Abstract
Cardiovascular malformations (CVMs) are the most common birth defect, occurring in 1–5% of all live births. Genetic, epigenetic, and environmental factors all influence the development of CVMs, and an improved understanding of causation of CVMs is a prerequisite for prevention. Cardiac development is a complex, multi-step process of morphogenesis that is under genetic regulation. Multiple developmental pathways act independently or in combination to effect proper cardiac lineage specification, differentiation, and structure. Because of this complexity, there are numerous potential mechanisms by which genetic variation can impact both fetal cardiac development and latent cardiac disease. Although the genetic contribution to CVMs is well recognized, the genetic causes of human CVMs are still identified relatively infrequently. Mouse models are important tools to investigate the molecular mechanisms underpinning cardiac development as well as the complex genetics that characterize human CVMs. In this review we provide an overview of the key genetic concepts characterizing human CVMs, review their developmental basis, and provide examples to illustrate the critical developmental and genetic concepts underlying the pathogenesis of CVMs.
Keywords: Congenital heart defects, congenital heart disease, development, gene dosage
INTRODUCTION
The underlying causes of CVMs can include cytogenetic abnormalities, single gene disorders, environmental etiologies, or most commonly, multifactorial etiologies (Table 1). Chromosomal abnormalities account for 12–14% of all live born cases and 20–33% of fetal cases of congenital cardiovascular malformations (CVMs), indicating that the proper genetic control of cardiac development is essential 1–4. CVMs can occur as isolated findings, as part of a well-defined syndrome, or in conjunction with additional extracardiac anomalies not formally recognized as a syndrome 5. The designation of CVMs as isolated can be problematic since many important distinguishing features of syndromic conditions, such as developmental delay or dysmorphic features, may not be apparent at initial evaluation. As a result, syndromic cases of CVM may be underestimated. In addition, the traditionally cited incidence for CVMs of ~1% of live births likely also underestimates the scope and impact of disease. Taking into account very high rates of CVMs in spontaneous abortuses, common malformations such as BAV (present in 1.2% of the population) and latent cardiac diseases such as aortic dilation which are not included in the birth incidence of CVMs, genetically mediated CVMs are likely much more common than previously thought. When considering the etiology of CVMs, as opposed to the proportion of CVM cases that manifest as disease at birth, the incidence increases to approximately 5%.
Table 1.
Causes of Cardiovascular Malformations
| Cause | Example | Characteristic CVMs |
|---|---|---|
| Environmental/teratogenic | Lithium Chloride | Ebstein’s anomaly |
| Genetic | ||
| Chromosomal | Trisomy 21 | Atrioventricular canal defect |
| Contiguous gene/CNV | 22q11.2 deletion syndrome | Conotruncal malformations |
| Single gene | Noonan syndrome | Pulmonary valve stenosis |
| Epigenetic | ||
| DNA methylation | De novo SMAD2 mutations | Conotruncal malformations LV obstructions Heterotaxy |
Recently, we summarized the overall progress in the molecular genetic analyses of CVMs and current recommendations for clinical application of genetic testing. In particular, we reviewed the utility and limitations of chromosomal microarray analyses (CMAs) and the emerging clinical roles for whole exome sequencing (WES) and other next-generation sequencing (NGS) technologies 6. Readers with an interest in the current clinical testing approaches for CVMs are referred there. Here, we focus on common genetic and developmental themes across the wide variety of CVMs and the ability of animal models and knowledge of cardiac developmental biology to impact our understanding and approach to CVMs.
The genetic basis of CVMs
Epidemiologic studies suggest that a syndromic form of CVM is identifiable in approximately 20% to 30% of cases 4. Known genetic causes are extremely heterogeneous, encompassing not only mutations in cardiac relevant genes but also more complex chromosomal abnormalities, submicroscopic duplications/deletions, and whole-chromosome aneuploidies (Table 1). As noted above, CVMs can be isolated or can occur as part of a well-recognized genetic syndrome, and this distinction may be subtle.
Inheritance patterns for many CVM-associated genetic conditions are well characterized (reviewed in 6) (Table 2). Genetic syndromic conditions associated with CVMs are most commonly de novo or autosomal dominant. For dominantly inherited conditions, such as Noonan or Holt-Oram syndromes, individual recurrence risks for offspring with the syndrome is 50%. Importantly, not all patients with a particular syndrome have associated heart defects and the proportion can vary by syndrome. Furthermore, the presence or severity of a CVM in the parent does not predict the severity in the child.
Table 2.
Examples of common syndromes with CVMs caused by single gene mutations
| Gene | Syndromes | Common cardiac anomalies |
|---|---|---|
| CHD7, SEMA3E | CHARGE syndrome | ASD, VSD, TOF |
| FBN1 | Marfan syndrome | Aortic dilation |
| JAG1, NOTCH2 | Alagille syndrome | PS, peripheral PS, TOF |
| KMT2D | Kabuki syndrome | ASD, VSD, TOF, CoA |
|
PTPN11, KRAS, NRAS, HRAS, RAF1, SOS1, NF1, CBL, BRAF, SHOC2, MAP2K1, MAP2K2 |
Rasopathies: Noonan, Cardiofaciocutaneous, Costello syndromes |
PS, HCM |
| SKI | Shprintzen-Goldberg syndrome | Aortic dilation |
| TBX5 | Holt-Oram syndrome | ASD, VSD, AVSD, conduction system disease |
| TFAP2b | Char syndrome | PDA |
|
TGFB2, TGFBR1, TGFBR2, SMAD3 |
Loeys-Dietz syndrome types 1–4 | Aortic dilation |
| TGFB3 | Rheinhoff syndrome | Aortic dilation |
| ZIC3 | X-linked heterotaxy syndrome | heterotaxy |
ASD, atrial septal defect; AVSD, atrioventricular septal defect; CoA, coarctation of the aorta; PDA, patent ductus arteriousus; PS, pulmonic stenosis; TOF, tetralogy of Fallot; VSD, ventricular septal defect
Isolated CVMs may be inherited as autosomal dominant, autosomal recessive, or X-linked conditions, but are most commonly sporadic with multifactorial etiology (Table 3). Like other conditions inherited as a complex trait, isolated CVMs may show familial clustering with reduced penetrance7. For these reasons, recurrence risks for isolated CVMs can be difficult to assign. Consistent evidence of high heritability of isolated CVMs indicate that a strong genetic component exists, even for defects occurring without an obvious mode of inheritance 8.
Table 3.
