.1005436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Urbina JA, Docampo R. Specific chemotherapy of Chagas disease: controversies and advances. Trends Parasitol. 2003;19:495–501. doi: 10.1016/j.pt.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 52.Molina I, et al. Randomized trial of posaconazole and benznidazole for chronic Chagas’ disease. N Engl J Med. 2014;370:1899–1908. doi: 10.1056/NEJMoa1313122. [DOI] [PubMed] [Google Scholar]
- 53.Morillo CA, et al. Randomized trial of benznidazole for chronic Chagas’ cardiomyopathy. N Engl J Med. 2015;373:1295–1306. doi: 10.1056/NEJMoa1507574. [DOI] [PubMed] [Google Scholar]
- 54.Chatelain E. Chagas disease drug discovery: toward a new era. J Biomol Screen. 2015;20:22–35. doi: 10.1177/1087057114550585. [DOI] [PubMed] [Google Scholar]
- 55.Urbina JA. Recent clinical trials for the etiological treatment of chronic chagas disease: advances, challenges and perspectives. J Euk Microbiol. 2015;62:149–156. doi: 10.1111/jeu.12184. [DOI] [PubMed] [Google Scholar]
- 56.McGwire BS. Satoskar AR Leishmaniasis: clinical syndromes and treatment. QJM. 2014;107:7–14. doi: 10.1093/qjmed/hct116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sundar S, Chakravarty J. An update on pharmacotherapy for leishmaniasis. Expert Opin Pharmacother. 2015;16:237–252. doi: 10.1517/14656566.2015.973850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.World Health Organization. Control of the Leishmaniases. Vol. 949. World Health Organization; 2010. pp. 1–186. Technical Report Series. [PubMed] [Google Scholar]
- 59.Olliaro PL, et al. Treatment options for visceral leishmaniasis: a systematic review of clinical studies done in India, 1980–2004. Lancet Infect Dis. 2005;5:763–774. doi: 10.1016/S1473-3099(05)70296-6. [DOI] [PubMed] [Google Scholar]
- 60.Perry MR, et al. Visceral leishmaniasis and arsenic: an ancient poison contributing to antimonial treatment failure in the Indian subcontinent? PLoS Negl Trop Dis. 2011;5:e1227. doi: 10.1371/journal.pntd.0001227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dey S, et al. High level arsenite resistance in Leishmania tarentolae is mediated by an active extrusion system. Mol Biochem Parasitol. 1994;67:49–57. doi: 10.1016/0166-6851(94)90095-7. [DOI] [PubMed] [Google Scholar]
- 62.Perry MR, Wyllie S, Raab A, Feldmann J, Fairlamb AH. Chronic exposure to arsenic in drinking water can lead to resistance to antimonial drugs in a mouse model of visceral leishmaniasis. Proc Natl Acad Sci USA. 2013;110:19932–19937. doi: 10.1073/pnas.1311535110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Perry MR, et al. Arsenic exposure and outcomes of antimonial treatment in visceral leishmaniasis patients in Bihar, India: a retrospective cohort study. PLoS Negl Trop Dis. 2015;9:e0003518. doi: 10.1371/journal.pntd.0003518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shaked-Mishan P, Ulrich N, Ephros M, Zilberstein D. Novel intracellular Sb-V reducing activity correlates with antimony susceptibility in Leishmania donovani. J Biol Chem. 2001;276:3971–3976. doi: 10.1074/jbc.M005423200. [DOI] [PubMed] [Google Scholar]
- 65.Decuypere S, et al. Gene expression analysis of the mechanism of natural Sb(V) resistance in Leishmania donovani isolates from Nepal. Antimicrob Agents Chemother. 2005;49:4616–4621. doi: 10.1128/AAC.49.11.4616-4621.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Decuypere S, et al. Molecular mechanisms of drug resistance in natural Leishmania populations vary with genetic background. PLoS Negl Trop Dis. 2012;6:e1514. doi: 10.1371/journal.pntd.0001514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brotherton MC, et al. Proteomic and genomic analyses of antimony resistant Leishmania infantum mutant. J Parasitol Res. 2013;8:e81899. doi: 10.1371/journal.pone.0081899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Biyani N, Singh AK, Mandal S, Chawla B, Madhubala R. Differential expression of proteins in antimony-susceptible and -resistant isolates of Leishmania donovani. Mol Biochem Parasitol. 2011;179:91–99. doi: 10.1016/j.molbiopara.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 69.Fyfe PK, Westrop GD, Silva AM, Coombs GH, Hunter WN. Leishmania TDR1 structure, a unique trimeric glutathione transferase capable of deglutathionylation and antimonial prodrug activation. Proc Natl Acad Sci USA. 2012;109:11693–11698. doi: 10.1073/pnas.1202593109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou Y, Messier N, Ouellette M, Rosen BP, Mukhopadhyay R. Leishmania major LmACR2 is a pentavalent antimony reductase that confers sensitivity to the drug Pentostam. J Biol Chem. 2004;279:37445–37451. doi: 10.1074/jbc.M404383200. [DOI] [PubMed] [Google Scholar]
