Abstract
Monosomy of chromosome 7 is the most frequent autosomal monosomy in acute myeloid leukemia (AML), where it associates with poor clinical outcomes. However, molecular features associated with this sole monosomy subtype (-7 AML) which may give insights into the basis for its poor prognosis have not been characterized. In this study, we analyzed 36 cases of -7 AML for mutations in 81 leukemia/cancer-associated genes using a customized targeted next-generation sequencing panel (Miseq). Global gene and microRNA expression profiles were also determined using paired RNA and small RNA sequencing data. Notably, gene mutations were detected in all the major AML-associated functional groups, which include activated signaling, chromatin remodeling, cohesin complex, methylation, NPM1, spliceosome, transcription factors and tumor suppressors. Gene mutations in the activated signaling and chromatin remodeling groups were relatively more frequent in patients <60 years of age, who also had more mutations in the methylation and spliceosome groups compared to patients {greater than or equal to} 60 years of age. Novel recurrent mutational events in AML were identified in the SMARCA2 gene. In patients {greater than or equal to} 60 years of age, the presence of spliceosome mutations associated with a lower complete remission rate (p=0.03). RNA sequencing revealed distinct gene and microRNA expression patterns between the sole -7 and non-7 AML cases, with reduced expression as expected of many genes and microRNAs mapped to chromosome 7, and overexpression of ID1, MECOM, and PTPRM, among others. Overall, our findings illuminate a number of molecular features of the underlying aggressive pathobiology in -7 AML patients.
Keywords: acute myeloid leukemia, monosomy 7, next-generation sequencing, gene mutation
Introduction
The loss of chromosome 7 (monosomy 7, -7) is the most frequent autosomal monosomy in acute myeloid leukemia (AML), and the most common chromosomal abnormality observed in high-risk AML patients (1). However, it has been rarely studied as a sole abnormality. The adverse prognostic impact of -7, both in combination with other chromosomal aberrations and as a sole abnormality, has been well known for decades. Patients with -7 are characterized by low initial complete remission (CR) rates, short remissions, and inferior survival compared with those of patients in most other cytogenetic groups (1–5). The elucidation of the underlying biology and identification of candidate genes responsible for the adverse clinical outcome of -7 AML have been the focus of numerous studies (6–14), but a comprehensive study using next-generation sequencing of a cohort of sole -7 AML patients without (possibly confounding) other cytogenetic abnormalities has not been reported to date.
The aim of our study was to characterize the mutational landscape and the gene- and microRNA-expression profiles of adult AML patients with -7 as a sole cytogenetic abnormality using next-generation sequencing techniques. Since, to our knowledge, all recent molecular studies characterizing -7 AML also included patients with additional chromosomal aberrations (15, 16), we hypothesized that restricting our study to sole -7 patients would enable us to find the molecular features associated with chromosome 7 loss, without the potential bias introduced by the presence of additional cytogenetic abnormalities. A more complete understanding of molecular events could provide insight into the pathobiology of chromosome 7 loss in AML.
Materials and Methods
Patients, treatment, and cytogenetic studies
Pretreatment bone marrow (BM) or peripheral blood (PB) samples containing ≥20% of leukemic blasts were obtained from 36 adult AML patients with -7 as a sole chromosomal abnormality. Cytogenetic analyses of pretreatment BM and/or PB samples were performed by institutional laboratories approved by Cancer and Leukemia Group B (CALGB)/Alliance for Clinical Trials in Oncology (Alliance). Cytogenetic results were confirmed by central karyotype review (17). All patients were treated with cytarabine and an anthracycline-based cytotoxic chemotherapy on CALGB/Alliance trials, the details of which are provided in the Supplementary data. Study protocols were in accordance with the Declaration of Helsinki and approved by the institutional review boards at each center, and all patients provided written informed consent.
Statistical analysis
Baseline characteristics were compared between younger and older -7 AML patients using Fisher’s exact test for categorical variables and the Wilcoxon rank-sum test for continuous variables (18). Definitions of the clinical endpoints–CR, disease-free (DFS) and overall survival (OS)–are provided in the Supplementary data. For time-to-event analyses, we calculated survival estimates using the Kaplan-Meier method, and compared groups using the Cox proportional hazard regression models. The dataset was locked on June 10th, 2015. Data collection and statistical analyses were performed by the Alliance Statistics and Data Center.
Molecular analyses
Patients provided written informed consent to participate in protocols CALGB 8461 (cytogenetic studies), CALGB 9665 (leukemia tissue bank) and CALGB 20202 (molecular studies), which involved collection of pretreatment BM aspirates and PB samples. Mononuclear cells were enriched through Ficoll-Hypaque gradient centrifugation and cryopreserved until use. Genomic DNA was extracted using the DNeasy Blood and Tissue Kit (QIAGEN, Hilden, Germany). The mutational status of 80 protein coding genes was determined centrally at The Ohio State University by targeted amplicon sequencing using the MiSeq platform (Illumina, San Diego, CA; see Supplementary data for details). Briefly, DNA library preparations were performed according to the manufacturer’s instructions. Samples were pooled and run on the MiSeq machine using the Illumina MiSeq Reagent Kit v3. Sequenced reads were aligned to the hg19 genome build using the Illumina Isis Banded Smith-Waterman aligner. Single nucleotide variant and indel calling were performed using MuTect and VarScan, respectively (19, 20). All called variants underwent visual inspection of the aligned reads using the Integrative Genomics Viewer (Broad Institute, Cambridge, MA; ref. 21). Testing for the presence or absence of FLT3 internal tandem duplication (FLT3-ITD) and CEBPA mutations were performed as previously described (22, 23). Thus, since FLT3 was also included in the 80-gene Miseq panel, the mutation status of 81 genes (80 genes plus CEBPA) was assessed in our study. Gene mutations were assigned to functional groups as previously described by Ley et al. (activated signaling, chromatin remodeling, cohesin complex, methylation, NPM1, spliceosome, transcription factors, tumor suppressors; ref. 24). Mutations detected in the targeted sequencing approach at the DNA level were also analyzed for their presence at the RNA level by visual inspection of the BAM files using the same criteria as for DNA variant calls (Supplementary Table S1). Of the 140 detected mutations, 99 were also evaluable at the RNA level. RNAseq analysis identified 93 (94%) of those 99 mutations. All six mutations not seen in the RNAseq data had Variant Allele Fractions (VAFs) <15%. Ten patients with available buccal swab material (n=9) or BM obtained during morphologic remission (n=1) were additionally analyzed for the germline status of the genes found mutated in leukemic samples using the same 80 gene targeted amplicon sequencing panel (Supplementary Table S2).
Gene- and microRNA-expression profiling
Total RNA was extracted from BM or PB cells from 31 patients using the TRIzol extraction method kit (QIAGEN). For five patients, the quantity and/or quality of RNA were insufficient to perform expression analyses. The RNA was used to make two sets of libraries. Gene expression libraries were constructed with the Illumina TruSeq stranded mRNA library preparation kit, while small RNA libraries were constructed with the NEBNext small RNA library prep kit (both New England Biolabs, Ipswich, MA). Both libraries were sequenced on an Illumina HiSeq 2500 targeting 40×106 and 25×106 reads for mRNA and small RNA libraries, respectively.
Quantification of polyA+ RNA sequences was performed as reported by TCGA (24). The ‘rnaseqv2’ University of North Carolina mRNA-seq Pipeline described at the TCGA Data Coordinating Center (DCC) was implemented in order to compare the -7 AML samples to the TCGA cohort (24). Essentially, the fastq files were aligned against hg19 using MAPSPLICE (25) and quantified with RSEM (26). The quantification of miRNAseq data was obtained using NovoAlign and miRBase 21 (www.mirbase.org). Batch effects were removed using the ComBat function (27) within the sva package in R (28, 29). The unsupervised clustering of sole -7 and TCGA samples (including patients with chromosome 7 abnormalities and/or complex karyotype) is depicted in Supplementary Figure S1. TCGA samples with chromosome 7 abnormalities and/or complex karyotype preferentially clustered with our sole -7 AML cohort.
Results
Clinical characteristics and outcome of AML patients with sole -7
Clinical characteristics of patients with sole -7, both as a total cohort and separated into younger (aged <60 years) and older (≥60 years) patients, are provided in Table 1. Only 21% of the sole -7 patients had extramedullary involvement of their leukemia, and there were no significant differences in the clinical characteristics between the age groups (Table 1).