Genes causing isolated heart defects
| Gene | Protein |
|---|---|
| ETS1 | V-Ets avian erythroblastosis virus E26 oncogene homolog 1 |
| TGFB2, TGFB3 | Transforming growth factor ligand 2, 3 |
| TGFBR1, TGFBR2 | Transforming growth factor receptor 1, 2 |
| SMAD2, SMAD3 | Mothers against decapentaplegic, drosophila, homologs 2, 3 |
| HAND1, HAND2 | Heart and neural crest derivatives expressed 1, 2 |
|
GATA4, GATA5, GATA6 |
GATA binding protein 4–6 |
| TBX1 | T-box 1 |
| TBX20 | T-box 20 |
| CITED2 | Cbp/P300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
| MESP1 | Mesoderm posterior 1 homolog |
| IRX4 | Iroquois homeobox 4 |
| MYOCD | Myocardin |
| Nkx2-5, Nkx2-6 | NK2 homeobox 5, 6 |
| NFATC1 | Nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 1 |
| NOTCH1 | Notch1 |
| ELN | Elastin |
Gene dosage as a mechanism for CVM
Gene dosage is an important concept underlying genetic disease, including birth defects. For many genes, a missing (deletion) or extra (duplication) copy of that gene results in no phenotypic consequences. In contrast, dosage sensitive genes produce abnormal phenotypes in the absence of two functional genes. Aneuploidies such as Trisomy 21 and Turner syndrome demonstrate that proper chromosome number is required for normal development. CVMs, including AVSD, are seen in approximately 50% of individuals with Down syndrome. Likewise, up to 50% of patients with Turner syndrome will have a CVM, most commonly a defect in the left ventricular outflow tract. Because of the large number of genes with abnormal dosage in these conditions, identifying the causal genes for the cardiac features has proven difficult. Furthermore, the decreased penetrance of the CVMs suggests that genetic modifiers interact with dosage-sensitive gene(s) on the same chromosome (in the case of Trisomy 21) or other chromosomes to cause CVM. Thus, a threshold exists in both aneuploid and euploid populations for the number of genetic perturbations that can be tolerated before CVM results. For example, Creld1 and Hey2 were recently identified as potential modifier genes in Trisomy 219. Mice with mutant forms of these potential modifiers were intercrossed to the Ts65Dn mouse model of Down syndrome. Breeding loss-of-function alleles of either Creld1 or Hey2 onto the trisomic background causes a significant increase in the frequency of CVM. This supports a threshold hypothesis for additive effects of genetic modifiers in the sensitized trisomic population.
Submicroscopic chromosome deletions and duplications also underlie many genetic syndromes, and the term genomic disorder is used to refer to these conditions. Two classic genomic disorders, 22q11.2 deletion syndrome and Williams-Beuren syndrome, are discussed in further detail below.
Understanding the genetic basis of syndromic CVM can identify important genes for isolated CVM
Williams-Beuren syndrome (WBS) is a relatively common genetic syndrome associated with CVM caused by deletion at 7q11.23, resulting in haploinsufficiency of multiple genes, including elastin, ELN. Supravalvar aortic stenosis (SVAS) is the most classic cardiac finding in WBS, although other defects, including peripheral pulmonic stenosis, occur. Subsequent to the description of WBS as a deletion at 7q11.23 in 199310, Ewart et al. showed close linkage of ELN and supravalvular aortic stenosis (SVAS) in two families11. Deletions or point mutations limited to the ELN gene appear to result in nonsyndromic SVAS, whereas larger deletions spanning multiple genes lead to the WBS. Studies in a mouse model with elastin deficiency have successfully corroborated the genetic findings with regard to SVAS and latent aortic disease 12, 13.
22q11.2 deletion syndrome provides a second example of a genomic disorder that led to the identification of a single gene causing CVM. CVMs occur in approximately 75% of patients with 22q11.2 deletion syndrome, with conotruncal defects predominating. After the identification of 22q11.2 deletion syndrome, significant effort was put forth to delineate the dosage sensitive gene(s) responsible for the CVMs using mouse development and genetics, ultimately identifying Tbx114, 15. Yagi et al. investigated TBX1 mutations in families who had 22q11.2 deletion syndrome phenotype but no detectable deletion and found that TBX1 mutations are responsible for many major phenotypes of the syndrome, including CVMs. In much the same way that modifiers for the Ts65Dn mouse model of Down syndrome were identified, sonic hedgehog and retinoic acid developmental signaling pathways modify the phenotypes of a 22q11.2 deletion syndrome mouse model, suggesting that mice with reduced gene dosage are sensitized to these morphogens16. These disorders illustrate the concept that genes which cause CVMs may be associated with syndromic or isolated presentations. Comprehensive identification of dosage sensitive candidate CVM genes and integration into an understanding about the genetic and developmental origins of CVM would facilitate the development of therapies to rescue the CVMs associated with both syndromic and isolated CVM.
Blurring the boundaries: single gene defects that can cause both syndromic and isolated CVMs
Genetic testing technologies have identified several genes that cause both syndromic and nonsyndromic CVMs (Table 2–3) 6. For example, CVMs, including aortic aneurysm, are reported in syndromic patients (i.e., Marfan syndrome (MFS) and Loeys-Dietz syndrome (LDS)) with mutations affecting the TGFβ pathway (TGFB2, TGFBR1, TGFBR2, SMAD3, FBN1) (Table 2)17, 18. Non-syndromic aortic disease is a frequently asymptomatic but potentially lethal disease characterized by familial cases of thoracic aortic aneurysm and dissection (FTAAD). This monogenic but genetically heterogeneous condition is primarily inherited as an autosomal dominant disorder with variable penetrance and expressivity. Mutations in TGFB genes have also been described in nonsyndromic patients with isolated CVM or aortic aneurysm (Table 3) 18–21. These facts complicate the clinical approach to patients with CVMs and the choice of genetic testing. Furthermore, patients with mutations in ACTA2, a gene known to cause isolated FTAAD, can have a syndromic presentation 22 and pediatric patients with FTAAD frequently have subtle signs of a connective tissue disorder 23. As the ability to identify genetic etiology improves, boundaries between syndromic and nonsyndromic disease often become less distinct. Careful phenotyping and improved interpretation of genetic variation are important to better refine our understanding of the spectrum of clinical effects of specific genetic variation.