- 71.Plourde M, et al. Generation of an aquaglyceroporin AQP1 null mutant in Leishmania major. Mol Biochem Parasitol. 2015;201:108–111. doi: 10.1016/j.molbiopara.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 72.Marquis N, Gourbal B, Rosen BP, Mukhopadhyay R, Ouellette M. Modulation in aquaglyceroporin AQP1 gene transcript levels in drug-resistant Leishmania. Mol Microbiol. 2005;57:1690–1699. doi: 10.1111/j.1365-2958.2005.04782.x. [DOI] [PubMed] [Google Scholar]
- 73.Gourbal B, et al. Drug uptake and modulation of drug resistance in Leishmania by an aquaglyceroporin. J Biol Chem. 2004;279:31010–31017. doi: 10.1074/jbc.M403959200. [DOI] [PubMed] [Google Scholar]
- 74.Mandal S, Maharjan M, Singh S, Chatterjee M, Madhubala R. Assessing aquaglyceroporin gene status and expression profile in antimony-susceptible and -resistant clinical isolates of Leishmania donovani from India. J Antimicrob Chemother. 2010;65:496–507. doi: 10.1093/jac/dkp468. [DOI] [PubMed] [Google Scholar]
- 75.Maharjan M, Singh S, Chatterjee M, Madhubala R. Role of aquaglyceroporin (AQP1) gene and drug uptake in antimony-resistant clinical isolates of Leishmania donovani. Am J Trop Med Hyg. 2008;79:69–75. [PubMed] [Google Scholar]
- 76.Rogers MB, et al. Chromosome and gene copy number variation allow major structural change between species and strains of Leishmania. Genome Res. 2011;21:2129–2142. doi: 10.1101/gr.122945.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sterkers Y, Crobu L, Lachaud L, Pages M, Bastien P. Parasexuality and mosaic aneuploidy in Leishmania: alternative genetics. Trends Parasitol. 2014;30:429–435. doi: 10.1016/j.pt.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 78.Rojo D, et al. A multiplatform metabolomic approach to the basis of antimonial action and resistance in Leishmania infantum. J Parasitol Res. 2015;10:e0130675. doi: 10.1371/journal.pone.0130675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Berg M, et al. Metabolic adaptations of Leishmania donovani in relation to differentiation, drug resistance, and drug pressure. Mol Microbiol. 2013;90:428–442. doi: 10.1111/mmi.12374. [DOI] [PubMed] [Google Scholar]
- 80.Dey S, Ouellette M, Lightbody J, Papadopoulou B, Rosen BP. An ATP-dependent As(III)-glutathione transport system in membrane vesicles of Leishmania tarentolae. Proc Natl Acad Sci USA. 1996;93:2192–2197. doi: 10.1073/pnas.93.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Manzano JI, Garcia-Hernandez R, Castanys S, Gamarro F. A new ABC half-transporter in Leishmania major is involved in resistance to antimony. Antimicrob Agents Chemother. 2013;57:3719–3730. doi: 10.1128/AAC.00211-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sundar S, et al. Efficacy of miltefosine in the treatment of visceral leishmaniasis in India after a decade of use. Clin Infect Dis. 2012;55:543–550. doi: 10.1093/cid/cis474. [DOI] [PubMed] [Google Scholar]
- 83.Rijal S, et al. Increasing failure of miltefosine in the treatment of Kala-azar in Nepal and the potential role of parasite drug resistance, reinfection, or noncompliance. Clin Infect Dis. 2013;56:1530–1538. doi: 10.1093/cid/cit102. [DOI] [PubMed] [Google Scholar]
- 84.Seifert K, et al. Characterisation of Leishmania donovani promastigotes resistant to hexadecylphosphocholine (miltefosine) Int J Antimicrob Ag. 2003;22:380–387. doi: 10.1016/s0924-8579(03)00125-0. [DOI] [PubMed] [Google Scholar]
- 85.Kulshrestha A, Sharma V, Singh R, Salotra P. Comparative transcript expression analysis of miltefosine-sensitive and miltefosine-resistant Leishmania donovani. Parasitol Res. 2014;113:1171–1184. doi: 10.1007/s00436-014-3755-6. [DOI] [PubMed] [Google Scholar]
- 86.Perlin DS, Shor E, Zhao Y. Update on antifungal drug resistance. Curr Clin Microbiol Rep. 2015;2:84–95. doi: 10.1007/s40588-015-0015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Denning DW, Bromley MJ. Infectious disease. How to bolster the antifungal pipeline. Science. 2015;347:1414–1416. doi: 10.1126/science.aaa6097. [DOI] [PubMed] [Google Scholar]
- 88.Epaulard O, et al. A multistep voriconazole-related phototoxic pathway may lead to skin carcinoma: results from a French nationwide study. Clin Infect Dis. 2013;57:e182–e188. doi: 10.1093/cid/cit600. [DOI] [PubMed] [Google Scholar]