Table 1.
Pretreatment clinical characteristics of patients with acute myeloid leukemia (AML) and sole -7, and comparison by age group (<60 years vs. ≥60 years)
| Characteristic | Sole -7 AML All patients (n=36) |
Sole -7 AML <60y (n=11) |
Sole -7 AML ≥60y (n=25) |
P valuea |
|---|---|---|---|---|
|
| ||||
| Age, years | -- | |||
| Median | 63 | 52 | 68 | |
| Range | 35–78 | 35–59 | 61–78 | |
|
| ||||
| Sex, n (%) of females | 15 (42) | 3 (37) | 12 (48) | 0.30 |
|
| ||||
| Race, n (%) | 0.29 | |||
| White | 34 (97) | 9 (90) | 25 (100) | |
| Non-white | 1 (3) | 1 (10) | 0 (0) | |
|
| ||||
| Hemoglobin, g/dl | 0.90 | |||
| Median | 9.1 | 9.1 | 9.1 | |
| Range | 7.3–12.3 | 7.9–10.6 | 7.3–12.3 | |
|
| ||||
| Platelet count, × 109/l | 0.27 | |||
| Median | 65 | 51 | 69 | |
| Range | 9–989 | 9–274 | 10–989 | |
|
| ||||
| WBC, × 109/l | 0.75 | |||
| Median | 12.3 | 12.2 | 13.2 | |
| Range | 1.6–212.7 | 5.2–111.0 | 1.6–212.7 | |
|
| ||||
| Bone marrow blasts, % | 0.47 | |||
| Median | 48 | 47 | 52 | |
| Range | 23–93 | 23–93 | 28–91 | |
|
| ||||
| Blood blasts, % | 0.31 | |||
| Median | 30 | 37 | 25 | |
| Range | 2–99 | 16–89 | 2–99 | |
|
| ||||
| s-AML, n (%) | 0.29 | |||
| Yes | 4 (11) | 0 (0) | 4 (16) | |
| No | 32 (89) | 11 (100) | 21 (84) | |
|
| ||||
| t-AML, n (%) | 1.00 | |||
| Yes | 1 (3) | 0 (0) | 1 (4) | |
| No | 35 (97) | 11 (100) | 24 (96) | |
|
| ||||
| Extramedullary involvement, n (%) | 1.00 | |||
| Total | 7 (21) | 2 (20) | 5 (22) | |
| Central nervous system | 0 (0) | 0 (0) | 0 (0) | |
| Gum hypertrophy | 0 (0) | 0 (0) | 0 (0) | |
| Hepatomegaly | 2 (6) | 1 (9) | 1 (4) | |
| Lymphadenopathy | 2 (6) | 1 (9) | 1 (4) | |
| Mediastinal mass | 0 (0) | 0 (0) | 0 (0) | |
| Skin Infiltrates | 0 (0) | 0 (0) | 0 (0) | |
| Splenomegaly | 5 (15) | 2 (18) | 3 (13) | |
|
| ||||
| Transplantation, n (%) | 0.52 | |||
| Allo in 1st CR | 2 (6) | 1 (9) | 1 (4) | |
| No Allo in 1st CR | 34 (94) | 10 (91) | 24 (96) | |
Abbreviations: Allo, allogeneic hematopoietic stem-cell transplantation (HSCT); CR, complete remission, n, number; s-AML, secondary AML; t-AML, therapy-related AML; WBC, white blood count; y, years.
P-values relate to the comparison of younger versus older sole -7 patients. P-values for categorical variables are from Fisher’s exact test, P-values for continuous variables are from the Wilcoxon rank sum test.
Consistent with previous reports (1–5), the outcomes of sole -7 AML patients were very poor, with only 42% of patients achieving a CR. The median DFS was 8.4 months and the median OS was 9.2 months (Table 2). Only two patients were alive three years after diagnosis; one of the surviving patients received allogeneic hematopoietic stem-cell transplantation (HSCT) in first CR. There were no significant differences in CR rates or OS between the younger and older patients (Table 2), despite differences in treatment intensity. Possible differences in DFS between the two age groups could not be statistically assessed due to the limited sample size.
Table 2.
Outcome of all patients with acute myeloid leukemia (AML) and sole -7, and comparison of outcomes by age group (<60 years vs. ≥60 years)
| Endpoint | Sole -7 AML All patients (n=36) |
Sole -7 AML <60 y (n=11) |
Sole -7 AML ≥60 y (n=25) |
P-valuea |
|---|---|---|---|---|
|
| ||||
| Complete remission, n (%) | 15 (42) | 5 (45) | 10 (40) | 1.00 |
|
| ||||
| Disease-free survivalb | -- | |||
| Median, months | 8.4 | 6 | 13.2 | |
| Disease-free at 12 months, % (95% CI) | 33 (10–59) | 0 | 50 (15–77) | |
| Disease-free at 36 months, % (95% CI) | 8 (1–31) | 0 | 13 (1–42) | |
|
| ||||
| Overall survival | 0.63 | |||
| Median, months | 9.2 | 9.6 | 8.4 | |
| Alive at 12 months, % (95% CI) | 44 (28–60) | 45 (17–71) | 44 (24–62) | |
| Alive at 36 months, % (95% CI) | 7 (1–19) | 9 (1–33) | 5 (0–21) | |
Abbreviations: CI, confidence interval, y, years.
P-values relate to the comparison of younger versus older sole -7 patients. P-value for complete remission is from Fisher’s exact test. P-value for overall survival is from the log-rank test.
Patients who received an allogeneic hematopoietic stem-cell transplantation in first complete remission were not included in the analysis of disease-free survival (DFS). The P-value for DFS could not be determined due to the limited sample size.
The mutational landscape of AML patients with sole -7
Results of mutational analyses performed in our patient cohort are shown in Table 3, Figure 1 and Supplementary Table S1. A total of 140 mutations were found in 41 of the 81 tested genes, with a median of four mutations detected per patient (range: 0–7 mutations). Six genes had a mutation frequency of >20%: the most frequently mutated gene was RUNX1 (n=10 patients, 28% of the cohort), followed by ASXL1 (n=9, 25%), NRAS (n=9, 25%), TET2 (n=9, 25%), DNMT3A (n=8, 22%) and SRSF2 (n=8, 22%). Other recurrently mutated genes included SETBP1 (n=4, 11%), mutations of which have been previously reported to be enriched in -7 patients (30), and NOTCH1 (n=4, 11%), a gene frequently mutated in chronic lymphocytic leukemia (CLL; ref. 31). Three patients had mutations in GATA2, which have been implicated in familial -7 syndromes (32, 33). Some of the detected mutational features were in line with previous findings in myelodysplastic syndromes (MDS) with loss of chromosome 7 material, including the detection of four mutations in the EZH2 gene, which is located on chromosome 7 and has been suggested to act as a tumor suppressor gene (10, 34). Interestingly, we did not detect any mutations in some of the major AML-associated genes, including CEBPA, FLT3-ITD, and TP53.
Table 3.