The developmental basis for CVMs: genes and pathways required for critical stages of heart formation
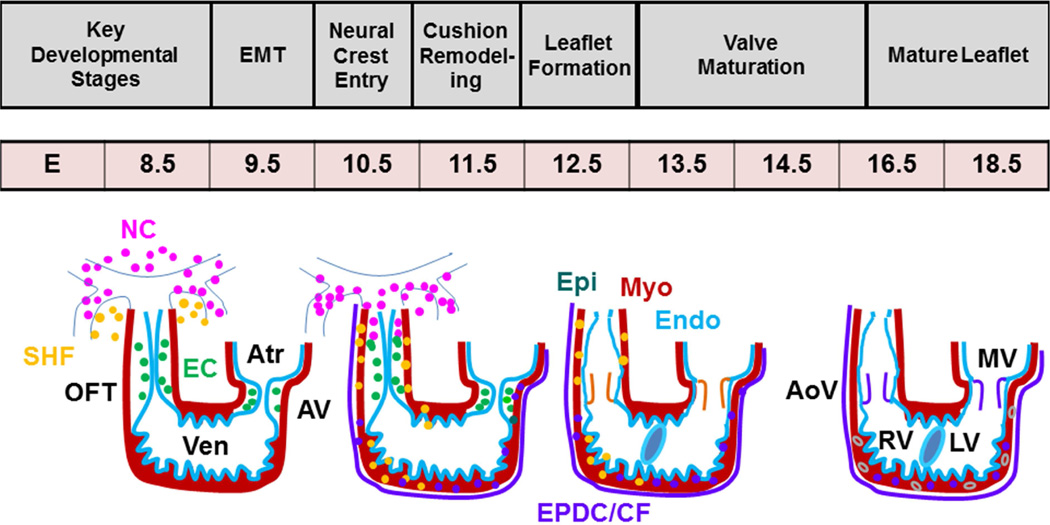
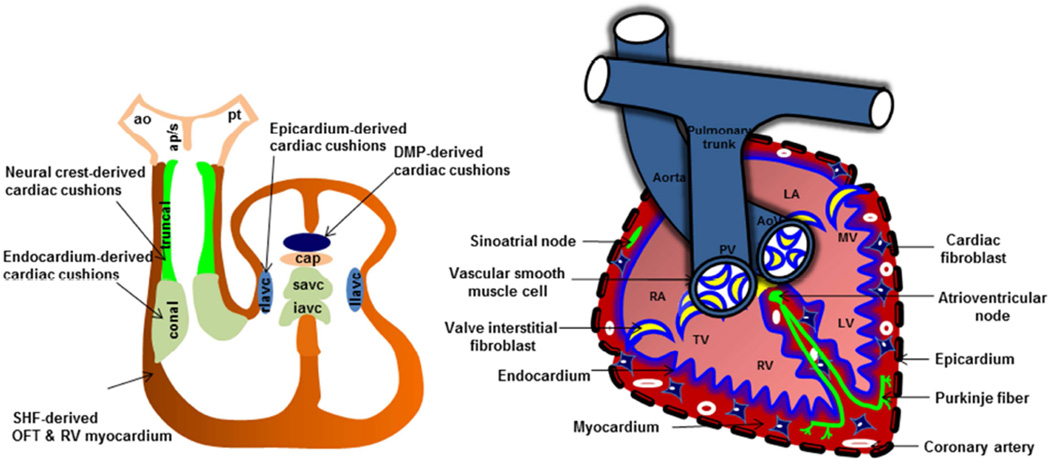
Genetically engineered mice are extensively used in CVM research and have contributed greatly to an understanding of the genetic control and mechanistic basis of CVMs. Multiple cell types contribute to the development of a fully septated four-chambered heart, including the first heart field (FHF) and second heart field (SHF), cardiac neural crest (NC), epicardial (Epi), and endocardial (EC) cell lineages (Fig. 1) 24. Both cardiac NC and endocardium-derived cushion mesenchyme are important precursors of the outflow tract (OFT) septa and semilunar valves25. At embryonic day 9.5 (E9.5), endocardium in the proximal OFT region gives rise to cushion mesenchyme via epithelial-mesenchymal transition (EMT) (Fig. 1) 25, 26. NC cells enter the distal OFT (E10), proliferate, and eventually colonize the endocardial ridges of the proximal cushions27. NC cells undergo apoptosis (E11.5 – 13.5), and are necessary for aorticopulmonary septation and OFT alignment28. Endocardium-derived OFT cells also undergo proliferation but remain restricted to the endocardial ridges of the proximal OFT cushions (Fig. 2). Remodeling and fusion of the endocardial ridges of the proximal cushions results in the formation of fibrous OFT septum that undergoes differentiation (E13.5–15.5) and eventually becomes a completely muscular structure (E16.5–18.5) through a process called myocardialization26. Despite abundant contributions of NC to the OFT mesenchyme, few NC derivatives are present in the mature semilunar valves27. Although the precise role of NC cells or their interaction with endocardial EMT-derived OFT cells remains unclear, more recent studies have suggested that NC cells are also important for the remodeling of the semilunar valves 27. Abnormal OFT cushion remodeling often results in semilunar valve thickening, defective OFT septation (persistent truncus arteriosus, PTA) or alignment defects such as double-outlet right ventricle (DORV) and ventricular septal defect (VSD) 25. The signals and cellular events that mediate valve remodeling are poorly characterized, although apoptosis and alterations in extracellular matrix production have been described 29, 30. Similar developmental events are noted in AV cushion formation and remodeling except that the cardiac NC are absent in the AV cushions and both dorsal mesocardium and epicardium provides additional cushion components of the AV cushion mesenchymal complex (Fig. 2) 31.
Figure 1. Diagrammatic representation of heart development.
Endocardium (EC) (aqua blue line) forms the endocardial cushions (green circle) via cushion EMT (E9.5). Neural crest (NC) (pink circle) cells migrate into the OFT during E10.5-12.5. Valve leaflets undergoing differentiation (orange color) and maturation (purple color) (E13.5-18.5) are clearly indicated. Only one semilunar valve is shown. SV, semilunar valves; AV, atrioventricular canal; RV, right ventricle; LV, left ventricle
Figure 2. Cardiac remodeling and septation.
Myocardium, endocardium, cardiac fibroblasts, and epicardium are major cell types in the heart. Left side, Components of OFT and AV cushions are indicated. OFT contains well demarcated conal (endocardium-derived) and truncal (NC-derived) cushions. AV cushion mesenchymal complex contains superior and inferior AV cushions (predominantly EC-derived), right and left lateral AV cushions (rlavc, llavc) (with contributions from both epicardium and EC), mesenchymal cap (cap) and dorsal mesenchymal protrusion (dmp) (dorsal mesocardium-derived). Right side, fully septated 4-chambered heart. The valve annulus and vascular wall of the aorta and pulmonary trunk are predominantly comprised of smooth muscle cells. Heart valves predominantly contain valve interstitial cells. Endocardium/endothelium is the innermost layer, whereas epicardium is the outermost layer. Both ventricular and atrial regions contain myocardium and cardiac fibroblasts along with coronary vasculature. Purkinje fiber and atrioventricular and sinoatrial nodes constitute the cardiac conduction system. AoV, aortic valve; PV, pulmonary valve, RA, right atrium; TV, tricuspid valve; RV, right ventricle; LV, left ventricle; MV, mitral valve; LA, left atrium.