- 89.Shor E, Perlin DS. Coping with stress and the emergence of multidrug resistance in fungi. PLoS Pathog. 2015;11:e1004668. doi: 10.1371/journal.ppat.1004668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kidd SE, Goeman E, Meis JF, Slavin MA, Verweij PE. Multi-triazole-resistant Aspergillus fumigatus infections in Australia. Mycoses. 2015;58:350–355. doi: 10.1111/myc.12324. [DOI] [PubMed] [Google Scholar]
- 91.van der Linden JW, et al. Prospective multicenter international surveillance of azole resistance in Aspergillus fumigatus . Emerg Infect Dis. 2015;21:1041–1044. doi: 10.3201/eid2106.140717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang E, et al. The ever-evolving landscape of candidaemia in patients with acute leukaemia: non-susceptibility to caspofungin and multidrug resistance are associated with increased mortality. J Antimicrob Chemother. 2015;70:2362–2368. doi: 10.1093/jac/dkv087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chowdhary A, Kathuria S, Xu J, Meis JF. Emergence of azole-resistant Aspergillus fumigatus strains due to agricultural azole use creates an increasing threat to human health. PLoS Pathog. 2013;9:e1003633. doi: 10.1371/journal.ppat.1003633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vermeulen E, Lagrou K, Verweij PE. Azole resistance in Aspergillus fumigatus: a growing public health concern. Curr Opin Infect Dis. 2013;26:493–500. doi: 10.1097/QCO.0000000000000005. [DOI] [PubMed] [Google Scholar]
- 95.Snelders E, et al. Triazole fungicides can induce cross-resistance to medical triazoles in Aspergillus fumigatus. J Parasitol Res. 2012;7:e31801. doi: 10.1371/journal.pone.0031801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Snelders E, Melchers WJ, Verweij PE. Azole resistance in Aspergillus fumigatus: a new challenge in the management of invasive aspergillosis? Future Microbiol. 2011;6:335–347. doi: 10.2217/fmb.11.4. [DOI] [PubMed] [Google Scholar]
- 97.Abdolrasouli A, et al. Genomic context of azole resistance mutations in Aspergillus fumigatus determined using whole-genome sequencing. mBio. 2015;6:e00536. doi: 10.1128/mBio.00536-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Katiyar SK, et al. Fks1 and Fks2 are functionally redundant but differentially regulated in Candida glabrata: implications for echinocandin resistance. Antimicrob Agents Chemother. 2012;56:6304–6309. doi: 10.1128/AAC.00813-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Singh SD, et al. Hsp90 governs echinocandin resistance in the pathogenic yeast Candida albicans via calcineurin. PLoS Pathog. 2009;5:e1000532. doi: 10.1371/journal.ppat.1000532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pfaller MA, Diekema DJ. Progress in antifungal susceptibility testing of Candida spp. by use of Clinical and Laboratory Standards Institute broth microdilution methods, 2010 to 2012. J Clin Microbiol. 2012;50:2846–2856. doi: 10.1128/JCM.00937-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Alexander BD, et al. Increasing echinocandin resistance in Candida glabrata: clinical failure correlates with presence of FKS mutations and elevated minimum inhibitory concentrations. Clin Infect Dis. 2013;56:1724–1732. doi: 10.1093/cid/cit136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Denoeud-Ndam L, et al. A multi-center, open-label trial to compare the efficacy and pharmacokinetics of Artemether-Lumefantrine in children with severe acute malnutrition versus children without severe acute malnutrition: study protocol for the MAL-NUT study. BMC Infect Dis. 2015;15:228. doi: 10.1186/s12879-015-0963-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kinga MK, et al. Quantitative genome re-sequencing defines multiple mutations conferring chloroquine resistance in rodent malaria. BMC Genomics. 2012;13:106. doi: 10.1186/1471-2164-13-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Manske M, et al. Analysis of Plasmodium falciparum diversity in natural infections by deep sequencing. Nature. 2012;487:375–379. doi: 10.1038/nature11174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cheeseman IH, et al. A major genome region underlying artemisinin resistance in malaria. Science. 2012;336:79–82. doi: 10.1126/science.1215966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yuan J, et al. Chemical genomic profiling for antimalarial therapies, response signatures, and molecular targets. Science. 2011;333:724–729. doi: 10.1126/science.1205216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Glover L, et al. Genome-scale RNAi screens for high-throughput phenotyping in bloodstream-form African trypanosomes. Nature Protoc. 2015;10:106–133. doi: 10.1038/nprot.2015.005. [DOI] [PubMed] [Google Scholar]