Frequencies of mutations in single genes observed in patients with acute myeloid leukemia (AML) and sole -7
| Genea | Sole -7 AML All patients (n=36) |
Sole -7 AML <60y (n=11) |
Sole -7 AML ≥60y (n=25) |
P valueb |
|---|---|---|---|---|
|
| ||||
| AKT1, n (%) | 0.31 | |||
| Mutated | 1 (3) | 1 (9) | 0 (0) | |
| Wild type | 35 (97) | 10 (91) | 25 (100) | |
|
| ||||
| ASXL1, n (%) | 0.69 | |||
| Mutated | 9 (25) | 3 (30) | 6 (24) | |
| Wild type | 27 (74) | 8 (70) | 19 (76) | |
|
| ||||
| BCOR, n (%) | 1.00 | |||
| Mutated | 2 (6) | 0 (0) | 2 (8) | |
| Wild type | 34 (94) | 11 (100) | 23 (92) | |
|
| ||||
| BCORL1, n (%) | 0.52 | |||
| Mutated | 2 (6) | 1 (9) | 1 (4) | |
| Wild type | 34 (94) | 10 (91) | 24 (96) | |
|
| ||||
| BRAF, n (%) | 1.00 | |||
| Mutated | 1 (3) | 0 (0) | 1 (4) | |
| Wild type | 35 (97) | 11 (100) | 24 (96) | |
|
| ||||
| BRD4, n (%) | 0.31 | |||
| Mutated | 1 (3) | 1 (9) | 0 (0) | |
| Wild type | 35 (97) | 10 (91) | 25 (100) | |
|
| ||||
| BRINP3, n (%) | 1.00 | |||
| Mutated | 2 (6) | 0 (0) | 2 (8) | |
| Wild type | 34 (94) | 11 (100) | 23 (92) | |
|
| ||||
| BTK, n (%) | 0.31 | |||
| Mutated | 1 (3) | 1 (9) | 0 (0) | |
| Wild type | 35 (97) | 10 (91) | 25 (100) | |
|
| ||||
| CBL, n (%) | 0.22 | |||
| Mutated | 3 (8) | 2 (18) | 1 (4) | |
| Wild type | 33 (92) | 9 (82) | 24 (96) | |
|
| ||||
| DNMT3A, n (%) | 1.00 | |||
| Mutated | 8 (22) | 2 (18) | 6 (24) | |
| Wild type | 28 (78) | 9 (82) | 19 (76) | |
|
| ||||
| ETV6, n (%) | 0.54 | |||
| Mutated | 3 (8) | 0 (0) | 3 (12) | |
| Wild type | 33 (92) | 11 (100) | 22 (88) | |
|
| ||||
| EZH2, n (%) | 0.08 | |||
| Mutated | 4 (11) | 3 (27) | 1 (4) | |
| Wild type | 32 (89) | 8 (73) | 24 (96) | |
|
| ||||
| FLT3-TKD, n (%) | 2 unknown | 2 unknown | 1.00 | |
| Present | 1 (3) | 0 (0) | 1 (4) | |
| Absent | 33 (97) | 11 (100) | 22 (96) | |
|
| ||||
| GATA2, n (%) | 1.00 | |||
| Mutated | 3 (8) | 1 (9) | 2 (8) | |
| Wild type | 33 (92) | 10 (91) | 23 (92) | |
|
| ||||
| HIST1H1E, n (%) | 1.00 | |||
| Mutated | 1 (3) | 0 (0) | 1 (4) | |
| Wild type | 35 (97) | 11 (100) | 24 (96) | |
|
| ||||
| IDH1, n (%) | 1.00 | |||
| Mutated | 1 (3) | 0 (0) | 1 (4) | |
| Wild type | 35 (97) | 11 (100) | 24 (96) | |
|
| ||||
| IDH2, n (%) | 0.64 | |||
| Mutated | 6 (17) | 1 (9) | 5 (20) | |
| Wild type | 30 (83) | 10 (91) | 20 (80) | |
|
| ||||
| KIT, n (%) | 1.00 | |||
| Mutated | 1 (3) | 0 (0) | 1 (4) | |
| Wild type | 35 (97) | 11 (100) | 24 (96) | |
|
| ||||
| KMT2A, n (%) | 1.00 | |||
| Mutated | 1 (3) | 0 (0) | 1 (4) | |
| Wild type | 35 (97) | 11 (100) | 24 (96) | |
|
| ||||
| KRAS, n (%) | 1.00 | |||
| Mutated | 1 (3) | 0 (0) | 1 (4) | |
| Wild type | 35 (97) | 11 (100) | 24 (96) | |
|
| ||||
| NOTCH1, n (%) | 0.57 | |||
| Mutated | 4 (11) | 2 (18) | 2 (8) | |
| Wild type | 32 (89) | 9 (82) | 23 (92) | |
|
| ||||
| NPM1, n (%) | 1.00 | |||
| Mutated | 1 (3) | 0 (0) | 1 (4) | |
| Wild type | 35 (97) | 11 (100) | 24 (96) | |
|
| ||||
| NRAS, n (%) | 0.41 | |||
| Mutated | 9 (25) | 4 (36) | 5 (20) | |
| Wild type | 27 (75) | 7 (64) | 20 (80) | |
|
| ||||
| PLEKHG5, n (%) | 1.00 | |||
| Mutated | 1 (3) | 0 (0) | 1 (4) | |
| Wild type | 35 (97) | 11 (100) | 24 (96) | |
|
| ||||
| PTPN11, n (%) | 0.08 | |||
| Mutated | 4 (11) | 3 (27) | 1 (4) | |
| Wild type | 32 (89) | 8 (73) | 24 (96) | |
|
| ||||
| RAF1, n (%) | 0.31 | |||
| Mutated | 1 (3) | 1 (9) | 0 (0) | |
| Wild type | 35 (97) | 10 (91) | 25 (100) | |
|
| ||||
| RUNX1, n (%) | 1.00 | |||
| Mutated | 10 (28) | 3 (27) | 7 (28) | |
| Wild type | 26 (72) | 8 (73) | 18 (72) | |
|
| ||||
| SETBP1, n (%) | 0.57 | |||
| Mutated | 4 (11) | 2 (18) | 2 (8) | |
| Wild type | 32 (89) | 9 (82) | 23 (92) | |
|
| ||||
| SF1, n (%) | 1.00 | |||
| Mutated | 1 (3) | 0 (0) | 1 (4) | |
| Wild type | 35 (97) | 11 (100) | 24 (96) | |
|
| ||||
| SF3B1, n (%) | 0.31 | |||
| Mutated | 1 (3) | 1 (9) | 0 (0) | |
| Wild type | 35 (97) | 10 (91) | 25 (100) | |
|
| ||||
| SMARCA2, n (%) | 0.63 | |||
| Mutated | 5 (14) | 2 (18) | 3 (12) | |
| Wild type | 31 (86) | 9 (82) | 22 (88) | |
|
| ||||
| SMC1A, n (%) | 1.00 | |||
| Mutated | 3 (8) | 1 (9) | 2 (8) | |
| Wild type | 33 (92) | 10 (91) | 23 (92) | |
|
| ||||
| SMC3, n (%) | 0.54 | |||
| Mutated | 3 (8) | 0 (0) | 3 (12) | |
| Wild type | 33 (92) | 11 (100) | 22 (88) | |
|
| ||||
| SRSF2, n (%) | 0.08 | |||
| Mutated | 8 (22) | 0 (0) | 8 (32) | |
| Wild type | 28 (78) | 11 (100) | 17 (68) | |
|
| ||||
| STAG2, n (%) | 1.00 | |||
| Mutated | 1 (3) | 0 (0) | 1 (4) | |
| Wild type | 35 (97) | 11 (100) | 24 (96) | |
|
| ||||
| TET2, n (%) | 0.03 | |||
| Mutated | 9 (25) | 0 (0) | 9 (36) | |
| Wild type | 27 (75) | 11 (100) | 16 (64) | |
|
| ||||
| TGM7, n (%) | 1.00 | |||
| Mutated | 2 (6) | 0 (0) | 2 (8) | |
| Wild-type | 34 (94) | 11 (100) | 23 (92) | |
|
| ||||
| TYK2, n (%) | 1.00 | |||
| Mutated | 1 (3) | 0 (0) | 1 (4) | |
| Wild type | 35 (97) | 11 (100) | 24 (96) | |
|
| ||||
| U2AF1, n (%) | 0.30 | |||
| Mutated | 5 (14) | 0 (0) | 5 (20) | |
| Wild type | 31 (86) | 11 (100) | 20 (80) | |
|
| ||||
| WT1, n (%) | 1.00 | |||
| Mutated | 4 (11) | 1 (9) | 3 (12) | |
| Wild type | 32 (89) | 10 (91) | 22 (88) | |
|
| ||||
| ZRSR2, n (%) | 0.31 | |||
| Mutated | 1 (3) | 1 (9) | 0 (0) | |
| Wild type | 35 (97) | 10 (91) | 25 (100) | |
|
| ||||
| Number of mutations per patient | 0.43 | |||
| Median (range) | 4 (0–7) | 3 (1–7) | 4 (0–7) | |
Abbreviations: FLT3-TKD, tyrosine kinase domain mutation in the FLT3 gene; y, years.