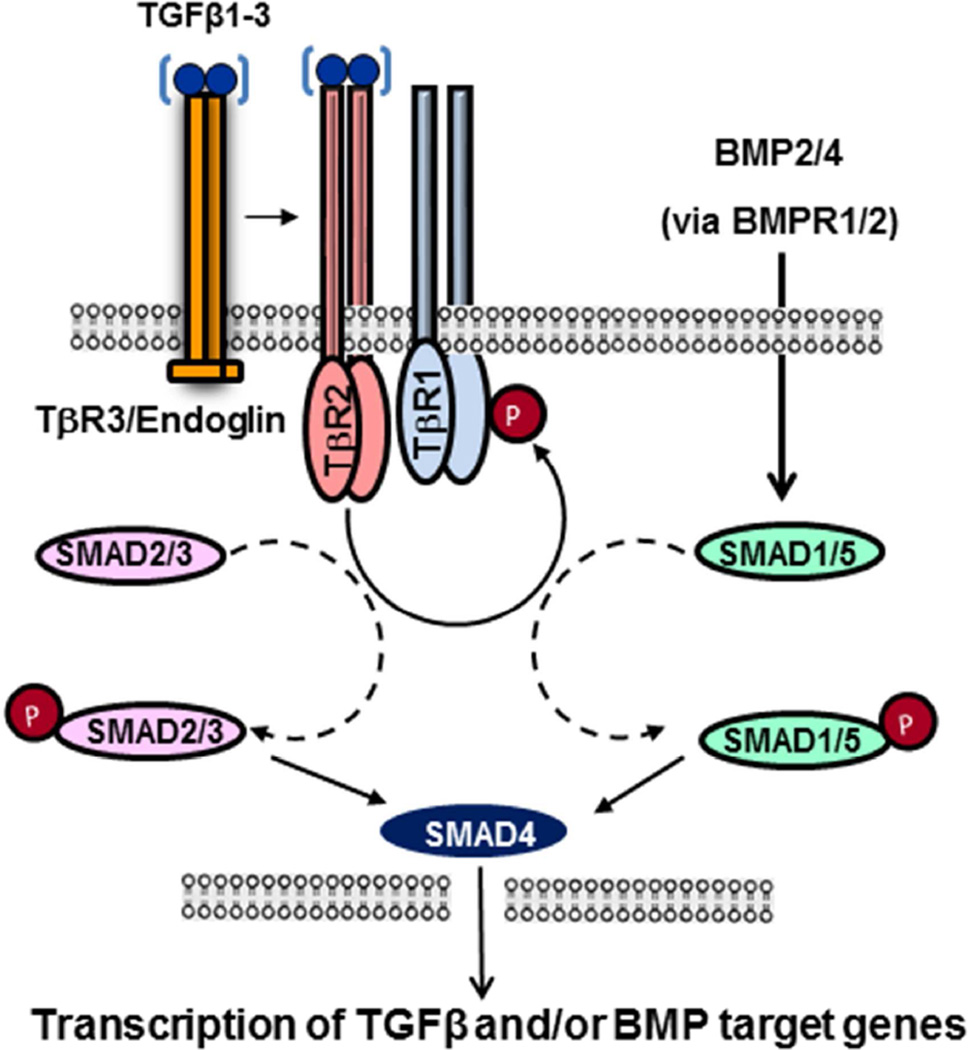
Developmental pathways acting independently or in combination contribute to heart development and have been reviewed recently 6, 32–34. For example, TGFβ and BMP family members play different roles during cardiac development (Fig. 3) (reviewed in 26, 35), and mutations in these genes result in distinct phenotypes 35–38. In Loeys-Dietz syndrome (LDS), mutations in the TGFβ pathway genes cause thoracic aortic aneurysm (TAA; Table 3) and are also highly associated with BAV39. Paradoxically, elevated levels of TGFβ1 are seen in these patients. Similar increases in TGFβ1 activity is also associated with BAV in Turner syndrome 40. Overall, it remains unclear whether the loss-of-TGFB function and/or gain-of-TGFB function is the primary cause of CVM. On the other hand, BMP signaling is required to induce differentiation of early cardiac progenitors, but BMP signaling is inhibited at later stages by Smad6a to permit chamber development mediated by Tbx2 and Tbx20. Importantly, the role of a particular signaling pathway can vary as development proceeds. For example, Wnt signals are critical for early cardiac precursor induction and proliferation, but later become inhibitory. Combinatorial interactions are the rule. Notch signaling interacts with both BMP and TGFβ pathways 6, 41. The cardiac transcription factor Nkx2.5 physically and functionally interacts with Gata4, Tbx5, and Mef2c, each of which forms additional unique and shared connections with other molecular, genetic, and signaling components 6, 33, 34. Such signaling and transcriptional networks hint at the possibility that some CVMs may result from additive effects of multiple low-effect susceptibility alleles.
Figure 3. Schematic diagram illustrating the TGFβ signaling pathway.
TGFβs binds to a common TGFβ receptor complex, and signals through phosphorylation of the canonical TGFβ-specific SMADs (i.e., pSMAD2/3). The pSMAD2/3 forms a complex with SMAD4, which accumulates in the nucleus and can regulate target gene expression. SMAD4 also binds to BMP-specific SMADs (SMAD1/5/8), and therefore regulates BMP-target gene expression in heart development.
Phenotypic heterogeneity and locus heterogeneity
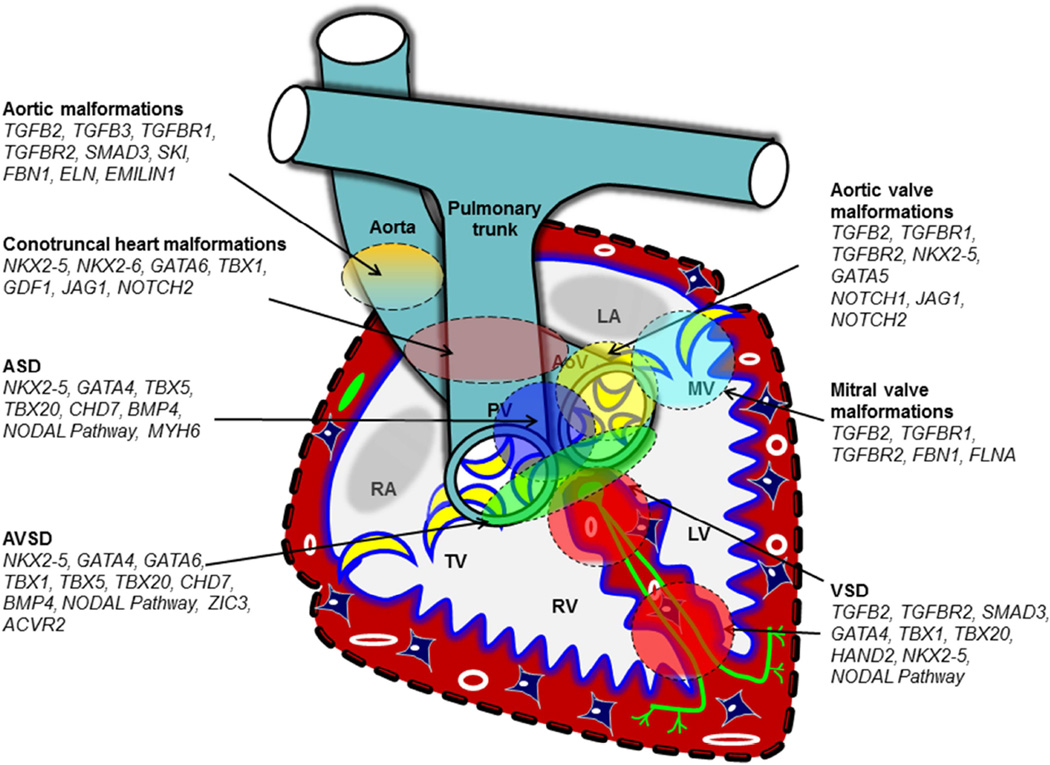
Studies of gene targeted mouse models indicate that loss of a single gene can result in a spectrum of CVMs (Table 4–5, Fig. 4; non-comprehensive examples). For example, the OFT malformations of the TGFβ2-deficient fetuses include DORV, PTA, abnormal morphology and thickening of aortic and/or pulmonary valves, aortic arch artery malformations (i.e. IAA), DILV and/or overriding of tricuspid valves orifice via a perimembranous inlet VSD, and abnormal morphology and thickening of tricuspid and mitral valves. Similarly, a range of CVM phenotype is seen in mice which lack Tbx1, Nkx2-5, Tbx20, Tbx5, and Gata4.