- 108.Alsford S, et al. High-throughput decoding of antitrypanosomal drug efficacy and resistance. Nature. 2012;482:232–236. doi: 10.1038/nature10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Begolo D, Erben E, Clayton C. Drug target identification using a trypanosome overexpression library. Antimicrob Agents Chemother. 2014;58:6260–6264. doi: 10.1128/AAC.03338-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cowman AF. Functional analysis of drug resistance in Plasmodium falciparum in the post-genomic era. Int J Parasitol. 2001;31:871–878. doi: 10.1016/s0020-7519(01)00201-6. [DOI] [PubMed] [Google Scholar]
- 111.Vincent IM, Barrett MP. Metabolomic-based strategies for anti-parasite drug discovery. J Biomol Screen. 2015;20:44–55. doi: 10.1177/1087057114551519. [DOI] [PubMed] [Google Scholar]
- 112.Goodman CD, et al. Parasites resistant to the antimalarial atovaquone fail to transmit by mosquitoes. Science. 2016;352:349–353. doi: 10.1126/science.aad9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Guttery DS, Roques M, Holder AA, Tewari R. Commit and transmit: molecular players in Plasmodium sexual development and zygote differentiation. Trends Parasitol. 2015;31:676–685. doi: 10.1016/j.pt.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 114.Hemingway J. The role of vector control in stopping the transmission of malaria: threats and opportunities. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130431. doi: 10.1098/rstb.2013.0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dias JC. Southern Cone Initiative for the elimination of domestic populations of Triatoma infestans and the interruption of transfusional Chagas disease. Historical aspects, present situation, and perspectives. Mem Inst Oswaldo Cruz. 2007;102(suppl. 1):11–18. doi: 10.1590/s0074-02762007005000092. [DOI] [PubMed] [Google Scholar]
- 116.Welburn SC, Maudlin I. Priorities for the elimination of sleeping sickness. Adv Parasitol. 2012;79:299–337. doi: 10.1016/B978-0-12-398457-9.00004-4. [DOI] [PubMed] [Google Scholar]
- 117.Dias JC, Silveira AC, Schofield CJ. The impact of Chagas disease control in Latin America: a review. Mem Inst Oswaldo Cruz. 2002;97:603–612. doi: 10.1590/s0074-02762002000500002. [DOI] [PubMed] [Google Scholar]
- 118.Abd-Alla AM, et al. Improving Sterile Insect Technique (SIT) for tsetse flies through research on their symbionts and pathogens. J Invertebr Pathol. 2013;112(suppl.):S2–S10. doi: 10.1016/j.jip.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Oliva CF, et al. Current status and future challenges for controlling malaria with the sterile insect technique: technical and social perspectives. Acta Trop. 2014;132(suppl.):S130–S139. doi: 10.1016/j.actatropica.2013.11.019. [DOI] [PubMed] [Google Scholar]
- 120.Dantzler KW, Ravel DB, Brancucci NM, Marti M. Ensuring transmission through dynamic host environments: host-pathogen interactions in Plasmodium sexual development. Curr Opin Microbiol. 2015;26:17–23. doi: 10.1016/j.mib.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Imhof S, Roditi I. The social life of African trypanosomes. Trends Parasitol. 2015;31:490–498. doi: 10.1016/j.pt.2015.06.012. [DOI] [PubMed] [Google Scholar]
- 122.Mony BM, et al. Genome-wide dissection of the quorum sensing signalling pathway in Trypanosoma brucei. Nature. 2014;505:681–685. doi: 10.1038/nature12864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mantel PY, et al. Malaria-infected erythrocyte-derived microvesicles mediate cellular communication within the parasite population and with the host immune system. Cell Host Microbe. 2013;13:521–534. doi: 10.1016/j.chom.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Regev-Rudzki N, et al. Cell-cell communication between malaria-infected red blood cells via exosome-like vesicles. Cell. 2013;153:1120–1133. doi: 10.1016/j.cell.2013.04.029. [DOI] [PubMed] [Google Scholar]
- 125.World Health Organization. Guidelines for the Treatment of Malaria. 3rd edn. World Health Organization; 2015. pp. 1–316. [Google Scholar]
- 126.World Health Organization. WHO Model List of Essential Medicines. 19th edn. World Health Organization; 2015. pp. 1–51. [Google Scholar]