Only genes found mutated in at least one patient are listed in alphabetical order in this Table. No mutations were detected in the ARAF, ATM, AXL, BCL2, BIRC6, CCND1, CCND2, CEBPA (biallelic), CSNKN1A, CTNNB1, FBXW7, GATA1, GSK3B, HNRNPK, IKZF1, IKZF3, IL7R, JAK1, JAK2, JAK3, KLHL6, MAPK1, MAPK3, MED12, MYD88, PHF6, PIK3CD, PIK3CG, PIKD3, PLCG2, PRKCB, PTEN, RAD21, SAMHD1, SF3A1, SYK, TP53, U2AF2, XPO1, or ZMYM3 genes. No patient was found to harbor FLT3-ITD.
P-values relate to the comparison of younger versus older sole -7 patients. P-values are from Fisher’s exact test.
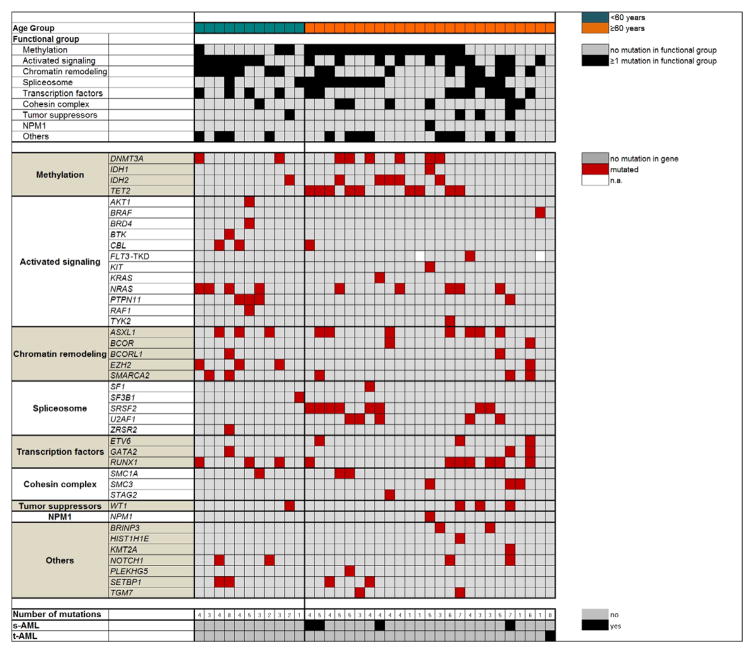
Figure 1.
Oncoprint of mutations found in functional groups (listed by descending frequencies; upper panel) and single gene mutations (lower panel) in patients with AML and sole -7. Patients (one per column) are depicted separately by age group (<60 years, green and ≥60 years, orange). Black highlights in the upper panels indicate the presence of ≥1 mutation in ≥1 gene assigned to the functional group. The lower panels list the respective gene mutations. Red highlights indicate the presence of a gene mutation, grey highlights indicate wild-type status (see Patients and Methods for details), white highlights indicate that the mutation status is not available (n.a.). s-AML denotes secondary AML; t-AML, therapy-related AML.
Five patients harbored mutations in the gene SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 (SMARCA2, Table 3, Figure 1), which encodes one of the two alternative catalytic subunits of the BAF chromatin remodeling complex (35). Non-truncating mutations in the ATPase region of SMARCA2 were previously identified in the rare Nicolaides-Baraitser syndrome (35, 36), which is characterized by intellectual disability and multiple congenital anomalies, but not increased risk of cancer. Recurrent SMARCA2 mutations are found in several different solid tumors, but in leukemia, SMARCA2 mutations were previously reported only in single cases of adult and pediatric AML (24) and in a patient with progressing CLL (37). In our cohort of sole -7 patients, four patients had a single SMARCA2 mutation and one patient had two SMARCA2 mutations. Some of the locations of mutations we discovered were in close proximity to mutation locations previously reported in Nicolaides-Baraitser syndrome patients (35, 36; Supplementary Figure S2). Interestingly, all three patients with a GATA2 mutation also had a mutation in SMARCA2 (Figure 1), suggesting there may be an undiscovered functional relationship between these genes. Four of the six SMARCA2 mutations had VAFs of <20%, suggesting that they likely represent later mutational events. While the mutation with the highest VAF (52%) could also be detected in the paired RNAseq data (37%, Supplementary Table S1), four of the remaining five SMARCA2 mutations could not be evaluated at the RNA level due to low SMARCA2 expression and thus inadequate coverage. The evaluable SMARCA2 mutation with a VAF of 13% at the DNA level was not detected at the RNA level (Supplementary Table S1).
Since -7 AML has been previously associated with familial AML syndromes (32, 33), we also tested germline samples of patients with available material (n=10) for the presence or absence of the variants detected in the leukemic samples (Supplementary Table S2). The analysis of the germline material of seven patients validated all variants detected in the diagnostic samples as somatic changes, whereas the germline material of two patients revealed one variant in each patient to be of germline origin. The CBL P510A variant found in patient no. 6 was detected with a VAF of 56% in the leukemic sample and with a VAF of 44% in the paired germline DNA (Supplementary Table S2). Further data mining revealed that this variant was also detected in a single individual in the 1000 genome project [minor allele frequency <0.0001%, amino acid change predicted not to affect the protein function (Scale-invariant feature transform; SIFT score: −0.62)], suggesting that CBL P510A represents an ultra-rare and probably benign germline variant. In contrast, the RAF1 R318W variant in patient no. 9, detected in both leukemic and germline material (with VAFs of 47% and 46%, respectively; Supplementary Table S2), is listed in the COSMIC database as a validated somatic variant, which was previously detected in a patient with endometrial cancer (COSM1037798, TCGA-B5-A0K6-01). Furthermore, the resulting amino acid change is predicted to be deleterious for RAF1’s protein function (SIFT scores: −3.84 to −4.52). Thus, RAF1 R318W could indeed represent a novel variant predisposing to -7 AML. Finally, the remission material of patient no. 10 could not be evaluated for possible germline variants, because all mutations in the remission sample had high VAFs suggesting the presence of residual disease (Supplementary Table S2).
Functional groups of gene mutations in AML patients with sole -7
To detect mutation patterns in sole -7 AML, we next assigned mutations to previously reported functional groups (24). All but a single patient had one or more mutations in at least one of the major functional groups comprised of the genes included in our mutation panel (median, 2 affected groups/patient; range, 0–5; Figure 1). The most frequently observed mutations were those in genes involved in methylation (DNMT3A, IDH1, IDH2 and TET2) that were found in 19 (53%) patients, and in genes leading to activated signaling (AKT1, BRAF, BRD4, BTK, CBL, FLT3-TKD, KIT, KRAS, NRAS, PTPN11, RAF1 and TYK2) that were present in 18 (50%) patients (Table 4 and Figure 1). Notably, the group of genes leading to activated signaling was dominated by mutations of genes affecting the RAS pathway, namely CBL, KRAS, NRAS, and PTPN11, which were detected in 15 (42%) patients. This is consistent with previous reports of the frequent involvement of RAS pathway members in myeloid malignancies, including AML, with unbalanced abnormalities involving chromosome 7 (15, 38). Other functional groups comprising frequently mutated genes were: chromatin remodeling (ASXL1, BCOR, BCORL1, EZH2 and SMARCA2; n=15 patients, 42%), spliceosome (SF1, SF3B1, SRSF2, U2AF1 and ZRSR2; n=14 patients, 39%) and transcription factors (ETV6, GATA2 and RUNX1; n=13 patients, 36%). In contrast, fewer mutations were detected in cohesin complex genes (SMC1A, SMC3 and STAG2; n=7 patients, 19%), and tumor suppressors (n=4 patients, 11%), with WT1 being the only tumor suppressor gene that was found mutated in the sole -7 cohort. Only one patient (3%) harbored an NPM1 mutation (Table 4 and Figure 1).
Table 4.