Table 4.
Phenotypic heterogeneity: the same genetic abnormality causes different heart defects
| Gene | CVMs in genetic mouse models |
|---|---|
| TGFB2 | VSD, TAA, BAV |
| NKX2-5 | VSD, AVSD, ASD |
| GATA4 | ASD, VSD, AVSD |
| TBX1 | AVSD, VSD, |
| TBX20 | VSD, AVSD, ASD |
| BMP4 | VSD, AVSD, ASD |
| GATA6 | VSD, AVSD, ASD |
| ZIC3 | Heterotaxy, d-TGA, DORV, AVSD, other heterotaxy spectrum heart defects |
| JAG1 | PS, ASD, TOF |
| GDF1 | DORV, TOF, d-TGA |
| TBX5 | ASD, VSD |
| Trisomy 21 | ASD, VSD, PDA |
| 45, X | BAV, HLHS, CoA |
| 22q11.2 deletion | TOF, VSD, IAA type B |
ASD, atrial septal defect; AVSD, atrioventricular septal defect; BAV, bicuspid aortic valve; CoA, coarctation of the aorta; DORV, double outlet right ventricle; d-TGA, d-transposition of the great arteries; HLHS, hypoplastic left heart syndrome; IAA, interrupted aortic arch type A; PDA, patent ductus arteriousus; PS, pulmonic stenosis; TAA, thoracic aortic aneurysm; TOF, tetralogy of Fallot; VSD, ventricular septal defect
Table 5.
Locus heterogeneity: the same CVM results from distinct genetic loci
| CVM Type | Examples of genetic etiologies |
|---|---|
| BAV | TGFB signaling pathway single gene mutations, aneuploidy (45,X) |
| VSD | TGFB and BMP signaling pathway single gene mutations, aneuploidy (45,X; Trisomy 21), 22q11.2 deletion |
| DORV | GDF1, TBX1, 22q11.2 deletion |
| AVSD | ACVR2, NKX2-5, GATA4, 22q11.2 deletion, aneuploidy (Trisomy 21) |
| PS | JAG1, NOTCH2 |
| TOF | JAG1, NOTCH2, 22q11.2 deletion |
| MVP | TGFB2, FBN1, FLNA |
| TAA | TGFB signaling pathway single gene mutations, MYH11, ACTA2, MYLK1, FBN1, |
| HCM | Rasopathy gene mutations Sarcomeric gene mutation |
Figure 4. Phenotypic and genetic heterogeneity in CVMs.
A single genetic abnormality can cause multiply types of CVMs. In addition, the same genetic abnormality can result in different CVMs. CVMs are indicated with colored circles.
Different CVM phenotypes are noted in patients with identical mutations, even among members of the same family (Table 4). Null mutation in several genes can cause AVSD in mice, including Nkx2-5, Gata4, and Tbx1 31. Additional examples are presented in Fig. 4. Developmental mechanisms that cause different CVMs in response to a mutation in a single gene remain incompletely understood.
Mutations in developmental pathways may result in latent disease
There are numerous potential mechanisms through which genetic mutations could affect the complex differentiation and morphogenetic processes in heart development. Once developed, the cardiovascular system must undergo homeostasis to maintain function throughout life. This ability to repair and remodel following stress and injury uses many of the same mechanisms involved in the original development and remodeling of those tissues38. Failure of these processes can result in late onset disease. Genes associated with CVMs (Fig. 4) 35, 38 are ideal candidates for these homeostatic, stress response and repair processes. Improvement in outcomes requires a better understanding of mechanisms underlying CVMs and dysregulated homeostatic/repair processes.
There are many interesting examples of genes in which homozygous gene deletion in mice results in CVMs in embryos and latent cardiac disease in adult mice, including elastin, emilin 1, periostin, and fibrillin 1. Elastin null (Eln−/−) mice die perinatally secondary to severe arterial obstruction reminiscent of SVAS12, whereas arteriopathy in the Eln+/− mouse manifests as systemic hypertension42. Juvenile Eln+/− mice demonstrate normal valve function, but progressive valve disease (predominantly aortic regurgitation) is identified in 17% of adult and 70% of aged adult Eln+/− mice by echocardiography13. Thus Eln+/− mice are a model of latent aortic valve disease and reduced elastin leads to dysregulation in valve pathogenesis. Other good examples of mouse models of latent aortic disease include Emilin1, Fibrillin-1+/C1039G, and Fibrillin1mgR/mgR (Fbn1mgR/mgR), the latter two being mouse models of Marfan syndrome (MFS). The Fbn1mgR/mgR mice die spontaneously from rupture of the thoracic aorta between 2 to 4 months of age, and are useful in testing therapeutic strategies for aortic aneurysm. On the other hand, Fibrillin-1+/C1039G mice, where a point mutation seen in MFS has been made, represent a viable mouse model to study the development and progression of aortic aneurysm. The early manifestation of elastic fiber fragmentation and aberrant TGFβ signaling suggests that these processes are crucial intermediate factors which provide novel information for diagnosis and treatment of patients with aortic disease 23, 43.
Decreased penetrance, variable expressivity and complex inheritance: lessons from mouse models
Genetically engineered mice can serve as a useful example of modifying genetic influences that affect phenotype. A good example is the different phenotypes seen in Tgfb2 null mice on mixed (129/Bl-Swiss) and inbred (C57BL/6) genetic backgrounds. The OFT malformations of the inbred null fetuses included DORV (100% cases), PTA (27.2% cases), and semilunar valve defects (100% cases). In addition, the null fetuses developed DILV and/or overriding of tricuspid valves orifice via a perimembranous inlet VSD (100% cases), and tricuspid/mitral valve defects (100%). Notably, the overall penetrance of the observed cardiac valve and septal defects was significantly higher in C57BL/6 inbred null fetuses compared to Tgfb2 null fetuses on the mixed genetic background. 44 This difference is attributed to the differences in genetic modifiers between the strains.