- 127.World Health Organization. Rapid Advice: Diagnosis, Prevention and Management of Cryptococcal Disease in HIV-Infected Adults, Adolescents and Children. World Health Organization; 2011. pp. 1–37. [PubMed] [Google Scholar]
- 128.Leroux S, Ullmann AJ. Management and diagnostic guidelines for fungal diseases in infectious diseases and clinical microbiology: critical appraisal. Clin Microbiol Infect. 2013;19:1115–1121. doi: 10.1111/1469-0691.12426. [DOI] [PubMed] [Google Scholar]
- 129.Kullberg BJ, Arendrup MC. Invasive candidiasis. N Engl J Med. 2015;373:1445–1456. doi: 10.1056/NEJMra1315399. [DOI] [PubMed] [Google Scholar]
- 130.Ecker A, Lehane AM, Clain J, Fidock DA. PfCRT and its role in antimalarial drug resistance. Trends Parasitol. 2012;28:504–514. doi: 10.1016/j.pt.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wootton JC, et al. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature. 2002;418:320–323. doi: 10.1038/nature00813. [DOI] [PubMed] [Google Scholar]
- 132.Sanchez CP, Stein W, Lanzer M. Trans stimulation provides evidence for a drug efflux carrier as the mechanism of chloroquine resistance in Plasmodium falciparum. Biochemistry. 2003;42:9383–9394. doi: 10.1021/bi034269h. [DOI] [PubMed] [Google Scholar]
- 133.Sanchez CP, Dave A, Stein WD, Lanzer M. Transporters as mediators of drug resistance in Plasmodium falciparum. Int J Parasitol. 2010;40:1109–1118. doi: 10.1016/j.ijpara.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 134.Ferdig MT, et al. Dissecting the loci of low-level quinine resistance in malaria parasites. Mol Microbiol. 2004;52:985–997. doi: 10.1111/j.1365-2958.2004.04035.x. [DOI] [PubMed] [Google Scholar]
- 135.Valderramos SG, et al. Identification of a mutant PfCRT-mediated chloroquine tolerance phenotype in Plasmodium falciparum. PLoS Pathog. 2010;6:e1000887. doi: 10.1371/journal.ppat.1000887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Pelleau S, et al. Adaptive evolution of malaria parasites in French Guiana: reversal of chloroquine resistance by acquisition of a mutation in pfcrt. Proc Natl Acad Sci USA. 2015;112:11672–11677. doi: 10.1073/pnas.1507142112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wellems TE, Walker-Jonah A, Panton LJ. Genetic mapping of the chloroquine-resistance locus on Plasmodium falciparum chromosome 7. Proc Natl Acad Sci USA. 1991;88:3382–3386. doi: 10.1073/pnas.88.8.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Duraisingh MT, et al. Linkage disequilibrium between two chromosomally distinct loci associated with increased resistance to chloroquine in Plasmodium falciparum. Parasitology. 2000;121:1–7. doi: 10.1017/s0031182099006022. [DOI] [PubMed] [Google Scholar]
- 139.Meshnick SR, Taylor TE, Kamchonwongpaisan S. Artemisinin and the antimalarial endoperoxides — from herbal remedy to targeted chemotherapy. Microbiol Rev. 1996;60:301. doi: 10.1128/mr.60.2.301-315.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wang P, Read M, Sims PFG, Hyde JE. Sulfadoxine resistance in the human malaria parasite Plasmodium falciparum is determined by mutations in dihydropteroate synthetase and an additional factor associated with folate utilization. Mol Microbiol. 1997;23:979–986. doi: 10.1046/j.1365-2958.1997.2821646.x. [DOI] [PubMed] [Google Scholar]
- 141.Heinberg A, Kirkman L. The molecular basis of antifolate resistance in Plasmodium falciparum: looking beyond point mutations. Ann N Y Acad Sci. 2015;1342:10–18. doi: 10.1111/nyas.12662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Cowman AF. In: Malaria: Parasite Biology, Pathogenesis, and Protection. Sherman IW, editor. 1998. pp. 317–330. [Google Scholar]
- 143.Rose GW, Suh KN, Kain KC, Le SN, McCarthy AE. Atovaquone-proguanil resistance in imported falciparum malaria in a young child. Pediatr Infect Dis J. 2008;27:567–569. doi: 10.1097/INF.0b013e318167918d. [DOI] [PubMed] [Google Scholar]
- 144.Carter NS, Fairlamb AH. Arsenical-resistant trypanosomes lack an unusual adenosine transporter. Nature. 1993;361:173–176. doi: 10.1038/361173a0. [DOI] [PubMed] [Google Scholar]
- 145.Maser P, Sutterlin C, Kralli A, Kaminsky R. A nucleoside transporter from Trypanosoma brucei involved in drug resistance. Science. 1999;285:242–244. doi: 10.1126/science.285.5425.242. [DOI] [PubMed] [Google Scholar]