Frequencies of gene mutations in functional groups observed in patients with acute myeloid leukemia (AML) and sole -7
| Functional groupa | Sole -7 AML All patients (n=36) |
Sole -7 AML <60y (n=11) |
Sole -7 AML ≥60y (n=25) |
P valueb |
|---|---|---|---|---|
|
| ||||
| Methylation, n (%) | 0.07 | |||
| Mutated | 19 (53) | 3 (27) | 16 (64) | |
| Wild type | 17 (47) | 8 (73) | 9 (36) | |
|
| ||||
| Activated signaling, n (%) | 0.47 | |||
| Mutated | 18 (50) | 7 (64) | 11 (44) | |
| Wild type | 18 (50) | 4 (36) | 14 (56) | |
|
| ||||
| Chromatin remodeling, n (%) | 0.14 | |||
| Mutated | 15 (42) | 7 (64) | 8 (32) | |
| Wild type | 21 (58) | 4 (36) | 17 (68) | |
|
| ||||
| Spliceosome, n (%) | 0.14 | |||
| Mutated | 14 (39) | 2 (18) | 12 (48) | |
| Wild type | 22 (61) | 9 (82) | 13 (52) | |
|
| ||||
| Transcription factors, n (%) | 1.00 | |||
| Mutated | 13 (36) | 4 (36) | 9 (36) | |
| Wild type | 23 (64) | 7 (64) | 16 (64) | |
|
| ||||
| Cohesin complex, n (%) | 0.40 | |||
| Mutated | 7 (19) | 1 (9) | 6 (24) | |
| Wild type | 29 (81) | 10 (91) | 19 (76) | |
|
| ||||
| Tumor suppressors, n (%) | 1.00 | |||
| Mutated | 4 (11) | 1 (9) | 3 (12) | |
| Wild type | 32 (89) | 10 (91) | 22 (88) | |
|
| ||||
| NPM1, n (%) | 1.00 | |||
| Mutated | 1 (3) | 0 (0) | 1 (4) | |
| Wild type | 35 (97) | 11 (100) | 24 (96) | |
|
| ||||
| Methylation and/or spliceosome and/or cohesin complex, n (%) | 0.04 | |||
| Mutated | 28 (78) | 6 (55) | 22 (88) | |
| Wild type | 8 (22) | 5 (45) | 3 (12) | |
Abbreviation: y, years.
The functional groups (ref. 24; ≥1 mutation in ≥1 gene assigned to the functional group, listed are only the genes which were found mutated in our cohort) are listed by descending mutation frequencies in all -7 patients. They include: Methylation, DNMT3A, IDH1/2 or TET2; Activated signaling, AKT1, BRAF, BRD4, BTK, CBL, FLT3-TKD, KIT, KRAS, NRAS, PTPN11, RAF1 or TYK2; Chromatin remodeling, ASXL1, BCOR, BCORL1, EZH2 or SMARCA2; Spliceosome, SF1, SF3B1, SRSF2, U2AF1 or ZRSR2; Transcription factors, ETV6, GATA2 or RUNX1; Cohesin complex, SMC1A, SMC3 or STAG2; Tumor suppressors, WT1; NPM1, NPM1.
P-values relate to the comparison of younger versus older sole -7 patients. P-values are from Fisher’s exact test.
Age-associated mutation patterns in AML patients with sole -7
For cytogenetically normal AML (CN-AML), it is known that certain mutations have age-related differences in their frequencies (39, 40). Our comparison of the frequencies of single gene mutations (Table 3) and functional groups (Table 4) between patients younger than 60 years and those aged 60 years and older revealed preferential mutation patterns in both age groups. Whereas EZH2 and PTPN11 mutations tended to occur more frequently in younger patients with sole -7 AML (27% of younger vs. 4% of older patients), TET2 and SRSF2 mutations were detected exclusively in older patients (TET2 in 36% and SRSF2 in 32%). However, with the exception of TET2 (P=0.03), none of the observed differences in the mutation frequencies between the two age groups reached statistical significance (Table 3). The fact that TET2 mutations were found exclusively in sole -7 AML patients who are aged ≥60 years differs somewhat from the previous reports on both CN-AML (40) and an unselected patient cohort comprised of patients with various cytogenetic findings (41). Although the incidence of TET2 mutations was also the highest among older patients in both CN-AML (29%, ref. 40) and an unselected AML patient population (24%, ref. 41), in contrast to younger sole -7 AML patients who did not harbor any TET2 mutations, considerable proportions of younger patients with CN-AML (15%, ref. 40) and those with AML with various karyotypes (7%, ref. 41) carried TET2 mutations. Because of a relatively low number of younger patients with sole -7 AML in our study, the lack of TET2 mutations in sole -7 AML patients under the age of 60 years should be corroborated.
We also observed a preferred involvement of functional groups depending on age (Table 4). Mutations in genes leading to activated signaling were frequently found in both age groups (64% of younger and 44% of older patients). However, while younger patients frequently harbored mutations in chromatin remodeling genes (64%, compared with 32% of older patients), older patients were characterized by mutations in methylation genes, which were found in 64% of patients (compared with 27% of younger patients), followed by mutations in spliceosome genes (48%, compared with 18% of younger patients). Eighty-eight percent of older patients harbored at least one mutation in genes involved in methylation, the spliceosome and/or the cohesin complex (compared with only 55% of younger patients, P=0.04). Even though the patient cohorts are small, this is the first report to suggest possible age-related differences in single genes and functional groups affected by mutations in sole -7 AML.
Effects of gene mutations on the outcome of AML patients with sole -7
To test whether any of the clinical parameters, single gene mutations or mutational groups (as defined by the TCGA, ref. 24) impact on the outcome of sole -7 patients, we performed univariable analyses for associations with CR achievement, DFS and OS in the whole patient cohort. None of the tested parameters was significantly associated with the achievement of CR, DFS or OS (Table 5).
Table 5.
Univariable outcome analyses in patients with acute myeloid leukemia and sole -7
| Endpoint | Variablesa | P valueb |
|---|---|---|
| Total cohort (n=36) | ||
| Complete remission | Spliceosome mutationc, ≥1 mutation vs. no mutation | 0.06 |
| SMARCA2, mutated vs. wild-type | 0.09 | |
| Overall survival | SMARCA2, mutated vs. wild-type | 0.09 |
| U2AF1, mutated vs. wild-type | 0.06 | |
| WT1, mutated vs. wild-type | 0.06 | |
| Patients ≥60 years (n=25) | ||
| Complete remission | Spliceosome mutationc, ≥1 mutation vs. no mutation | 0.03d |
| Overall survival | U2AF1, mutated vs. wild-type | 0.07 |
Variables tested in the univariable outcome analyses are listed in the Supplementary data. Listed in Table 5 are all variables with a P-value ≤0.10. Variables written in bold-type letters are those associated with unfavorable outcome. No variable tested for association with DFS in all patients or in patients ≥60 years had a P-value ≤0.10.
P-values for achievement of CR using logistic regression; P-values for disease-free and overall survival using Cox proportional hazards regression.
Spliceosome mutations include mutations in the SF1, SF3B1, SRSF2, U2AF1 and/or ZRSR2 genes.
Odds ratio for achievement of CR ≥1 spliceosome mutation versus no mutation: 0.13, 95% confidence interval: 0.02 to 0.82.
We next tested the older and younger patient groups separately for associations between clinical and molecular parameters and outcome. Again, no parameter was significantly associated with any of the outcome endpoints in younger sole -7 patients, which may be due to the small sample size. However, older patients with sole -7 harboring a mutation in a spliceosome gene had a lower CR rate compared with that of patients without a spliceosome mutation (17% vs. 62%, P=0.03; Table 5). No parameter was significantly associated with DFS or OS.
Gene- and microRNA-expression profiling of AML patients with sole -7
The reduced expression of some genes mapped to chromosome 7 caused by the chromosome loss has been suggested as a major factor in the pathogenesis of -7 AML (6, 7), and several promising haploinsufficient genes located on chromosome 7 have already been identified as candidates contributing to the phenotype of -7 AML (8–13). Using our, the largest to date, series of patients with -7 as a sole abnormality, we comprehensively characterized the gene- and microRNA-(miR) expression changes caused by chromosome 7 loss. We compared the gene- and miR-expression profiles between sole -7 patients and AML patients with other cytogenetic findings in the TCGA AML cohort (24), excluding patients with chromosome 7 abnormalities and/or complex karyotype. We derived a signature comprising 284 genes that were significantly downregulated and 42 genes significantly upregulated in sole -7 AML (adjusted P-value <0.001; Supplementary Figure S3 and Supplementary Tables S3 and S4). Consistent with a gene dosage effect, 94% of the genes significantly downregulated in -7 AML were mapped to chromosome 7.