Epigenetic factors in CVM
An increasing recognition of epigenetic factors has revealed an unanticipated breadth to the causes of CVMs45. Epigenetics refers to functionally relevant changes to the genome that do not involve change in the DNA sequence. DNA methylation and histone modification are major epigenetic mechanisms which alter chromatin remodeling and gene expression without altering the underlying genetic information 5. A recent study by Pediatric Cardiac Genomics Consortium of the NHLBI identified de novo point mutations in several histone modifying genes that collectively contribute to approximately 10% of severe CVM5, 21. Refinements in technologies such as ChIP-seq and systems biology approaches will aid the understanding of global regulation and functional redundancies in cardiac transcription factors in CVMs33, 34. MicroRNAs (miRNAs), a class of "small" non-coding RNAs, negatively regulate the expression of their target genes through post-transcriptional processes and also interact with epigenetic machinery46. Regulation of gene expression via mechanisms that affect epigenetic machinery will identify novel etiologies for CVMs.
Future developments
The effect of gene variation on the assembly of distinct cardiac and extracardiac cell lineages during heart development is an important area that warrants future investigation. The relative importance and role of different cell types in cardiac morphogenesis and remodeling remains to be fully understood. Defining gene function in specific cell types in mouse models at high resolution will enable predictions to be made about the phenotypic consequences of variants in humans that currently lack functional interpretation. Experiments that delineate fundamental differences/similarities in loss-of-function and gain-of-function genetic backgrounds in mice will provide insight into the consequences of gene dosage perturbation in humans and mechanisms of genetic disease. Finally, incorporation of new technologies such as next generation sequencing, gene expression profiling (i.e., RNA seq), and CRISPR/Cas9 -based methodologies to discover and validate novel genes involved in CVMs will significantly enhance the understanding of cardiac genetics and development.
SUMMARY
Cardiac development is a complex, multi-step process under genetic regulation. A detailed understanding of the molecular basis of cardiac development is necessary to understand disease causation. The field of cardiovascular genetics is progressing at a rapid pace, leading to novel diagnostic genetic testing for CVMs. Recent efforts to integrate developmental studies from animal models with systems biology approaches offers significant promise for future CVM research. Understanding how genetic mutations affect the integration of multiple signal transduction pathways to cause CVM is an active area of research. The information gained from these developmental and genetic investigations should generate novel hypotheses for future experimentation and to provide diagnostic and therapeutic avenues for CVM patients.
KEY POINTS.
There is a strong genetic contribution to cardiovascular malformations (CVMs).
Genes important for syndromic CVM may also cause nonsyndromic CVM.
An understanding of the genes and pathways required for critical stages of heart formation informs the approach to genetic testing and diagnosis.
The same gene or genetic locus may cause different types of CVMs (phenotypic heterogeneity)
The same CVM may result from mutations in different genes (locus heterogeneity)
Mouse models are important tools to investigate the complex genetics of CVMs.
Best Practices Box.
What is the current practice?
Genetic Testing in Cardiovascular Malformations
Genetic testing practices for congenital heart defects have yet to be standardized in many centers and testing is frequently underutilized.
Guidelines for cardiac imaging and genetic testing for thoracic aortic aneurysm and cardiomyopathy
A genetic diagnosis has important implications for patient management, screening recommendations for family members, and recurrence risk counseling
Genetic Testing Options
Chromosome analysis is the gold standard for diagnosis of aneuploidies and other large chromosomal abnormalities.
Chromosomal microarray (CMA) and fluorescence in situ hybridization (FISH) permit identification of microdeletion and duplication syndromes resulting from abnormalities too small to be detected by conventional chromosome analyses.
Next generation sequencing (NGS) panels are the test of choice for some syndromic congenital heart defects, thoracic aortic aneurysm and cardiomyopathy.
CMA and targeted NGS are non-redundant tests. Consulting a geneticist is important for establishing a differential, ordering the appropriate test(s), and interpreting results.
Implications for family members
First degree relatives of patients with specific cardiovascular malformations (i.e. left ventricular outflow tract obstructive defects, thoracic aortic aneurysm) should undergo cardiac screening
Family based risk assessment and recurrence risk information differs by type of cardiovascular malformation and should be provided to the family be a knowledgeable genetics professional.
What changes in current practice are likely to improve outcomes?
Continued integration of genetic testing services into cardiovascular practice will improve diagnostic and prognostic accuracy and will support risk assessment and family planning initiatives.
NGS technologies promise to greatly benefit patient diagnosis and gene discovery efforts.
Appropriate cardiac screening and surveillance in at risk relatives will identify latent disease.
Is there a Clinical Algorithm?
An algorithm for congenital heart defects has been proposed recently. 6
Guidelines summarize genetic testing for syndromic and non-syndromic thoracic aortic aneurysm as well as cardiac screening in first degree relatives.47
Major Recommendations
Genetic testing and referral decisions should be determined based on the nature of the cardiac defect.
A detailed pedigree should be obtained in all cases of CVM.
Chromosome analysis is recommended for patients with suspected aneuploidy.
Patients with multiple congenital anomalies, neurological findings, developmental delay, and/or dysmorphic features should be referred for genetic evaluation.
CMA and/or FISH should be used in patients with conotruncal defects.
Patients with apparently non-syndromic LVOTO, RVOTO, AVSD, heterotaxy, or other complex defects should have CMA.
Patients with TAA should have a genetics evaluation and appropriate genetic testing.
Specific CVMs should trigger cardiac screening of first degree relatives.
Rating for the Strength of the Evidence
C Recommendation based on consensus, usual practice, expert opinion, disease-oriented evidence, and case series for studies of diagnosis, treatment, prevention, or screening
References
Pierpont ME, Basson CT, Benson DW, Jr., et al. Genetic basis for congenital heart defects: current knowledge. Circulation 2007;115:3015–38 (American Heart Association Position Statement).
Hiratzka LF, Bakris GL, Beckman JA, et al. 2010 ACCF/AHA/AATS/ACR/ASA/SCA/SCAI/SIR/STS/SVM guidelines for the diagnosis and management of patients with Thoracic Aortic Disease: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines, American Association for Thoracic Surgery, American College of Radiology, American Stroke Association, Society of Cardiovascular Anesthesiologists, Society for Cardiovascular Angiography and Interventions, Society of Interventional Radiology, Society of Thoracic Surgeons, and Society for Vascular Medicine. Circulation. 2010;121(13):e266–369.
Cowan JR, Ware SM. Genetics and genetic testing in congenital heart disease. Clinics in Perinatology 2015; 42:373–93.
Summary
Genetic evaluation and testing are increasingly important for cardiovascular malformations. Improvements in genetic testing technologies have assisted gene discovery and helped to reshape standards of patient care. Risk assessment of family members and recurrence risk counseling are important components of management and care.