- 146.Baker N, et al. Aquaglyceroporin 2 controls susceptibility to melarsoprol and pentamidine in African trypanosomes. Proc Natl Acad Sci USA. 2012;109:10996–11001. doi: 10.1073/pnas.1202885109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Graf FE, et al. Chimerization at the AQP2-AQP3 locus is the genetic basis of melarsoprol-pentamidine cross-resistance in clinical Trypanosoma brucei gambiense isolates. Int J Parasitol Drugs Drug Resist. 2015;5:65–68. doi: 10.1016/j.ijpddr.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Pyana PP, et al. Melarsoprol sensitivity profile of Trypanosoma brucei gambiense isolates from cured and relapsed sleeping sickness patients from the Democratic Republic of the Congo. PLoS Negl Trop Dis. 2014;8:e3212. doi: 10.1371/journal.pntd.0003212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Graf FE, et al. Aquaporin 2 mutations in Trypanosoma brucei gambiense field isolates correlate with decreased susceptibility to pentamidine and melarsoprol. PLoS Negl Trop Dis. 2013;7:e2475. doi: 10.1371/journal.pntd.0002475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kazibwe AJ, et al. Genotypic status of the TbAT1/P2 adenosine transporter of Trypanosoma brucei gambiense isolates from Northwestern Uganda following melarsoprol withdrawal. PLoS Negl Trop Dis. 2009;3:e523. doi: 10.1371/journal.pntd.0000523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Vincent IM, et al. A molecular mechanism for eflornithine resistance in African trypanosomes. PLoS Pathog. 2010;6:e1001204. doi: 10.1371/journal.ppat.1001204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Mathieu C, et al. Trypanosoma brucei eflornithine transporter AAT6 is a low-affinity low-selective transporter for neutral amino acids. Biochem J. 2014;463:9–18. doi: 10.1042/BJ20140719. [DOI] [PubMed] [Google Scholar]
- 153.Wilkinson SR, Taylor MC, Horn D, Kelly JM, Cheeseman I. A mechanism for cross-resistance to nifurtimox and benznidazole in trypanosomes. Proc Natl Acad Sci USA. 2008;105:5022–5027. doi: 10.1073/pnas.0711014105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hall BS, Bot C, Wilkinson SR. Nifurtimox activation by trypanosomal type I nitroreductases generates cytotoxic nitrile metabolites. J Biol Chem. 2011;286:13088–13095. doi: 10.1074/jbc.M111.230847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Patterson S, Wyllie S. Nitro drugs for the treatment of trypanosomatid diseases: past, present, and future prospects. Trends Parasitol. 2014;30:289–298. doi: 10.1016/j.pt.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Sokolova AY, et al. Cross-resistance to nitro drugs and implications for treatment of human African trypanosomiasis. Antimicrob Agents Chemother. 2010;54:2893–2900. doi: 10.1128/AAC.00332-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wyllie S, et al. Nitroheterocyclic drug resistance mechanisms in Trypanosoma brucei . J Antimicrob Chemother. 2015;71:625–634. doi: 10.1093/jac/dkv376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Mejia AM, et al. Benznidazole-resistance in Trypanosoma cruzi is a readily acquired trait that can arise independently in a single population. J Infect Dis. 2012;206:220–228. doi: 10.1093/infdis/jis331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Trochine A, Creek DJ, Faral-Tello P, Barrett MP, Robello C. Benznidazole biotransformation and multiple targets in Trypanosoma cruzi revealed by metabolomics. PLoS Negl Trop Dis. 2014;8:e2844. doi: 10.1371/journal.pntd.0002844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hall BS, Wilkinson SR. Activation of benznidazole by trypanosomal type I nitroreductases results in glyoxal formation. Antimicrob Agents Chemother. 2012;56:115–123. doi: 10.1128/AAC.05135-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zingales B, et al. A novel ABCG-like transporter of Trypanosoma cruzi is involved in natural resistance to benznidazole. Mem Inst Oswaldo Cruz. 2015;110:433–444. doi: 10.1590/0074-02760140407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Murta SMF, et al. Deletion of copies of the gene encoding old yellow enzyme (TcOYE), a NAD(P)H flavin oxidoreductase, associates with in vitro-induced benznidazole resistance in Trypanosoma cruzi. Mol Biochem Parasitol. 2006;146:151–162. doi: 10.1016/j.molbiopara.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 163.Andrade HM, et al. Proteomic analysis of Trypanosoma cruzi resistance to benznidazole. J Proteome Res. 2008;7:2357–2367. doi: 10.1021/pr700659m. [DOI] [PubMed] [Google Scholar]