The most significant downregulation was observed for an open-reading-frame on chromosome 7 which likely encodes a transmembrane spanning protein C7orf42 (TMEM248). The mismatch repair gene PMS2 (42) was the second most significantly downregulated gene. We also confirmed downregulation of CUX1 (8), EZH2 (10) and MLL3 (11), all of which have been previously identified as haploinsufficient genes of chromosome 7 (Supplementary Table S3).
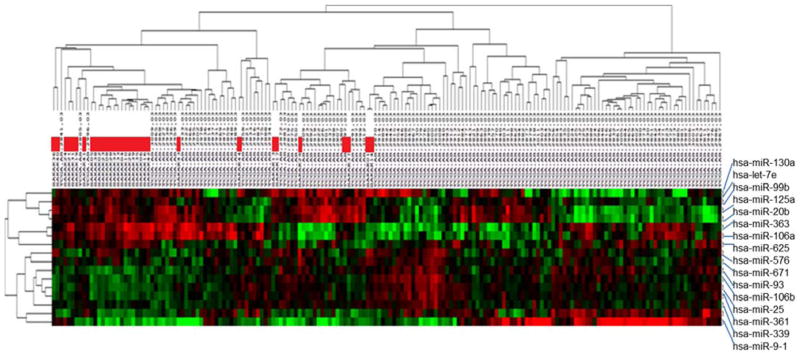
The most significantly upregulated gene in sole -7 AML was the protein tyrosine phosphatase receptor type M gene (PTPRM), which is an important regulator of cell growth, differentiation and oncogenic transformation (43). Other highly overexpressed genes in sole -7 AML included ID1, which is a common downstream target of oncogenic tyrosine kinases in leukemic cells (44), and MECOM, the MDS1/EVI1 complex locus, which has been previously associated with inv(3)(q21q26.2)/t(3;3)(q21;q26.2) and chromosome 7 abnormalities (38, 45) (Supplementary Table S4). The miR-expression profile consisted of 16 differentially expressed miRs, six of which were significantly downregulated and 10 significantly upregulated in sole -7 patients compared with non-sole -7 AML (Figure 2, Supplementary Tables S5 and S6). All but one of the six downregulated miRs are mapped to chromosome 7 (miR-25, miR-93, miR-106b, miR-339, and miR-671). MiR-25, miR-93 and miR-106b belong to a miR cluster located in different introns of the MCM7 gene, which itself did not belong to the most downregulated mRNAs in sole -7 AML. The most downregulated miR with respect to the observed fold-change was miR-9-1, which is mapped to chromosome 1 (Supplementary Table S5).
Figure 2.
Heatmap depicting the differential miR expression of AML patients with sole -7 (n=31, indicated by red bars) and AML patients with other cytogenetics of the TCGA AML cohort19 (excluding patients with chromosome 7 abnormalities and/or complex karyotype, n=136). Lower expression is shown in green, while higher expression is shown in red. The derived miR-expression signature comprised six miRs downregulated and 10 miRs upregulated in sole -7 AML, with an adjusted P-value of <0.001. The significantly down- and upregulated miRs and the corresponding P-values and fold changes can be found in Supplementary Tables S5 and S6, respectively.
The miRs upregulated in sole -7 patients mainly belong to two miR-clusters: the cluster comprising miR-20b, miR-363 and miR-106a located on the X chromosome and the cluster of miR-99b, miR-125a and miR-let7e located on chromosome 19 (Supplementary Table S6), suggesting a shared regulatory mechanism.
It has been recently shown that large-scale loss of chromosome material in cancer cells can disturb multiple signaling pathways and produce phenotypes distinct from those arising through loss of a single gene (46). We performed gene ontology (GO) analyses of the differentially expressed mRNAs and of the mRNA targets of the differentially expressed miRs to test whether genes involved in specific biologic processes are enriched in signatures associated with loss of the entire chromosome 7. Indeed, our GO analysis of downregulated genes located on chromosome 7 revealed a significant enrichment of genes involved in the molecular function of RNA binding (both Poly-A and non-Poly-A), and the biological process of small protein conjugation (Supplementary Table S7).
In addition, an analysis of the mRNA targets of the miRs which were upregulated in -7 AML showed a striking enrichment of target mRNAs involved in metabolic processes, especially in the cellular nitrogen compound (Supplementary Figure S4). In contrast, GO analysis of the mRNA targets of the downregulated miRs only resulted in weak clustering (Supplementary Figure S5). Taken together, the observed enrichments suggest that multiple gene networks might also be important in the pathogenesis of -7 AML.
Discussion
In this study, we analyzed the hitherto largest, to our knowledge, homogeneous group of adult AML patients with -7 as the sole abnormality to provide insights into the molecular characteristics associated with chromosome 7 loss using a comprehensive panel of 81 cancer- and/or leukemia-associated genes. The relatively large number of gene mutations detected in virtually all major functional groups indicates that -7 AML is molecularly heterogeneous. However, our data also suggest that mutation patterns we observed are partly associated with the patients’ age. Although both the younger and older patients frequently harbored mutations in genes leading to activated signaling that comprise kinases and RAS pathway members, younger patients with -7 AML often had mutations in chromatin modifiers. In contrast, older -7 AML patients often harbored mutations in genes involved in methylation and/or the spliceosome. Our finding that spliceosome mutations may confer a reduced probability of achieving a CR in patients receiving cytarabine and an anthracycline (7+3)-based induction chemotherapy might provide a first rationale for testing of spliceosome mutations at diagnosis in older -7 AML patients. Whether this molecular feature will also be predictive of CR attainment in patients receiving other therapeutic approaches, such as a demethylating agent therapy, should be assessed by future studies, as thus far, to our knowledge, no published data about molecular prognosticators in -7 AML in patients treated with demethylating agents are available.
Concordant with our results, enrichment in mutations affecting the RAS pathway has been previously suggested as an important biologic feature of -7 AML by McNerney et al. (15). However, while that study suggested a paucity of mutations in methylation genes (15), our data found the methylation group to be the most frequently affected functional group of sole -7 AML. This discrepancy may be related to the limited sample sizes in both studies, but also may stem from the fact that McNerney et al. (15) studied not only patients with -7 but also patients with del(7q) (~18%), and that two-thirds of patients analyzed in their study harbored chromosomal aberrations in addition to -7 or del(7q). Moreover, a recent study by Gröschel et al. (38) on 30 patients with AML, five patients with myelodysplastic syndromes, four cell lines and two patients with chronic myelogenous leukemia in blast crisis that harbored inv(3) or t(3;3), which in two thirds of the cases was accompanied by -7 as an additional cytogenetic abnormality, detected mutations in genes leading to activated signaling (kinases and RAS pathway members) in 98% of samples studied, but only 10% of their samples had mutations in methylation genes. We also frequently detected activated signaling mutations in our patient cohort, but our sole -7 AML patients had frequent mutations in methylation, chromatin remodeling and spliceosome genes. These apparently discordant results may be attributed not only to the fact that one-third of cases studied by Gröschel et al. (38) did not harbor -7, but also to the age-related differences in mutation distribution we report herein, since the majority of our patients with sole -7 were older than 60 years, whereas patients with -7 that accompanies inv(3) or t(3;3) are usually younger.
The use of large-scale next-generation sequencing for molecular profiling studies of leukemia/cancer patients is becoming increasingly common. However, the diversity of the analyzed cohorts makes it difficult to detect novel variants with potential importance in specific patient subgroups. The identification of six SMARCA2 mutations in five sole -7 AML patients in our cohort underlines the discovery potential of sequencing efforts in small but homogeneous patient populations. While a SMARCA2 mutation has been previously reported in a single adult patient with AML and a translocation involving the KMT2A (MLL) gene (24), the prevalence of SMARCA2 mutations and the potential of their enrichment in certain subsets of AML patients have likely been underestimated. It seems noteworthy that four of the six SMARCA2 mutations had VAFs <20%, providing a possible reason why they were not more frequently discovered in other sequencing studies. Efforts to assess the frequency of SMARCA2 mutations in other cytogenetic subgroups and to evaluate their possible functional consequences are already ongoing. Mutations in other genes belonging to the ATP-dependent mSWI/SNF chromatin remodeling complex have also been detected recently in solid tumors. Those studies and the detected mutation diversity not only highlight the pathophysiologic importance of these genes, but also suggest that specific genes encoding subunits of the mSWI/SNF complex appear to be mutated in specific cancers (47).