Acknowledgments
FUNDING SOURCES:
Supported, in part, by Riley Children’s Foundation, Showalter Trust, Center of Excellence in Cardiovascular Research Fund, and R01HL126705-01 (M.A.) and by an American Heart Association Established Investigator Award, Burroughs Wellcome Foundation, March of Dimes, and Indiana University Health - Indiana University School of Medicine Strategic Research Initiative (S.M.W.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DISCLOSURE STATEMENT:
The authors have nothing to disclose.
Reference List
- 1.Gillum RF. Epidemiology of congenital heart disease in the United States. Am Heart J. 1994 Apr 127;(4 Pt 1):919–927. doi: 10.1016/0002-8703(94)90562-2. [DOI] [PubMed] [Google Scholar]
- 2.Chaoui R, Korner H, Bommer C, Goldner B, Bierlich A, Bollmann R. [Prenatal diagnosis of heart defects and associated chromosomal aberrations] Ultraschall Med. 1999 Oct;20(5):177–184. doi: 10.1055/s-1999-8912. [DOI] [PubMed] [Google Scholar]
- 3.Tennstedt C, Chaoui R, Korner H, Dietel M. Spectrum of congenital heart defects and extracardiac malformations associated with chromosomal abnormalities: results of a seven year necropsy study. Heart. 1999 Jul;82(1):34–39. doi: 10.1136/hrt.82.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferencz C, Boughman JA, Neill CA, Brenner JI, Perry LW. Congenital cardiovascular malformations: questions on inheritance. Baltimore-Washington Infant Study Group. J Am Coll Cardiol. 1989 Sep;14(3):756–763. doi: 10.1016/0735-1097(89)90122-8. [DOI] [PubMed] [Google Scholar]
- 5.Bruneau BG, Srivastava D. Congenital heart disease: entering a new era of human genetics. Circ Res. 2014 Feb 14;114(4):598–599. doi: 10.1161/CIRCRESAHA.113.303060. [DOI] [PubMed] [Google Scholar]
- 6.Cowan JR, Ware SM. Genetics and genetic testing in congenital heart disease. Clin Perinatol. 2015 Jun;42(2):373–393. doi: 10.1016/j.clp.2015.02.009. ix. [DOI] [PubMed] [Google Scholar]
- 7.Lalani SR, Belmont JW. Genetic basis of congenital cardiovascular malformations. Eur J Med Genet. 2014 Aug;57(8):402–413. doi: 10.1016/j.ejmg.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oyen N, Poulsen G, Boyd HA, Wohlfahrt J, Jensen PK, Melbye M. Recurrence of congenital heart defects in families. Circulation. 2009 Jul 28;120(4):295–301. doi: 10.1161/CIRCULATIONAHA.109.857987. [DOI] [PubMed] [Google Scholar]
- 9.Li H, Cherry S, Klinedinst D, et al. Genetic modifiers predisposing to congenital heart disease in the sensitized Down syndrome population. Circ Cardiovasc Genet. 2012 Jun;5(3):301–308. doi: 10.1161/CIRCGENETICS.111.960872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curran ME, Atkinson DL, Ewart AK, Morris CA, Leppert MF, Keating MT. The elastin gene is disrupted by a translocation associated with supravalvular aortic stenosis. Cell. 1993 Apr 9;73(1):159–168. doi: 10.1016/0092-8674(93)90168-p. [DOI] [PubMed] [Google Scholar]
- 11.Ewart AK, Jin W, Atkinson D, Morris CA, Keating MT. Supravalvular aortic stenosis associated with a deletion disrupting the elastin gene. J Clin Invest. 1994 Mar;93(3):1071–1077. doi: 10.1172/JCI117057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li DY, Brooke B, Davis EC, et al. Elastin is an essential determinant of arterial morphogenesis. Nature. 1998 May 21;393(6682):276–280. doi: 10.1038/30522. [DOI] [PubMed] [Google Scholar]
- 13.Hinton RB, Adelman-Brown J, Witt S, et al. Elastin haploinsufficiency results in progressive aortic valve malformation and latent valve disease in a mouse model. Circ Res. 2010 Aug;107(4):549–557. doi: 10.1161/CIRCRESAHA.110.221358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindsay EA, Vitelli F, Su H, et al. Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature. 2001 Mar 1;410(6824):97–101. doi: 10.1038/35065105. [DOI] [PubMed] [Google Scholar]
- 15.Merscher S, Funke B, Epstein JA, et al. TBX1 is responsible for cardiovascular defects in velo-cardio-facial/DiGeorge syndrome. Cell. 2001 Feb 23;104(4):619–629. doi: 10.1016/s0092-8674(01)00247-1. [DOI] [PubMed] [Google Scholar]
- 16.Maynard TM, Gopalakrishna D, Meechan DW, Paronett EM, Newbern JM, LaMantia AS. 22q11 Gene dosage establishes an adaptive range for sonic hedgehog and retinoic acid signaling during early development. Hum Mol Genet. 2013 Jan 15;22(2):300–312. doi: 10.1093/hmg/dds429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Braverman AC. Heritable thoracic aortic aneurysm disease: recognizing phenotypes, exploring genotypes. J Am Coll Cardiol. 2015 Apr 7;65(13):1337–1339. doi: 10.1016/j.jacc.2014.12.056. [DOI] [PubMed] [Google Scholar]
- 18.Micha D, Guo DC, Hilhorst-Hofstee Y, et al. SMAD2 Mutations are Associated With Arterial Aneurysms and Dissections. Hum Mutat. 2015 Aug 6;10 doi: 10.1002/humu.22854. [DOI] [PubMed] [Google Scholar]
- 19.Gago-Diaz M, Blanco-Verea A, Teixido-Tura G, et al. Whole exome sequencing for the identification of a new mutation in TGFB2 involved in a familial case of non-syndromic aortic disease. Clin Chim Acta. 2014 Nov 1;437:88–92. doi: 10.1016/j.cca.2014.07.016. Epub@2014 Jul@19.:88–92. [DOI] [PubMed] [Google Scholar]
- 20.Matyas G, Naef P, Tollens M, Oexle K. De novo mutation of the latency-associated peptide domain of TGFB3 in a patient with overgrowth and Loeys-Dietz syndrome features. Am J Med Genet A. 2014 May 5;10 doi: 10.1002/ajmg.a.36593. [DOI] [PubMed] [Google Scholar]
- 21.Zaidi S, Choi M, Wakimoto H, et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013 Jun 13;498(7453):220–223. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo DC, Pannu H, Tran-Fadulu V, et al. Mutations in smooth muscle alpha-actin (ACTA2) lead to thoracic aortic aneurysms and dissections. Nat Genet. 2007 Dec;39(12):1488–1493. doi: 10.1038/ng.2007.6. [DOI] [PubMed] [Google Scholar]
- 23.Landis BJ, Ware SM, James J, Shikany AR, Martin LJ, Hinton RB. Clinical Stratification of Pediatric Patients with Idiopathic Thoracic Aortic Aneurysm. J Pediatr. 2015 Jul;167(1):131–137. doi: 10.1016/j.jpeds.2015.02.042. [DOI] [PubMed] [Google Scholar]