- 164.Kubata BK, et al. A key role for old yellow enzyme in the metabolism of drugs by Trypanosoma cruzi. J Exp Med. 2002;196:1241–1251. doi: 10.1084/jem.20020885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wyllie S, Cunningham ML, Fairlamb AH. Dual action of antimonial drugs on thiol redox metabolism in the human pathogen Leishmania donovani. J Biol Chem. 2004;279:39925–39932. doi: 10.1074/jbc.M405635200. [DOI] [PubMed] [Google Scholar]
- 166.Baiocco P, Colotti G, Franceschini S, Ilari A. Molecular basis of antimony treatment in leishmaniasis. J Med Chem. 2009;52:2603–2612. doi: 10.1021/jm900185q. [DOI] [PubMed] [Google Scholar]
- 167.Wyllie S, et al. Elevated levels of tryparedoxin peroxidase in antimony unresponsive Leishmania donovani field isolates. Mol Biochem Parasitol. 2010;173:162–164. doi: 10.1016/j.molbiopara.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Mukhopadhyay R, et al. Trypanothione overproduction and resistance to antimonials and arsenicals in Leishmania. Proc Natl Acad Sci USA. 1996;93:10383–10387. doi: 10.1073/pnas.93.19.10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wyllie S, Vickers TJ, Fairlamb AH. Roles of trypanothione S-transferase and tryparedoxin peroxidase in resistance to antimonials. Antimicrob Agents Chemother. 2008;52:1359–1365. doi: 10.1128/AAC.01563-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Callahan HL, Beverley SM. Heavy metal resistance: a new role for P-glycoproteins in Leishmania. J Biol Chem. 1991;266:18427–18430. [PubMed] [Google Scholar]
- 171.Mukherjee A, et al. Role of ABC transporter MRPA, γ-glutamylcysteine synthetase and ornithine decarboxylase in natural antimony-resistant isolates of Leishmania donovani. J Antimicrob Chemother. 2007;59:204–211. doi: 10.1093/jac/dkl494. [DOI] [PubMed] [Google Scholar]
- 172.El Fadili K, et al. Role of the ABC transporter MRPA (PGPA) in antimony resistance in Leishmania infantum axenic and intracellular amastigotes. Antimicrob Agents Chemother. 2005;49:1988–1993. doi: 10.1128/AAC.49.5.1988-1993.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Brochu C, Haimeur A, Ouellette M. The heat shock protein HSP70 and heat shock cognate protein HSC70 contribute to antimony tolerance in the protozoan parasite Leishmania. Cell Stress Chaperones. 2004;9:294–303. doi: 10.1379/CSC-15R1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Chawla B, Jhingran A, Panigrahi A, Stuart KD, Madhubala R. Paromomycin affects translation and vesicle-mediated trafficking as revealed by proteomics of paromomycin–susceptible–resistant Leishmania donovani. J Parasitol Res. 2011;6:e26660. doi: 10.1371/journal.pone.0026660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Rakotomanga M, Saint-Pierre-Chazalet M, Loiseau PM. Alteration of fatty acid and sterol metabolism in miltefosine-resistant Leishmania donovani promastigotes and consequences for drug-membrane interactions. Antimicrob Agents Chemother. 2005;49:2677–2686. doi: 10.1128/AAC.49.7.2677-2686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Rakotomanga M, Blanc S, Gaudin K, Chaminade P, Loiseau PA. Miltefosine afects lipid metabolism in Leishmania donovani promastigotes. Antimicrob Agents Chemother. 2007;51:1425–1430. doi: 10.1128/AAC.01123-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Vincent IM, et al. Untargeted metabolomic analysis of miltefosine action in Leishmania infantum reveals changes to the internal lipid metabolism. Int J Parasitol Drugs Drug Resist. 2014;4:20–27. doi: 10.1016/j.ijpddr.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Dorlo TPC, Balasegaram M, Beijnen JH, de Vries PJ. Miltefosine: a review of its pharmacology and therapeutic efficacy in the treatment of leishmaniasis. J Antimicrob Chemother. 2012;67:2576–2597. doi: 10.1093/jac/dks275. [DOI] [PubMed] [Google Scholar]
- 179.Perez-Victoria FJ, Gamarro F, Ouellette M, Castanys S. Functional cloning of the miltefosine transporter: a novel P-type phospholipid translocase from Leishmania involved in drug resistance. J Biol Chem. 2003;278:49965–49971. doi: 10.1074/jbc.M308352200. [DOI] [PubMed] [Google Scholar]
- 180.Perez-Victoria FJ, Castanys S, Gamarro F. Leishmania donovani resistance to miltefosine involves a defective inward translocation of the drug. Antimicrob Agents Chemother. 2003;47:2397–2403. doi: 10.1128/AAC.47.8.2397-2403.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Coelho AC, et al. Multiple mutations in heterogeneous miltefosine-resistant Leishmania major population as determined by whole genome sequencing. PLoS Negl Trop Dis. 2012;6:e1512. doi: 10.1371/journal.pntd.0001512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Perez-Victoria FJ, Sanchez-Canete MP, Castanys S, Gamarro F. Phospholipid translocation and miltefosine potency require both L. donovani miltefosine transporter and the new protein LdRos3 in Leishmania parasites. J Biol Chem. 2006;281:23766–23775. doi: 10.1074/jbc.M605214200. [DOI] [PubMed] [Google Scholar]