In addition, our finding of the RAF1 variant as a germline change, which may be a leukemia-predisposing mutation in the affected patient, highlights the usefulness of paired germline-leukemic DNA analyses, especially in AML with -7, which has been previously associated with familial AML syndromes. In fact, two other RAF1 germline variants have been previously detected both in the primary tumors and in the leukemic blasts of two patients with therapy-related AML after treatment for, respectively, embryonal cell carcinoma of the testis and colorectal cancer (48), suggesting that these RAF1 mutations may constitute possible cancer- and AML-predisposing events.
Furthermore, the results of our gene- and microRNA-expression analyses provide important insights into the biology of -7 AML. Besides the expected and previously described largely homogeneous downregulation of genes located on chromosome 7 (49), we observed an overexpression of the MECOM (EVI1/MDS1) locus in most sole -7 AML samples. A possible mechanistic link between MECOM overexpression and subsequent loss of chromosome 7 has been previously suggested in a case report of two patients with genomic instability and myelodysplasia with -7 consequent to MECOM activation after gene therapy for chronic granulomatous disease (50). Moreover, MECOM overexpression has been reported as a consistent feature of AML with the chromosome 3q rearrangements, most often inv(3) and t(3;3), which in most, but not all, patients occur together with -7 as a secondary abnormality (14, 38). Our study shows that high MECOM expression is a molecular characteristic of AML with -7 also in the absence of inv(3) or t(3;3). While high expression of MECOM is known to be associated with very poor outcome of AML patients, little is known how to target this deregulated expression. The observation that additional mechanisms other than the super-enhancer formation may lead to aberrantly increased MECOM may provide an additional challenge to target this important oncogene. However, the first in vitro data about the possible efficacy of the BET inhibitor JQ1 in cell lines with t(3;3) or inv(3;3) might provide a rationale to test this therapeutic approach also in patients with sole -7 AML with aberrantly expressed MECOM (51).
Although this is the largest study characterizing the mutational landscape of adult AML with sole -7 published to date, our findings should be interpreted with caution because of the still limited sample size of 36 patients. Furthermore, while the targeted sequencing approach enabled us to also identify subclonal mutations, it did not permit discovery of novel mutations in genes that have not hitherto been associated with leukemia and/or solid tumors. These limitations may also explain the paucity of mutations detected in the only patient with t-AML, who might harbor mutation(s) in less frequently affected gene(s).
In summary, the identification of age-associated gene mutation patterns and commonly deregulated gene- and microRNA-expression profiles sheds light on the molecular events in sole -7 AML.
Acknowledgments
Financial support: This work was supported in part by the National Cancer Institute (grants CA101140, CA140158, CA180861, CA196171, CA016058, CA180821, CA180882, and CA077658), the Coleman Leukemia Research Foundation, the Pelotonia Fellowship Program (A-KE), and by an allocation of computing resources from The Ohio Supercomputer Center.
The authors are grateful to the patients who consented to participate in these clinical trials and the families who supported them; to Donna Bucci and the CALGB/Alliance Leukemia Tissue Bank at The Ohio State University Comprehensive Cancer Center, Columbus, OH, for sample processing and storage services and Lisa J. Sterling and Chris Finks for data management.
Footnotes
Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest: The authors declare no conflict of interest.
References
- 1.Grimwade D, Hills RK, Moorman AV, Walker H, Chatters S, Goldstone AH, et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 2010;116:354–65. doi: 10.1182/blood-2009-11-254441. [DOI] [PubMed] [Google Scholar]
- 2.Byrd JC, Mrózek K, Dodge RK, Carroll AJ, Edwards CG, Arthur DC, et al. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461) Blood. 2002;100:4325–36. doi: 10.1182/blood-2002-03-0772. [DOI] [PubMed] [Google Scholar]
- 3.Slovak ML, Kopecky KJ, Cassileth PA, Harrington DH, Theil KS, Mohamed A, et al. Karyotypic analysis predicts outcome of preremission and postremission therapy in adult acute myeloid leukemia: a Southwest Oncology Group/Eastern Cooperative Oncology Group study. Blood. 2000;96:4075–83. [PubMed] [Google Scholar]
- 4.Mrózek K, Heerema NA, Bloomfield CD. Cytogenetics in acute leukemia. Blood Rev. 2004;18:115–36. doi: 10.1016/S0268-960X(03)00040-7. [DOI] [PubMed] [Google Scholar]
- 5.Hussain FT, Nguyen EP, Raza S, Knudson R, Pardanani A, Hanson CA, et al. Sole abnormalities of chromosome 7 in myeloid malignancies: spectrum, histopathologic correlates, and prognostic implications. Am J Hematol. 2012;87:684–6. doi: 10.1002/ajh.23230. [DOI] [PubMed] [Google Scholar]
- 6.Kere J, Ruutu T, Lahtinen R, de la Chapelle A. Molecular characterization of chromosome 7 long arm deletions in myeloid disorders. Blood. 1987;70:1349–53. [PubMed] [Google Scholar]
- 7.Le Beau MM, Espinosa R, 3rd, Davis EM, Eisenbart JD, Larson RA, Green ED. Cytogenetic and molecular delineation of a region of chromosome 7 commonly deleted in malignant myeloid diseases. Blood. 1996;88:1930–5. [PubMed] [Google Scholar]
- 8.McNerney ME, Brown CD, Wang X, Bartom ET, Karmakar S, Bandlamudi C, et al. CUX1 is a haploinsufficient tumor suppressor gene on chromosome 7 frequently inactivated in acute myeloid leukemia. Blood. 2013;121:975–83. doi: 10.1182/blood-2012-04-426965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou L, Opalinska J, Sohal D, Yu Y, Mo Y, Bhagat T, et al. Aberrant epigenetic and genetic marks are seen in myelodysplastic leukocytes and reveal Dock4 as a candidate pathogenic gene on chromosome 7q. J Biol Chem. 2011;286:25211–23. doi: 10.1074/jbc.M111.235028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Dai H, Wang Q, Wang Q, Xu Y, Wang Y, et al. EZH2 mutations are related to low blast percentage in bone marrow and -7/del(7q) in de novo acute myeloid leukemia. PLoS One. 2013;8:e61341. doi: 10.1371/journal.pone.0061341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen C, Liu Y, Rappaport AR, Kitzing T, Schultz N, Zhao Z, et al. MLL3 is a haploinsufficient 7q tumor suppressor in acute myeloid leukaemia. Cancer Cell. 2014;25:652–65. doi: 10.1016/j.ccr.2014.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagamachi A, Matsui H, Asou H, Ozaki Y, Aki D, Kanai A, et al. Haploinsufficiency of SAMD9L, an endosome fusion facilitator, causes myeloid malignancies in mice mimicking human diseases with monosomy 7. Cancer Cell. 2013;24:305–17. doi: 10.1016/j.ccr.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 13.Kratz CP, Emerling BM, Bonifas J, Wang W, Green ED, Le Beau MM, et al. Genomic structure of the PIK3CG gene on chromosome band 7q22 and evaluation as a candidate myeloid tumor suppressor. Blood. 2002;99:372–4. doi: 10.1182/blood.v99.1.372. [DOI] [PubMed] [Google Scholar]
- 14.Gröschel S, Lugthart S, Schlenk RF, Valk PJM, Eiwen K, Goudswaard C, et al. High EVI1 expression predicts outcome in younger adult patients with acute myeloid leukemia and is associated with distinct cytogenetic abnormalities. J Clin Oncol. 2010;28:2101–7. doi: 10.1200/JCO.2009.26.0646. [DOI] [PubMed] [Google Scholar]
- 15.McNerney ME, Brown CD, Peterson AL, Banerjee M, Larson RA, Anastasi J, et al. The spectrum of somatic mutations in high-risk acute myeloid leukemia with -7/del(7q) Br J Haematol. 2014;166:550–6. doi: 10.1111/bjh.12964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Honda H, Nagamachi A, Inaba T. -7/7q- syndrome in myeloid-lineage hematopoietic malignancies: attempts to understand this complex disease entity. Oncogene. 2015;34:2413–25. doi: 10.1038/onc.2014.196. [DOI] [PubMed] [Google Scholar]
- 17.Mrózek K, Carroll AJ, Maharry K, Rao KW, Patil SR, Pettenati MJ, et al. Central review of cytogenetics is necessary for cooperative group correlative and clinical studies of adult acute leukemia: the Cancer and Leukemia Group B experience. Int J Oncol. 2008;33:239–44. [PMC free article] [PubMed] [Google Scholar]
- 18.Vittinghoff E, Glidden DV, Shiboski SC, McCulloch CE. Regression methods in biostatistics: linear, logistic, survival and repeated measures models. New York: Springer; 2005. [Google Scholar]
- 19.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–9. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–6. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thiede C, Steudel C, Mohr B, Schaich M, Schäkel U, Platzbecker U, et al. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: association with FAB subtypes and identification of subgroups with poor prognosis. Blood. 2002;99:4326–35. doi: 10.1182/blood.v99.12.4326. [DOI] [PubMed] [Google Scholar]
- 23.Marcucci G, Maharry K, Radmacher MD, Mrózek K, Vukosavljevic T, Paschka P, et al. Prognostic significance of, and gene and microRNA expression signatures associated with, CEBPA mutations in cytogenetically normal acute myeloid leukemia with high-risk molecular features: a Cancer and Leukemia Group B study. J Clin Oncol. 2008;26:5078–87. doi: 10.1200/JCO.2008.17.5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo myeloid leukemia. N Engl J Med. 2013;368:2059–74. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang K, Singh D, Zeng Z, Coleman SJ, Huang Y, Savich GL, et al. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res. 2010;38:e178. doi: 10.1093/nar/gkq622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–27. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 28.Leek JT, Johnson WE, Parker HS, Fertig EJ, Jaffe AE, Storey JD. Sva: Surrogate Variable Analysis. 2015 R package version 3.14.0. Available from: http://www.bioconductor.org/packages/release/bioc/html/sva.html.