- 24.Kelly RG, Buckingham ME, Moorman AF. Heart fields and cardiac morphogenesis. Cold Spring Harb Perspect Med. 2014 Oct 1;4(10):a015750. doi: 10.1101/cshperspect.a015750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin CJ, Lin CY, Chen CH, Zhou B, Chang CP. Partitioning the heart: mechanisms of cardiac septation and valve development. Development. 2012 Sep;139(18):3277–3299. doi: 10.1242/dev.063495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Azhar M, Schultz JE, Grupp I, et al. Transforming growth factor beta in cardiovascular development and function. Cytokine Growth Factor Rev. 2003 Oct;14(5):391–407. doi: 10.1016/s1359-6101(03)00044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jain R, Engleka KA, Rentschler SL, et al. Cardiac neural crest orchestrates remodeling and functional maturation of mouse semilunar valves. J Clin Invest. 2011 Jan;121(1):422–430. doi: 10.1172/JCI44244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kirby ML, Gale TF, Stewart DE. Neural crest cells contribute to normal aorticopulmonary septation. Science. 1983 Jun 3;220(4601):1059–1061. doi: 10.1126/science.6844926. [DOI] [PubMed] [Google Scholar]
- 29.Markwald RR, Norris RA, Moreno-Rodriguez R, Levine RA. Developmental basis of adult cardiovascular diseases: valvular heart diseases. Ann N Y Acad Sci. 2010 Feb;1188:177–183. doi: 10.1111/j.1749-6632.2009.05098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hinton RB, Jr, Lincoln J, Deutsch GH, et al. Extracellular matrix remodeling and organization in developing and diseased aortic valves. Circ Res. 2006 Jun 9;98(11):1431–1438. doi: 10.1161/01.RES.0000224114.65109.4e. [DOI] [PubMed] [Google Scholar]
- 31.Briggs LE, Kakarla J, Wessels A. The pathogenesis of atrial and atrioventricular septal defects with special emphasis on the role of the dorsal mesenchymal protrusion. Differentiation. 2012 Jul;84(1):117–130. doi: 10.1016/j.diff.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rana MS, Christoffels VM, Moorman AF. A molecular and genetic outline of cardiac morphogenesis. Acta Physiol (Oxf) 2013 Apr;207(4):588–615. doi: 10.1111/apha.12061. [DOI] [PubMed] [Google Scholar]
- 33.Chong JJ, Forte E, Harvey RP. Developmental origins and lineage descendants of endogenous adult cardiac progenitor cells. Stem Cell Res. 2014 Nov 13;(3 Pt B):592–614. doi: 10.1016/j.scr.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 34.Kathiriya IS, Nora EP, Bruneau BG. Investigating the transcriptional control of cardiovascular development. Circ Res. 2015 Feb 13;116(4):700–714. doi: 10.1161/CIRCRESAHA.116.302832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arthur HM, Bamforth SD. TGFbeta signaling and congenital heart disease: Insights from mouse studies. Birth Defects Res A Clin Mol Teratol. 2011 Jun;91(6):423–434. doi: 10.1002/bdra.20794. [DOI] [PubMed] [Google Scholar]
- 36.Hinck AP. Structural studies of the TGF-betas and their receptors - insights into evolution of the TGF-beta superfamily. FEBS Lett. 2012 Jul 4;586(14):1860–1870. doi: 10.1016/j.febslet.2012.05.028. [DOI] [PubMed] [Google Scholar]
- 37.Akhurst RJ, Hata A. Targeting the TGFbeta signalling pathway in disease. Nat Rev Drug Discov. 2012 Oct;11(10):790–811. doi: 10.1038/nrd3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doetschman T, Barnett JV, Runyan RB, et al. Transforming growth factor beta signaling in adult cardiovascular diseases and repair. Cell Tissue Res. 2012 Jan;347(1):203–223. doi: 10.1007/s00441-011-1241-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maccarrick G, Black JH, III, Bowdin S, et al. Loeys-Dietz syndrome: a primer for diagnosis and management. Genet Med. 2014 Feb 27;10 doi: 10.1038/gim.2014.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou J, Arepalli S, Cheng CM, Bakalov VK, Bondy CA. Perturbation of the transforming growth factor beta system in Turner syndrome. Beijing Da Xue Xue Bao. 2012 Oct 18;44(5):720–724. [PMC free article] [PubMed] [Google Scholar]
- 41.de la Pompa JL, Epstein JA. Coordinating tissue interactions: Notch signaling in cardiac development and disease. Dev Cell. 2012 Feb 14;22(2):244–254. doi: 10.1016/j.devcel.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carta L, Wagenseil JE, Knutsen RH, et al. Discrete Contributions of Elastic Fiber Components to Arterial Development and Mechanical Compliance. Arterioscler Thromb Vasc Biol. 2009 Oct 22; doi: 10.1161/ATVBAHA.109.193227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pierpont ME, Lacro RV. Children with Thoracic Aortic Aneurysm: Challenges in Diagnosis and Therapy. J Pediatr. 2015 Jul;167(1):14–16. doi: 10.1016/j.jpeds.2015.03.056. [DOI] [PubMed] [Google Scholar]
- 44.Azhar M, Brown K, Gard C, et al. Transforming growth factor Beta2 is required for valve remodeling during heart development. Dev Dyn. 2011 Sep;240(9):2127–2141. doi: 10.1002/dvdy.22702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang CP, Bruneau BG. Epigenetics and cardiovascular development. Annu Rev Physiol. 2012;74:41–68. doi: 10.1146/annurev-physiol-020911-153242. Epub@2011 Oct 24.:41–68. [DOI] [PubMed] [Google Scholar]
- 46.Kataoka M, Wang DZ. Non-Coding RNAs Including miRNAs and lncRNAs in Cardiovascular Biology and Disease. Cells. 2014 Aug 22;3(3):883–898. doi: 10.3390/cells3030883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hiratzka LF, Bakris GL, Beckman JA, Bersin RM, Carr VF, Casey DE, Jr, et al. 2010 ACCF/AHA/AATS/ACR/ASA/SCA/SCAI/SIR/STS/SVM guidelines for the diagnosis and management of patients with Thoracic Aortic Disease: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines, American Association for Thoracic Surgery, American College of Radiology, American Stroke Association, Society of Cardiovascular Anesthesiologists, Society for Cardiovascular Angiography and Interventions, Society of Interventional Radiology, Society of Thoracic Surgeons, and Society for Vascular Medicine. Circulation. 2010;121(13):e266–e369. doi: 10.1161/CIR.0b013e3181d4739e. [DOI] [PubMed] [Google Scholar]