- 183.Perez-Victoria JM, et al. Alkyl-lysophospholipid resistance in multidrug-resistant Leishmania tropica and chemosensitization by a novel P-glycoprotein-like transporter modulator. Antimicrob Agents Chemother. 2001;45:2468–2474. doi: 10.1128/AAC.45.9.2468-2474.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Castanys-Munoz E, Perez-Victoria JM, Gamarro F, Castanys S. Characterization of an ABCG-like transporter from the protozoan parasite Leishmania with a role in drug resistance and transbilayer lipid movement. Antimicrob Agents Chemother. 2008;52:3573–3579. doi: 10.1128/AAC.00587-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Canuto GA, et al. Multi-analytical platform metabolomic approach to study miltefosine mechanism of action and resistance in Leishmania. Anal Bioanal Chem. 2014;406:3459–3476. doi: 10.1007/s00216-014-7772-1. [DOI] [PubMed] [Google Scholar]
- 186.Blum G, et al. New insight into amphotericin B resistance in Aspergillus terreus. Antimicrob Agents Chemother. 2013;57:1583–1588. doi: 10.1128/AAC.01283-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Gray KC, et al. Amphotericin primarily kills yeast by simply binding ergosterol. Proc Natl Acad Sci USA. 2012;109:2234–2239. doi: 10.1073/pnas.1117280109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Odds FC, Brown AJ, Gow NA. Antifungal agents: mechanisms of action. Trends Microbiol. 2003;11:272–279. doi: 10.1016/s0966-842x(03)00117-3. [DOI] [PubMed] [Google Scholar]
- 189.Prasad R, Rawal MK. Efflux pump proteins in antifungal resistance. Front Pharmacol. 2014;5:202. doi: 10.3389/fphar.2014.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Costa C, Dias PJ, Sa-Correia I, Teixeira MC. MFS multidrug transporters in pathogenic fungi: do they have real clinical impact? Front Physiol. 2014;5:197. doi: 10.3389/fphys.2014.00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Cowen LE, Sanglard D, Howard SJ, Rogers PD, Perlin DS. Mechanisms of antifungal drug resistance. Cold Spring Harb Perspect Med. 2015;5:a019752. doi: 10.1101/cshperspect.a019752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Flowers SA, et al. Gain-of-function mutations in UPC2 are a frequent cause of ERG11 upregulation in azole-resistant clinical isolates of Candida albicans. Eukaryot Cell. 2012;11:1289–1299. doi: 10.1128/EC.00215-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Cannon RD, et al. Efflux-mediated antifungal drug resistance. Clin Microbiol Rev. 2009;22:291–321. doi: 10.1128/CMR.00051-08. Table. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Kwon-Chung KJ, Chang YC. Aneuploidy and drug resistance in pathogenic fungi. PLoS Pathog. 2012;8:e1003022. doi: 10.1371/journal.ppat.1003022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Selmecki A, Gerami-Nejad M, Paulson C, Forche A, Berman J. An isochromosome confers drug resistance in vivo by amplification of two genes, ERG11 and TAC1. Mol Microbiol. 2008;68:624–641. doi: 10.1111/j.1365-2958.2008.06176.x. [DOI] [PubMed] [Google Scholar]
- 196.Selmecki A, Forche A, Berman J. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science. 2006;313:367–370. doi: 10.1126/science.1128242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Nishikawa JL, et al. Inhibiting fungal multidrug resistance by disrupting an activator-mediator interaction. Nature. 2016;530:485–489. doi: 10.1038/nature16963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Cowen LE. The evolution of fungal drug resistance: modulating the trajectory from genotype to phenotype. Nature Rev Microbiol. 2008;6:187–198. doi: 10.1038/nrmicro1835. [DOI] [PubMed] [Google Scholar]
- 199.Sun HY, Singh N. Characterisation of breakthrough invasive mycoses in echinocandin recipients: an evidence-based review. Int J Antimicrob Agents. 2010;35:211–218. doi: 10.1016/j.ijantimicag.2009.09.020. [DOI] [PubMed] [Google Scholar]
- 200.Vandeputte P, Ferrari S, Coste AT. Antifungal resistance and new strategies to control fungal infections. Int J Microbiol. 2012;2012:713687. doi: 10.1155/2012/713687. [DOI] [PMC free article] [PubMed] [Google Scholar]