- 29.Ruffalo M, LaFramboise T, Koyutürk M. Comparative analysis of algorithms for next-generation sequencing read alignment. Bioinformatics. 2011;27:2790–6. doi: 10.1093/bioinformatics/btr477. [DOI] [PubMed] [Google Scholar]
- 30.Makishima H, Yoshida K, Nguyen N, Przychodzen B, Sanada M, Okuno Y, et al. Somatic SETBP1 mutations in myeloid malignancies. Nat Genet. 2013;45:942–6. doi: 10.1038/ng.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Puente XS, Pinyol M, Quesada V, Conde L, Ordóñez GR, Villamor N, et al. Whole-exome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–5. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hahn CN, Chong C-E, Carmichael CL, Wilkins EJ, Brautigan PJ, Li X-C, et al. Heritable GATA2 mutations associated with familial myelodysplastic syndrome and acute myeloid leukemia. Nat Genet. 2011;43:1012–7. doi: 10.1038/ng.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsu AP, Sampaio EP, Khan J, Calvo KR, Lemieux JE, Patel SY, et al. Mutations in GATA2 are associated with the autosomal dominant and sporadic monocytopenia and mycobacterial infection (MonoMAC) syndrome. Blood. 2011;118:2653–5. doi: 10.1182/blood-2011-05-356352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42:722–6. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 35.Van Houdt JKJ, Nowakowska BA, Sousa SB, van Schaik BDC, Seuntjens E, Avonce N, et al. Heterozygous missense mutations in SMARCA2 cause Nicolaides-Baraitser syndrome. Nat Genet. 2012;44:445–9. doi: 10.1038/ng.1105. [DOI] [PubMed] [Google Scholar]
- 36.Wolff D, Endele S, Azzarello-Burri S, Hoyer J, Zweier M, Schanze I, et al. In-frame deletion and missense mutations of the C-terminal helicase domain of SMARCA2 in three patients with Nicolaides-Baraitser syndrome. Mol Syndromol. 2011;2:237–44. doi: 10.1159/000337323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doménech E, Gómez-López G, Gzlez-Peña D, López M, Herreros B, Menezes J, et al. New mutations in chronic lymphocytic leukemia identified by target enrichment and deep sequencing. PLoS One. 2012;7:e38158. doi: 10.1371/journal.pone.0038158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gröschel S, Sanders MA, Hoogenboezem R, Zeilemaker A, Havermans M, Erpelinck C, et al. Mutational spectrum of myeloid malignancies with inv(3)/t(3;3) reveals a predominant involvement of RAS/RTK signaling pathways. Blood. 2015;125:133–9. doi: 10.1182/blood-2014-07-591461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schneider F, Hoster E, Schneider S, Dufour A, Benthaus T, Kakadia PM, et al. Age-dependent frequencies of NPM1 mutations and FLT3-ITD in patients with normal karyotype AML (NK-AML) Ann Hematol. 2012;91:9–18. doi: 10.1007/s00277-011-1280-6. [DOI] [PubMed] [Google Scholar]
- 40.Metzeler KH, Maharry K, Radmacher MD, Mrózek K, Margeson D, Becker H, et al. TET2 mutations improve the new European LeukemiaNet risk classification of acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2011;29:1373–81. doi: 10.1200/JCO.2010.32.7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chou W-C, Chou S-C, Liu C-Y, Chen C-Y, Hou H-A, Kuo Y-Y, et al. TET2 mutation is an unfavorable prognostic factor in acute myeloid leukemia patients with intermediate-risk cytogenetics. Blood. 2011;118:3803–10. doi: 10.1182/blood-2011-02-339747. [DOI] [PubMed] [Google Scholar]
- 42.Peltomäki P, de la Chapelle A. Mutation predisposing to hereditary nonpolyposis colorectal cancer. Adv Cancer Res. 1997;71:93–119. doi: 10.1016/s0065-230x(08)60097-4. [DOI] [PubMed] [Google Scholar]
- 43.Craig SEL, Brady-Kalnay SM. Regulation of development and cancer by the R2B subfamily of RPTPs and the implications of proteolysis. Semin Cell Dev Biol. 2015;37:108–18. doi: 10.1016/j.semcdb.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tam WF, Gu T-L, Chen J, Lee BH, Bullinger L, Fröhling S, et al. Id1 is a common downstream target of oncogenic tyrosine kinases in leukemic cells. Blood. 2008;112:1981–92. doi: 10.1182/blood-2007-07-103010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lavallée VP, Gendron P, Lemieux S, D’Angelo G, Hébert J, Sauvageau G. EVI1-rearranged acute myeloid leukemias are characterized by distinct molecular alterations. Blood. 2015;125:140–3. doi: 10.1182/blood-2014-07-591529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cai Y, Crowther J, Pastor T, Abbasi Asbagh L, Baietti MF, De Troyer M, et al. Loss of chromosome 8p governs tumor progression and drug response by altering lipid metabolism. Cancer Cell. 2016;29:751–66. doi: 10.1016/j.ccell.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 47.Kadoch C, Crabtree GR. Mammalian SWI/SNF chromatin remodeling complexes and cancer: mechanistic insights gained from human genomics. Sci Adv. 2015;1:e1500447. doi: 10.1126/sciadv.1500447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zebisch A, Staber PB, Delavar A, Bodner C, Hiden K, Fischereder K, et al. Two transforming C-RAF germ-line mutations identified in patients with therapy-related acute myeloid leukemia. Cancer Res. 2006;66:3401–8. doi: 10.1158/0008-5472.CAN-05-0115. [DOI] [PubMed] [Google Scholar]
- 49.Schoch C, Kohlmann A, Dugas M, Kern W, Hiddemann W, Schnittger S, et al. Genomic gains and losses influence expression levels of genes located within the affected regions: a study on acute myeloid leukemias with trisomy 8, 11, or 13, monosomy 7, or deletion 5q. Leukemia. 2005;19:1224–8. doi: 10.1038/sj.leu.2403810. [DOI] [PubMed] [Google Scholar]
- 50.Stein S, Ott MG, Schultze-Strasser S, Jauch A, Burwinkel B, Kinner A, et al. Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease. Nat Genet. 2010;16:198–204. doi: 10.1038/nm.2088. [DOI] [PubMed] [Google Scholar]
- 51.Dawson MA, Gudgin EJ, Horton SJ, Giotopoulos G, Meduri E, Robson S, et al. Recurrent mutations, including NPM1c, activate a BRD4-dependent core transcriptional program in acute myeloid leukemia. Leukemia. 2014;28:311–20. doi: 10.1038/leu.2013.338. [DOI] [PMC free article] [PubMed] [Google Scholar]